Abstract
The 3′ nontranslated region (NTR) of the hepatitis C virus (HCV) genome is highly conserved and contains specific cis-acting RNA motifs that are essential in directing the viral replication machinery to initiate at the correct 3′ end of the viral genome. Since the ends of viral genomes may be damaged by cellular RNases, preventing the initiation of viral RNA replication, stable RNA hairpin structures in the 3′ NTR may also be essential in host defense against exoribonucleases. During 3′-terminal sequence analysis of serum samples of a patient with chronic hepatitis related to an HCV1b infection, a number of clones were obtained that were several nucleotides shorter at the extreme 3′ end of the genome. These shorter 3′ ends were engineered in selectable HCV replicons in order to enable the study of RNA replication in cell culture. When in vitro-transcribed subgenomic RNAs, containing shorter 3′ ends, were introduced into Huh-7 cells, a few selectable colonies were obtained, and the 3′ terminus of these subgenomic RNAs was sequenced. Interestingly, most genomes recovered from these colonies had regained the wild-type 3′ ends, showing that HCV, like several other positive-stranded RNA viruses, has developed a strategy to repair deleted 3′ end nucleotides. Furthermore, we found several genomes in these replicon colonies that contained a poly(A) tail and a short linker sequence preceding the poly(A) tail. After recloning and subsequent passage in Huh-7 cells, these poly(A) tails persisted and varied in length. In addition, the connecting linker became highly diverse in sequence and length, suggesting that these tails are actively replicated. The possible terminal repair mechanisms, including roles for the poly(A) tail addition, are discussed.
Following entry into a host cell, positive-strand RNA viruses produce several viral proteins, including an RNA-dependent RNA polymerase (RdRp), which is responsible for the production of progeny genomic RNA. RNA viruses have developed several strategies to ensure that sufficient amounts of each template are synthesized and that replication starts at the extreme ends. Specific cis-acting RNA motifs on the 3′ end of both the plus and minus strands play critical roles in directing the viral replication machinery to initiate at the 3′ ends (9, 11, 17, 34).
For hepatitis C virus (HCV), a positive-strand RNA virus of the Flaviviridae family, the 3′ nontranslated region of the genome (3′NTR) consist of three subregions, i.e., the variable region, the poly(U/UC) region, and the 3′ X-tail (Fig. 1A). First, directly following the open reading frame, the 3′NTR commences with a short variable region (approximately 40 nucleotides [nt]) that is poorly conserved between different HCV genotypes (40). Next to this 3′ variable region is a polyhomopolymeric (U/UC) tract of variable length and composition containing mainly uridines with occasionally interspersed cytidine residues. Directly downstream of the internal poly(U/UC) tract is the extreme 3′ end of 98 highly conserved bases, which is designated 3′ X-tail or core element (19, 35). The secondary structure of this X-tail was defined by enzymatic and chemical approaches and consists of a 46-nt long stem-loop structure at the extreme 3′ end (SL1) (2). The upstream 52 nt of the X-tail appear to be less ordered both in secondary structure predictions as well as in chemical probing experiments (2, 40), suggesting that there is no obvious RNA structure. Deletion of the 3′ X-tail renders the genomic RNA noninfectious in naive chimpanzees, demonstrating that this RNA element is critical for productive in vivo replication(20, 41). Also, in the tissue culture HCV replicon system, deletion studies demonstrated that each element in the X-tail is required for efficient RNA replication (8, 42).
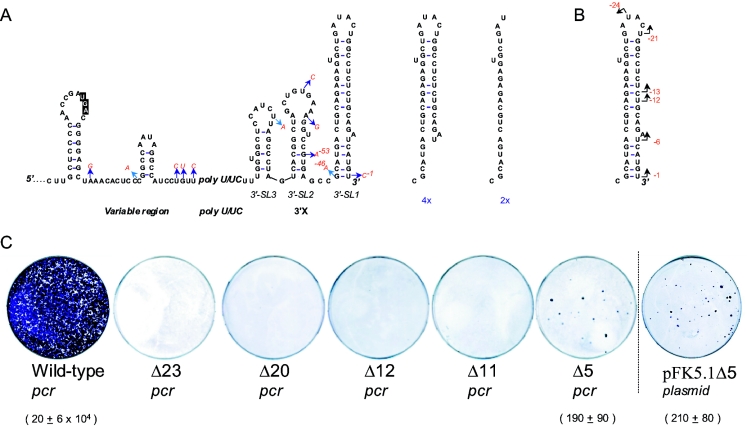
FIG. 1.
