Abstract
Hydrogen peroxide (H2O2) is the major reactive oxygen species (ROS) produced in sperm. High concentrations of H2O2 in sperm induce nuclear DNA fragmentation and lipid peroxidation and result in cell death. The respiratory chain of the mitochondrion is one of the most productive ROS generating systems in sperm, and thus the destruction of ROS in mitochondria is critical for the cell. It was recently reported that H2O2 generated by the respiratory chain of the mitochondrion can be efficiently destroyed by the cytochrome c-mediated electron-leak pathway where the electron of ferrocytochrome c migrates directly to H2O2 instead of to cytochrome c oxidase. In our studies, we found that mouse testis-specific cytochrome c (T-Cc) can catalyze the reduction of H2O2 three times faster than its counterpart in somatic cells (S-Cc) and that the T-Cc heme has the greater resistance to being degraded by H2O2. Together, these findings strongly imply that T-Cc can protect sperm from the damages caused by H2O2. Moreover, the apoptotic activity of T-Cc is three to five times greater than that of S-Cc in a well established apoptosis measurement system using Xenopus egg extract. The dramatically stronger apoptotic activity of T-Cc might be important for the suicide of male germ cells, considered a physiological mechanism that regulates the number of sperm produced and eliminates those with damaged DNA. Thus, it is very likely that T-Cc has evolved to guarantee the biological integrity of sperm produced in mammalian testis.
Keywords: mouse testis, antioxidation
Roughly half of infertility cases are caused by male infertility (1). One of the mechanisms of male infertility is the excess production of reactive oxygen species (ROS) in sperm, which can induce nuclear DNA fragmentation, lipid peroxidation, and protein–protein cross links (2–4). The primary source of ROS in sperm is the respiratory chain of the mitochondrion (5–8). The main ROS form in sperm is hydrogen peroxide (H2O2), the concentration of which in sperm has not been measured (2–4), whereas in liver cells, the H2O2 concentration (10−7 to 10−9 mol/liter) is known to be 100- to 10,000-fold higher than that of superoxide (O2·−) (≈10−11 mol/liter) (6, 9, 10). O2·−, as the precursor of H2O2 and a short-lived ion, can be rapidly converted to H2O2 by several pathways and does not accumulate in the cell (10, 11). The downstream ROS product of H2O2, namely hydroxyl radical (·OH), is highly reactive in directly inducing DNA single strand break and lipid peroxidation (12, 13). Because H2O2 can also diffuse freely among the membrane systems of the cell, making it the most prevalent ROS form (14), its reduction becomes a major challenge for the normal functions of the cell. Recently, it was demonstrated that the cytochrome c (Cc)-mediated electron-leak pathway can strongly suppress the formation of H2O2 by the respiratory chain and efficiently destroy preexisting H2O2 (10, 15, 16). Cc is thus considered a significant physiological contributor for H2O2 detoxification in the cell (10, 15, 17).
The reduction of H2O2 is more critical for sperm than for somatic cells because of their high content of polyunsaturated fatty acids within the plasma membrane and a low concentration of ROS scavenging enzymes in the cytoplasm (4, 18). During the development of male mammalian germ cells, a testis-specific form of Cc (T-Cc) that differs from its counterpart in somatic cells (S-Cc) is expressed. In the mouse, the genes coding for T-Cc and S-Cc are located at chromosomes 2 and 6, respectively (19). Although the primary structures of these two proteins are 86.5% identical, T-Cc and S-Cc are different not only in the intron–exon genomic organizations but also in expression patterns (20). S-Cc in germ cells gradually declines as the cells undergo meiosis, with pachytene spermatocytes harboring low levels of S-Cc at an approximate S-Cc to T-Cc ratio of 1:4, whereas T-Cc is the predominant form in sperm (21). Thus, the biochemical property changes of T-Cc would have great impact on the Cc-mediated biological processes in male germ cells rich of T-Cc, which include electron transfer chain operation, antioxidation, and probably apoptosis.
