Abstract
The reiterated nature of histone genes has hampered genetic approach to dissect the role of histones in chromatin dynamics. We here report isolation of three temperature-sensitive (ts) Schizosaccharomyces pombe strains, containing amino-acid substitutions in the sole histone H2B gene (htb1+). The mutation sites reside in the highly conserved, non-helical residues of H2B, which are implicated in DNA–protein or protein–protein interactions in the nucleosome. In the allele of htb1-72, the substitution (G52D) occurs at the DNA binding loop L1, causing disruption of the gene silencing in heterochromatic regions and lagging chromosomes in anaphase. In another allele htb1-223 (P102L) locating in the junction between α3 and αC, the mutant residue is in contact with H2A and other histones, leading to structural aberrations in the central centromere chromatin and unequal chromosome segregation in anaphase. The third allele htb1-442 (E34K) near α1 displayed little defect. Evidence is provided that monoubiquitinated H2B is greatly unstable in P102L mutant, possibly owing to proteasome-independent destruction or enhanced deubiquitination. Histone H2B thus plays an important role in centromere/kinetochore formation.
Keywords: centromere, chromatin, gene silencing, histone H2B, monoubiquitination
Introduction
In eukaryotic cells, DNA is packaged repetitively into nucleosomes by means of interactions among four classes of histones, H2A, H2B, H3 and H4 (Richmond et al, 1984; van Holde, 1988; Kornberg and Lorch, 1999; Elgin and Workman, 2002). The crystal structure of the nucleosome core particle consists of approximately 147 bp of DNA organized into a superhelix around an octamer of two of each of these four histones (Luger et al, 1997; Luger, 2003; Richmond and Davey, 2003). Histones are phosphorylated, acetylated, methylated, ubiquitinated and sumoylated (Peterson and Laniel, 2004). Extensive post-translational modifications mainly occur in the tail regions of histones, and affect transcriptional activation, repression, chromatin assembly, heterochromatin gene silencing, centromere chromatin, response to DNA damages, histone deposition, chromosome segregation, mitosis, meiosis and spermatogenesis: histone tails may be viewed as complex protein–protein interaction surface that are regulated by numerous modifications (e.g., Grewal and Elgin, 2002; Fischle et al, 2003a, 2003b; Hayashi et al, 2004; Korber and Horz, 2004; Tagami et al, 2004).
Histone tails contribute significantly neither to the structure of individual nucleosomes nor to the stability, but they are thought to strongly affect the folding of nucleosomal arrays into higher-order structure. What then would be the role of inner histone structure in cellular physiological regulations? Does it solely contribute toward the assembly to nucleosomal structure? Post-translational modifications infrequently occur in the inner structure, which is often not exposed to the outer surface of nucleosome. However, K79 in the L1 region between α1 and α2 of H3 (L and α represent, respectively, loop and α-helix; Luger et al, 1997) is methylated by a methylase Dot1 and this methylation has been shown to be implicated in meiotic checkpoint control and telomeric silencing (San-Segundo and Roeder, 2000; Feng et al, 2002; Ng et al, 2002; van Leeuwen et al, 2002). Amino-acid substitution mutants in the inner histone structure have been scarce, as genetic analyses of histone functions in metazoan cells are hampered because of the high copy numbers of the histone genes in the genome.
In this study, we characterized the temperature-sensitive (ts) mutants of the fission yeast Schizosaccharomyces pombe histone H2B gene. The mutation sites reside in the highly conserved, non-helical, inner regions of histone H2B. Our results suggested that the inner structure of H2B is essential for proper ubiquitination, centromere function, gene silencing and chromosome segregation.
Results
Isolation of S. pombe histone H2B mutants
To isolate histone H2B mutants, ts mutants were randomly isolated, followed by characterization of cytological mutant phenotypes (Hayashi et al, 2004). Then, mass transformation using plasmid pH2AH2B that carried the paired genes of histone H2A–H2B (Yuasa et al, 2004) was performed by introducing plasmid into ∼500 individual strains. The resulting transformants were screened for their ability to produce colonies at the restrictive temperature (36°C). Three mutant strains (72, 223, 442), whose ts phenotype was fully suppressed by plasmid pH2AH2B at 36°C, were obtained. Subcloning established that the histone H2B gene (designated htb1+: Matsumoto and Yanagida, 1985), but not H2A gene, was capable of suppressing the ts phenotype (Figure 1A). S. pombe has the sole gene of H2B, but the two H2A and three paired H3–H4 genes are present in the genome (Matsumoto and Yanagida, 1985); one may expect that only htb1+ can generate ts mutations.
Figure 1.
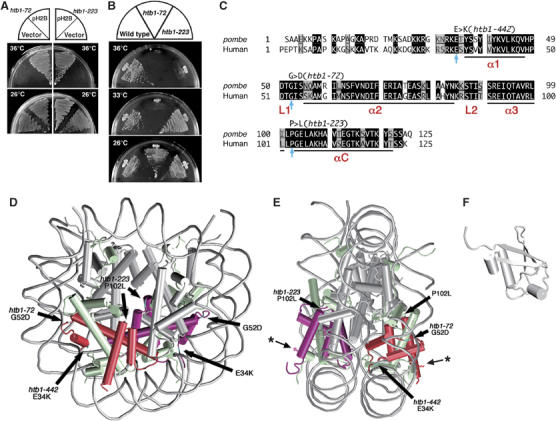
S. pombe H2B mutations reside in the conserved, non-helical regions. (A) Plasmid pH2B suppresses the ts phenotype of htb1-72 and htb1-223. (B) htb1-72 and htb1-223 mutants fail to produce colonies at 36°C on the complete YPD plates. (C) Amino-acid sequences of fission yeast and human histone H2B. Identical and similar amino acids are indicated in black and gray boxes, respectively. Substituted amino acids in the three htb1 mutants are indicated by arrows (see text). The α-helices are labeled as α1, α2, α3 and αC, whereas L1 and L2 represent the DNA binding loops. (D, E) The nucleosome structure (D, front view; E, side view; White et al, 2001) is depicted with the positions of altered amino acids. The atomic coordinates file (ID; 1ID3) was obtained from Brookhaven Protein Databank (PDB) and visualized with VMD (version 1.8.3). The DNA phosphodiester backbone is shown in gray. The octamer histone chains are schematized by the helical rods and the interhelical loops. Histone H2B subunits are indicated in red. The asterisks indicate the position of K that is associated with monoubiquitin. (F) The 3D structure of ubiquitin (PDB ID; 1UBQ) (Vijay-Kumar et al, 1987).
