Abstract
The negative effect of permanent contamination of populations because of spontaneous mutations does not appear to be very high if judged from the relatively good health of humans or many wild and domesticated species. This is partly explained by the fact that, in diploids, the new mutations are usually located in heterozygous loci and therefore are masked by wild-type alleles. The expression of mutations at the phenotypic level may also strongly depend on environmental factors if, for example, deleterious alleles are more easily compensated under favorable conditions. The present experiment uses diploid strains of yeast in which mutations arise at high rates because a mismatch-repair protein is missing. This mutagenesis resulted in a number of new alleles that were in heterozygous loci. They had no detectable effect on fitness when the environment was benign. A very different outcome was seen when thermal shock was applied, where fitness of the mutation-contaminated clones was lower and more diverse than that of the nonmutagenized clones. This shows that the genetic load conferred by spontaneous mutations can be underestimated or even overlooked in favorable conditions. Therefore, genetic variation can be higher and natural selection more intense when environmental conditions are getting poorer. These conclusions apply, at least, to that component of variation that directly originates from spontaneous mutations (as opposed to the variation resulting from the history of selection).
Most of the spontaneous alterations in the DNA sequence are expected to be either neutral or deleterious (1, 2). The actual impact of new alleles on an organism's fitness and thus their role in evolution is likely to depend on environment (3–5). This problem directly relates to the current debate on the rate at which mutations affecting fitness appear and can be experimentally detected (1, 2). Research on spontaneous mutations is difficult because often neither their molecular basis is known nor can they be completely protected from natural selection during prolonged accumulation experiments (6). The present work aims at overcoming these difficulties by using a yeast strain deficient in a major DNA mismatch repair system such that mutations were collected rapidly and their nature was known. A former study showed that such an approach could be fruitful because a haploid yeast that accumulated a large number of mutations had significantly reduced fitness under optimal conditions, which deteriorated even more when the environment worsened (7, 8). However, from the perspective of human health and conservation biology, it is particularly interesting to know what would be the phenotypic effects of a relatively small burden of heterozygous mutations in diploids (9).
The present experiment uses a diploid strain of yeast and a simple system of turning on and off the mismatch repair system. One of the genes essential for the majority of mismatch repair, PMS1, was deleted from its chromosomal locus. However, a functional copy was present on a plasmid. The mutation rate was normal in the presence of the plasmid, but, when it was absent, frame shifts and substitutions increased in frequency by about two orders of magnitude (10). The starting strain was strictly homozygous, with two identical alleles in all loci (except for the mating-type locus). The newly generated alleles, however, were expected to be heterozygous because it was highly improbable that both copies of a gene would be mutated. The accumulation of mutations began when a cell lost its plasmid containing the wild-type PMS1 gene. Mutagenesis continued for about 35–40 generations of vegetative growth and was terminated when the wild-type gene was reintroduced by transformation. A large number of replicate lines were established in which the losses, and hence mutation accumulation, were independent. These experimental clones were compared with a control group, that is, clones that were started from single cells of the same strain as the experimental group, but which were never deprived of the plasmid and therefore had not experienced an elevated mutation rate. The performance of both groups was estimated by measuring the maximum growth rate and the density of stationary phase cultures. The first trait revealed whether a clone was able to propagate with a normal doubling time; the second determined whether it exploited all resources before growth ceased. The two temperatures tested were a benign 30°C and a stressful 38°C.
Materials and Methods
Strain.
The strain used to initiate this study was originally derived from a natural isolate, Y55 (11). Introduction of genetic markers and other manipulations yielded the clone Y55 2231 Δpms1 ΔHO MATa his4 leu2 lys2 ade1 ura3. This strain was transformed with the plasmid pWBK3 PMS1 URA3 (12) and subsequently propagated on media lacking uracil to maintain the plasmid, thus ensuring that the mutation rate was low. The Y55 2231/pWBK3-PMS1 clone was then transformed with a plasmid containing the HO gene, whose action helped to obtain a homozygous (except for the MAT locus) diploid clone with the genotype of Y55 2231 (13). This diploid, in which the HO plasmid was lost but the PMS1 plasmid was still maintained, will be denoted in the following sections by F because it is considered free of new mutations.
Mutation Rate.
