Abstract
To identify molecular mechanisms controlling vein patterns, we analyzed scarface (sfc) mutants. sfc cotyledon and leaf veins are largely fragmented, unlike the interconnected networks in wild-type plants. SFC encodes an ADP ribosylation factor GTPase activating protein (ARF-GAP), a class with well-established roles in vesicle trafficking regulation. Quadruple mutants of SCF and three homologs (ARF-GAP DOMAIN1, 2, and 4) showed a modestly enhanced vascular phenotype. Genetic interactions between sfc and pinoid and between sfc and gnom suggest a possible function for SFC in trafficking of auxin efflux regulators. Genetic analyses also revealed interaction with cotyledon vascular pattern2, suggesting that lipid-based signals may underlie some SFC ARF-GAP functions. To assess possible roles for SFC in auxin transport, we analyzed sfc roots, which showed exaggerated responses to exogenous auxin and higher auxin transport capacity. To determine whether PIN1 intracellular trafficking was affected, we analyzed PIN1:green fluorescent protein (GFP) dynamics using confocal microscopy in sfc roots. We found normal PIN1:GFP localization at the apical membrane of root cells, but treatment with brefeldin A resulted in PIN1 accumulating in smaller and more numerous compartments than in the wild type. These data suggest that SFC is required for normal intracellular transport of PIN1 from the plasma membrane to the endosome.
INTRODUCTION
Vascular development provides a useful model for studying patterning. Vascular tissues arise early in organogenesis, and organ-specific patterns allow investigation of vein patterning in a variety of developmental contexts. In broad, flat organs, such as leaves and cotyledons, which are typically primary sites of photosynthesis, veins are dispersed to provide physiological support (water supply and sugar transport). Veins arise early in leaf development, and the first morphological indication of vascular fate specification is the appearance of elongated procambial cells (Esau, 1953). In the leaf, consecutive rounds of vascular recruitment lead to stereotypical patterns of veins of different size, with primary veins being large (diameter) veins that are recruited early, secondary veins recruited next (slightly narrower than primary veins and generally branching from the primary vein), followed by tertiary and quaternary veins (Turner and Sieburth, 2002). However, mechanisms that control leaf vein patterning are poorly understood.
One hypothesis for establishment of veins is canalization (Sachs 1981, 1989), which proposes that auxin acts as a self-amplifying transported signal that induces vein formation. The polar transport of auxin, which is controlled by directional efflux from cells (Blakeslee et al., 2005), was initially linked to xylem regeneration in studies of wound repair in stems (Jacobs, 1952). Since then, numerous studies using a variety of techniques have continued to link auxin to vascular development (Aloni, 2004). For example, in Arabidopsis thaliana, seedling growth on polar auxin transport inhibitors results in significant modification of leaf vein patterns (Mattsson et al., 1999; Sieburth, 1999). Furthermore, auxin-responsive reporter genes are expressed early in leaf development, in both morphologically unspecialized cells and in elongated procambial cells (Aloni et al., 2003; Mattsson et al., 2003). This observation is consistent with auxin flux occurring through cells destined to differentiate into procambium. Positions of auxin sources have been proposed to be both dispersed and dynamic to facilitate production of spatially distributed leaf veins via a canalization mechanism (Aloni, 2001). Nevertheless, how sites of leaf auxin synthesis and pathways for auxin flux are selected remains poorly understood.
Studies of auxin efflux have indicated a pivotal role for PIN proteins. There are eight members of the PIN gene family in Arabidopsis, five of which have defined functional roles in auxin-related processes (reviewed in Paponov et al., 2005). PIN1 and PIN2 were first implicated in auxin transport because pin1 and pin2/eir1/agr1 showed phenotypes consistent with tissue-specific defects in polar auxin transport (Bell and Maher, 1989; Okada et al., 1991; Luschnig et al., 1998), and both PIN1 and PIN2/AGR1/EIR1 expression and localization patterns are consistent with roles in polar auxin transport (Chen et al., 1998; Gälweiler et al., 1998; Luschnig et al., 1998). Auxin efflux also requires Arabidopsis Multidrug Resistance-like/P-glycoproteins (MDR/PGPs) (Noh et al., 2001; Blakeslee et al., 2005). PGP1 is sufficient to transport auxin in a heterologous system (Geisler et al., 2005), but precise roles for PIN and MDR/PGP gene products in polar auxin transport in planta remain to be elucidated. Nevertheless, localization of auxin efflux–related proteins, such as PINs and PGPs, is clearly critical for directional auxin movement
An important regulator of PIN protein localization is GN/EMB30, which is necessary for embryogenesis and later events in plant development (Mayer et al., 1993; Shevell et al., 1994; Geldner et al., 2003a). The gn/emb30 mutants have pleiotropic defects that have been linked to mislocalization of PIN1 (Steinmann et al., 1999). Treatment of plants with brefeldin A (BFA) causes PIN1 to accumulate in endosomes and revealed constitutive cycling of PIN1 between the plasma membrane and endosome (Geldner et al., 2001). GN/EMB30 was later shown to be a target of BFA and to function specifically in the movement of proteins (including PIN1) from the endosome to the plasma membrane (Geldner et al., 2003b). Another protein participating in PIN dynamics is BIG/TIR3/DOC1; mutants (tir3 and doc1) also show pleiotropic defects (albeit milder than gn/emb30) and mislocalize PIN1 following treatment with polar auxin transport inhibitors (Gil et al., 2001). PINOID (PID), which encodes a Ser-Thr protein kinase, has also been implicated in control of auxin efflux (Christensen et al., 2000; Benjamins et al., 2001). pid mutants produce phenotypes resembling pin1 (Bennett et al., 1995), and PIN1 localizes to incorrect cellular positions in pid mutants and in plants overexpressing PID (Friml et al., 2004). However, we do not know the targets of PID kinase nor how it controls PIN1 localization. In addition, the complex cell biology of vesicle trafficking suggests that many additional components required for PIN trafficking remain to be identified.
GN/EMB30 performs its role in PIN1 cycling through regulation of an ADP ribosylation factor (ARF). ARFs are small GTP binding proteins that function as molecular switches to regulate vesicle traffic. ARF-GTP is the active form, and it participates in formation of transport vesicles. ARF-GDP is the inactive form of ARF, and conversion of ARF-GTP to ARF-GDP is necessary for vesicle uncoating, a prelude for vesicle fusion with specific target membranes. ARF-GTP/ARF-GDP interconversion is regulated by two classes of proteins. ARF guanine exchange factors (GEFs) are positive regulators that cause ARF activation (ARF-GDP conversion to ARF-GTP). ARF GTPase activating proteins (GAPs) are negative regulators that activate latent GTPase activity of ARF, which then hydrolyzes the bound GTP (converting ARF-GTP to the inactive ARF-GDP). The large number of ARF, ARF-like, ARF-GAPs, and ARF-GEFs in the Arabidopsis genome (Vernoud et al., 2003) provides the possibility for highly regulated vesicle trafficking.
To gain insight into leaf vein patterning, we used genetic approaches to identify Arabidopsis leaf vein pattern mutants. One of these mutants, scarface (sfc), produces isolated vascular islands in the place of continuous and interconnected secondary and higher-order leaf and cotyledon veins (Deyholos et al., 2000). Disruption of auxin processes in sfc mutants was implicated because sfc mutant root elongation was hypersensitive to exogenous auxin, and we proposed that SFC functions as a negative regulator of auxin signaling. Here, we report identification of SFC as an ARF-GAP and present genetic, biochemical, and cell biological data implicating a role for SFC in membrane trafficking–based control of auxin efflux.
RESULTS
Intermediate and Weak sfc Alleles
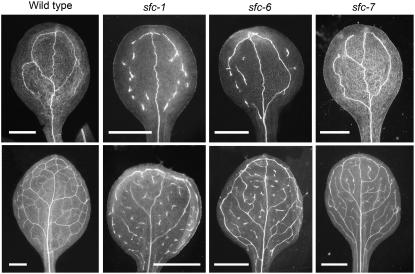
We previously described five phenotypically identical sfc alleles (sfc-1 through sfc-5) (Deyholos et al., 2000). sfc mutants are small, with epinastic cotyledons and leaves that also show distinct vein pattern defects. In contrast with the wild type, which produces organs with continuous veins (each vein connected to at least one additional vein), sfc-1 mutant leaves and cotyledons contain vascular islands (VIs), which are isolated fragments of vascular tissue (Figure 1; Deyholos et al., 2000). We found two new sfc alleles in our mutant collection (sfc-6 and sfc-7). Complementation tests indicated that these mutants were allelic to sfc-1, but they showed weaker vein pattern phenotypes (Figure 1).