To determine whether the ts mutations actually resided in the htb1 locus, an integration vector pYC11 carrying the htb1+ gene and the Saccharomyces cerevisiae LEU2 was chromosomally integrated in the leu1 mutant strain by homologous recombination (because of the low sequence homology between LEU2 and leu1+, homologous integration occurred at the htb1+ locus). The resulting Leu+ integrant strain was crossed with ts mutants, and tetrad dissection was performed. The Ts− and Leu+ phenotypes were tightly linked. With the finding that pH2B, but not pH2A, suppressed the ts phenotype (Figure 1A), we concluded that the ts mutants were derived from the htb1 locus.
After several rounds of crossing with the wild-type strain for removing extra silent mutations, if any, the ts phenotypes of three ts strains (htb1-72, -223, -442) at 36°C were characterized. The Ts+ and Ts− phenotypes were segregated 2:2 for the strains 72 and 223, but not for htb1-442. The 442 strain appeared to contain a leaky ts and an additional silent mutation (data not shown). We hence employed mostly the two strains 72 and 223 for subsequent physiological experiments. In Figure 1B, the failure of htb1-72 and htb1-223 to form colonies on the complete YPD plate at 36°C is shown. The strain htb1-72 showed the ts phenotype severer than htb1-223 as it did not form colonies at 33°C.
Mutation sites are located in the distinct, but in the conserved and non-helical regions
Mutant genes of histone H2B were obtained from the isolated ts strains by the PCR method, and their nucleotide sequences were determined. It was thus established that single-nucleotide substitutions existed for each of the three htb1 mutant strains. For htb1-72, only the 52nd amino-acid residue showed the nucleotide change from GGT to GAT in comparison with the wild-type sequence. For htb1-223, the 102nd residue was altered from CCC to CTC. The resulting amino-acid substitutions in htb1-72 and htb1-223, respectively, were 52Gly to Asp (G52D) and 102Pro to Leu (P102L). The 442 segregant that showed the weak ts phenotype contained a nucleotide change in the 34th residue (from GAA to AAA), causing the change from Glu to Lys (E34K). These three mutations occurred at the highly conserved residues in histone H2B. The amino acids of the mutation sites were conserved between fission yeast (Pombe) and human histone H2B, as shown in Figure 1C.
One of them (72) resides in the middle of histone fold, α1–L1–α2–L2–α3, whereas the location of the two others (223 and 442) is not within but nearby the histone fold. Residue G52D (72) is positioned in the loop L1 that exists between α1 and α2 helices (Figure 1C). L1 and L2 are the sites to be bound to DNA. In contrast, the P102L residue (223) locates in the short linker between the two helices, α3 and the αC extension. The long αC extension is unique to H2B, and defines the outer limit of the protein surface of nucleosome core particle. P102L is thus close in contact with H2Aα2 by extensive hydrophobic interaction (Luger et al, 1997). The E34K (442) leaky mutation is in the conserved, amino-terminal tail that is connected to the amino-terminal edge of α1 helix. The physical locations of mutant residues are indicated by the thick arrows in Figure 1D (front view) and Figure 1E (side view) in the nucleosome core structure determined by crystallography (Luger et al, 1997; White et al, 2001; Richmond and Davey, 2003). The thin arrows with asterisk represent the K119 site to bind to monoubiquitin (Figure 1E and F). The ts mutations identified in this study are thus all located in the conserved and non-helical regions. The G52D substitution is in contact with the minor groove of DNA, whereas P102L is in the central region of the histone octamer in the front view (Figure 1D) and in the middle of the outer sides in the side view (Figure 1E). P102L hence interacts with the histone H2A in the same nucleosome, or also with the histones of adjacent nucleosomes (Schalch et al, 2005). The E34K substitution is close in contact with DNA.
Missegregation and chromatin shrinkage occur in htb1 mutants
We investigated whether these htb1 mutations affected the cell division cycle of S. pombe. Exponentially growing cultures of wild-type, htb1-72 and htb1-223 strains at 26°C in the complete YPD were shifted to 36°C for 12 h. Cells were scored for viability and observed by DAPI staining after glutaraldehyde fixation. Interestingly, htb1-223, but not htb1-72, showed frequent unequal nuclear division after 8 h at 36°C (arrowheads in Figure 2A, lower right panel), the characteristic phenotype seen in the centromere/kinetochore protein mutants (Takahashi et al, 1994, 2000; Saitoh et al, 1997; Goshima et al, 1999; Hayashi et al, 2004). Quantitative data (Figure 2B, red line with rectangles in the middle panel) showed that the frequency of cells with unequal daughter nuclei increased after 4 h at 36°C in coincidence with the gradual decrease of cell viability (bottom panel), and reached 38% of binucleated cells. Typical centromere/kinetochore protein mutants mis6 and mis12 produced ∼80% unequal nuclei in binucleate cells so that the unequal segregation phenotype of htb1-223 was not severe as mis6 and mis12. The unequal nuclear division phenotype was scarce (a few %) in htb1-72. Instead, the nuclear chromatin observed by DAPI staining appeared to be shrunk (12 h; Figure 2A, middle right panel). Cell viability of htb1-72 decreased at the time of nuclear chromatin shrinkage. The gross chromatin organization in htb1-72 appeared to be altered. Additionally, the thick septa indicative of cytokinesis delay were observed in htb1-72.
Figure 2.
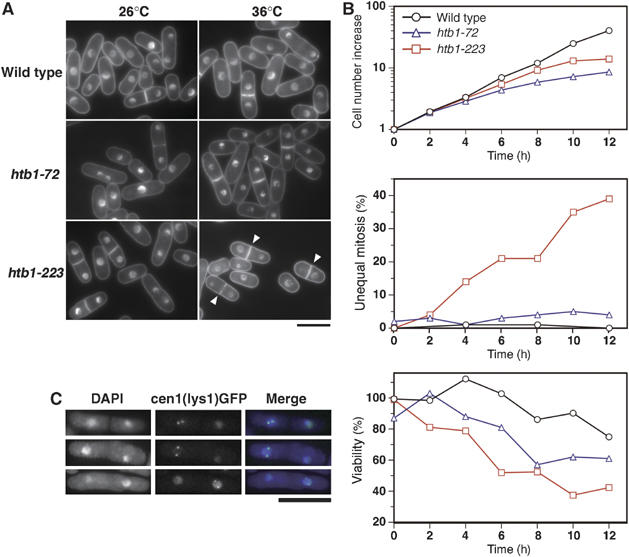
Cellular phenotypes of histone H2B mutants htb1-72 and htb1-223. Asynchronously growing htb1 mutant cells in YPD medium were shifted to 36°C for 0–12 h. (A) Mutant cells cultured at 26°C (left panels) or 36°C for 8 or 12 h (right panels) were stained by DAPI. Upper panels, wild type; middle panels, htb1-72; lower panels, htb1-223. Arrowheads indicate binucleate cells displaying unequal nuclear division. Bar, 10 μm. (B) Top, cell number increase; middle, the frequencies of unequal nuclear division; bottom, cell viability. (C) Unequal chromosome segregation was examined by the cen1(lys1)-GFP method (see text). Three examples of htb1-223 mutant cells cultured at 36°C for 10 h displaying unequal segregation of cen1(lys1)-GFP signals are shown. Bar, 10 μm.