The rate of spontaneous mutation was estimated by finding how often resistance to canavanine appears. Haploid and not diploid strains were used because this marker is recessive. Both Y55 2231 and Y55 2231/pWBK3-PMS1 were brought to stationary phase cultures in a synthetic minimal medium (SM) enriched with relevant amino acids and nucleotides. They were then diluted 1:10,000 in a fresh medium of the same composition, each dispensed to 30 aliquots of 1 ml, and allowed to reach the stationary phase density. Samples of these replicate cultures were transferred on plates with the SM medium containing relevant enrichments and 60 mg/liter of canavanine. Colonies were scored after 48 h of incubation. The mutation rates were determined by a maximum likelihood method (14), followed by approximate estimation of confidence limits (15).
Mutation Accumulation.
Mutations were accumulated in a period of propagation in which mismatch repair was absent. Mutagenesis began with the loss of pWBK3-PMS1 from the F strain and ended with its reacquisition. The procedure involved four steps. First, the F strain was streaked to single cells on nutrient-rich YEPD plates. During the growth of colonies, plasmid was spontaneously lost in some cells. To obtain clones in which the plasmid had been lost, the colonies from YEPD were restreaked on a synthetic complete medium (SC) containing 5-fluro orotic acid (5-FOA). This killed the cells still containing the URA3 gene, and thus the plasmid (16). Next, the very small colonies that appeared on the 5-FOA medium after 24 h of incubation were transferred with a toothpick on the YEPD plates for another 48 h. Finally, the resulting relatively large colonies were retransformed with the wild-type PMS1. These steps were done independently for each clone reported in the following sections as “mutation contaminated” (abbreviated by M).
The control clones were essentially identical to the initial F clone because they were obtained directly from the latter by streaking it to single cells on plates lacking uracil and then maintained and propagated independently from each other on this medium. Because the wild-type PMS1 plasmid was maintained, they did not experience a period of propagation with an enhanced mutation rate.
Fitness Assays.
To measure the growth rate at 30°C, the F and M replicate clones were first grown overnight to stationary phase in SC without uracil. These were used to inoculate new cultures at 1% of the stationary phase density and then incubated with vigorous shaking. After the cultures reached “exponential” growth phase (7), their growth was monitored by measuring optical density at five time points every 30 min. The assay at 38°C was somewhat different. The initial stationary phase cultures were those obtained at 30°C in SC without uracil. From this, aliquots of 100 μl were transferred to 3 ml of preheated fresh medium and kept at 38°C with vigorous agitation. Measurements of optical density were taken every 1 h for up to 11 h. The analysis of growth rate was based on five measurements. For slow growing cultures, the last five time points were used. For rapidly growing cultures, the last five measurements lower than a fixed value were used. This fixed density, after which the rate of growth was likely to decrease because of the effect of saturation, was determined in a pilot experiment. The measurements gathered in both 30°C and 38°C were converted by using a log-normal transformation, and a regression analysis was performed.
Another trait measured for the F and M clones was the optical density (OD600) of cultures at stationary phase. At 30°C, the density was measured after 24 h of incubation in SC without uracil. These cultures were used to inoculate the 38°C cultures in the same way as described in the preceding paragraph. Measurements were then taken after 24 and 48 h.
Statistical Analysis.
Two statistics were calculated for the F and M clones at 30°C and 38°C. One of them was mean, x̄, and it served as a measure of central tendency. The second, opportunity for selection, s2/x̄2 where s2 stands for variance, was used to estimate variation (17). The latter parameter was chosen over variance because the mean population densities, and especially growth rates, were much lower at the stressful than at the benign temperature (see Results). Standard errors of both statistics were found by bootstrapping. Testing for differences between the F and M clones and between different temperatures involved several planned comparisons. Therefore, the experiment-wise type I error was adjusted by using the Dunn-Šidak procedure (18). Finally, the Welch's approximate t′ was used instead of the standard Student's t because both the sizes of samples and their standard errors often differed substantially (19).
Results
Mutation Rate.
The rate of spontaneous mutation to canavanine resistance in the mutator strain (Y55 2231) was 9.64 × 10−6, with the lower and upper 95% confidence limits at 6.79 × 10−6 and 1.27 × 10−5, respectively. When the same strain contained the pWBK3-PMS1 plasmid, the mutation rate decreased to 2.63 × 10−7, with the analogous confidence limits equal to 1.50 × 10−7 and 3.86 × 10−7.
Growth Rate.