(A) Secondary structure of the 3′NTR and quasispecies therein derived from the serum of an HCV1b-infected patient. A total of 40 cDNA clones were sequenced: 29 cDNA clones corresponded to the WT sequence, and 11 individual cDNA clones contained single nucleotide changes indicated by arrows, followed by the nucleotide substitution (red). On the right side of the panel, four independent clones lacked five extreme 3′-end nucleotides, and two clones lacked 23 extreme 3′-end nucleotides. Note that the numbering starts at the last 3′ nucleotide, annotated with “−1,” and counts back. (B) The location of reported potential initiation sites is indicated by an arrow in SL1 (17, 28; the present study). (C) Effect of 3′-end deletions on cell colony formation using selectable replicons. WT is the pFK5.1 (Con1) construct amplified using T7 primer and reverse primer corresponding to the 3′ end (see the text for further details). Numbers below the plates refer to the CFU per microgram of in vitro-transcribed replicon RNA.
Like other Flaviviridae, the HCV RdRp, NS5b, is able to initiate RNA synthesis without the need for a primer in a de novo RNA synthesis assay and requires only a single nucleotide for priming (25, 28, 44). A possible function for the extreme 3′ stem-loop structure of HCV may be to recruit the replication complex and to ensure de novo initiation at the very 3′ terminal end (28, 32). Incorrect initiation of minus strands will lead to inadvertent loss of genetic information. Furthermore, if viral genome 3′ ends are exposed to cellular exonucleases, deletion of 3′ nucleotides could prevent viral RNA replication. Stable 3′ stem-loop structures, therefore, may be essential in viral defense against host nucleases, by acting as a barrier against exoribonucleolytic trimming (30). Alternatively, RNA viruses could escape nuclease degradation via sequestering replicating RNA in virus-induced membranes structures (36), covalent linkage of amino acids (7), or polyadenylation of the 3′ end (6, 29).
We describe here an HCV1b variant, with a deletion of 5 nt in the 3′-terminal end, that is repaired in subgenomic HCV replicons when introduced in human hepatoma Huh-7 cells. This suggests that there is a selective pressure for the restoration of truncated ends. Analysis of the restored 3′ ends further revealed that in the repair process a subpopulation of the genomic RNA was polyadenylated at the 3′end and contained an additional linker sequence between the 3′NTR and poly(A), which is atypical for Flaviviridae but reminiscent of Sindbis virus genome termini.
MATERIALS AND METHODS
Cell culture.
Huh-7 hepatocellular carcinoma cells were maintained in Dulbecco modified Eagle medium (Cambrex, Belgium) supplemented with 10% heat-inactivated fetal calf serum (PAA Laboratories), 2 mM l-glutamine, and 1% nonessential amino acids (Invitrogen). For cell lines carrying HCV replicons, 500 μg of G418 ml−1 was added (Geneticin; Life Technologies).
HCV 3′NTR amplification.
3′-End determination of serum extracted HCV RNA was performed as previously described (19). In brief, a 3′-blocked and 5′-phosphorylated oligonucleotide 760 (GACTGTTGTGGCCTGCAGGGCCGAATT) was ligated to the 3′ end of the HCV genome by using T4 RNA ligase (Invitrogen). From this ligation reaction, cDNA was made by using an antisense primer (TAATTCGGCCCTGCAGGCCACAACA) partially complementary to the ligated oligonucleotide 760 using Thermoscript reverse transcriptase (Invitrogen). First-strand cDNA was amplified by using the primers 799 (9246′-TGGTTCGTTGCTGGTTACAGC′-9266; forward primer) and 758 (AATTCGGCCCTGCAGGCCACAACAGTC; reverse primer), complementary to the ligated oligonucleotide 760, in the Expand High Fidelity PCR system (Roche). The resulting band of approximately 360 bp was cloned into the pCR2.1-Topo vector (Invitrogen) and then sequenced using T7forward and M13rev primers; Invitrogen) in both directions with an ABI Prism 310 Genetic Analyzer (Applied Biosystems) and the Dye Dideoxy terminator sequencing kit (Applied Biosystems). For determination of 3′ ends from replicon cells extracted RNA, an identical procedure was followed except the forward primer 799 was replaced with 800 (9504′-CTTTGGTGGCTCCATCTTAG′-9523).
Plasmid constructions.
The replicon vector used to test 3′NTR variants was pFK-I389neo/NS3-5/5.1 (pFK5.1 [21]). Shorter and longer 3′NTR replicons were generated by using the cloned 3′NTR cDNA pCR2.1-Topo plasmids (see above) as a template for PCR amplification. The forward primer for all of these PCR products was primer 800 (see above). The reverse primers were as follows: for construct pFK5.1Δ5, primer 1158 (CCACTAGTGATATCTGCAGAGAGGCCAGTATC′-9580), for pFK5.1C-5U, primer 1222 (GGGACTAGTAGTACTTAATCTGCAGAGAGGCCAGC′-9584); and for pFK5.1pA57 and pFK5.1pA65, primer 1250 (CGGACTAGTTTCGGCCCTGCAGGCCACAACAGTACTTTTTTTTTTTT). The resulting 3′NTR PCR products were BmtI (nt 9536 in the HCV genome) × SpeI (underlined in the oligonucleotides) cloned into a shuttle vector consisting of pBluescript(−) (Stratagene) containing XhoI (nt 7185)-SpeI fragment of pFK5.1. The BmtI-SpeI fragments generated by PCR were sequenced upon cloning. Next, the XhoI-SpeI fragment of the pBluescript(−) shuttle vector, containing the new 3′NTR, was cloned into pFK5.1 digested with XhoI-SpeI.