Here, we compare the biological functions of T-Cc with those of S-Cc from various aspects. First, measurements of the electron transfer activities with Cc oxidase (CcO) in beef mitochondrial particles show that T-Cc is the better substrate in an alkaline environment. Secondly, the apoptotic activity of T-Cc is shown to be three to five times larger than that of S-Cc in the Xenopus egg extract apoptosis measurement system. Furthermore, we demonstrate that T-Cc catalyzes the reduction of H2O2 three times faster than S-Cc and that the T-Cc heme has the greater resistance to being degraded by H2O2. Finally, in attempting to illustrate these differences of T-Cc from S-Cc, a 1.6-Å crystal structure of T-Cc is resolved and provides a structural interpretation to the increased H2O2 detoxification activity of T-Cc.
Results
For further details, see Supporting Text, Figs. 6–9, and Table 3, which are published as supporting information on the PNAS web site.
Overall Structure of Mouse T-Cc.
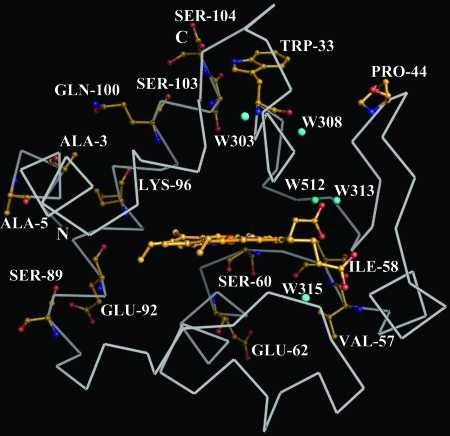
The final structure of T-Cc was refined at 1.6-Å resolution to an Rfactor of 18.2% and Rfree of 20.9%. The (ϕ, ψ) angles of all residues were evaluated by Ramachandran plot (data not shown) and, except for several glycines and for Lys-27, which takes part in forming an unusual γ-turn (22), all other residues are within the permitted region. The skeleton of T-Cc is similar to that of horse heart Cc (hh-Cc; Protein Data Bank ID 1HRC) with the greatest rms deviations (rmsd) at the residue regions 1–5, 21–23, 37–46, 55–58, 85–87, and 96–104. These differences largely occur at the regions with the differences of the primary structures of the two proteins. All of the residues in T-Cc differing from those in S-Cc are located at the distal side (back side as shown in Fig. 1) of T-Cc, whereas there are few changes at the proximal side. The prosthetic group, the heme, bonded covalently to Cys-14 and Cys-17, is buried in the hydrophobic pocket surrounded by the upper part (residues 1–58) and bottom part (residues 59–104) of the protein. The mouth of the pocket is formed by two loop regions from the upper part (residues 14–18, 28–30) and bottom part (residues 78–83). The inner part of the protoporphyrin ring of the heme is involved in a network of hydrogen bonds with the rest of the protein (Table 1). In T-Cc, the hydrogen bond interactions around the posterior propionate of the heme show significant variations compared to those of hh-Cc (Table 1).
Fig. 1.
Schematic diagram of T-Cc illustrated from the proximal side, as viewed from the heme opening of the molecule. Backbones are drawn with the ribbon model and colored white. The 14 residues different to those in S-Cc are shown in the ball and stick model and colored orange. Five water molecules located in the internal part of the protein are in cyan.
Table 1.
Heme propionate hydrogen bond interactions
| Interaction | Distances, Å |
|
|---|---|---|
| T-Cc (Protein Data Bank ID 2AIU) | hh-Cc (Protein Data Bank ID 1hrc) | |
| Heme O1A-Try-48 OH* | 2.60 | 2.69 |
| Heme O1A-Water A OH* | 2.59 | 2.91 |
| Heme O1A-Water B OH* | 3.53† (3.05) | |
| Heme O2A-Trp-59 NE1* | 2.91 | 2.67 |
| Heme O2A-Asn-52 ND2* | 2.95 | 3.31 |
| Heme O2A-Gly-41 N* | 2.83 | 3.18 |
| Heme O1D-Thr-49 N | 2.99 | 2.97 |
| Heme O2D-Thr-49 OG1 | 2.63 | 2.69 |
| Heme O2D-Lys-79 N | 2.99 | 2.73 |
| Heme O2D-Thr-78 OG1 | 3.22 | 2.98 |
Value in parentheses is averaged mean for rice, horse, iso-1, and iso-2 Cc. Waters A and B are short for equivalents to Water 125 (Water 313 in T-Cc) and Water 139 in hh-Cc, respectively.