The shape of the growth curves indicated that the increase in population density was truly exponential at 30°C, but not always at 38°C (data not shown). In many clones, and especially those contaminated with mutations, the rate of growth tended to slow down gradually when cultures were maintained in the higher temperature. Thus, the parameter used to quantify the rate of growth, the coefficient of regression, should be interpreted differently for the two temperature regimes. At 30°C, it was equal to the maximum growth rate, which was constant in time. At 38°C, however, it should be seen as an average rate of growth in the tested period of population expansion (see Materials and Methods for more details).
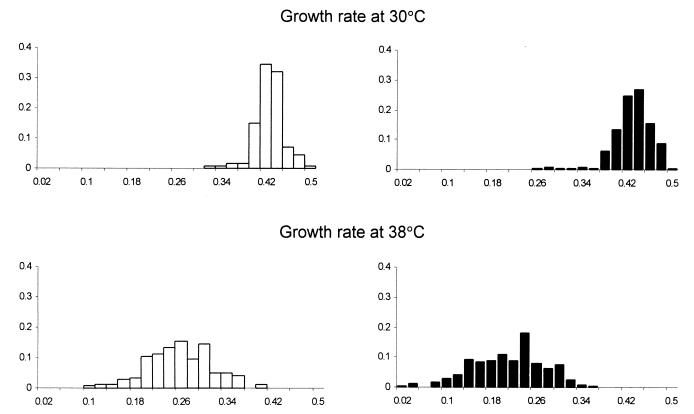
Fig. 1 presents the frequency distributions of the growth rate for the clones free of mutations (F) and those contaminated with them (M) at both temperatures. Statistical comparisons are provided in Table 1. The data show that the growth rate of the F and M clones was practically the same at 30°C. At 38°C, however, the mutation load of the M clones manifested itself as about a 20% decline in the growth rate when compared with that of the F clones. Both the F and M clones were approximately twice as slow at the unfavorable temperature than at the favorable one, indicating that the higher temperature was indeed stressful. Variation among F and among M clones was similar at the mild temperature, but the latter became more variable when compared at the stressful temperature as indicated by the increase in “opportunity for selection” (see Materials and Methods for explanations and Table 1 for statistics).
Figure 1.
Frequency distributions of growth rates (1/h) of the control F clones (open bars) and the mutation-contaminated M clones (filled bars).
Table 1.
Means and opportunities for selection calculated for growth rates of clones free of mutations (F) and contaminated with mutations (M)
| Pair compared | Sample
sizes
|
Mean growth rates,
1/h
|
df† | t′(B−A) | Opportunity
for selection,
×100
|
df† | t′(B−A) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A* vs. B* | A | B | A | B | A | B | ||||
| F30 vs. M30 | 127 | 229 | 0.418 | 0.417 | 254.1 | −0.083 | 0.380 | 0.714 | 353.9 | 2.211 |
| F38 vs. M38 | 143 | 266 | 0.247 | 0.200 | 244.9 | −7.448** | 5.266 | 9.926 | 386.7 | 3.884** |
| F30 vs. F38 | 127 | 143 | 0.418 | 0.247 | 210.1 | −31.84** | 0.380 | 5.266 | 145.9 | 7.238** |
| M30 vs. M38 | 229 | 266 | 0.417 | 0.200 | 433.9 | −47.74** | 0.714 | 9.926 | 275.7 | 9.178** |
indicates probability of the type I error lower than 0.001. Critical values of t′ were calculated for four planned comparisons.
A and B serve as variables that should be substituted within each row for appropriate F or M assayed at 30°C or 38°C.
Degrees of freedom are not natural numbers because they were calculated according to Welsh's procedure to compensate for unequal sizes and variances of the compared samples (19).
Stationary Phase Density.