To confirm that the unequal nuclear division phenotype of htb1-223 was due to chromosome missegregation, the cen1-GFP method (Nabeshima et al, 1998) was employed. After 10 h at 36°C, 25% of the binucleate htb1-223 cells (50/204) exhibited unequal nuclear division. As shown in Figure 2C, the cen1-GFP signals failed to segregate in these cells. Missegregation of cen1-GFP signals was observed in 22% (11/50) of cells showing unequal nuclear division. We thus concluded that the unequal nuclear division phenotype in htb1-223 mutant was owing to chromosome missegregation.
Histone H2B mutants were sensitive to a protein synthesis inhibitor
To understand the causes for the histone H2B mutants to produce distinct phenotypes, we examined whether these mutants might be differentially sensitive to drugs and irradiation. Effects of hydroxyurea (DNA replication inhibitor) and UV irradiation were tested, but their sensitivities were similar to that of wild type (data not shown). Cycloheximide (Cyh, a protein synthesis inhibitor), however, showed different sensitivities as shown in Figure 3A. At 30 and 33°C (semi-restrictive temperature), both htb1-72 and htb1-223 were sensitive to Cyh. At 30°C, htb1-223 was more sensitive than htb1-72. Introducing pH2B plasmid into the mutants restored the hypersensitivity (data not shown), indicating that the htb1 mutations caused both ts and Cyh phenotypes. The addition of Cyh thus produced the synthetic lethal effect on htb1 mutations.
Figure 3.
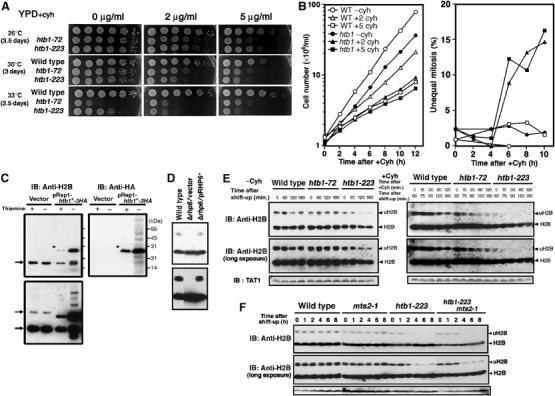
Cyh sensitivity and turnover of Rhp6-dependent ubiquitination of H2B in htb1 mutants. (A) The htb1 mutants are hypersensitive to Cyh, a protein synthesis inhibitor. Both htb1-72 and htb1-223 were slower in growth than wild type in the presence of Cyh. Cells were serially diluted (1:5) and spotted onto the YPD plates containing Cyh (0, 2 and 5 μg/ml). The highest-density spots contained 5 × 104 cells. (B) The cell number increase (left panel) and the frequencies of unequal nuclear division (right) are shown for wild type (open) and htb1-223 (closed) in the absence (circles) or presence (rectangles, triangles) of Cyh at 33°C. (C) Immunoblot patterns using polyclonal antibodies against histone H2B (anti-H2B) and monoclonal antibodies against HA (anti-HA). Extracts of cells expressing endogenous H2B (Rep1 vector) and ectopically expressed H2B tagged with HA (pRep1-htb1HA) in the presence of thiamine (promoter off) and absence of thiamine (14 h after removal of thiamine; promoter on) were used for immunoblot. The 17 kDa H2B band and the 24–25 kDa uH2B bands (indicated by the arrows) were detected in cells carrying vector. In cells carrying pRep1-htb1HA, additional bands for H2B-3HA (asterisks) were detected by anti-H2B and anti-HA antibodies. (D) Immunoblot was performed for extracts of wild type, Δrhp6 carrying vector or plasmid pRHP6. The presumed monoubiquitinated H2B band was missing in Δrhp6. (E) Wild type, htb1-72 and htb1-223 were cultured in the absence (−) or presence of (+) of Cyh (100 μg/ml) at 36°C for 0–180 min. In the case of +Cyh, strains were first cultured at 36°C for 60 min and then followed by the addition of Cyh. Immunoblot patterns by short and long exposure are shown. The positions for H2B and uH2B are indicated by arrowheads. (F) Wild type, mts2-1, htb1-223 and the double mutant mts2-1 htb1-223 were cultured first at 26°C, and then shifted to 36°C for 8 h. Immunoblot patterns using anti-H2B antibodies indicated that the level of decrease of uH2B was not arrested in the proteasome mutant mts2-1 background.
We then tested whether Cyh affected the cellular phenotypes of the htb1-223 mutant at 33°C, the semi-restrictive temperature. To this end, the culture of htb1-223 grown at 26°C was shifted to 33°C for 2 h in the absence of Cyh followed by the addition of Cyh (2 and 5 μg/ml) for the continued culture at 33°C for 12 h (Figure 3B). For control, wild type without or with Cyh (5 μg/ml) was tested. In the absence of Cyh, the frequency of unequal nuclear division was negligible in htb1-223 at 33°C. In the presence of Cyh, however, the cell number increase (left panel) was considerably retarded and the frequency of unequal nuclear division (right panel) strikingly increased after 4 h. These results suggested that the level of functional H2B protein might decrease in the presence of Cyh in htb1-223 mutant at the semi-restrictive temperature (see below).
Rhp6-mediated monoubiquitination of H2B
The above results suggested that mutant histone H2B might be unstable or faster in turnover than that of wild-type histone H2B. The inhibition of protein synthesis might reduce the level of mutant histone H2B. To test this hypothesis, polyclonal antibodies against a 16 amino-acid peptide of the amino-terminal tail sequence SAAEKKPASKAPAGKA of S. pombe histone H2B were raised. Immunoblot of cell extracts were carried out using affinity-purified anti-H2B antibodies. An intense ∼17 kDa band (corresponding to histone H2B) was seen in wild-type cells carrying vector plasmid pREP1 (Figure 3C, left). A weak ∼25 kDa band (indicated by the arrows) was observed in the long exposed immunoblot patterns. This 25 kDa band corresponds to monoubiquitinated histone H2B (designated uH2B; see below).
The assignment of the bands was confirmed using extracts of cells that expressed the HA-tagged histone H2B under the control of the inducible nmt1 promoter (Figure 3C). Antibodies against H2B (left panel) and against HA (right panel) were used to detect HA-tagged H2B that was overproduced in the absence of thiamine (promoter, on) for 14 h after the removal of thiamine. The HA-tagged H2B band (indicated by asterisks) was dramatically increased in the absence of thiamine. Polyclonal antibodies against H2B thus detected both endogenous and ectopically expressed H2B.