Figure 1.
sfc Phenotypes.
Vein patterns in wild-type and representative sfc alleles. Cotyledons are shown at the top and one of the first leaf pair at the bottom. Bars = 1 mm.
We compared cotyledon vein pattern and root development in sfc-1, sfc-6, and sfc-7 mutants (Table 1). Root length, lateral root numbers, and cotyledon vein pattern features (numbers of areoles, VIs, and branch points) showed a clear trend, with sfc-1 having the most severe defects, sfc-6 intermediate defects, and sfc-7 the weakest defects. These data indicate that sfc-7, sfc-6, and sfc-1 represent weak, intermediate, and strong alleles, respectively. Furthermore, the correspondence between root phenotype severity and vein pattern severity suggests that SFC, either directly or indirectly, controls root development.
Table 1.
Cotyledon and Root Defects in sfc Alleles
| Cotyledon Vein Pattern
|
Root Growth and Development
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Areole Numbera | VIsb | Branch Pointsc | n | Root Length (day 5)d | Root Length (day 10)d | Lateral Roots (day 11) | n |
| sfc-1 | 0.0 | 13.4 | 2.3 | 29 | 5.2 | 11.1 | 0.95 | 22 |
| sfc-6 | 0.4 | 8.6 | 5.5 | 27 | 6.6 | 16.7 | 6.40 | 20 |
| sfc-7 | 1.4 | 2.3 | 4.4 | 28 | 10.2 | 40.1 | 10.80 | 27 |
| Ler | 3.1 | 0.0 | 6.4 | 20 | 16.5 | 69.0 | 21.60 | 27 |
Regions delimited by veins.
Vein segments isolated from other veins.
Sites where two or more veins intersect.
Primary root (mm).
SFC Encodes an ARF-GAP Protein
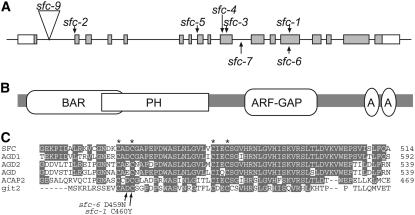
To identify the SFC gene, we mapped SFC to the upper arm of chromosome 5 (within BACs T31B5 and T22N19) and sequenced the genes within the mapping interval from our sfc mutants. Only gene At5g13300 contained mutations, and we identified a different lesion for each of our seven sfc alleles. We also obtained a line containing a T-DNA insertion within this gene (Alonso et al., 2003); this insertion resulted in a strong sfc phenotype, and the mutant (sfc-9) failed to complement sfc-1, confirming that we identified the correct gene (Figure 2A, Table 2). Strong alleles included two with splice site defects (sfc-2 and sfc-5), one nonsense allele (sfc-4), and a deletion leading to a frame shift (sfc-3). Two alleles had missense mutations; one missense mutation led to a strong phenotype (sfc-1), and the other missense mutation led to an intermediate phenotype (sfc-6). Finally, the weak sfc-7 allele had a lesion within intron 10 (a C-to-T at nucleotide 160 within this 335-nucleotide intron). At5g13300 was recently identified in the cloning of VAN3, which was also identified as an Arabidopsis mutant with vein pattern defects (Koizumi et al., 2000; Koizumi et al., 2005). This correspondence indicates that sfc is allelic to van3 and corroborates that we have identified the correct gene.
Figure 2.
SFC Molecular Characterization.
(A) Diagram of the SFC gene structure and positions of mutations. The 5′ end of this gene is at the left. Boxes indicate exons, gray portions of boxes correspond to translated regions, and white boxes are untranslated regions. Lines indicate either introns or 5′ and 3′ regions.
(B) Illustration of SFC domain organization. A, Ankyrin domain.
(C) Alignment of AGDs. The four critical Cys residues are indicated by an asterisk above the SFC amino acid sequence. Alignment includes the three Arabidopsis SFC-related ARF-GAP proteins, the human ACAP2 (centaurin β2), and a more distantly related mammalian GIT2 ARF-GAP. The changes in sfc missense mutations are shown below the alignment.
Table 2.
Lesions in sfc Alleles
| Allele | Lesion | Allele Strength | Lesion Site Relative to Protein Domains |
|---|---|---|---|
| sfc-1 | C460Y | Strong | ARF-GAP |
| sfc-2 | AG to AA, 3′ splice site intron 1 | Strong | BAR |
| sfc-3 | Deletion at codon 225a | Strong | Between BAR and PH |
| sfc-4 | Q209STOP | Strong | Between BAR and PH |
| sfc-5 | AG to AA, 3′ splice site intron 7 | Strong | BAR |
| sfc-6 | D459N | Intermediate | ARF-GAP |
| sfc-7 | C to T, intron 10 (160th nucleotide of a 335-nucleotide intron) | Weak | Between BAR and PH |
| sfc-9 | T-DNA insertion in intron 1 Salk_069166 | Strong | BAR |
Inserted amino acids: CRSTKDRWIERAGGVEMVLMDHRMEMAYKESVEALTKStop.
The SFC gene encompasses 17 exons, and it encodes a predicted 1344–amino acid protein. The protein contains four domains: a BAR domain, a PH domain, an ARF-GAP domain, and two Anykrin repeats (Figure 2B). This is the same domain organization as β-centaurin, an ACAP-type ARF-GAP (Randazzo and Hirsch, 2004). SFC and the human centaurin β2 share 27% amino acid identity and 50.5% similarity and are aligned over 85% of their length. BAR domains, especially in conjunction with an adjacent PH domain, have been implicated as membrane curvature sensors (Peter et al., 2004). PH domains bind to inositol phosphates and can confer binding to specific membranes (Lemmon et al., 1995; Falasca et al., 1998). The ARF-GAP domain activates latent ATPase activity of ARFs, which then leads to vesicle uncoating. Ankyrin domains confer binding to other proteins and as components of ARF-GAP proteins have been shown to bind actin (Randazzo et al., 2000).
A previous analysis of Arabidopsis small GTPase genes identified 15 ARF-GAP domain (AGD) proteins (AGD1 to AGD15) that fell into four classes (Vernoud et al., 2003). SFC corresponds to AGD3 and is one of four proteins belonging to class 1. We aligned the AGDs of SFC with those of the three other Arabidopsis class 1 AGD proteins and two animal AGDs (Figure 2C). AGDs are characterized by four invariant Cys residues, all four of which are present in SFC. Our two missense alleles have lesions within the AGD. In sfc-1, a Tyr is substituted for the second conserved Cys (C460Y). This residue is essential for function; thus, the strong sfc-1 phenotype is consistent with SFC functioning as an ARF-GAP. In sfc-6, an Asn is substituted for an Asp (D459N). This position shows less conservation than the Cys (Figure 2C); thus, this lesion is also consistent with the sfc-6 allele's intermediate phenotype.
Expression Patterns of Class 1 AGD Genes of Arabidopsis
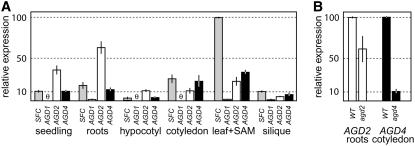
The Arabidopsis genome contains three additional genes encoding AGD proteins highly related to SFC. Amino acid alignments showed 63% amino acid identity (79% similarity) between SFC and AGD1 and 45% identity (67% similarity) between both SFC and AGD2, and SFC and AGD4. The similarity of these four genes raised the possibility that they could carry out partially redundant functions. To determine whether expression patterns of these genes overlapped, we compared expression using real-time RT-PCR and RNA isolated from a variety of tissues (Figure 3A). We detected expression of each of these four genes. In roots, RNA corresponding to all four genes was present; however, AGD2 transcript levels were much higher than SFC, AGD1, or AGD4. By contrast, in hypocotyl tissue, only low transcript levels for SFC, AGD2, and AGD4 were detected, and no AGD1 RNA was detected. AGD1 RNA was also not detected in cotyledons; however, for this tissue, SFC and AGD4 RNA levels were intermediate, and AGD2 RNA levels were low. We also detected RNA from all four genes in the leaf + shoot apical meristem samples; here, SFC RNA predominated, AGD1 was low, and AGD2 and AGD4 were intermediate. RNA extracted from mixed-age siliques also showed expression of all four AGD genes, but in this tissue, all four RNAs were present at low levels. The 12-d-old seedlings reflected the combined RNA levels detected in each 8-d-old tissue. The relatively high level of AGD2 probably reflects the abundant roots on these seedlings, and the absence of AGD1 is likely to reflect this gene's expression in only a few tissues or cell types.
Figure 3.
Real-Time RT-PCR Analysis of AGD1, AGD2, AGD4, and SFC Expression.