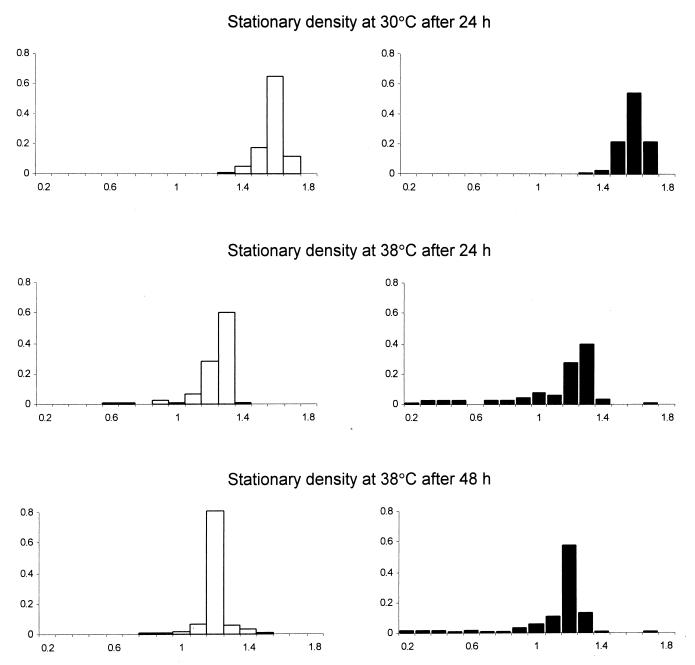
The optical density of the cultures was measured after 24 h of incubation at 30°C, and after 24 and 48 h of incubation at 38°C. An additional measurement in the higher temperature was necessary because some cultures did not finish their growth after the first day. The results are presented in Fig. 2 and Table 2. The densities of the F and M cultures were equal at 30°C. At 38°C, the cultures of M clones were less dense than that of F clones by 8% or 6% (measured after 24 or 48 h, respectively). This difference was smaller than in the case of the growth rate but remained statistically significant. Furthermore, the densities at the benign temperature were clearly higher than at the high one, indicating that the latter environment was indeed stressful. An interesting result was found when the opportunities for selection were compared. This measure of variation indicated that the F clones were not more variable than the M clones in the benign environment. Nor did the variation among F clones increase significantly in the harsh environment. The variation among M clones at 38°C did increase, when compared with the F clones at 38°C or with the M clones at 30°C. This effect was visible after both 24 and 48 h of incubation (Table 2). This indicates that the impact of high temperature was not restricted to postponing initiation of growth or slowing down its rate, but also halted growth of different mutation-contaminated clones at different densities.
Figure 2.
Frequency distributions of optical densities (OD600) of the stationary phase cultures of the control F clones (open bars) and the mutation-contaminated M clones (filled bars).
Table 2.
Means and opportunities for selection calculated for stationary phase densities of clones free of mutations (F) and contaminated with mutations (M)
| Pair
compared
|
Sample sizes
|
Mean stationary
densities,
OD600
|
df† | t′(B−A) | Opportunity
for selection,
×100
|
df† | t′(B−A) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A* vs. B* | A | B | A | B | A | B | ||||
| F30/24h vs. M30/24h | 127 | 229 | 1.537 | 1.545 | 204.9 | 1.119 | 0.198 | 0.227 | 291.3 | −0.475 |
| F38/24h vs. M38/24h | 127 | 228 | 1.179 | 1.081 | 350.6 | −4.907** | 0.773 | 7.184 | 246.5 | 3.621*** |
| F38/48h vs. M38/48h | 127 | 222 | 1.150 | 1.077 | 327.2 | −4.420** | 0.394 | 5.399 | 226.7 | 3.363*** |
| F30/24h vs. F38/24h | 127 | 127 | 1.537 | 1.179 | 219.4 | −33.04** | 0.198 | 0.773 | 131.1 | 2.075 |
| F30/24h vs. F38/48h | 127 | 127 | 1.537 | 1.150 | 250.3 | −43.65** | 0.198 | 0.394 | 149.4 | 1.463 |
| M30/24h vs. M38/24h | 229 | 228 | 1.545 | 1.081 | 255.7 | −25.37** | 0.227 | 7.184 | 227.3 | 3.975** |
| M30/24h vs. M38/48h | 229 | 222 | 1.545 | 1.077 | 258.2 | −29.65** | 0.227 | 5.399 | 221.4 | 3.486*** |
and *** indicate probability of the type I error lower than 0.001 and 0.01, respectively. Critical values of t′ were calculated for seven planned comparisons.
A and B serve as variables that should be substituted within each row for appropriate F or M, assayed at 30°C or 38°C, after 24 or 48 h. Note that all measurements at 30°C were done only after 24 h, whereas those at 38°C were measured also after 48 h due to slow growth at this temperature.
Degrees of freedom are not natural numbers because they were calculated according to Welsh's procedure to compensate for unequal sizes and variances of the compared samples (19).
Added New or Magnified Existing Effects?