The 25 kDa upper H2B band was produced in an Rhp6-dependent manner. As shown in Figure 3D, the upper band disappeared in the deletion mutant Δrhp6 (Kitamura et al, 2001). This band was restored in Δrhp6 carrying plasmid pRHP6 (constructed by PCR method in this study). Rhp6 is a homolog of budding yeast Rad6, a ubiquitin conjugating enzyme, which is known to be responsible for monoubiquitination of histone H2B (Robzyk et al, 2000). We hence supposed that the 25 kDa band was an Rhp6-mediated monoubiquitinated band. The band intensity of uH2B in the absence of Cyh was approximately 10% of total cellular histone H2B in wild type.
Monoubiquitinated H2B-223 mutant protein is unstable
We then tested whether mutant H2B was less stable than wild type in the presence or absence of Cyh. Cyh (100 μg/ml) was added to the cultures of wild type, htb1-72 and htb1-223, 60 min after the shift to 36°C, and cell extracts were prepared. Immunoblot patterns of wild-type and mutant extracts using antibodies against histone H2B are shown in Figure 3E. Although the levels of H2B remained high and did not alter significantly, the uH2B level in htb1-223 mutant was already low before the shift (roughly 50% compared with that in wild-type cells), and decreased after the shift to 36°C in the absence of Cyh. In the presence of Cyh, the level of uH2B-223 rapidly decreased. The levels of wild-type uH2B and other mutant uH2B-72 also decreased in the presence of Cyh.
The quantitative densitometric analyses of immunoblot patterns indicated that the half-life of uH2B-223 in the presence of Cyh at 36°C was only 15 min, whereas those in wild type and htb1-72 were 88 and 86 min, respectively. In the absence of Cyh, mutant H2B-223 became unstable by the temperature shift to 36°C, and the apparent half-life of uH2B-223 was 92 min. The level of H2B in wild type and htb1-72 did not alter in the absence of Cyh. We therefore concluded that the half-life of uH2B was specifically short in htb1-223.
Instability of uH2B in htb1-223 is not due to 26S proteasome-dependent proteolysis
A question was raised whether instability of ubiquitinated H2B was due to proteolysis. Immunoblot of H2B protein was carried out for extracts in a proteasome mutant mts2-1 and the double mutant mts2-1 htb1-223. As shown in Figure 3F, the level of uH2B was not affected at all in the absence of 26S proteasome function for wild-type H2B, but mutant H2B-223 protein destruction seemed to be slightly suppressed so that mutant uH2B might go through the proteasome for degradation. However, the decay in the level of uH2B might be principally owing to deubiquitination rather than 26S-dependent proteolysis. In any case, uH2B became highly dynamic in htb1-223 cells in the presence of Cyh so that chromatin would be strongly altered if uH2B was indispensable.
Ubiquitination-deficient H2B does not support cell viability
To investigate whether instability of uH2B has any physiological consequence, we constructed a plasmid containing htb1-K119R mutant gene (pH2B-K119R) that had a lysine-to-arginine substitution at the conserved monoubiquitination site K119 of histone H2B to R by site-directed mutagenesis. Plasmid pH2B-K119R tagged with FLAG and under the inducible promoter nmt1 failed to produce the normal band of uH2B (Supplementary Figure 1). We first tested whether high-copy plasmid pH2B-K119R could complement the ts phenotype of htb1 mutants. As shown in Figure 4A, pH2B-K119R suppressed neither the ts phenotype of htb1-72 nor htb1-223. Secondly, we examined whether htb1-223 mutant carrying plasmid pH2B-K119R displayed the missegregation phenotype at 36°C. Although the appearance was delayed, the missegregation phenotype was still observed (closed diamonds in the middle panel in Figure 4B). In addition, the septation index increased, indicating that cytokinesis was delayed, resembling the phenotype of htb1-72. In control htb1-223 cells carrying wild-type plasmid pH2B, no missegregation phenotype was observed.
Figure 4.
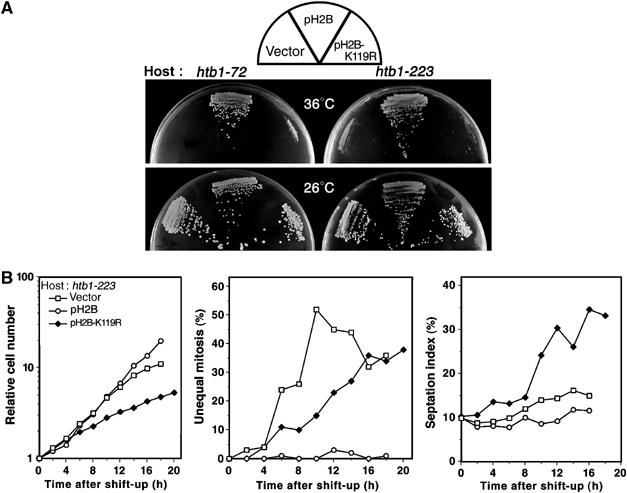
H2B K119R mutant substituted at monoubiquitination site fails to suppress the phenotype of htb1 mutant. (A) Plasmid carrying the htb1-K119R gene failed to suppress the ts phenotype of htb1-72 and htb1-223 mutants. (B) The cultures of htb1-223 mutant carrying vector, pH2B, or pH2B-K119R were asynchronously grown in the EMM2 medium and shifted to 36°C. The cell number increase (left panel), the frequencies of unequal nuclear division phenotype (middle) and the septation index (right) after the shift-up are shown.
CENP-A localization was partly diminished in htb1-223
To examine whether Cnp1, the centromere-specific histone H3 variant CENP-A in fission yeast (Takahashi et al, 2000), could be normally localized to the centromere in htb1-223 mutant, the cnp1+ gene was tagged with GFP and integrated onto the chromosome and expressed under the native promoter. Cnp1-GFP signal was observed after methanol fixation. In control htb1+ cells, Cnp1-GFP signals were observed as dots signals in nuclei throughout the cell cycle. After shifting-up to 36°C, the frequencies of cells with dots signals in htb1+ were 83% (0 h), 76% (4 h) and 76% (8 h) (Figure 5A, bottom panel). However, in htb1-223 cells cultured at 36°C for 8 h, the frequencies of Cnp1-GFP signals were significantly diminished from 82% at 26°C (0 h) to 53% at 36°C (6 h). Examples of the dot and dispersed signals of Cnp1-GFP are shown in the upper panels of Figure 5A. Quantitative data are indicated in the lower panel. The blue portions represent the fractions of cells that displayed the dispersed Cnp1-GFP signals. The frequencies of centromeric Cnp1-GFP in htb1-72 cells, however, are similar as those in htb1+. These results suggested that Cnp1 was not properly loaded onto centromere in htb1-223 or that centromeric chromatin architecture was structurally spread whereas Cnp1 binding to centromeres remained. We examined whether Rhp6 has any influence over Cnp1 localization, and found that Cnp1 localization was normal in Δrhp6 deletion mutant cells (data not shown), suggesting that monoubiquitinated H2B did not seem to be implicated in Cnp1 localization.