(A) Relative expression of class 1 ARF-GAP domain genes in RNA isolated from roots, hypocotyl, cotyledons, leaves plus meristem of 8-d-old (22°C-grown) seedlings, 12-d-old whole seedlings, and mixed-age siliques of mature Landsberg erecta (Ler) plants. Expression was normalized to β-tubulin and shown relative to the expression of SFC in RNA isolated from leaf and shoot apical meristem combined.
(B) Relative transcript levels in agd2 and agd4 mutants. RNA was isolated from 8-d-old roots (of the wild type and agd2 mutants) and 8-d-old cotyledons (of the wild type and agd4 mutants). Transcript levels were measured using real-time RT-PCR, normalized to β-tubulin, and are shown relative to expression of that gene (AGD2 or AGD4) in the wild-type tissue.
Light-gray bars represent SFC, dark-gray bars represent AGD1, white bars represent AGD2, and black bars represent AGD4. θ indicates no expression detected. Error bars represent sd (two to six experiments).
Functional Analysis of Class 1 AGD Genes of Arabidopsis
To assess functions of AGD1, AGD2, and AGD4, we characterized T-DNA insertion alleles (agd1, agd2, and agd4) (Alonso et al., 2003). The T-DNA insertions for agd1 and agd2 mutants fall in exon 3 and intron 11, respectively, while the T-DNA insertion for agd4 is 68 nucleotides upstream of the translational start site but within the 145-nucleotide 5′ untranslated region (cDNA accession number AJ130878) (see Supplemental Figure 1 online). To determine whether the T-DNA insertions into the AGD2 and AGD4 genes affected RNA abundance, we isolated RNA from agd2 and agd4 mutants and performed real-time RT-PCR analysis (Figure 3B). For analysis of agd2, we used RNA prepared from roots, where AGD2 expression is high. Primers used for this analysis were located 5′ of the T-DNA insertion, and we found reduced transcript levels (63%). For analysis of agd4, we used RNA prepared from cotyledons, where AGD4 expression is high. In this case, primers were located 3′ of the insertion. The agd4 mutant had strongly reduced RNA levels (10.4%). These analyses indicated that the agd2 and agd4 insertion alleles were at least hypomorphic, so we proceeded with our phenotypic analyses.
We used PCR to identify heterozygous and homozygous mutants and followed these plants through two generations. We identified no defects in agd1, agd2, or agd4 single mutants and no enhancement of sfc-9 in each of the three sfc agd double mutant combinations (data not shown). Additional crosses were performed to make multiple mutant plants, and enhancement of the sfc-9 phenotype was not observed until we had constructed the quadruple mutant.
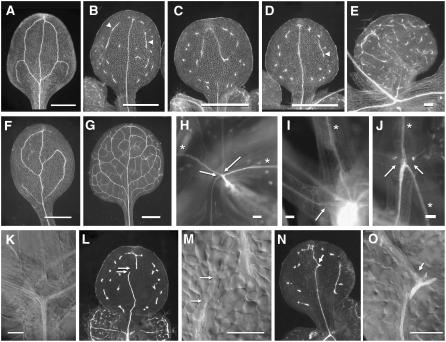
The agd1 agd2 agd4 sfc-9 quadruple mutant seedlings appeared similar to sfc single mutants (short roots, epinastic cotyledons and leaves, and accumulation of anthocyanins on the abaxial face of the cotyledon). However, quadruple mutant seedlings were slightly smaller and had shorter roots than sfc-9 single mutants (see Supplemental Figure 2 online; data not shown), and cotyledon and leaf vein pattern defects were modestly enhanced. agd1 agd2 agd4 sfc-9 mutant cotyledons contained many small VIs (Figures 4C and 4D) and fewer of the elongated VIs that are typical of sfc mutants (Figure 4B, arrowheads), although elongated VIs were not completely eliminated (Figure 4D, arrowhead). The most notable feature of the quadruple mutant, however, was the discontinuous cotyledon primary vein (Figure 4C). We quantified the cotyledon primary vein defect and considered the vein to be intact if it extended through the proximal 80% of the organ. We observed discontinuous primary veins in 36% of quadruple mutant cotyledons (n = 88), and many quadruple mutants showed a discontinuous primary vein in one cotyledon, while the other cotyledon's primary vein was intact. Among the quadruple mutants with intact primary veins, 4.5% showed regions where the primary vein was very thin, sometimes narrowed to a single file of tracheary elements (data not shown). In addition, 12.5% showed a zigzag like pattern (Figure 4N). Although included with the intact primary veins, close examination of these zigzag primary veins showed that the nonlinear portions were often not continuous (Figure 4O). In addition to the quadruple mutant phenotype, the agd1 agd2 agd4 triple mutants also showed a low penetrance (∼12%) cotyledon vein pattern phenotype (e.g., Figure 4F).
Figure 4.
Loss of AGD1, AGD2, and AGD4 Enhances sfc Vein Pattern Defects.
(A) Wild-type cotyledon vein pattern.
(B) sfc-9 cotyledon vein pattern. Arrowheads point to elongated VIs.
(C) agd1 agd2 agd4 sfc-9 quadruple mutant cotyledon. Note the loss of primary vein and small sized VI.
(D) agd1 agd2 agd4 sfc-9 quadruple mutant cotyledon; approximately half retain an intact primary vein. Arrowhead points to uncommon elongated VI.
(E) agd1 agd2 agd4 sfc-9 quadruple mutant leaf; compare with sfc leaf vein patterns in Figure 1. This leaf shows greater interruption of marginal vein network and smaller VIs than in sfc single mutants.
(F) agd1 agd2 adg4 triple mutant cotyledons occasionally had incomplete proximal and distal areoles.
(G) agd1 agd2 agd4 triple mutant leaf vein pattern is similar to the wild type.
(H) to (J) Connection of cotyledon and leaf primary veins with the stem/hypocotyl vasculature. Asterisks indicate cotyledon veins, and arrows point to the junctions between leaf primary veins and the stem.
(H) sfc-9 leaf veins always connect with the stem's vascular system.
(I) agd1 agd2 agd4 triple mutant leaf veins always connect to the stem's vascular system.
(J) agd1 agd2 agd4 sfc-9 quadruple mutant leaf veins failed to connect to the stem's vascular system.
(K) DIC image of agd1 ad2 agd4 sfc-9 quadruple mutant leaf vein base shows neither vascular nor procambial connections between the leaf primary vein and the hypocotyl.
(L) and (M) agd1 agd2 agd4 sfc-9 quadruple mutant with interrupted primary vein shown in dark field (L) and the region of primary vein disruption (M). Arrows indicate the extent of procambium that extended above the lower segment of the primary vein.
(N) and (O) Another agd1 agd2 agd4 sfc-9 quadruple mutant showing a zigzag primary vein (arrows).
Bars = 1 mm ([A] to [D], [F], and [G]) and 100 μm ([E], [H] to [K], [M], and [O]).
To determine whether the discontinuous primary veins resulted from defects in vascular differentiation or patterning, we used differential interference contrast (DIC) to examine areas of primary vein discontinuity for the presence of procambial cells. Among 32 cotyledons, 26 (81%) contained only one or two procambial cells extending beyond the differentiated tracheary element cells (e.g., Figure 4M). Six cotyledons contained longer procambial strands, but only one had procambium that was continuous with other vascular tissue (data not shown). These results strongly suggest that these four AGD genes are required to pattern the cotyledon primary vein. A defect in patterning, rather than differentiation, is also consistent with the expression (low) of all four class 1 AGD genes in mixed-age siliques, which would include developing embryos, but not in mature cotyledons (Figure 3A).
Leaves of the quadruple mutant also showed slightly less vein connectivity than sfc-9 single mutants, which was especially noticeable at the leaf margin (Figures 4E and 4L). In addition, the quadruple mutant's leaf primary vein failed to connect with the vascular tissue of the stem (Figures 4J and 4K). This lack of connectivity was only observed in quadruple mutants (observed in 88 of 88 leaves examined) and was never observed in sfc single mutants (Figure 4H) nor in agd1 agd2 agd4 triple mutants (Figure 4I). This phenotype has previously only been observed in plants grown in media containing polar auxin transport inhibitors (Mattsson et al., 1999; Sieburth, 1999). The agd1 agd2 agd4 triple mutant leaf vein pattern appeared mostly normal, although the leaf shape was occasionally asymmetric (Figure 4G). The similarity between the quadruple mutant's leaf vein pattern phenotype and leaves from polar auxin transport inhibitor–grown plants (Figure 3) suggests that these four genes may perform partially redundant functions in polar auxin transport during leaf development.