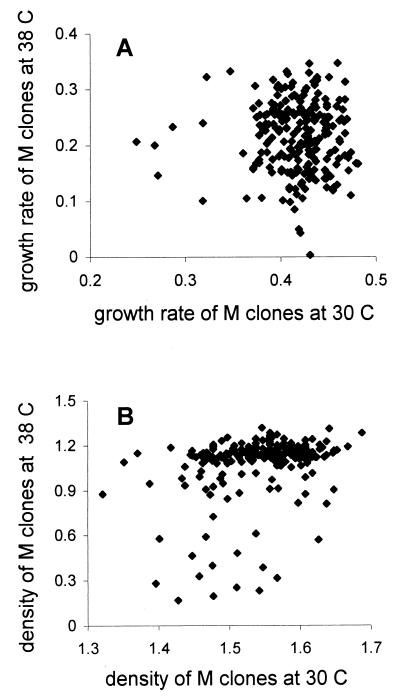
The higher decrease in fitness visible at 38°C can mean that either more mutations become deleterious or that the same mutations become more harmful under stress. If the latter were true, clones that were inferior under benign conditions would be even more so under stress. Fig. 3 shows how the fitness of mutation-contaminated (M) clones correlated between the benign and stressful environment. There was no correlation in growth rates between 30°C and 38°C (r = −0.011, t = −0.172, df = 227, P = 0.863) whereas such a correlation was statistically significant for stationary phase densities (r = 0.383, t = 6.152, df = 220, P < 0.0001). The correlation of the stationary densities was brought about by relatively few clones (around 10%), which were moderately affected at 30°C but became clearly inferior at 38°C (Fig. 3B). This is evidence that environmental stress magnified some deleterious effects. No such relation was visible in the case of growth rate (Fig. 3A) where many clones of average performance at 30°C became much more affected at 38°C. This suggests that these were mutations that were neutral under benign conditions but became deleterious under stressful ones. There may also have been clones that grew slower in both environments, but, if there were, there were too few to produce a significant correlation.
Figure 3.
Correlations of the growth rates (A) and stationary phase densities (B) for the M clones (bearing mutations) when tested in the benign and stressful environment.
Discussion
The constitution of organisms may be such that they are not visibly affected by a moderate load of small alterations in DNA. The present experiment indicates that this can indeed be the case when mutations occur in diploids, and therefore are masked by the wild-type alleles, and the environment is favorable. The performance of mutation-contaminated clones deteriorated, however, when they were exposed to a strong stress. A genetic load was evident in the unfavorable environment because the mean fitness decreased and its variation increased. This effect was observed both when the rate of growth after transferring the cultures to a stressful temperature was determined and when the final density in this environment was measured. The general decrease in fitness under stress was probably caused both by an increase in the number of mutations that were deleterious and a magnification of deleterious effect of the mutations.
It has been estimated that the mutation rate in wild-type yeast is about 0.003 per haploid genome per cell division (20). Data based on several mutational targets suggest that the mutation rate in mismatch repair-deficient strains can be enhanced by several hundred times (10). A similar factor applies here because the increase in mutation rate observed when the PMS1 plasmid was absent was indistinguishable from estimates obtained with other mismatch repair-deficient strains (21–23). However, it is another question how many of those mutations were non-neutral. A recent study suggests that, when mismatch repair is absent, the visibly harmful mutations arise at a rate of 0.06 per diploid cell division (D. M. Wloch, K.S., R.H.B., and R.K., unpublished data), of which 40% were lethal. Mutations with selection coefficients lower than 0.01 could not have been individually scored, but they were probably even more numerous. These estimates were done only at 30°C. In the present experiment, diploid cells underwent about 35–40 generations before the repair defect that facilitated mutation accumulation was restored. Thus, the strains were burdened with about one lethal and several deleterious mutations. It is remarkable how efficiently the deleterious and especially the lethal alleles were masked by their wild-type counterparts when environment was benign. A similar finding was made when viability of heterozygous mutants was analyzed in Drosophila (24).