T7 PCR runoff transcription.
In order to create a transcription runoff sites for the generation of selectable replicon RNAs with various 3′ ends, we used a T7 PCR runoff strategy using the pFK5.1 plasmid as a template for PCR amplification. After heating at 94°C for 2 min, 5 ng of pFK5.1 was amplified by 30 cycles (30 s at 94°C, 30 s at 58°C, and 8 min at 68°C) of PCR by using Expand Long Template PCR System 3 (Roche Applied Science). Reaction mixtures (50 μl) contained 300 nM T7for primer (GCTAAGCTTCGTAATACGACTC) and 3′ runoff reverse primer corresponding to the last nucleotide, wild type (9605-′ACTTGATCTGCAGAGAGGCCAGTATCAGCACTC′-9573), Δ23 (9582-′ATCAGCACTCTCTGCAGTCAAGCGG′-9558), Δ20 (9585-′AGTATCAGCACTCTCTGCAGTCAAG′-9561), Δ12 (9593-′GAGAGGCCAGTATCAGCACTCTCTG′-9569), Δ11 (9594-′AGAGAGGCCAGTATCAGCACTCTC′-9571), and Δ5 (9600-′ATCTGCAGAGAGGCCAGTATCAGCAC′-9575).
Reactions typically yielded between 4 and 8 μg of an 8-kb PCR product. After PCR amplification, 10 U of DpnI restriction enzyme was added to the PCR, followed by incubation for 1 h to digest the methylated parental pFK5.1 plasmid. After digestion DpnI was heat inactivated for 20 min at 80°C, and the reaction mixture was loaded onto a 0.7% agarose gel (0.5× TAE) and run for 1 h at 10 V/cm. The 8-kb PCR product was subsequently extracted by using the QIAEX II agarose gel extraction protocol (QIAGEN). Eluted DNA was subsequently used in a T7 in vitro transcription reaction.
In vitro transcription, electroporation, and selection of selectable replicon cells.
In vitro transcription, electroporation, and selection of G418-resistant cell lines was done as described by Lohman et al. (24). The only adjustments were the restriction sites used for runoff transcription (see Results).
RESULTS
Determination of 3′NTR quasispecies variants from an HCV genotype 1b clinical isolate.
We have determined the 3′-terminal sequence of an HCV1b genotype derived from the serum of a patient with chronic hepatitis (37) by ligation of a synthetic oligonucleotide to the 3′ end, followed by reverse transcription-PCR (RT-PCR), cDNA cloning, and subsequent sequencing analysis as described in Materials and Methods. This procedure was performed in duplicate on separate serum RNA extractions, and 20 cDNA clones were analyzed from each sample. When the consensus sequence of the 40 clones was compared to the 3′NTR of other HCV type 1 genomes characterized, no differences were found outside the poly(U/UC) stretch. In 11 individual cDNA clones, single nucleotide changes were observed differing from the consensus (Fig. 1A). Most of these mutations (8 of 11) were within the predicted single-stranded regions of the 3′NTR. Of the remaining three mutations, one variant, A-53, abolished a predicted base pair, while the other two still retained base pairing to the adjacent residue (C-1 and A-46, Fig. 1A). RNA synthesis of HCV has been shown to initiate de novo from the terminal U nucleotide (−1) of SL1, but also a C residue at the ultimate position could direct correct RNA synthesis (4, 18).
Replication of truncated 3′ ends in selectable HCV replicons.