SFC and Other Leaf Vein Pattern Mutants
To determine the relationship between SFC and other genes implicated as playing a role in vein patterning, we characterized sfc-1 lop1-1 double mutants. lop1 mutants have misshapen leaves with greatly reduced vein patterns and a twisted root phenotype but a normal cotyledon vein pattern (Carland and McHale, 1996; see Supplemental Figure 3 online). sfc-1 lop1-1 double mutants produced cotyledons that resembled sfc-1 and leaves that showed a lop1-1 shape with the epinasty of sfc. The cotyledon vein pattern also resembled the sfc-1 single mutant. The leaf showed the reduced and disorganized vein pattern of lop1 with the fragmentation into the VI of sfc (see Supplemental Figure 3 online). This additive phenotype suggests that LOP1 and SFC function in independent pathways.
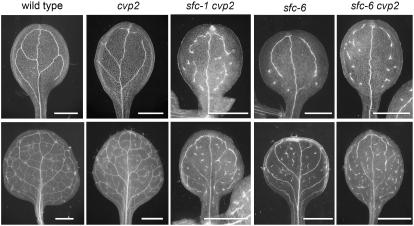
We also analyzed double mutants with cotyledon vein pattern2 (cvp2). CVP2 encodes an inositol polyphosphate 5′ phosphatase, and cvp2 mutants produce cotyledons and leaves lacking vascular connections at organ margins and between higher-order veins (Carland et al., 1999). An F3 line homozygous for cvp2 and segregating sfc-1 produced double mutants that were slightly smaller than sfc-1 single mutants alone (data not shown) and produced a vein pattern that was essentially the same as sfc-1 (Figure 5). To further explore possible sfc cvp2 interactions, we also characterized double mutants using the intermediate sfc-6 allele. The sfc-6 cvp2 double mutants were smaller than sfc-6 single mutants, and they showed a more severe vein pattern phenotype. The sfc-6 single mutant leaves contain intact primary and secondary veins, and higher-order veins are fragmented, whereas the sfc-6 cvp2 double mutant had fragmented secondary veins and many VIs (Figure 5). This enhancement of sfc-6 by the loss of cvp2 suggests that the products of these two genes might function in the same pathway.
Figure 5.
Vein Patterns in sfc cvp2 Double Mutants Show Enhanced Fragmentation.
Vein patterns of cotyledons (top row) and leaves (bottom row) of the genotype listed above the panel. Bars = 1 mm.
Auxin Response Is Intact in sfc Mutants
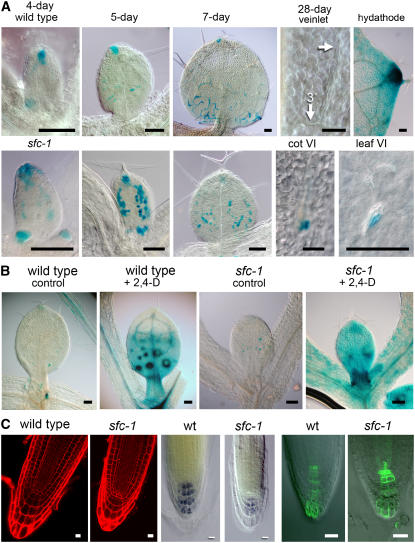
The major hypothesis to explain vein patterning, canalization of auxin flow, proposes that vein position is specified by paths of high auxin flux (Sachs, 1981). Mindful of this hypothesis, we considered that sfc's VIs could arise due to defects in auxin transport or defects in response to transported auxin. To sort out these possibilities, we characterized DR5:β-glucuronidase (GUS) expression patterns in sfc mutants. DR5:GUS contains a synthetic auxin-inducible promoter (Ulmasov et al., 1997), and expression is most commonly interpreted as indicating patterns of auxin accumulation in auxin-responsive cells, although induction by brassinolide has also been observed (Nakamura et al., 2003). Patterns of GUS staining conferred by DR5 in wild-type leaves and cotyledons have been well characterized (Aloni et al., 2003; Mattsson et al., 2003); DR5 confers GUS staining in leaf provascular and procambial cells, consistent with a positive role for auxin flux as an inductive signal for vascular identity. In sfc leaves, DR5 conferred GUS staining at the same developmental stage as the wild type; however, the sfc GUS staining was largely confined to isolated spots that corresponded to developing VIs (Figure 6A). Even in older organs (e.g., 24 d old), when DR5 no longer conferred GUS staining to veins of wild-type leaves but instead only to hydathodes, most sfc VIs (both cotyledon and leaf) continued to show strong GUS staining (Figure 6A). In these older sfc organs, cotyledon GUS staining was consistently at proximal positions of VIs, while leaf VI GUS staining was typically at one end of the VI but with no consistent proximal/distal or medial/lateral polarity.
Figure 6.
DR5:GUS Expression and Root Development in sfc Mutants.
(A) DR5:GUS expression in developing leaves of the wild type (top row) and sfc-1 mutant (bottom row). From the left are representative day 4, day 5, and day 7 leaves. The right two images of both rows show tissue from a 28-d-old plant. In the wild type (top), a leaf from a 28-d-old plant no longer has GUS staining associated with differentiated veins (arrow shows veinlet; 3° refers to a tertiary vein), but the same leaf shows strong GUS staining associated with hydathodes. In 28-d-old sfc-1 mutants, DR5:GUS staining is maintained in cells associated with VIs of both cotyledons and leaves. Bars = 0.1 mm.
(B) A 5-h treatment with 5 μm 2,4-D induces DR5:GUS staining in both the wild type and sfc mutants. Large spots in the wild type correspond to differentiating trichomes. Bars = 0.1 mm.
(C) Roots of the wild type and sfc mutants have similar cellular organization (propidium iodide–stained 6-d-old tissue to the far left), although the sfc cells are smaller and the root has a more blunt shape. Starch staining shows similar patterns of cell types (center, 6-d-old seedling), and DR5(rev):GFP (right, 6-d-old seedling) shows similar expression patterns. Bars = 10 μm.
The prolonged DR5 expression associated with sfc VIs suggested that auxin responses were intact. To test this, we examined auxin inducibility of DR5 in sfc and the wild type. For both the wild type and sfc mutants, exogenous auxin treatment induced strong expression in roots and leaves (Figure 6B). These data, combined with previous results indicating exaggerated auxin-induced inhibition of root elongation in sfc mutants (Deyholos et al., 2000), indicated that auxin responses are intact in sfc mutants.
SFC and Auxin Efflux Regulatory Components
To examine possible roles for SFC in auxin transport, we analyzed double mutants using pid-2. PID is a Ser-Thr protein kinase that affects polar positioning of the PIN1 auxin efflux regulator (Christensen et al., 2000; Benjamins et al., 2001; Friml et al., 2004). pid mutants show defects in cotyledon separation, flower morphology, and inflorescence structure (Bennett et al., 1995). We found that cotyledons of pid-2 plants showed occasional incomplete or ectopic areoles (Figures 7A, 7E, and 7F). We analyzed seedling and vein pattern phenotypes of F3 seedlings from homozygous pid-2 parents that segregated sfc-1. The double mutant seedlings and their vein patterns were variable; most double mutants resembled sfc-1 single mutants (68%, n = 76), but some showed additional defects in leaf initiation (21%; Figures 7C and 7H) or both root development and leaf initiation defects (11%; Figures 7D and 7I). We never observed these leaf or root phenotypes in either single mutant. Double mutants with more severe morphological defects also showed greater vascular interruptions. These observations suggest that PID and SFC might function in the same pathway.
Figure 7.
The sfc Phenotype Is Affected by Loss of Either PID or GN.
(A) to (I) sfc pid-2 double mutant analysis of 11-d-old seedlings ([A] to [D]) and cotyledon vein patterns ([E] to [I]).
(A) pid-2 homozygous mutant (note the three cotyledons).
(B) sfc-1 homozygous mutant.
(C) and (D) Two pid-2 sfc double mutants with enhanced phenotypes (reduced leaf development and root defects).
(E) and (F) Cotyledon vein patterns of the pid-2 homozygote shown in (A).
(G) sfc-1 cotyledon vein pattern.
(H) and (I) Cotyledon vein patterns of the seedlings shown in (C) and (D), respectively. Arrows in (D) and (I) point to the short, blunt root apex.
(J) to (Q) sfc gn4577 double mutant analysis of 11-d-old seedlings ([J] to [M]) and cotyledon vein patterns ([N] to [Q]).
(J) Wild-type control (Ler).
(K) sfc-1 single mutant.
(L) gn4577 single mutant.
(M) sfc-1 gn4577 double mutant.
(N) Wild-type cotyledon vein pattern.
(O) sfc-1 cotyledon vein pattern.
(P) gn4577 cotyledon vein pattern.