Figure 5.
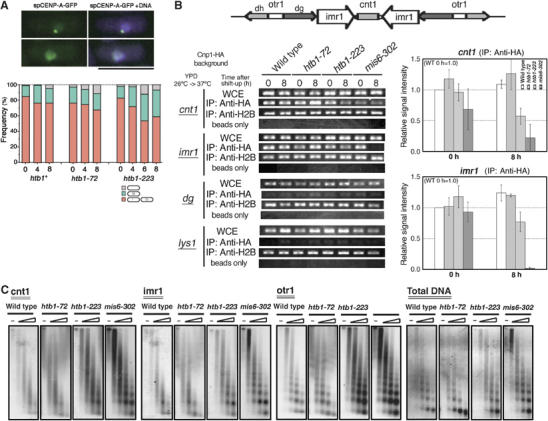
Centromere-specific chromatin structure is partly disrupted in htb1-223 mutant cells. (A) Cnp1-GFP signals (green) were observed in the htb1 mutant background. DAPI was used to stain DNA (purple). Bar, 10 μm. For quantitative measurements, cells were grouped into three classes based on the localization patterns of Cnp1: the dots signals (orange), the dispersed signal (blue) and no detected signal (gray). (B) CHIP experiment was performed using the central centromere probes of cnt1, imr1, the outer heterochromatic repeat dg and pericentric lys1 (lower left). The organization of cen1 is schematized (top). Relative signal intensities were determined and quantitative data are shown (lower right, the value for wild type 0 h is 1.0). The amount of cnt1 and imr1 DNAs co-precipitated with the Cnp1-HA fusion was reduced in htb1-223 mutant cells at 37°C (anti-HA for cnt1 and imr1). (C) Micrococcal nuclease experiments. Nuclear chromatin were prepared from wild-type, htb1-72, htb1-223 and mis6-302 cells that were cultured at 37°C for 7 h, and digested with micrococcal nuclease for 0, 1, 2, 4 and 8 min, followed by agarose gel electrophoresis and Southern hybridization using the three centromeric DNA probes cnt1, imr1 and otr1. The ethidium bromide staining patterns of the gel are also shown.
To further substantiate the above localization result, chromatin immunoprecipitation (CHIP) was performed under the htb1 mutant background. The cnp1+ gene was tagged with HA and integrated onto the chromosome under the native promoter (Takahashi et al, 2000). Cnp1-HA was immunoprecipitated after formaldehyde fixation. The levels of PCR signals in the htb1-223 mutant background were estimated, using the central centromere primers (cnt and imr). As shown in Figure 5B, the CHIP patterns (lower left) and their quantitative estimations (lower right) indicated that the levels of the central centromere sequences bound to Cnp1 were reduced after the shift-up to 37°C. Wild-type and mis6-302 are also shown as a control: it is known that Cnp1 centromere localization is greatly reduced in mis6-302 (Takahashi et al, 2000). These results supported the possibility that htb1-223 mutation had an effect on Cnp1 centromere localization, and showed that the degree of association between centromeric DNA and Cnp1 was diminished. The t-test was conducted and the P-values obtained were 0.024 and 0.013 for cnt and imr, respectively. A possibility was raised that, if nucleosome density was altered by the H2B mutation, then the measurements of nucleosome modifications and variants are not informative without correction for nucleosome content. We therefore performed CHIP using anti-H2B antibodies. No significant change in the nucleosome density was found in the centromeric regions (Figure 5B).
Partial disruption of specialized centromere chromatin in htb1-223
The above experiments could not establish that centromeric chromatin was disrupted in htb1-223 mutant cells. To examine whether centromeric structure was defective in htb1-223, we performed a micrococcal nuclease digestion experiment. When nuclear chromatin of wild-type S. pombe is digested with micrococcal nuclease, Southern blot using the probes of inner centromere DNAs displays a specialized chromatin with smeared digestion pattern (Polizzi and Clarke, 1991; Takahashi et al, 1992). In cnp1, mis6 and mis12 centromere-defective mutants (Saitoh et al, 1997; Goshima et al, 1999; Takahashi et al, 2000), the specialized chromatin was abolished and this was the cause for unequal chromosome segregation. We tested whether the centromere-specific chromatin digestion pattern was impaired in htb1-223 mutant. As shown in Figure 5C, the smeared patterns were significantly diminished, and the more regular-ladder like patterns were obtained for the imr centromere probes in htb1-223 mutant at 37°C for 7 h (Figure 5C, imr1). The smeared pattern was maintained in the cnt region, however. The digestion pattern of htb1-72 was smeared as that in wild type. The pattern for mis6-302 is also shown as a control of the loss of specialized chromatin. These results showed that the specialized centromere chromatin was disrupted in the imr region of htb1-223 mutant cells. The imr regions are inverted repeats that are 100% identical in each centromere and contain a number of tRNA genes (Takahashi et al, 1991).
We tested whether Rhp6 has any effect on centromere chromatin. To this end, micrococcal nuclease digestion experiment was carried out for Δrhp6 deletion mutant. As shown in Supplementary Figure 2, the smeared digestion patterns were observed in the central centromere chromatin in Δrhp6 mutant cells, suggesting that the reduced monoubiquitination of H2B did not significantly affect centromere chromatin at the level of micrococcal digestion pattern.
Genetic interactions of htb1-223 with segregation-defective mutants
To identify genetic interactions between htb1 and various mutations, we constructed the double mutants. The results are shown in Table I. Synthetic growth defects at the permissive temperature were found for the double mutants of htb1-223 with cnp1-1, mis6-302, mis12-537 or dis1-288. These mutants were all defective in centromere/kinetochore functions (Nakaseko et al, 1996; Saitoh et al, 1997; Nabeshima et al, 1998; Goshima et al, 1999; Takahashi et al, 2000). Such synthetic phenotypes were not observed at all for the double mutants employing the allele of htb1-72 (Supplementary Figure 3). These results confirmed that centromere/kinetochore functions were defective in htb1-223.
Table 1.