In addition to single nucleotide changes, four cDNA clones were obtained that were 5 nt shorter at the extreme 3′ end and two clones that lacked 23 nt at the 3′ end (Fig. 1A). It has been reported that in vitro minus-strand RNA synthesis can initiate internally either within the single-stranded loop of SL1 at position −21 (28) or within the stem of SL1 at position −12 or −13 (18). These internal initiation products, and the shorter 3′ termini that we observe, all have in common that they end with a pyrimidine base (Fig. 1B). This is consistent with observations that HCV RdRp preferentially initiates opposite uridine and cytidine residues and can use either ATP or GTP for de novo initiation of RNA synthesis (23, 25). Because the −12, −13, and −21 internal initiation products resulted from in vitro replication assays and the −23 and −5 variants we found in serum could be aberrant replication products, we tested these shorter variants in selectable HCV replicons, which enable the study of HCV RNA replication in cell culture (24). Since it was impossible to introduce correct restriction sites to create unique transcription runoff sites for the generation of selectable replicon RNAs, we used a T7 PCR runoff strategy using the HCV1b con1 replicon construct (pFK-I389neo5.1) (21). In short, a PCR amplification product was generated by using this replicon plasmid as a template, starting with a forward oligonucleotide primer complementary to the T7 transcription promoter and ending with an oligonucleotide reverse primer opposite to the last nucleotide of the truncated 3′NTR. After PCR amplification, the template DNA was removed with DpnI endonuclease, which specifically digests methylated DNA. The unmethylated ∼8-kb PCR product was subsequently purified by agarose gel electrophoresis and gel extraction. This purified PCR product was then used for T7 in vitro transcription reactions, producing selectable replicon RNAs with 3′ ends corresponding to the runoff site determined by the reverse primer. Human hepatoma Huh-7 cells were then electroporated with the different in vitro-synthesized RNAs and incubated for 3 weeks in the presence of 0.5 mg of G418/ml in the growth medium. Subgenomic RNAs missing 23, 20, 12, or 11 nucleotides at the 3′ end were unable to generate Geneticin-resistant colonies (Fig. 1C). However, colonies were observed for the 5-nt truncated RNAs, suggesting that this shorter variant could replicate (Fig. 1C). Despite the use of a high-fidelity polymerase during PCR amplification, extra unwanted mutations might have been introduced by the polymerase during production of the PCR template that consequently suppressed the actual efficiency of replication. Therefore, the same truncation was created by introducing a unique EcoRV restriction site at this position (GAT|ATC) to linearize the plasmid, circumventing the need for PCR amplification for this particular mutant. In vitro transcribed subgenomic RNA from the resulting plasmid (pFK5.1Δ5) was generated, and introduced into Huh-7 cells (Fig. 1C). Similar numbers of selectable colonies were obtained compared to the T7 PCR-runoff generated transcript (Δ5 PCR, Fig. 1C), showing that possible errors introduced during PCR amplification had little effect on colony formation.
Repair of truncated 3′ ends in selectable HCV replicons.
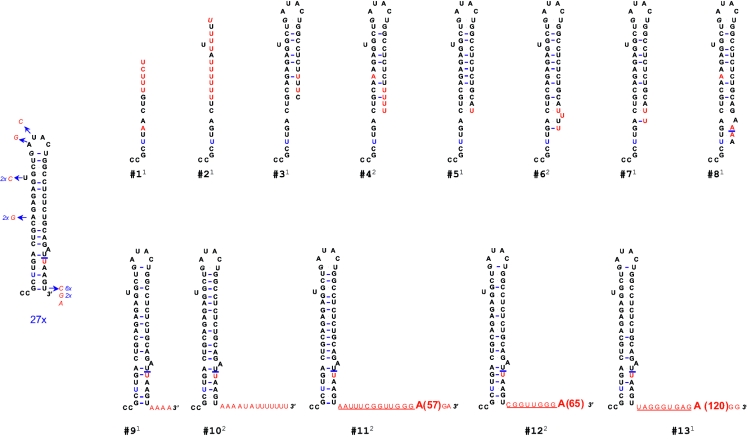
The surprising ability of the Δ5 replicon to form a small number of selectable colonies suggests that RNA replicated at low efficiency or that mutations have arisen to compensate for the defect. To address the latter possibility, we expanded two independent pFK5.1Δ5 replicon colonies and isolated total RNA, and the 3′ ends of replicon RNA were cloned and sequenced (Fig. 2). Interestingly, most replicons recovered from these pFK5.1Δ5 colonies (27 of 40, Fig. 2) had regained the missing 5 nt and thus restored base pairing in SL1 (Fig. 2). Restoration of these 5 nt indicates the existence of a repair pathway for truncated genomes in HCV selectable replicons. Compared to the wild type (WT) (pFK5.1) sequence, a U base instead of a C residue was found at position −5 in the two replicon colonies analyzed. Other cDNA clones retrieved showed truncated 3′ ends (sequences 1 to 8) or contained extra sequences (sequences 9 to 13, Fig. 2). Most shorter genomes (varying from 2 to 32 bases shorter) contained several U residues at their 3′ recessed ends. Of the genomes with an extended 3′ end, three contained an extremely long poly(A) tail (57, 65, and 120 A's, respectively) linked to the correct 3′ end with a short linker sequence (underlined in Fig. 2). How these U and A residues are added to the 3′ end is unclear, but these sequences must be added either by the viral polymerase (nontemplated terminal transferase activity) or by the cellular cytoplasmic polyadenylation machinery (see Discussion).
FIG. 2.