(Q) sfc-1 gn4577 double mutant cotyledon vein pattern.
Bars = 1 mm.
We also analyzed sfc gnom/emb30 double mutants. GNOM/EMB30 encodes an ARF-GEF that is necessary for vesicle trafficking between the endosome and the plasma membrane (Steinmann et al., 1999; Geldner et al., 2001). We first analyzed crosses between sfc-1 and the strong emb30-2 allele. F2 plants showed segregation at a 9:4:3 (wild type:gn/emb30:sfc) ratio. This ratio is consistent with the double mutant resembling gn/emb30, indicating epistasis of gn/emb30. To gain additional insight, we also characterized double mutants using gn4577, a weak allele identified in our mutant screen (L.E. Sieburth and M.K. Deyholos, unpublished data) that was similar to previously described weak gn alleles (Geldner et al., 2003b). gn4577 single mutants had a long unbranched root and generally a single large cotyledon with too many very thick and highly interconnected veins (Figures 7L and 7P). The gn4577 sfc-1 double mutant typically had two epinastic cotyledons (similar to sfc-1) and a long unbranched root similar to gn4577 (Figure 7M; data not shown). The gn4577 sfc-1 cotyledon vein pattern resembled that of sfc-1, except the sfc phenotype was modestly suppressed in that there were fewer VIs (Figure 7Q). The sfc-1 gn4577 double mutant also resembled that of gn4577, although the gn4577 phenotype was modestly suppressed in that veins were less thick, and had fewer ectopic tracheary elements, than in the gn4577 single mutant. A similar gn van3 double mutant phenotype was also reported recently (Koizumi et al., 2005). This double mutant phenotype suggested mutual suppression of each single mutant phenotype (including vein pattern, cotyledon fusion, and root length), and it suggested that the SFC ARF-GAP might function in a pathway opposing that of the GN/EMB30 ARF-GEF.
sfc Roots Show Altered Auxin Transport and Response
Because methods for measuring auxin transport in Arabidopsis leaves and cotyledons have not been developed, we used sfc-1 roots to analyze auxin transport and PIN1:green fluorescent protein (GFP) localization. The roots of sfc are shorter than the wild type (Table 1), and to explore whether this tissue was suitable for these analyses, we characterized sfc root cellular organization (Figure 6C). The arrangement of cells within the root meristem of sfc mutants was the same as the wild type, although the zone of starch-staining cells was often smaller. sfc roots also showed normal patterns of DR5:GUS and DR5(rev):GFP expression, indicating that there were no gross defects in the sfc mutants (Figure 6C; data not shown). In addition, SFC is expressed within the protoxylem of the stele, the same cells that show strong expression of PIN1 (Birnbaum et al., 2003). We also observed a modest vascular phenotype in sfc roots, tracheary elements differentiated much closer to the root apex than in wild-type roots (see Supplemental Figure 4 online), and sfc pid double mutants showed occasional severe root development defects. All these observations suggested that SFC might be required in roots, so we performed additional experiments to measure auxin transport.
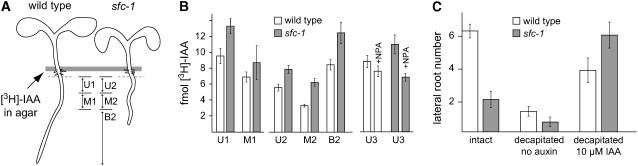
Our strategy for measuring auxin transport is outlined in Figure 8A. [3H]-indole-3-acetic acid (IAA) was applied at the root/shoot junction, and after 18 h, the amount transported was measured in 3-mm segments adjacent to the application site or further down the root. Surprisingly, all sfc root segments contained higher amounts of [3H]-IAA than the control (Figure 8B). These reported experiments used plants of slightly different ages (5 d old for Ler and 6 d old for sfc-1) to better match the root length; similar trends were observed with roots that were age matched but had a greater difference in root length (data not shown). We also examined the effect of napthylphthalamic acid (NPA) on polar IAA transport in the wild type and sfc-1. In the wild type, IAA transport was reduced by 10 μM NPA to the same extent as previously reported (Figure 8B; data not shown) (Rashotte et al., 2001). NPA also inhibited IAA transport in sfc. These data indicate that sfc mutant roots have a higher capacity for IAA uptake and carrier-mediated transport than roots of the wild type.
Figure 8.
sfc Roots Transport and Respond to Exogenous Auxin.
(A) Schematic diagram of auxin transport assays. The gray bar represents an agar block containing [3H]-IAA, which was placed at the root-shoot junction. The dashed line indicates that portion of the root that was discarded (2 mm). Lines between the two plants indicate the portions of the roots where [3H]-IAA was measured.
(B) Bar graph depicting acropetally transported [3H]-IAA. Tissue measured is diagramed in (A). The set to the left shows measurements of two root segments, while the center set depicts an independent measurement of three root segments. The measurements to the right show the influence of 10 μM NPA on acropetally transported [3H]-IAA in the upper segment of the wild type and sfc mutants. Similar results were obtained in the other segments. Bars indicate averages ± sd.
(C) Lateral root numbers of sfc and wild-type decapitated roots. The controls are nondecapitated (intact) plants; bars to the right are numbers for plants where an agar block containing no [3H]-IAA or 10 μM was applied at the cut site (root-shoot junction). Lateral root counts were made 3 d after auxin application. Bars indicate averages ± sd.
To determine whether the reduced numbers of sfc lateral roots was related to shoot-derived auxin, we quantified lateral root development on intact roots, roots decapitated to block auxin flow from the shoots, and decapitated roots supplied with exogenous auxin (Figure 8C). sfc-1 mutants produced fewer lateral roots than control plants, and removal of the shoot resulted in a decrease in lateral root production on both sfc-1 and Ler roots. However, application of exogenous auxin to the decapitated root resulted in a dramatic increase in lateral root production in sfc-1, more so than the wild-type control. These data are consistent with the lateral root phenotype resulting from decreased auxin movement from the shoot to the root in intact sfc-1. Moreover, these data indicate that the ability to deliver auxin is not impaired in sfc roots, thereby corroborating the measurements of auxin transport. Taken together, these data are consistent with reduced delivery of auxin to the roots of sfc-1 mutants but greater than wild-type capacity of roots to transport available auxin.
sfc Roots Show Altered PIN1 Cycling
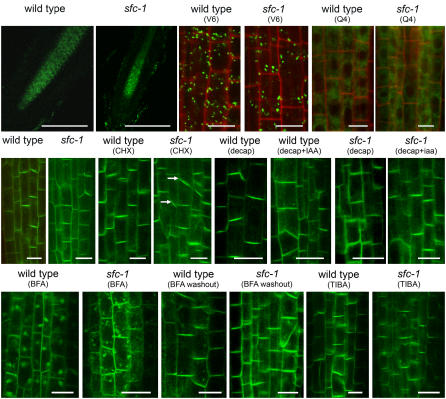
The molecular identification of SFC as an ARF-GAP, combined with observations suggesting possible roles for SFC in polar auxin transport and observations of sfc gn4577 genetic interactions, suggested that SFC might participate in PIN1 cycling. To test this, we characterized PIN1:GFP fusion protein dynamics in sfc roots using PIN1:GFP (Heisler et al., 2005). In the wild type, PIN1:GFP is abundant within the vascular stele from the root apex to the differentiation zone. By contrast, in sfc mutants, the PIN1:GFP signal was restricted to a narrow zone near the root apex (Figure 9). We do not believe this reflects gross defects in sfc roots, as DIC, confocal, and DR5 expression patterns were normal (Figure 6C). Furthermore, the cell tag markers V6 (streaming dots) and Q4 (endoplasmic reticulum surface) (Cutler et al., 2000) showed similar intracellular localization patterns in the wild type and sfc (Figure 9), suggesting that organization within sfc root cells was largely normal. Alternatively, the small domain of PIN1-GFP–expressing cells in sfc mutant roots could reflect altered regulation of PIN1 gene expression (e.g., due to different auxin levels; Peer et al., 2004) or premature differentiation of PIN1-expressing cells (see Supplemental Figure 3 online).
Figure 9.
Confocal Analysis of PIN1:GFP and Controls Reveals Cycling Defects in sfc Mutants.
The wild type and sfc mutants show similar patterns of V6 and Q4 controls (streaming dots and endoplasmic reticulum surface) and similar patterns of PIN1:GFP except following treatment with BFA. In sfc mutants, BFA treatment results in PIN1:GFP accumulation in many small compartments in contrast with the two to three large compartments in the wild type. Bars = 10 μm, except for the two top left low-magnification images of PIN1:GFP, in which bars = 100 mm.