The two environments applied in this study are much more different from each other than a mere difference of 8°C would suggest. The stressful temperature induces intense transcription of various shock response proteins that protect existing enzymes and membranes or participate in the repair or degradation of damaged proteins (25). The response is rapid because transcription of some heat shock-induced mRNAs peaks between 10 and 20 min after temperature shift (26) and is accompanied by a transient decrease in synthesis of many other proteins (27). It seems likely that an intense metabolic stress could uncover the damages caused by spontaneous mutations that were hidden when the cell was maintained in an optimal environment. This effect should be most visible when the cells were most challenged. This is probably directly after transferring stationary phase cells from 30°C to a fresh medium of 38°C, when the cells would have had to cope both with the thermal stress and the need to restart the metabolism required for growth. This relatively short period was not monitored. However, even in the following interval of a few hours, the difference in the rate of growth between the control and the mutation-contaminated clones was conspicuous. Fig. 1 shows that many mutation-contaminated clones grew considerably slower at the high temperature. There were mutations that were neutral for this trait in the benign conditions, but they became deleterious in the stressful ones, which resulted in a lack of correlation between the two environments. Some clones did not reach the stationary phase density even after 48 h, when any growth ceased (Fig. 2). Thus, the stress not only slowed down the startup of many clones but also prevented reaching maximum density by some of them. The minority of clones that were inferior even at the final measurement probably carried some serious defects that did harm even in the benign environment, although much smaller (Fig. 3B).
The general conclusion is that fitness of mutation-contaminated organisms could seriously decrease if environmental stress were frequently encountered. This is expected to be true not only for thermal stress but also for chemical agents, osmotic pressure, radiation, or starvation because the physiological response to those stresses has been shown to be similar to that of heat shock (28). The present work shows that a harsh environment can enhance genetic load conferred by preexisting mutations. Another recent study suggests that environmental stress may also enhance the rate at which mutations arise (29).
Hoffmann and Merilä (30) argue that understanding whether and how genetic variation changes under environmental stress is difficult because the history of the studied population is often unsure. For example, one cannot rule out the possibility that alleles deleterious to fitness in unfavorable conditions have been maintained in a population because of their hypothetical advantages in other habitats. In addition to antagonistic pleiotropy (31), purging selection (32, 33) and the ascendance of compensatory mutations (34, 35) can modify the spectrum of deleterious alleles when they are either accumulated in long-term experiments (36–40) or sampled from extant populations (41–44). This criticism can be applied, at least partly, to those of former experiments that did report enlargement of deleterious effects by harsh environmental conditions or intraspecific competition (40, 45, 46). In this experiment, propagation was only the number of generations to go from a single cell to a colony. This resulted in the accumulation of mutations that were “new” and represented a relatively unbiased sample of deleterious alleles. Thus, the present work shows unambiguously that stress can help to differentiate between genetically loaded and unloaded individuals. This was shown in the noncompetitive fitness assays, although also for yeast it has been shown that competition can help in uncovering differences in growth rate (47). Enlargement of genetic variation resulting from spontaneous mutation would lead to more efficient action of natural selection. Because the mutational pressure is omnipresent and the physiological response to environmental stress is similar in many organisms (48), the results of this study may be of general significance.
Acknowledgments
We thank A. S. Kondrashov and two anonymous reviewers for helpful comments, and W. Kramer for providing the pWBK3 plasmid. This study was supported by The Wellcome Trust.