Terminal repair of a 5-nt 3′-truncated HCVcon1 replicon. The secondary structure of 3′-SL1 cDNA clones derived from two separate replicon colonies selected from the pFK5.1Δ5 plate (Fig. 1C) is shown. A total of 40 clones were sequenced (20 per colony). Arrows followed by the nucleotide substitution depict mutations found. Shown in red are nucleotides different from the consensus 3′-SL1 (pFK5.1). Underlined sequences indicate the linkers connecting the 3′ end and the poly(A) tail. Clone numbers are indicated under each sequence, and the annotations in superscript (1 or 2) indicate the colony number where it was found.
Effects of a 3′ terminal poly(A) tail on replicon cell colony formation.
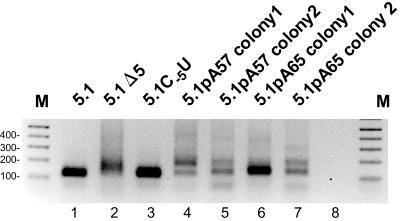
The fact that HCV replicon RNAs with truncated and extended 3′ ends exist in selectable replicons colonies (Fig. 2) suggests that they are either replication competent or complemented in trans by the restored WT replicons present in the same cell. To determine whether the HCV replicons benefit from the poly(A) tails gained in the repair process and to confirm that the replicons with a poly(A) tail are actively replicated, we tested two of these longer genomes in the selectable replicon system (pFK5.1_pA57 and pFK5.1_pA65; Fig. 3). Since a reverse oligo(dT) primer would not have the opportunity to bind to the extreme end of a long A stretch, the use of the T7 PCR runoff strategy to create in vitro transcripts with a long poly(A) tail was not possible. Therefore, we introduced one mutation or added two nucleotides for the pFK5.1_pA57 and pFK5.1_pA65 constructs, respectively, in order to create a ScaI restriction runoff site (AGT|ACT) (Table 1). Upon transfection of Huh-7 cells, both poly(A)-containing replicons formed selectable colonies, albeit with somewhat lower efficiency compared to the parental replicon (pFK5.1C-5U, Fig. 3). To determine further whether these replicon RNAs still contained their A-tail, two independent colonies were again expanded from each replicon. When the 3′ ends from these poly(A) replicon colonies were analyzed by RT-PCR and gel electrophoresis, two different-length PCR products could be observed for three of the four colonies: one corresponding to the length of the parental 3′ end and one approximately 50 bp larger (Fig. 4). One replicon colony showed only a single band corresponding to the parental 3′ end, suggesting that the poly(A) tail was lost (Fig. 4, pFK5.1_pA65 colony 1). As expected, both the pFK5.1 and the pFK5.1C-5U replicons only showed one band corresponding to the WT 3′ end. When cDNA clones of these colonies were sequenced, a surprising high sequence variability was observed (Table 1). The poly(A) stretch was shown to vary in length from 26 to 69 A residues and was thus, in some cases, longer than the original poly(A) tail. In a few clones, the poly(A) structure is interrupted by a single G residue (clones 57.19 and 57.24). Furthermore, the linker sequence connecting the 3′NTR and poly(A) tail appeared to be hypervariable in sequence and length. Only in a small number of clones the original linker sequence was retrieved (clones 57.2 to 57.5). Most newly formed linker domains appear GU-rich and varied in length by 5 to 16 nt. Consistent with the results of the RT-PCR analysis (Fig. 4), no poly(A) tail was observed for pFK5.1_pA65 colony 1 (Table 1), whereas for the other colonies only a small amount of the cDNA clones had lost their A-tails. Of these, several contained one to three extra U's at their 3′ ends (e.g., clones 57.32 and 57.33). These extra U's were also observed when cDNA clones derived from the WT 3′-end replicon colony were sequenced (pFK5.1C-5U, Table 1), and it is, therefore, unlikely that these added U's are remnants of the linker.
FIG. 3.
Colony formation of selectable replicons carrying a poly(A) tail. The top images show the secondary structures of 3′ SL1 structures including the poly(A) tail; the bottom images show colonies formed using replicons containing poly(A) tails (pFK5.1_pA57 and pFK5.1_pA65) and the WT 3′ end (pFK5.1C-5U). The numbers below the plates refer to the CFU per microgram of in vitro-transcribed replicon RNA.
TABLE 1.
Variants found in the 3′NTR poly(A)-tail of poly(A)-containing repliconsa
Twenty cDNA clones derived from two separate colonies selected from the pFK5.1_pA57, pFK5.1_pA65, and pFK5.1_C-5U plates (Fig. 3) were sequenced. Nucleotides different compared to the input selectable replicon RNA are indicated in red. Nucleotides introduced to generate a transcription runoff transcription site (ScaI) are indicated in blue. Clone numbers (#no.) are indicated next to the sequence.