*The interactions around Arg-38.
†Listed for comparison.
In the later stage of refinement, 224 water molecules were located in the protein, of which five (Fig. 1) are buried in the internal part of T-Cc under the van der Waals surface of the protein. These five water molecules are believed to account for some specific biochemical properties of the protein and are discussed below. The locations of these five water molecules were confirmed by the 2Fo − Fc electron density map (Supporting Text and Fig. 8). The other 219 water molecules are all at the surface of the protein, and form a hydrogen-bond network stabilizing the surface area of T-Cc.
Kinetic Assays.
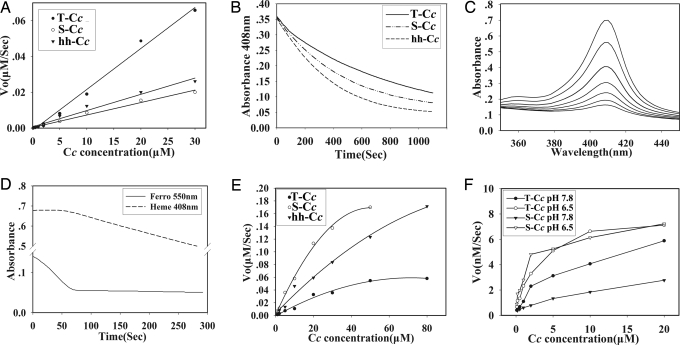
In the presence of 170 μM H2O2, ferroT-Cc reduced H2O2 3-fold faster than S-Cc and hh-Cc (Figs. 2A and 9). From 0 to 30 μM Cc, the reaction appeared as a first-order reaction where Vo = K × [Cc], where the reaction constants K (μM−2·s−1) for T-Cc, S-Cc, and hh-Cc are shown in Table 2, with the K value of T-Cc being about three times that of S-Cc and hh-Cc.
Fig. 2.
Kinetic studies of T-Cc. (A) The initial rates of H2O2 reduction by various concentrations of ferroT-Cc, ferroS-Cc, and ferrohh-Cc. (B) Degradation of the heme of ferriT-Cc, ferriS-Cc, and ferrihh-Cc in the presence of 3 mM H2O2. (C) Heme destruction of ferriT-Cc as indicated by dissipation of the Soret band measured at 408 nm. Spectra were scanned every 3 min. (D) Time course of ferroT-Cc oxidation (solid line) and inactivation (dashed line) in the presence of 3 mM H2O2. (E) Reduction titration of T-Cc, S-Cc, and hh-Cc with ascorbate. (F) Activities of purified CcO with T-Cc and S-Cc. Measurements were performed under both low pH (50 mM phosphate/Tris, pH 6.5) and high pH (25 mM acetate/Tris, pH 7.8) conditions.
Table 2.
Summary of kinetic assays
| T-Cc | S-Cc | hh-Cc | |
|---|---|---|---|
| Reducing activity, K, sec−1 × 10−3 | 2.3 | 0.7 | 0.9 |
| Degradation constant, KD, sec−1 × 10−4 | 10.29 | 13.67 | 18.26 |
| Half life of bleaching, T1/2, sec | 604 | 410 | 300 |
| Ascorbate reduction, KR, sec−1 × 10−4 | 16.08 | 55.53 | 35.92 |
As indicated by the decrease of the Soret band absorption at 408 nm (Fig. 2B), the heme of ferriT-Cc showed much more resistance to being degraded by excess H2O2 (3 mM) than ferriS-Cc and ferrihh-Cc. With the same initial concentration, the half lives of T-Cc, S-Cc, and hh-Cc were 604 s, 410 s, and 300 s, respectively. Confirming the decrease at 408 nm, the complete spectrum of the Soret band of T-Cc showed the expected changes with time (Fig. 2C). The similar data for S-Cc and hh-Cc are not shown. In contrast, under the same conditions, the heme of the three ferroCc’s used is not degraded by H2O2 as long as they remain in the ferrous state (Fig. 2D). The curves of heme degradation and of oxidation of the protein to the ferric state by H2O2 in all three ferroCc’s had biphasic behaviors. The first phase, a flat line at 408 nm, indicating no heme degradation, and a dropping curve at 550 nm, representing continuous oxidation of the protein to the ferric state, demonstrated that Cc can protect its heme from H2O2 degradation when the protein is not fully in the ferric form. In the second phase, the fully ferric Cc (as seen from the flat line at 550 nm) started to be destroyed by H2O2, generating a slope at 408 nm.