Abbreviations
- SC
synthetic complete medium
- M
mutation-contaminated
- F
new mutation-free
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021390798.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021390798
References
- 1.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch M, Blanchard J, Houle D, Kibota T, Schultz S, Vassilieva L, Willis J. Evolution (Lawrence, Kans) 1999;53:645–663. doi: 10.1111/j.1558-5646.1999.tb05361.x. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann A A, Parsons P A. Evolutionary Genetics and Environmental Stress. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 4.Kawecki T J. Evolution (Lawrence, Kans) 1997;51:1751–1763. doi: 10.1111/j.1558-5646.1997.tb05099.x. [DOI] [PubMed] [Google Scholar]
- 5.Kawecki T J, Barton N B, Fry J D. J Evol Biol. 1997;10:407–429. [Google Scholar]
- 6.Keightley P D, Eyre-Walker A. Genetics. 1999;153:515–523. doi: 10.1093/genetics/153.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korona R. Evolution. 1999;53:1966–1971. doi: 10.1111/j.1558-5646.1999.tb04577.x. [DOI] [PubMed] [Google Scholar]
- 8.Korona R. Genetics. 1999;151:77–85. doi: 10.1093/genetics/151.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crow J F. Proc Natl Acad Sci USA. 1997;94:8380–8386. doi: 10.1073/pnas.94.16.8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouse G F. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Totowa, NJ: Humana; 1988. [Google Scholar]
- 11.McCusker J H, Perlin D S, Haber J E. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer W, Kramer B, Williamson M S, Fogel S. J Bacteriol. 1989;171:5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herskowitz I, Jensen R E. In: Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. [PubMed] [Google Scholar]
- 14.Sarkar S, Ma W T, Sandri Gv H. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- 15.Stewart F M. Genetics. 1994;137:1139–1146. doi: 10.1093/genetics/137.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–354. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 17.Crow J F. Hum Biol. 1958;30:1–13. [PubMed] [Google Scholar]
- 18.Sokal R R, Rohlf F J. Biometry. 3rd Ed. New York: Freeman; 1995. [Google Scholar]
- 19.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice–Hall; 1999. [Google Scholar]
- 20.Drake J W. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson M S, Game J, Fogel S. Genetics. 1985;110:609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz B A, Ramachandran K, Vonarx E J. Genetics. 1998;148:1491–1505. doi: 10.1093/genetics/148.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 24.Crow J F. Genetics. 1979;92:s165–s172. [PubMed] [Google Scholar]
- 25.Craig E A, Gambill B D, Nelson R J. Microbiol Rev. 1993;57:402–411. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner-Washburne M, Becker J, Kosic-Smithers J, Craig E A. J Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller M J, Xuong N-H, Geiduschek E P. J Bacteriol. 1982;151:311–316. doi: 10.1128/jb.151.1.311-327.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig E A. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 29.Goho S, Bell G. Proc R Soc London Ser B. 2000;267:123–129. doi: 10.1098/rspb.2000.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann A A, Merillä A. Trends Ecol Evol. 1999;14:96–101. doi: 10.1016/s0169-5347(99)01595-5. [DOI] [PubMed] [Google Scholar]
- 31.Rose M R. Heredity. 1982;48:63–78. [Google Scholar]
- 32.Wang J L, Hill W G, Charlesworth D, Charlesworth B. Genet Res. 1999;74:165–178. doi: 10.1017/s0016672399003900. [DOI] [PubMed] [Google Scholar]
- 33.Willis J H. Evolution (Lawrence, Kans) 1999;53:1678–1691. doi: 10.1111/j.1558-5646.1999.tb04553.x. [DOI] [PubMed] [Google Scholar]
- 34.Crill W D, Wichman H A, Bull J J. Genetics. 2000;154:27–37. doi: 10.1093/genetics/154.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore F B G, Rozen D E, Lenski R E. Proc R Soc London Ser B. 2000;267:515–522. doi: 10.1098/rspb.2000.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukai T. Genetics. 1964;50:1–19. doi: 10.1093/genetics/50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kibota T T, Lynch M. Nature (London) 1996;381:694–696. doi: 10.1038/381694a0. [DOI] [PubMed] [Google Scholar]
- 38.Keightley P D, Caballero A. Proc Natl Acad Sci USA. 1997;94:3823–3827. doi: 10.1073/pnas.94.8.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilieva L L, Lynch M. Genetics. 1999;151:119–129. doi: 10.1093/genetics/151.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shabalina S A, Yampolsky L, Yu, Kondrashov A S. Proc Natl Acad Sci USA. 1997;94:13034–13039. doi: 10.1073/pnas.94.24.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlesworth B, Charlesworth D, Morgan M T. Nature (London) 1990;347:389–392. [Google Scholar]
- 42.Johnston M, Shoen D J. Science. 1995;267:226–229. doi: 10.1126/science.267.5195.226. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez J, Lopez-Fanjul S. Genetics. 1996;143:829–837. doi: 10.1093/genetics/143.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng H-W, Lynch M. Genetics. 1997;147:147–155. doi: 10.1093/genetics/147.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondrashov A S, Houle D. Proc R Soc London Ser B. 1994;258:221–227. doi: 10.1098/rspb.1994.0166. [DOI] [PubMed] [Google Scholar]
- 46.Vassilieva L L, Hook A M, Lynch M. Evolution (Lawrence, Kans) 2000;54:1234–1246. doi: 10.1111/j.0014-3820.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 47.Thather J W, Shaw J M, Dickinson W J. Proc Natl Acad Sci USA. 1998;95:253–257. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mager W H, de Kruijff A J J. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]