FIG. 4.
3′-NTR PCR amplification products obtained from RNA isolated from various selectable replicon cell lines (indicated above the lanes) by ligation of synthetic oligonucleotides, followed by RT-PCR analysis. The “M” lane shows a 100-bp ladder (Invitrogen). For contrast purposes, a negative image of the ethidium bromide-stained agarose gel is shown.
DISCUSSION
Repair of viral 3′-truncated products generated during RNA replication could be crucial for the production of infectious progeny. The 3′ truncations of the viral RNA may arise due to viral imperfection either by internal initiation or premature termination of replication of the viral RdRp (18, 28). Also, in a hostile cellular environment exoribonucleolytic trimming of the 3′ termini could occur by the action of RNases. Several viruses have developed a strategy in which truncated products can be repaired in order to maintain functional full-length viral genomes (see references 3, 27, and 29). In the present study we describe an HCV genome variant that has a truncation of 5 nt at the 3′ end and demonstrate repair of this deletion in selectable HCV replicons. This novel finding strongly suggests that HCV possesses a 3′-end repair mechanism.
Based on in vitro RNA transcription studies, a model for 3′-end repair of the HCV terminus was suggested by Oh et al. (28). Because the 3′ stem-loop 1 has internal complementary sequences (see Fig. 1A), it was proposed that missing nucleotides might be added to the 3′ end of this self-priming hairpin. The recovered SL1 3′ ends we observe, however, all contain two G-U base pairs (UAAGU, see Fig. 2) in the restored RNA hairpin, whereas copy back of the recessed stem-loop by the RNA polymerase would produce complementary G-C base pairs (CAAGC), making the model of Oh et al. unlikely for the observed repair in our system.
Another possibility for 3′-end repair could be effectuated by abortive synthesis of internally initiated RNA molecules, which subsequently realign to the recessed 3′ ends to prime minus-strand synthesis. Such a repair mechanism was suggested for turnip crinkle carmovirus, a positive-strand RNA virus (27). Whether this mechanism is used in repair of HCV 3′ ends is unknown. One genome terminus we found in the poly(A)65 HCV replicon colony 1 contained an overlapping repeat of the last 9 nt of 3′ end (Table 1, clone 65.1, UUAAGUUUA), a finding suggestive of an abortive product reinitiating at the 3′ end.
A third route for repair of 3′-terminal deletions may be the ability of the RdRp to add nontemplated nucleotides to the 3′ end of its genome. In this model the terminal transferase activity, which is either an intrinsic property of NS5b or a cellular enzyme binding to NS5b (1, 23, 30, 30, 30, 32, 38), would elongate defective 3′ ends, which should then allow sufficient replication for further evolution in the direction of fitter genomes, containing the original 3′ end (30, 38). The identity of the nontemplated nucleotides added at the 3′ end by the polymerase may be determined by the genome 3′-terminal structure and/or sequence (30, 38). For the shorter genomes remaining after repair, mostly U-stretches were added to the genome termini (Fig. 2, clones 1 to 7), while the longer genomes present contained extra A's (clones 8 to 13). Not only recessed termini contained extra U's; WT 3′ ends also occasionally obtained one or two additional U residues (see Table 1 and reference 40). Interestingly, in vitro replication experiments by Oh et al. showed that the addition of single-stranded U's to the 3′ end of the X-template generated RNA products that initiated RNA synthesis from the 3′ end most single-stranded nucleotide (28). Furthermore, binding assays showed that purified RdRp binds homopolymeric U's very efficiently (23, 28), suggesting that the recessed 3′-end genomes containing extra U's we observed (Fig. 2) could be used as a template for replication and provide a basis for further evolution in the direction of a more efficient WT-like terminus. Besides shorter 3′ ends, which could represent intermediates in the repair process, several longer and polyadenylated genome termini were observed (Fig. 2). Replicons containing these poly(A) tails are >10-fold more efficient in colony formation than the Δ5 replicon from which they originated, showing that they benefit from this poly(A) tail. As for cellular mRNAs, a poly(A) tail connected to the HCV genome could contribute to genome stability and help to protect the 3′ end against nuclease degradation. The finding of poly(A) tails on the HCV replicon RNA is somewhat surprising since, in contrast to most other positive-strand viruses, flaviviruses generally lack a poly(A) tail. One exception thus far are certain tick-borne encephalitis virus strains, which contain a poly(A) tract that varies in length from 20 to approximately 200 nt as an internal part of the 3′NTR (26). The origin and selective advantage of this internal poly(A) tract is unknown.