Fig. 2E and Table 2 show that the reduction rate of T-Cc by ascorbate was ≈1/3 and 1/2 that for S-Cc and hh-Cc, respectively.
The electron transfer activities of T-Cc and S-Cc with CcO in the beef mitochondrial particle preparation were compared by measuring the initial rates of oxidation of fully ferrous samples (Fig. 2F). The activities of T-Cc and S-Cc with CcO were similar in the pH 6.5 buffer, but T-Cc was more efficiently oxidized than S-Cc in a more alkaline buffer (pH 7.8). T-Cc had a more steady performance than S-Cc indicated by the lower response to pH changes (Fig. 2F).
Apoptotic Activity of Mouse T-Cc in Xenopus Egg Extracts.
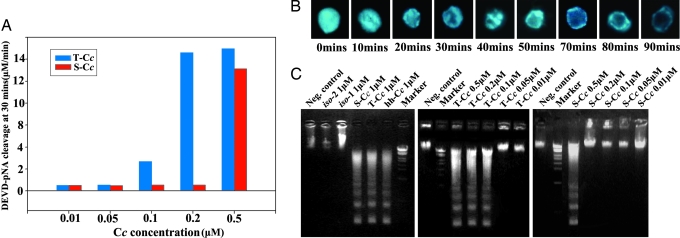
The threshold concentration for T-Cc to fully activate caspase-3, one of the downstream components from Cc in the intrinsic apoptosis pathway (23), was 0.1 μM and 3- to 5-fold lower than that for S-Cc (0.3–0.5 μM) (Fig. 3A), indicating that T-Cc has a much higher apoptotic activity than S-Cc. To further confirm this conclusion, two other independent hallmarks of apoptosis were used, namely chromatin condensation (Fig. 3B), which is caused by the loss of chromatin decondensation activity of nucleoplasmin, and DNA fragmentation (Fig. 3C), which is the result of digesting chromatin by caspase-activated DNase/DFF40 (DNA fragmentation factor, 40 kDa). The differences of activities between T-Cc and S-Cc in these two experiments were similar to those in the caspase-3 activation experiment.
Fig. 3.
Apoptotic activity of T-Cc. (A) Effect of T-Cc and S-Cc on the rate of DEVD-pNA cleavage. Initial rates of DEVD-pNA cleavage were determined by comparing to the standard curve for pNA. (B) Chromatin condensation induced by adding 1 μM T-Cc and erythrocyte nuclei into Xenopus egg extracts. Samples were fixed and stained by DAPI at the time indicated for fluorescence observation. (C) DNA fragmentation assays were carried out by adding various concentrations of T-Cc, S-Cc, and 105 chicken erythrocyte nuclei into Xenopus egg extracts, followed by electrophoresis (see Materials and Methods).
Differences of Internal Water Patterns Between Testicular and Somatic Cytochromes c.
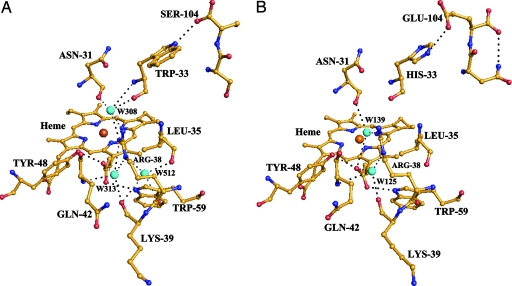
From the comparison of the crystal structures of rice, yeast iso-1, yeast iso-2, and horse heart cytochromes c, there are two internal water molecules, namely W139 and W313 (Fig. 4B), highly conserved in the vicinity of the heme. Surprisingly we found that W139 is missing in T-Cc (Fig. 4A), whereas two internal water molecules are added. One of them (W512) has a high level of vibration as indicated by the relatively large value of the calculated B factor. W313 forms three hydrogen bonds to Arg-38. W512, located just next to W313, also forms a hydrogen bond to Arg-38. The other new internal water molecule, W308, has a low level of vibration and is located just above the location of W139, namely the water molecule which is absent in T-Cc as compared to other cytochromes c. As shown in Fig. 4A, W308 forms hydrogen bonds to three residues, including two residues invariant in all cytochromes c (Arg-38, Asn-31) and Trp-33, which is substituted by His-33 in all currently known mammalian S-Cc.