Synthetic genetic interactions of htb1-72 and htb1-223 mutants with various mitotic, phosphatase, kinase and deacetylase mutants
| htb1-72 | htb1-223 | |
|---|---|---|
| cnp1-1 | ND | Lethal |
| mis6-302 | No effect | Synthetic growth defect |
| mis12-537 | No effect | Synthetic growth defect |
| dis1-288 | No effect | Synthetic growth defect |
| dis2-11 | Synthetic growth defect | Synthetic growth defect |
| Δpka1 | No effect | Suppression |
| clr6-1 | Synthetic growth defect | Weak synthetic growth defect |
| cut9-665 | No effect | No effect |
| mts2-1 | No effect | No effect |
| ND, not determined. | ||
For the double mutants using dis2-11 (defective in type 1 protein phosphatase) and clr6-1 (histone deacetylase, HDAC) (Ohkura et al, 1989; Grewal et al, 1998), both htb1-223 and htb1-72 showed the same synthetic effects, and for pka1 deletion mutants (cAMP-dependent kinase) (Yamashita et al, 1996) suppressed the ts phenotype of htb1-223. It may not be surprising for H2B to interact with PP1, PKA and HDAC as H2B is known to have the sites for phosphorylation and acetylation in the amino-terminal tail. The above results suggest that deacetylated and dephosphorylated H2B is appropriate for the central centromere chromatin, consistent with our current view for the assembly mechanism of central centromeric architecture (Hayashi et al, 2004). The phosphorylation site in the tail has the PKA consensus sequence. Opposite effects of Dis2 (PP1) and PKA on both htb1 mutants are consistent with a hypothesis that they may competitively act on the same phosphorylation site.
Chromatin silencing diminished in htb1 mutants
We examined whether transcriptional silencing was normal in the htb1-72 and htb1-223 mutant background. To this end, the ura4+ reporter gene chromosomally inserted in the heterochromatin regions was employed (Ekwall et al, 1996, 1999). In S. pombe, centromere, telomere and the mating-type locus are heterochromatic regions, and the common DNA sequences are present. Centromere consists of the central core domain (cnt), the inverted repeats domain flanking the central core (imr) and the outer heterochromatic repeats domain (otr), as shown in Figure 5B. To examine gene expression in htb1 mutants, expression of the inserted ura4+ was assayed by growth on non-selective (N/S) and counterselective (5-fluoroorotic acid (5-FOA), which inhibits the growth of Ura+-expressing cells) media. As shown in Figure 6A (top panel, 30°C in the YPD medium), derepression of ura4+ occurred in the whole centromeres (cnt1, otr1) and the mating-type locus (mat3) in the htb1-72 mutant background. However, in the htb1-223 background, silencing was maintained in these ura4+ inserted cells as in wild-type cells. The control of dicer deletion (Δdcr1; Volpe et al, 2003) that is desilenced is also shown. These results demonstrated that transcriptional silencing was generally abolished in htb1-72 but not in htb1-223 under this experimental condition (YPD, 30°C). When the synthetic medium EMM2 was used at 33°C for assaying the silencing in the htb1-223 background, it became possible to detect desilencing in the cnt1 but not in the otr region (Figure 6A, bottom panel).
Figure 6.

Effect of htb1 mutations on silencing of the inserted ura4+ marker gene and histone modification patterns in the centromere region. The ura4+ insertion sites within cen1: otr1R∷ura4+ and cnt1∷ura4+ are indicated by the short bar in the top panel of Figure 5B. (A) Cells were serially diluted (1:5) and spotted onto the YPD plates: non-selective (N/S) or containing 5-FOA. They were incubated at 30°C for 3 days, or they were spotted onto the EMM2 plates: nonselective (N/S), lacking uracil (−URA) or containing 5-FOA. The highest-density spots contained 1 × 104 cells. (B) Lagging chromosomes observed in anaphase of htb1-72 cells at 18°C. (C) Histone acetylation and methylation patterns in the centromere region were altered in htb1-223 mutant. The levels of histone H3 and H4 acetylation and methylation in the centromeric regions were examined by CHIP using antibodies against acetylated H3 and H4. Anti-AcH4 is against acetylated K5, K8, K12, K16 in H4, whereas anti-AcH3 is against acetylated K9 and K14 in H3. Antibodies against dimethylated K4 in H3 were also used. The centromere probes used were central cnt1, although pericentric probe lys1 was also used. Indicated strains were cultured at the restrictive temperature (37°C) for 0 or 8 h. (D) The levels of histone H2B and (di- or tri-) methylated histone H3 (Lys4) in htb1 mutant cells were examined by immunoblotting. The levels of Lys4 (di- or tri-) methylated histone H3 were diminished in htb1 mutants, particularly in htb1-223 cells.
Mutations such as Δswi6 that disrupt the formation of outer centromeric silent chromatin are known to cause lagging chromosomes in anaphase at low temperature (Ekwall et al, 1995, 1996). We examined whether the same was true for htb1-72 mutant cells by immunofluorescence microscopy using DNA and tubulin staining (Figure 6B). Quantitative data are indicated in the table. The htb1-72 cells were found to display a high incidence of lagging chromosomes on anaphase spindle (27%, n=100) at a low temperature, 18°C. On the other hand, htb1-223 cells showed much lower incidence of lagging chromosomes (7%, n=100) at 18°C. In wild-type cells, lagging chromosomes were scarce (1%, n=100) at 18°C. Although lagging chromosomes were observed in both Δswi6 and htb1-72 mutants, underlying defects might greatly differ, as htb1-72 was not sensitive to TBZ, a tubulin poison (Supplementary Figure 4). The cause of lagging chromosome in htb1-72 might be owing to chromatin defect rather than spindle–chromosome interaction defect.
Altered histone modifications in the centromere chromatin of htb1 mutant
To further substantiate alterations of histone modifications, we performed the CHIP experiments for htb1 mutants cultured at the restrictive temperature for 8 h, using the central centromere probe (cnt) and a pericentric probe for the regular gene lys1. Results using antibodies against acetylated histones H3 (AcH3) and H4 (AcH4), and Lys 4 di- or trimethylated H3 (MeH3K4) under the htb1 mutant background with the control of wild-type and mis16-53 mutant are shown in Figure 6C. It was previously demonstrated that histone H4 was hyperacetylated in control mis16 mutant, specifically in the central centromere regions (Hayashi et al, 2004). An extensive study using the CHIP-on-CHIP method showed that Lys 4-methylated histone H3 was present in the central centromere regions (Cam et al, 2005). In this experiment, it was found that methylation of histone H3 in the central centromere also increased in mis16. Both effects were limited in the central centromere regions in mis16 mutant. In the htb1-72 mutant background, however, such central centromere-specific hyperacetylation or hypermethylation was not found in comparison with the wild-type control. In htb1-223, however, acetylation and methylation slightly increased, but the degree of increase was much less than the case of mis16 centromere mutant cells. These CHIP experiments were performed twice, producing reproducible results, and for Figure 6C, the P-values obtained by the t-test for AcH4, AcH3 and MeH3 were 0.076, 0.100 and 0.022, respectively. A question was raised whether the nucleosome density might be altered by the H2B mutants. If so, the measurements of nucleosome modifications and variants require correction for nucleosome content. The H2B levels were thus examined by immunoblot using antibodies against H2B (Figure 6D), and it was found that there was no significant variation in the H2B contents among the wild type and mutants.