For poly(A) tail-containing positive-strand RNA viruses it is proposed that the poly(A) region is used as a template to initiate minus-strand RNA synthesis opposite one of the multiple adenylates. Due to the variation of initiation sites within the poly(A) region, short stretches of A's of different lengths occur at the 3′ end of the newly synthesized progeny RNA, which are subsequently polyadenylated to restore the integrity of the genomic RNA (6, 13, 15, 29). Several different mechanisms are proposed to explain polyadenylation at the viral plus strand. Analogous to the polyadenylation of eukaryotic mRNAs, some plus-strand RNA viruses of plants contain a cis-acting polyadenylation signal (AAUAAA) in the 3′ noncoding region essential for binding of the cytoplasmic polyadenylation factors (39) and subsequent polyadenylation of the RNA genomes (6, 12). Since such a requisite signal is not present in the 3′NTR of HCV, the mechanism of addition of the poly(A) tail is likely to be different. As for HCV, a canonical polyadenylation signal is absent in members of the plus-strand Togaviridae and Coronaviridae families, which contain poly(A) tails of approximately 10 to 150 nt, which have been shown to be essential for viral replication (5, 15, 22, 29, 33). For Sindbis virus as well as for mouse hepatitis virus, removal of the poly(A) tail resulted in severely delayed infectivity and replication appeared to be mediated by restoration of the missing poly(A) tail (14, 22, 29). The mechanism of poly(A) addition to the progeny of these tail-less virus genomes is still unknown, but for these viruses the poly(A) tail clearly represents an essential cis-acting signal for replication.
An alternative mechanism for polyadenylation occurs when short A-tails, which are added by the action of terminal transferase activity (see, for example, Fig. 2, clones 8 and 9), are subsequently recognized and extended by the cytoplasmic poly(A) polymerase that normally extends cellular mRNAs in the cytoplasm. RNAs containing such short preformed A-tails do not require the AAUAAA polyadenylation sequence to form long poly(A) tails (31).
A third possible mechanism to gain extra sequences and a poly(A) tail is nonhomologous recombination with cellular messenger RNAs. Pestiviral genomes, which are closely related to HCV, frequently include insertions of cellular mRNA through nonhomologous RNA recombination (10). Addition of host-derived poly(A) tail containing mRNAs to a defective HCV genome through breakage and rejoining could render a replication-incompetent genome viable.
Finally, many (viral) polymerases have the inherent ability to stutter at homopolymeric sequences during duplication (43). The length of the poly(A) tails we observed might be influenced by slippage of any of the polymerases used to identify 3′ ends, being either T7 polymerase, reverse transcriptase, Taq polymerase, or the HCV RdRp itself. Unmistakably, poly(A) tails observed for selectable replicons longer than the input poly(A) tail must be synthesized by a slippage mechanism of one of the polymerases (Table 1).
The occurrence of the poly(A) tails coincided with the presence of novel short linker sequences connecting the poly(A) tail and the correct 3′ end (Fig. 2). A further unexpected feature of this linker sequence was that this motif turned out to be hypervariable in length and composition when submitted to another round of selection in replicon colonies (Table 1). Intriguingly, similar concomitant heterogeneous sequences linking the poly(A) tail and the 3′ end were reported during the terminal repair process of Sindbis virus and beet yellow vein virus genomes (13, 16, 29). For Sindbis virus a model was proposed in which internal bases adjacent to the poly(A) tail could be lost due to polymerase jumping events during viral minus-strand synthesis (29). A polymerase-jumping event may account for the internal deletion observed when the poly(A) linkers of clones 11 and 12 (Fig. 2) are compared, in which the linker of clone 12 seems to have lost part of the linker of clone 11 (underlined in Fig. 2). Similarly, jumping could explain the subsequent shortening of the linkers of clones 57.8 and 57.9 compared to the parental sequence (AAUUUCGGUUGGG > UCGGUUGGG > GUUGGG; Table 1). The hypervariable nature of the linkers, both in length and composition, suggests that there is little selective pressure on this sequence other than connecting the poly(A) tails to the genome 3′ end. Remarkably, when we examined the various 3′ ends for possible alterations in RNA secondary structure folds, using the web-based Mfold server (45), the observed poly(A) tail was predicted to fold back onto the poly(U/UC) stretch, forming a large double-stranded RNA stretch. Whether such a structure is really formed remains to be established. The fact that the poly(A) tail is maintained in the selectable replicons for the duration of expansion of a single electroporated cell to approximately 10 million cells suggests that the A's are actively duplicated. Clearly, whenever initiation takes place internally at the original 3′ end of the virus genome, the poly(A) tail is irreversibly lost. Of the four selectable replicon colonies we analyzed, one had lost its poly(A) tail, while of the other three colonies approximately half of the genomes still contained a poly(A) tail (Table 1). An additional role for the poly(A) tail could be increased stability or translatability of the HCV genome, ensuring longer survival. Increased stability or translatability could compensate for lower replication efficiency, ensuring longer survival. The unresolved question remains whether the poly(A) tails we observe are a functional part of the HCV replication or a strategy for 3′-end repair. Nevertheless, we have shown that HCV may be able to repair the loss of genetic information and is able to use alternative strategies to maintain the integrity of the viral genome.