Consistent with previous characterizations, we found PIN1:GFP targeted to the apical (lower) membrane of procambial cells in the stele of wild-type roots (Figure 9). sfc roots showed a similar pattern of PIN1-GFP accumulation, indicating that pathways for PIN1 synthesis and apical membrane targeting do not require SFC. Because PIN1:GFP was found in a smaller zone in sfc mutants, we considered it possible that PIN1 was less stable in sfc mutants. However, after incubating sfc and the wild type in the protein synthesis inhibitor cycloheximide for 4 h, the sfc and wild-type signal intensities were similar (Figure 9). This observation indicates that, in sfc mutants, PIN1:GFP was not overtly destabilized. These analyses also revealed occasional misoriented end walls in the protoxylem (see arrows in sfc-1 [CHX] in Figure 9), supporting a role for SFC in root development.
To determine whether the strong influence of decapitation and auxin supplementation on lateral root development in sfc mutants affected PIN1:GFP localization, we also performed confocal analysis on decapitated plants and decapitated roots incubated in IAA for 24 h (Figure 9). For both the wild type and sfc-1, decapitation and decapitation followed by IAA incubation resulted in no strong effect on PIN1:GFP localization.
To explore whether SFC was required for subcellular movement of PIN1, we compared localization of PIN1:GFP following incubation in BFA. Although BFA inhibits several different plant ARF-GEFs (Nebenführ et al., 2002), it specifically inhibits movement of PIN1 from the endosome to the plasma membrane, without affecting movement from the plasma membrane to the endosome (Geldner et al., 2003a). Consistent with previous studies, the wild type incubated in BFA led to PIN1:GFP accumulation in two to three large organelles (Figure 9) that have previously been identified as endosomes (Geldner et al., 2003a). By contrast, BFA treatment of sfc mutants resulted in PIN1:GFP localization in 5 to 11 smaller organelles in each cell (Figure 9). This result suggests that SFC is required for normal cycling of PIN1:GFP, possibly in the delivery of endocytosed vesicles to the endosome. Consistent with previous studies, we also found that washing out BFA in the wild type resulted in return of PIN1:GFP to its normal apical membrane (Figure 9). In sfc, BFA washout also resulted in return of PIN1:GFP to the apical membrane. To further assess PIN1:GFP cycling in sfc mutants, we compared the wild type and sfc treated with tri-iodo benzoic acid (TIBA), a polar auxin transport inhibitor. It was shown previously that TIBA treatment prevented movement of PIN1:GFP in response to BFA or BFA washout, suggesting that TIBA prevents cycling (Geldner et al., 2001). In the wild type, TIBA treatment did not affect PIN1:GFP localization (Figure 9). In sfc, most of the PIN1:GFP signal also remained at the plasma membrane, but some signal also accumulated in perinuclear regions (Figure 9). This result is consistent with no gross defect in PIN1 localization, and together with the BFA analyses, these studies strongly suggest that PIN1 cycling is altered in sfc roots. Note that these experiments were all performed using the sfc-1 allele; identical BFA responses were observed in sfc mutants in 15 independent F2 lines, and we never observed the altered BFA response in wild-type plants (phenotypically wild-type plants from the 15 lines segregating for sfc and in 12 F2 lines that did not segregate for sfc). These observations indicate that the altered BFA response resulted from the loss of sfc rather than an unlinked mutation.
DISCUSSION
Axial patterning of leaf veins is most commonly attributed to cellular pathways of auxin flux that are established during early leaf development. Here, we showed that SFC, which is required for axial development of cotyledon and leaf veins, encodes an ARF-GAP. ARF-GAPs are negative regulators of ARF activity in that they are required for ARF GTPase activity, which converts ARF-GTP into its inactive ARF-GDP form. ARF-GTP conversion to ARF-GDP is classically linked to vesicle uncoating, a necessary prelude for vesicle fusion with target membranes, although studies also implicate additional roles in vesicle coat formation and cargo sorting (Randazzo and Hirsch, 2004).
SFC is closely related to the animal ARF-GAP proteins ACAP1 and ACAP2 (centaurins β1 and β2) and is the same gene as was identified earlier this year as VAN3 (Koizumi et al., 2005). In animal cells, ACAP1 and ACAP2 are activated by phosphatidylinositol 4,5-bisphosphate, they affect the actin cytoskeleton, and they regulate ARF6 activity (Jackson et al., 2000). ARF6 functions in receptor-mediated endocytosis (D'Souza-Schorey et al., 1995), and dominant-negative forms of ARF6 cause recycling defects in which proteins normally recycled to the plasma membrane instead became trapped in vesicles (Brown et al., 2001). ACAP1 has also recently been shown to play a specific role in sorting cargo for recycling from the endosome (Dai et al., 2004). Our studies suggest that SFC/VAN3, an Arabidopsis ACAP-type ARF-GAP, also functions in cycling of plasma membrane proteins.
SFC Is Required for PIN1 Internalization in Roots
The sfc and gn4577 mutants produce nearly opposite vascular phenotypes (too few/too many cotyledon veins that are underconnected/overconnected). They also encode proteins of opposite functions; ARF-GEFs are positive regulators of ARF proteins, whereas ARF-GAPs are negative regulators of ARF proteins. The partial suppression of each single mutant's vascular defects in the gn4577 sfc-1 double mutant provides genetic support for SFC and GN/EMB30 performing opposing biochemical activities. GN/EMB30 is required for some proteins, including PIN1, to move from the endosome to the plasma membrane (Geldner et al., 2003b), and in gn mutants, PIN1 appears to reach the plasma membrane but in somewhat random patterns (Steinmann et al., 1999). There are two general ways in which the SFC/VAN3 ARF-GAP could function in a pathway opposing the GN/EMB30 ARF-GEF: SFC/VAN3 could negatively regulate the same ARF that is regulated by GN/EMB30, or SFC/VAN3 could regulate vesicle trafficking in the opposing direction (i.e., plasma membrane to endosome).
The membrane trafficking inhibitor BFA inhibits GN/EMB30 activity and results in PIN1 accumulation in endosomes (Geldner et al., 2003b). Thus, BFA treatment of PIN1:GFP-containing plants allows observations of PIN1:GFP internalization. This effect is reversible, as washout of BFA allows PIN1:GFP to return to its normal localization on the apical surface of root protoxylem cells. In sfc, BFA treatment resulted in PIN1:GFP accumulation in novel small-sized compartments (Figure 9), while washout of BFA allowed PIN1:GFP to return to its normal distribution. This observation corroborated the genetic evidence that SFC/VAN3 functions in a pathway opposing GN/EMB30 and indicated a role for SFC/VAN3 in endocytosis. We note that our observations imply SFC/VAN3 localization in close proximity to the plasma membrane, but a recent study showed that in protoplasts, 35S:VAN3:GFP localized to the trans-Golgi network (Koizumi et al., 2005). To resolve this difference, we are currently developing PSFC:SFC:GFP lines using the native SFC promoter and assaying rescue; these fusion proteins should provide valuable insights into SFC localization and dynamics. The defect in PIN1 internalization in sfc mutants might also explain the higher auxin transport capacity in sfc roots. A defect in PIN1 endocytosis could result in greater amounts of PIN1 remaining on the apical surface of the plasma membrane.
SFC/VAN3 Is Required for Normal Auxin Efflux in Aerial Tissues
The striking vascular phenotype of sfc/van3 mutants features isolated VIs in all broad flat organs (Deyholos et al., 2000; Koizumi et al., 2000). The major hypothesis to explain vein patterning, canalization of auxin flow, could explain the sfc/van3 phenotype if SFC/VAN3 functioned either in the perception or in the transport of auxin. We found that the auxin-responsive DR5:GUS reporter gene was inducible by auxin in sfc leaves, indicating that auxin responses were intact and suggesting that VIs might arise due to defects in auxin efflux.
Additional support for decreased auxin efflux in aerial tissues of sfc mutants comes from the sfc root phenotype. Although development of the sfc primary root was largely normal, sfc mutants had greatly reduced lateral root numbers. The reduced lateral root numbers did not indicate a direct role for SFC in lateral root initiation, as exogenous auxin restored sfc lateral root production. Thus, the reduced numbers of lateral roots in sfc mutants indicated reduced auxin delivery from the sfc shoot, in support of SFC being required for normal auxin efflux in shoots.