Discussion
In this study, we report pleiotropic phenotypes of three htb1 mutants, which contained amino-acid substitutions at non-helical residues in the highly conserved inner structure of histone H2B. Each mutant showed quite distinct, characteristic cellular phenotypes that include Cyh hypersensitivity, defective monoubiquitination, gene silencing, centromere chromatin and chromosome segregation. One strain (E34K) was only slightly slow in the cell number increase (data not shown), whereas two other strains (G52D and P102L) showed the clear ts phenotypes. Isolation of these mutants appeared to be possible because of the sole H2B gene. The same situation may be applied if three other histone genes, H2A, H3 and H4, will be disrupted and the single remaining histone subtype gene is mutagenized randomly or in a site-directed manner. It was once thought that substitution-type ts mutants might be scarce for the histone genes, as the sequence was extremely conserved during evolution. This seemed to be, however, a misconception. In this study, three H2B substitution mutants could be obtained by screening only ∼500 ts strains. Further, the substitution of conserved E34 residue to K showed little phenotype. Curiously, all the mutated residues were located in the non-helical regions, probably because the structurally flexible loop and the linker regions between helices might be able to produce conditionally altered conformations at different temperatures. These mutants will provide a unique opportunity to experimentally dissect the roles of H2B in the formation of heterochromatin and complex centromeres, and the regulation of gene silencing by siRNA in S. pombe.
The htb1-72 mutation was substituted (G52D) in the DNA-contacting loop L1 of histone fold, and became lethal after 8 h at 36°C. In these mutant cells, broad gene desilencing occurred in the heterochromatin of centromere and the mating-type locus. Overall chromatin structure including the heterochromatin regions might be affected. In fact, the interphase nuclear chromatin stained by DAPI was clearly shrunk (compacted) after 12 h. Although no obvious mitotic defect was observed at the restrictive temperature (36°C), lagging chromosomes were abundantly seen in htb1-72 at a rather low temperature (18°C) after 12 h. As htb1-72 differed from Δswi6 in its insensitive nature to TBZ, further investigation is needed to clarify the nature of lagging chromosomes. Centromere architecture rather than spindle–kinetochore interaction might be defective in htb1-72 mutant at a low temperature, and lagging chromosomes observed were owing to the defect in centromere chromatin itself.
The htb1-223 mutation is substituted (P102L) in the short linker region between α3 and αC helices. The αC extension is specific to H2B and not an element of the histone fold. This extension forms a part of the outer protein surface that also contain antiparallel H2B α3 helix and H2A α2 helix, bound by the strong hydrophobic interaction (Luger et al, 1997). An attractive hypothesis is that htb1-223 affects the chromatin fiber formation by altering the interface interactions between the adjacent nucleosomes. This is consistent with the crystal structure of a tetra-nucleosome recently solved (Schalch et al, 2005). It reveals that linker DNA zigzags back and forth between two stacks of nucleosome cores and that H2B αC is present in the interface between two adjacent nucleosome cores in the chromatin fiber models. This interface might be structurally defective in htb1-223 mutant so that the central centromeric chromatin essential for equal chromosome segregation might be disrupted. Additionally, monoubiquitinated H2B in htb1-223 might become accessible to deubiquitinase or protease. Alternatively, monoubiquitination of H2B might become inefficient in htb1-223 owing to the inaccessibility of ubiquitin conjugating or ligating enzyme.
Two striking phenotypes, frequent unequal chromosome segregation and short-lived uH2B (half-life, 15 min), were found in htb1-223. A specific higher-order chromatin fiber in the central centromere region might be impaired in mutant cells. We suspect that monoubiquitin bound to mutant H2B is either inefficiently ligated, rapidly deubiquitinated or degraded by non-proteasomal protease. It is tempting to speculate that centromere/kinetochore function requires proper monoubiquitination and long half-life of uH2B. However, our results do not support such hypothesis. The centromere location of Cnp1 and the micrococcal nuclease digestion patterns appear to be normal in Δrhp6 deletion that is responsible for monoubiquitination. uH2B itself may be unstable in these mutant cells. The defect in Cnp1 loading is most likely not due to defective monoubiquitination of H2B but solely due to P102L mutation. In consistence with the results of synthetic genetic interactions, centromeric CENP-A localization determined by Cnp1-GFP and CHIP was diminished in htb1-223 cells at restrictive temperature (Figure 5A and B). Further, the innermost repeat (imr) regions appeared to lose the specialized chromatin detected by micrococcal nuclease digestion (Figure 5C). Taken together, chromatin in the central centromere was functionally and structurally impaired in htb1-223, probably owing to the change in protein conformation of mutant H2B protein.
Certain phenotypes of the H2B mutants isolated in this study might be caused by general defects in transcription, so that the expression of other genes involved in these processes are affected, thus causing the observed phenotypes. We measured the level of total RNAs before and after (8 h, 36°C) the temperature shift of H2B mutants, and found that the levels did not alter significantly (data not shown), indicating the lack of great reduction or increase of transcription in these mutants. In htb1-223, the smear-like pattern is changed to a regular ladder at imr1, but only a little change is observed at cnt1. While Cnp1 levels are generally supposed to correlate with smear-like digestion patterns, Cnp1 levels are more reduced at cnt1 than at imr1 in htb1-223. We have no satisfactory explanation for this apparent discrepancy. The imr regions contain a large number of tRNA genes, but the cnt regions do not. Recently, these tRNA genes were reported to play a role in chromatin boundary (Scott et al, 2006). The changes in the imr chromatin structure due to a slight decrease of Cnp1 enrichment might be amplified to a significant change in the micrococcal nuclease digestion pattern. Alternatively, the chromatin properties detected by the digestion and Cnp1 CHIP are different, so that an unknown defect in H2B mutant protein might cause these two phenotypes.
In the budding yeast S. cerevisiae, the substitutions in histone H4 caused the ts phenotype defective in mitotic chromosome transmission (Smith et al, 1996). Two mutations are needed within histone H4 to generate the ts phenotype: (1) lethal change at 82nd residue from T to I and (2) at 89th residue from A to V as an intragenic suppressor. The H4 82nd residue is located in the loop L2 that is bound to DNA. In consistent with the mutation site, transcription is affected in mutant cells. These mutant phenotypes are somewhat reminiscent to those of htb1-72, but quite different with regard to its strong genetic interactions with centromere-related gene CSE4 (similar to CENP-A). Apparently, the budding yeast H4 mutant displayed both phenotypes of htb1-72 and htb1-223. In the budding yeast H2A substitution mutant (the residues 20th from S to F, and at 30th from G to D), the phenotypes similar to H4 were found, resulting in G2–M arrest and an increased rate of chromosome loss (Pinto and Winston, 2000). The centromere-specific chromatin structure is disrupted in the H2A and H4 histone mutants (Meluh et al, 1998; Pinto and Winston, 2000). Taken together, the present and the previous results of budding yeast and fission yeast histone mutant analyses indicate that histones H2A, H2B (and uH2B) and H4 in addition to CENP-A, a centromere-specific histone H3, play important roles in centromere/kinetochore functions.