Fig. 4.
The region around the Arg-38 site in T-Cc (A) and hh-Cc (B). Water molecules are shown as cyan spheres. Possible hydrogen bond interactions are marked as black dashed lines. The ferric ion in the center of the heme is shown as an orange sphere. The carbon, oxygen, and nitrogen atoms are in yellow, red, and blue, respectively.
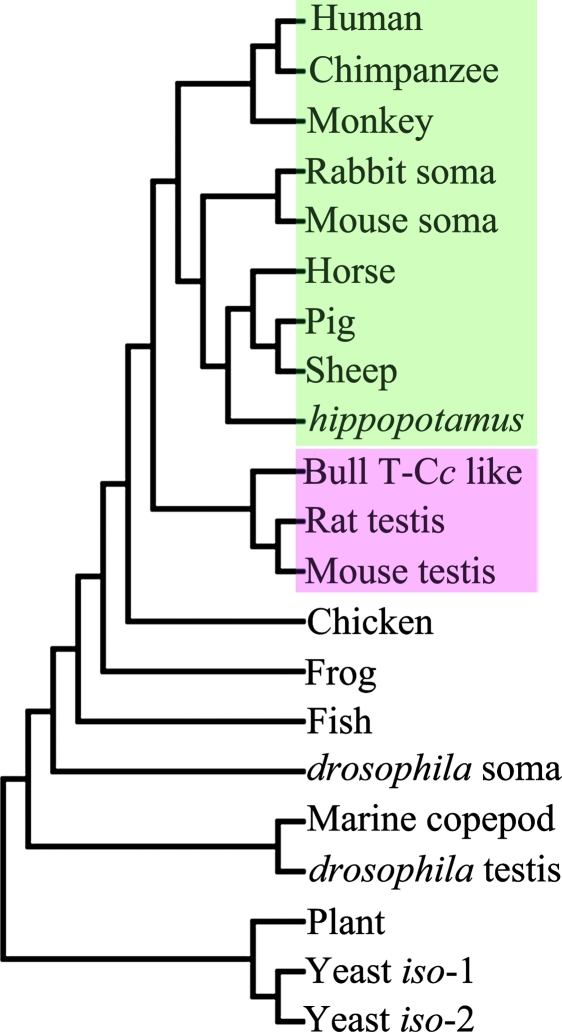
Evolutionary Analysis.
The amino acid sequences of 21 Cc genes were retrieved from the NCBI database. From the evolutionary analysis using the protein sequences of these cytochromes c to build a phylogenetic tree (Fig. 5), it turns out that the testicular and somatic cytochromes c are clustered into two classes (Fig. 5). These tissue-specific Cc genes are thought to evolve by duplication from a common ancestral Cc gene (24, 25). Applying a value of 11 million years for every residue difference along two divergent lines of Cc (26), the 14-aa residue differences between S-Cc and T-Cc implies that they separated from each other ≈150 million years ago.
Fig. 5.
Evolutionary analysis. Phylogenetic tree for Cc’s from various species, constructed as described in Materials and Methods. The clades for testicular and somatic Cc’s are marked pink and green, respectively.
Discussion
A fundamental question is why a testis-specific Cc has evolved in male mammalian germ cells. Because Cc has three major functions, namely, electron transfer, apoptosis and antioxidation (10, 23, 27), the question becomes what functions of these three have significant changes in T-Cc as compared to S-Cc.