While our data imply decreased auxin efflux in shoots, our measurements for roots indicated increased auxin transport capacity. These differing conclusions could reflect the different developmental contexts of auxin transport in roots and developing leaves. That is, roots are established during embryogenesis, and root auxin transport in seedlings occurs with this preestablished polarity. By contrast, pathways for auxin efflux must be established during both leaf and cotyledon organogenesis, and these organs appear to have a unique requirement for SFC. Furthermore, auxin levels can have opposite effects on expression of PIN genes in roots and shoots (Peer et al., 2004), and PID plays different roles in root and shoot tissues (Friml et al., 2004). PID is required for apical targeting of the auxin efflux machinery in the shoot, while in roots, it is required for basal (distal from the root apical meristem) targeting. Our genetic data suggest that SFC/VAN3 might function in the same pathway as PID, which supports the possibility for different shoot and root SFC/VAN3 functions. Analyzing the role of SFC in targeting of different PIN proteins in above-ground tissues is an important future goal.
SFC/VAN3 and Cotyledon and Leaf Vein Patterning
The canalization hypothesis for vein patterning proposes that sites initially selected for vascular differentiation are based on stochastic modulations in auxin efflux, followed by self-reinforcing networks. The recent demonstration that PIN1 localization is reinforced by auxin flux supports this possibility (Paciorek et al., 2005). Auxin also induces PID expression (Benjamins et al., 2001), and high levels of PID expression are sufficient to induce changes in PIN1 distribution (Friml et al., 2004). Thus, the self-reinforcing flux of auxin combined with PID-induced changes in PIN1 polarity may be sufficient to generate complex branched vein patterns. In this scenario, the VIs in the sfc mutant might arise if SFC/VAN3 is required for either normal PIN1 cycling or for PID-directed efflux machinery relocation.
Alternatively, directions of auxin transport could be specified by a (nonauxin) signal that directs intracellular trafficking of the auxin efflux machinery. A precedent for this type of regulation is provided by gravistimulation, which induces rapid relocation of PIN3 to lateral positions in root columella cells (Friml et al., 2002). What positional signals could serve as a primary signal directing movement of the auxin efflux machinery? Studies implicate a variety of signals, including small peptides (Casson et al., 2002), sterols (Jang et al., 2000; Carland et al., 2002; Souter et al., 2002; Willemsen et al., 2003), and phosphoinositides (Carland and Nelson, 2004). A phosphoinositide signal is particularly attractive, as phosphatidylinositol 4-monophosphate has been shown to bind strongly to the SFC/VAN3 PH domain (Koizumi et al., 2005) and is a possible product of CVP2. Furthermore, our genetic analyses suggested that SFC and CVP2 might function in the same pathway. In this scenario, the BAR and PH domains of SFC could confer binding to a specific lipid domain that, in turn, could be regulated by CVP2. Alternatively, the SFC PH domain could function as a regulatory domain that controls ARF-GAP activity, similar to the way the ASAP1 ARF-GAP protein's PH domain inhibits its ARF-GAP activity unless bound to a specific phosphoinositide (Kam et al., 2000). Development of tools for in planta localization of SFC/VAN3 may allow us to start distinguishing between these possibilities.
METHODS
Plant Growth
Most experiments used surface-sterilized seeds that were plated onto growth medium (GM) plates, which were composed of 0.5× Murashige and Skoog salts (Caisson Labs), 1% sucrose, 0.5 g/L MES (Sigma-Aldrich), and 0.8% phytoblend agar (Caisson Labs), pH 5.8. The auxin transport and root auxin response experiments used GM that was the same except Murashige and Skoog salts were used at 1×, and sucrose increased to 1.5%. Seeds were cold-stratified at 4°C for 3 d and then transferred to a TC-30 Conviron controlled environment chamber maintained at 22°C and constant light (100 to 120 μE m2 s). Seedling age refers to days after transfer from the cold.
Microscopy and Tissue Preparation
Seedling images were obtained using an Olympus SZX9 microscopy. Vascular patterns were observed using a dark-field base, and tissue was fixed in Carnoys (3:1 ethanol:acetic acid) and cleared by incubation in saturated chloral hydrate (Sigma-Aldrich). Details of cellular anatomy and GUS staining patterns were examined using an Olympus BX-50 microscopy. Images of leaf bases were obtained for tissue without cover slips to prevent breaking the tissue.
Mapping and Gene Identification
Mapping primers were developed from the Cereon database (Jander et al., 2002) and are available upon request. The sfc-9 allele corresponds to Salk_069116, and its insertion position within intron 1 was verified by sequencing.
AGD Mutant Analysis
Insertion alleles for members of the AGD gene family were identified in the Salk Insertion Sequence Database (Alonso et al., 2003). The insertion site for each mutant was characterized by sequencing. Our agd1 mutant corresponds to Salk_036034, and genotyping was performed using LBB1 and the following gene specific primers: forward, 5′-AGCTCGATGATTCTCCCA-3′, and reverse 5′-AAGCAACCGATCACTCAG-3′ (LBB1+F gives the agd1 insertion-specific product). The agd2 mutant corresponds to Salk_125988, and genotyping was performed using LBB1 and the following gene-specific primers: forward, 5′-GGAGAACTCGAAGTCAGC-3′, and reverse, 5′-TCCATGGTTGTCAAGCAC-3′ (LBB1+F produces the insertion-specific product). The agd4 mutant corresponds to Salk_024103, and genotyping was performed with LBB1 and the following gene-specific primers: forward, 5′-CATATTCATCACCAACCGA-3′, and reverse, 5′-AAAACGGGACACTCTGAAT-3′ (the insertion-specific reaction used LBB1+R).
Real-Time RT-PCR Analysis
RNA extraction and cDNA template preparation were performed as described previously (Deyholos et al., 2003). For real-time RT-PCR, we used the Roche LightCycler and Roche FastStart DNA Master SYBR Green 1 master mix. Each real-time RT-PCR run included, as an internal control, a reaction for β-tubulin. We tested each reaction's specificity using the machine's standard melt curve method, and products were also analyzed by gel electrophoresis. We determined the relative expression of each AGD gene for each tissue analyzed using the standard formula ΔCT = CT(gene analyzed) − CT(β-tubulin). We next calculated the average ΔCT value for each gene's expression in each tissue, and we calculated the relative expression using the equation 2[(average ΔCT (geneX, tissueX) − average ΔCT (SFC, leaf+shoot apical meristem)]. Oligonucleotides for AGD1 analysis were as follows: forward (exon5/exon6 junction), 5′-CCACGAAGTTAAGGAAGCTCGTA-3′, and reverse (exon7/exon8), 5′-CGCTATGTAGGTCCTCCTCAATT-3′. Oligonucleotides for AGD2 analysis were as follows: forward (exon6), 5′-TCGGCGTCGGTTCGATAA-3′, and reverse (exon 7/exon8), 5′-TTCTCCAAATCCTCTTCCAACTC-3′. Oligonucleotides for AGD4 analysis were as follows: forward (exon4/exon5), 5′-TTGCTCACAGGTGGAGCAT-3′, and reverse (exon7), 5′-CTCTTCCAACTCTGCCACAAT-3′. Oligonucleotides for SFC were as follows: forward (exon1), 5′-GACTCTCCCATGTTCCGCAA-3′, and reverse (exon2/exon3), 5′-CTCGCCTAGTCCTTCAGTGTATTT-3′. Approximate positions of these oligonucleotides are also shown in Supplemental Figure 1 online.
We also used real-time RT-PCR analysis to analyze transcript levels in agd2 and agd4 mutants. For the agd2 analysis, we isolated RNA from 8-d-old roots of wild-type and agd2 homozygous mutants, and for the agd4 analysis, we isolated RNA from 8-d-old cotyledons (wild-type and agd4 homozygotes). For these experiments, we used two independent RNA isolations for agd2 analysis and three independent RNA isolations for agd4, and we used the same procedures and primers as described above.
Auxin Transport and Root Auxin Responses
To measure acropetal auxin transport, a line of [3H]-IAA was applied at the root-shoot junction of wild-type and sfc-1 seedlings and incubated for 18 h. Experiments presented in Figure 6 used the 4- or 5-d-old wild type and 6-d-old sfc-1 to facilitate size-matching of the roots. Another experiment using 6-d-old plants of both genotypes produced the same results. Following the 18-h incubation, the 2-mm portion of root closest to the applied [3H]-IAA was discarded, and the remaining roots were divided into 3-mm upper and middle segments (upper being closest to the shoot) and a third variably sized bottom segment (Figure 8A). For NPA treatments, seedlings were transferred to either control media or media containing 10 μM NPA, and the agar line containing [3H]-IAA was applied 6 h after transfer to this media. For the NPA treatment, similar segments were used as described above, but only the results from the upper segment are reported. The amount of [3H]-IAA was determined using a scintillation counter (Beckman). To test auxin-induced lateral root formation, 5-d-old seedlings were decapitated at the root-shoot junction, and an agar block (±10 μM IAA) was placed over the cut surface. Lateral root numbers were determined 3 d later.