Materials and methods
Strains, media, PCR primers and antibodies
S. pombe histone H2B mutant strains used were described previously (Yuasa et al, 2004). The complete YPD, the minimal EMM2 and the sporulation medium SPA were used (Moreno et al, 1991). The cell number was measured by the Sysmex F-800 (Toa Med. Elec. Co.). For isolation of mutant htb1 gene and construction of plasmids pH2B and pRHP6, fragments of htb1 gene and rhp6 gene were amplified by the PCR method with the nucleotides 5′-CGCGGATCCAACGATGAAGCAGCGAATGG-3 ′ and 5′-CGCGGATCCAAACCACATCAACTTCGCAC-3 ′ (for htb1) and 5′-TCAATCACGTAGCAGCGC-3′ and 5′-TTGGGAACAGCCGTTGAG-3′ (for rhp6). Polyclonal antibodies against S. pombe histone H2B were prepared by immunizing rabbits using the amino-terminal peptide (SAAEKKPASKAPAGKA) as antigen. Anti-HA (12CA5) monoclonal antibodies (Boehringer) and antibodies against methylated histone H3 (Abcam; ab7766) and acetylated histones H3 and H4 (Upstate Inc., #06-598 and #06-599) were used.
Microscopy
Cells expressing the GFP-tagged genes integrated on the chromosome were fixed with methanol. For the missegregation assay, LacI-GFP-NLS bound near the cen1 locus was expressed in the presence of thiamine (Nabeshima et al, 1997). The procedures for immunofluorescence microscopy were described by Hagan and Hyams (1988). Cells were fixed with paraformaldehyde and stained by TAT1 antibodies (Woods et al, 1989).
Immunoblotting
One volume of S. pombe cell culture (containing 1 × 108 cells) was mixed with 1/4 volume of ice-chilled 100% trichloroacetic acid (TCA). The resulting mixture was collected by centrifugation, and the pellets were washed with 10% TCA, followed by cell disruption with glass beads in 10% TCA. After centrifugation at 8000 r.p.m. for 10 min at 4°C, the washed precipitates were resuspended in the SDS sample buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and boiled at 70°C for 30 min. After centrifugation at 14 000 r.p.m. for 10 min, the supernatant was used for SDS–PAGE and immunoblot.
Chromatin immunoprecipitation
CHIP was carried out as described (Saitoh et al, 1997; Takahashi et al, 2000). Cells cultured at 37°C were fixed by formaldehyde. For immunoprecipitation, cell extracts were prepared using the extraction buffer (25 mM HEPES–KOH at pH 7.5, containing 200 mM NaCl, 10% glycerol, 0.2% NP-40, protease inhibitors, 1 mM PMSF and 1% Trasylol). Antibodies conjugated with protein A–Sepharose were used.
Micrococcal nuclease digestion
The procedures for micrococcal nuclease digestion were according to Takahashi et al (1992). Cells cultured at 37°C for 7 h were used for preparing extracts and treated with micrococcal nuclease (250 U/ml; Worthington Biochem). Plasmids pKT110, pKT108 and pYC148 were used as hybridization probes for making the micrococcal digestion patterns.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Acknowledgments
We are grateful to Dr Kozo Ajiro for discussion and technical instruction. This study was supported by the CREST research grant from the Japan Science and Technology Agency (JST) and the COE research grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). TM, TH and TN were supported by the 21st Century COE grant of the Graduate School of Biostudies, from the MEXT. TN is a recipient of postdoctoral fellowships of the Japan Society for Promotion of Science (JSPS).
References
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC (1999) Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153: 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109 (Part 11): 2637–2648 [DOI] [PubMed] [Google Scholar]
- Elgin SC, Workman JL (2002) Chromosome and expression mechanisms: a year dominated by histone modifications, transitory and remembered. Curr Opin Genet Dev 12: 127–129 [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12: 1052–1058 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003a) Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003b) Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183 [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M (1999) Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev 13: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Bonaduce MJ, Klar AJ (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12: 178–187 [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS (1988) The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci 89: 343–357 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, Toda T (2001) Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev Cell 3: 389–399 [DOI] [PubMed] [Google Scholar]
- Korber P, Horz W (2004) SWRred not shaken; mixing the histones. Cell 117: 5–7 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- Luger K (2003) Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev 13: 127–135 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Yanagida M (1985) Histone gene organization of fission yeast: a common upstream sequence. EMBO J 4: 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M (1998) Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell 9: 3211–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Saitoh S, Yanagida M (1997) Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol 283: 459–471 [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Nabeshima K, Kinoshita K, Yanagida M (1996) Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells 1: 633–644 [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K (2002) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M (1989) The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell 57: 997–1007 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14: R546–R551 [DOI] [PubMed] [Google Scholar]
- Pinto I, Winston F (2000) Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J 19: 1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C, Clarke L (1991) The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol 112: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA (2003) The structure of DNA in the nucleosome core. Nature 423: 145–150 [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A (1984) Structure of the nucleosome core particle at 7 Å resolution. Nature 311: 532–537 [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M (1997) Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90: 131–143 [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell 11: 3601–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16: 119–129 [DOI] [PubMed] [Google Scholar]
- Smith MM, Yang P, Santisteban MS, Boone PW, Goldstein AT, Megee PC (1996) A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol 16: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M (1991) A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol 218: 13–17 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamada H, Yanagida M (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell 5: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- van Holde KE (1988) Chromatin. New York: Springer-Verlag [Google Scholar]
- Vijay-Kumar S, Bugg CE, Cook WJ (1987) Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol 194: 531–544 [DOI] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC (2003) RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146 [DOI] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K (1989) Definition of indivisiual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci 93: 491–500 [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M (1996) 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature 384: 276–279 [DOI] [PubMed] [Google Scholar]
- Yuasa T, Hayashi T, Ikai N, Katayama T, Aoki K, Obara T, Toyoda Y, Maruyama T, Kitagawa D, Takahashi K, Nagao K, Nakaseko Y, Yanagida M (2004) An interactive gene network for securin-separase, condensin, cohesin, Dis1/Mtc1 and histones constructed by mass transformation. Genes Cells 9: 1069–1082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4