Acknowledgments
This study was supported by EC grant QLK2-CT-2002-01329 to H.V.L. and Netherlands Organization for Scientific Research grant 901-02-099 from the Council for Medical and Health Research.
We thank Kelly Moran for critical reading of the manuscript and Fred Wassenaar for advice.
REFERENCES
- 1.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgyan, J., and F. Garcia-Arenal. 1998. Template-independent repair of the 3′ end of cucumber mosaic virus satellite RNA controlled by RNAs 1 and 2 of helper virus. J. Virol. 72:5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, Z. H., T. J. Liang, and G. X. Luo. 2004. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J. Virol. 78:3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, M. H., and T. K. Frey. 1999. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73:3386-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J. H., C. W. Peng, Y. H. Hsu, and C. H. Tsai. 2002. The synthesis of minus-strand RNA of bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. J. Virol. 76:6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreher, T. W., C. H. Tsai, and J. M. Skuzeski. 1996. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc. Natl. Acad. Sci. USA 93:12212-12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-Acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallei, A., M. Orlich, H. J. Thiel, and P. Becher. 2005. Noncytopathogenic pestivirus strains generated by nonhomologous RNA recombination: alterations in the NS4A/NS4B coding region. J. Virol. 79:14261-14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilford, P. J., D. L. Beck, and R. L. S. Forster. 1991. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of white clover mosaic potexvirus. Virology 182:61-67. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, R. W., and C. M. Rice. 2005. Requirements at the 3′ end of the Sindbis virus genome for efficient synthesis of minus-strand RNA. J. Virol. 79:4630-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, K. R., M. Hajjou, J. Y. Hu, and R. Raju. 1997. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 71:2693-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, M. A., and D. A. Brian. 1991. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J. Virol. 65:6331-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jupin, I., S. Bouzoubaa, K. Richards, G. Jonard, and H. Guilley. 1990. Multiplication of beet necrotic yellow vein virus RNA-3 lacking a 3′ poly(A) tail is accompanied by reappearance of the poly(A) tail and a novel short U-rich tract preceding it. Virology 178:281-284. [DOI] [PubMed] [Google Scholar]
- 17.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 18.Kim, M., H. Kim, S. P. Cho, and M. K. Min. 2002. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J. Virol. 76:6944-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, Y. J., C. L. Liao, and M. M. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 25.Luo, G. X., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandl, C. W., C. Kunz, and F. X. Heinz. 1991. Presence of poly(A) in a flavivirus: significant differences between the 3′ noncoding regions of the genomic RNAs of tick-borne encephalitis-virus strains. J. Virol. 65:4070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., C. D. Carpenter, and A. E. Simon. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. USA 94:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh, J. W., G. T. Sheu, and M. M. C. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 29.Raju, R., M. Hajjou, K. R. Hill, V. Botta, and S. Botta. 1999. In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of the Sindbis virus RNA genome: a novel 3′-end repair pathway. J. Virol. 73:2410-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheets, M. D., and M. Wickens. 1989. Two phases in the addition of a poly(A) tail. Genes Dev. 3:1401-1412. [DOI] [PubMed] [Google Scholar]
- 32.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, X. P., G. H. Zhang, and A. E. Simon. 2005. Short internal sequences involved in replication and virion accumulation in a subviral RNA of Turnip crinkle virus. J. Virol. 79:512-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchil, P. D., and V. Satchidanandam. 2003. Architecture of the flaviviral replication complex: protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388-24398. [DOI] [PubMed] [Google Scholar]
- 37.van Leeuwen, H. C., C. B. E. M. Reusken, M. Roeten, T. J. Dalebout, J. I. Riezu-Boj, J. Ruiz, and W. J. M. Spaan. 2004. Evolution of naturally occurring 5′ non-translated region variants of hepatitis C virus genotype 1b in selectable replicons. J. Gen. Virol. 85:1859-1866. [DOI] [PubMed] [Google Scholar]
- 38.Vo, N. V., J. R. Tuler, and M. M. C. Lai. 2004. Enzymatic characterization of the full-length and C-terminally truncated hepatitis C virus RNA polymerases: function of the last 21 amino acids of the C terminus in template binding and RNA synthesis. Biochemistry 43:10579-10591. [DOI] [PubMed] [Google Scholar]
- 39.Wahle, E., and U. Ruegsegger. 1999. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23:277-295. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 41.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi, M. K., and S. M. Lemon. 2003. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, H. Y., H. A. Lee, P. Palese, and A. Garcia-Sastre. 1999. Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J. Virol. 73:5240-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong, W. D., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]