GUS Staining
GUS staining reactions and auxin induction of DR5 expression were performed as described by Deyholos et al. (2003), except that auxin treatment lasted 5 h (instead of 8), and control incubations were performed in liquid GM. The GUS staining reactions were terminated after 3 to 4 h, and tissue was fixed and cleared as described above.
Double Mutant Analyses
The emb30-2 sfc double mutant analysis used multiple independent segregating lines. In one F2, the following phenotype ratio was obtained: 155(wild type):64(emb30-2):49(sfc-1), which is consistent with epistasis of emb30 (χ2 = 0.26, df = 2; critical value = 5.991 for 95% confidence). The gn4577 mutant was mapped to the top of chromosome 1 and then verified as an allele by noncomplementation with emb30-2. For double mutants from all crosses, single siliques were plated, and segregation was analyzed using >200 plants. For analyses where one mutant is homozygous viable (e.g., cvp2 and pid-2), F3 plants that were homozygous for the viable mutation were obtained. Segregation of sfc was found to be consistent with a 3:1 segregation ratio, and for these F3, all sfc mutants were assumed to be homozygous for both loci.
Confocal Analysis
Confocal analysis was performed using 5-d-old vertically grown seedlings and a Zeiss 510 Meta laser scanning confocal microscope. We used the metadetector to obtain spectra of wild-type (non-GFP) and GFP-expressing roots. Comparison of these spectra revealed no confounding autofluorescence, and GFP fluorescence signals were found in patterns consistent with their previously published patterns. BFA, cycloheximide, and TIBA treatments were used at concentrations and treatment durations matching those described by Geldner et al. (2001). Treatments were performed in 48-well microtiter plates in liquid GM, and measurements were made from live tissue. To allow direct comparison of PIN1:GFP in the wild type and sfc mutants, our analyses used the region just distal to the root meristem.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_121333 (SFC/VAN3), NM_125591 (AGD1), NM_104767 (AGD2), and NM_100962 (AGD4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genomic Structure and T-DNA Insertion Sites for the AGD1, AGD2, and AGD4 Genes.
Supplemental Figure 2. Seedling Phenotypes of sfc-9 and agd1 agd2 agd4 sfc-9 Quadruple Mutants.
Supplemental Figure 3. Additive Interaction in the lop1 sfc-1 Double Mutant.
Supplemental Figure 4. Root PIN1:GFP Expression and Tracheary Element Differentiation.
Supplementary Material
Acknowledgments
We are grateful for contributions from David Goeres, Jennifer Dickman, Nasheed Shaams, Nick Peters, and Louise Saw. We thank Marcus Babst for useful suggestions during the course of this work, Josh Steffen for assistance with real-time RT-PCR, Denise Dearing for use of the LightCycler, Elliot Meyerowitz and Marcus Heisler for use of PIN1:GFP, and the ABRC for various seed stocks. We are also grateful for financial support from the National Science Foundation (IBN-0344389 to L.E.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Leslie E. Sieburth (sieburth@biology.utah.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039008.
References
- Aloni, R. (2001). Foliar and axial aspects of vascular differentiation: Hypotheses and evidence. J. Plant Growth Regul. 20 22–34. [Google Scholar]
- Aloni, R. (2004). The induction of vascular tissues by auxin. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 471–492.
- Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Benjamins, R., Quint, A., Wiejers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Maher, E.P. (1989). Mutants of Arabidopsis thaliana with abnormal gravitropic responses. Mol. Gen. Genet. 220 289–293. [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8 505–520. [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Peer, W.A., and Murphy, A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8 494–500. [DOI] [PubMed] [Google Scholar]
- Brown, F.D., Rozelle, A.L., Yin, H.L., Balla, T., and Donaldson, J.G. (2001). Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Berg, B.L., FitzGerald, J.N., Jinamornphongs, S., Nelson, T., and Keith, B. (1999). Genetic regulation of vascular tissue patterning. Plant Cell 11 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., and McHale, N.A. (1996). LOP1: A gene involved in auxin transport and vascular patterning in Arabidopsis. Development 122 1811–1819. [DOI] [PubMed] [Google Scholar]
- Carland, F.M., and Nelson, T. (2004). COTYLEDON VASCULAR PATTERN2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson, S.A., Chilley, P.M., Topping, J.F., Evans, I.M., Souter, M.A., and Lindsey, K. (2002). The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14 1705–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J., Li, J., Bos, E., Porcionatto, M., Premont, R.T., Bourgoin, S., Peters, P.J., and Hsu, V.W. (2004). ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev. Cell 7 771–776. [DOI] [PubMed] [Google Scholar]
- Deyholos, M.K., Cavaness, G.F., Hall, B., King, E., Punwani, J., Van Norman, J., and Sieburth, L.E. (2003). VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130 6577–6588. [DOI] [PubMed] [Google Scholar]
- Deyholos, M.K., Cordner, G., Beebe, D., and Sieburth, L.E. (2000). The SCARFACE gene is required for cotyledon and leaf vein patterning. Development 127 3205–3213. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Li, G., Colombo, M.I., and Stahl, P.D. (1995). A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267 1175–1178. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1953). Plant Anatomy. (New York: John Wiley & Sons).
- Falasca, M., Logan, S.K., Lehto, V.P., Baccante, G., Lemmon, M.A., and Schlessinger, J. (1998). Activation of phospholipase Cγ by PI 3-kinase-induced PH domain-mediated membrane targeting. EMBO J. 17 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003. b). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torrest-Ruiz, R.A., Mayer, U., and Jürgens, G. (2003. a). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131 389–400. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44 179–194. [DOI] [PubMed] [Google Scholar]
- Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K.C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001). BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Jackson, T.R., Brown, F.D., Nie, Z., Miura, K., Foroni, L., Sun, J., Hsu, V.W., Donaldson, J.G., and Randazzo, P.A. (2000). ACAPs are Arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, W.P. (1952). The role of auxin in differentiation of xylem around a wound. Am. J. Bot. 39 301–309. [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genomic era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.-C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kam, J.L., Miura, K., Jackson, T.R., Gruschus, J., Roller, P., Stauffer, S., Clark, J., Aneja, R., and Randazzo, P.A. (2000). Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. J. Biol. Chem. 275 9653–9663. [DOI] [PubMed] [Google Scholar]
- Koizumi, K., Naramoto, S., Sawa, S., Yahara, N., Ueda, T., Nakano, A., Sugiyama, M., and Fukuda, H. (2005). VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vacular network formation. Development 132 1699–1711. [DOI] [PubMed] [Google Scholar]
- Koizumi, K., Sugiyama, M., and Fukuda, H. (2000). A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular networks: Calling the auxin signal flow canalization hypothesis into question. Development 127 3197–3204. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Ferguson, K.M., O'Brien, R., Sigler, P.B., and Schlessinger, J. (1995). Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 92 10472–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, C., Caxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thalilana. Genes Dev. 12 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2992. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Büttner, G., and Jürgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 117 149–162. [Google Scholar]
- Nakamura, A., Higuchi, K., Goda, H., Fujiwara, M.T., Sawa, S., Koshiba, T., Shimada, Y., and Yoshida, S. (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Ritzenthaler, C., and Robinson, D.G. (2002). Brefelding A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug Resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek, T., Zazímalová, E., Ruthardt, N., Petrásek, J., Stierhof, Y.D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Paponov, I.A., Teale, W.D., Trebar, M., Blilou, I., and Palme, K. (2005). The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 10 170–177. [DOI] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, B.J., Kent, H.M., Mills, I.G., Vallis, Y., Butler, P.J.G., Evans, P.R., and McMahon, H.T. (2004). BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303 495–499. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A., Andrade, J., Miura, K., Brown, M.T., Long, Y.Q., Stauffer, S., Roller, P., and Cooper, J.A. (2000). The Arf-GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 97 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P.A., and Hirsch, D.S. (2004). Arf GAPs: Multifunctional proteins that regulate membrane traffic and actin remodelling. Cell. Signal. 16 401–413. [DOI] [PubMed] [Google Scholar]
- Rashotte, A.M., DeLong, A., and Muday, G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response and lateral root elongation. Plant Cell 13 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. (1981). The control of the patterned differentiation of vascular tissues. Adv. Bot. Res. 9 151–162. [Google Scholar]
- Sachs, T. (1989). The development of vascular networks during leaf development. Curr. Topics Plant Biochem. Physiol. 8 168–183. [Google Scholar]
- Shevell, D.E., Leu, W.M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77 1051–1062. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E. (1999). Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 121 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Turner, S., and Sieburth, L.E. (2002). Vascular Patterning. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0073, http://www.aspb.org/publications/arabidopsis.
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.