Has T-Cc particularly evolved to fulfill the high energy requirement of sperm? Our studies demonstrated that the electron transfer activities of T-Cc and S-Cc with CcO in beef mitochondrial particle preparations are about the same in an acidic environment (pH 6.5), whereas T-Cc is the better substrate in an alkaline environment (pH 7.8). Assuming the absolute concentration of Cc in mitochondria of sperm is the same to that in somatic cells, the similar rates of reaction of T-Cc and S-Cc with CcO at pH 6.5 suggest that there is no significant difference of electron transfer activities between T-Cc and S-Cc. Likewise, the much higher energy need in motile sperm is satisfied by the increased concentration of mitochondria within the midpiece of sperm (28). The energy needs in sperm can be met even without T-Cc, because mice deficient of T-Cc are still fertile (20). Moreover, in mammalian cells, the cellular ATP concentration is sufficiently high (≈2 mM) to keep cultured cell alive for several days upon ATP synthase inhibition (29). Taken together, these observations argue strongly against the possibility that T-Cc has particularly evolved to increase the energy production of sperm. On the other hand, because Cc interacts with CcO by its proximal side (30), where the residues are the same in T-Cc and S-Cc (Fig. 1), the very different rates of oxidation of these two proteins by CcO at pH 7.8 would indicate that factors other than the proximal side structure must be involved. However, what these factors are and how they affect the reaction at different pH values need further investigation.
The apoptosis of male germ cells is proposed to be a mechanism regulating the quantity and quality of sperm produced in mammalian testis (20). Although, to date, there is still no direct evidence that Cc is the principal factor responsible for signaling apoptosis in male germ cells, it has recently been demonstrated that the mitochondria-dependent pathway of apoptosis plays a key role in the death processes of male germ cells (31). Moreover, infertile males show significant elevated levels of Cc, Caspase-9 (the direct downstream component from Cc in the intrinsic apoptosis pathway), and Caspase-3 in their semen samples (32). These two observations strongly underscore the importance of the Cc mediated apoptotic pathway in the apoptosis of male germ cells. The apoptotic activity of T-Cc is three to five times that of S-Cc in the Xenopus egg extract system. This dramatically stronger apoptotic activity of T-Cc might be important for the apoptosis of male germ cells. A rational indication is that T-Cc could make male germ cells, as compared to somatic cells, undergo apoptosis more easily in reacting to ROS challenges, and therefore play a much more stringent “quality control” role, as compared to S-Cc. T-Cc has 14 residues differing from those in S-Cc, almost all of which are located at the distal surface of the protein (Fig. 1) and might account for the enhanced apoptotic activity of T-Cc.
Imbalance between ROS production and antioxidant capacity in sperm would lead to increased production of oxidants, causing mitochondrial and genomic damages and finally resulting in cell death (2–4). Cc works as a “bodyguard” protecting cells from damages by H2O2 (10, 15, 16), the major form of ROS in sperm. The H2O2 reduction activity of T-Cc is 3-fold that of S-Cc. Therefore, in addition to the possible “quality control” role after ROS damages of mitochondria and nuclear DNA, T-Cc can also prevent these damages by much more efficiently destroying H2O2. Down-regulation of the H2O2 production in sperm can thus ensure the integrity of the genome transmitted by sperm.
Interestingly, what confers to T-Cc the much stronger H2O2 reduction activity than S-Cc? The crystal structure of T-Cc reveals that the environment around Arg-38 of T-Cc is significantly altered from that of hh-Cc (Fig. 4 A and B). Arg-38 affects the redox behaviors of Cc by charge-charge interactions with the heme posterior propionate and/or by hydrogen bonding to the heme posterior propionate mediated by surrounding water molecules, namely W125 and W139 (22, 27, 33, 34). The Arg38Ala mutant of yeast iso-1 Cc shows a significant increase of its reduction potential (≈40 mV) and a more stable oxidized form compared to the wild-type yeast iso-1 Cc (34). Our experiments also demonstrate that T-Cc has a much lower rate of reduction by ascorbate than S-Cc and hh-Cc (Table 2). The ascorbate-binding site is suggested to be at the crevice defined by Arg-38 (35, 36). Therefore, in the absence of significant changes elsewhere, the above observations jointly suggest that the changes of T-Cc from S-Cc around Arg-38, including the new pattern of the internal water molecules, cause the extraordinarily high activity of T-Cc in H2O2 reduction, and its resistance to ascorbate reduction.
Whether the testicular form of the Cc gene is present only in rodent mammals has not been examined directly. However, the evolutional divergence of T-Cc from S-Cc, occurred ≈150 million years ago as calculated above (Evolutionary Analysis) before the eutherian–mammalian radiation (≈100 million years ago) (37), and is close to the estimated divergence time of birds and mammals (38). In 2003, a pseudogene similar to T-Cc was recognized in the human genome (25). Recently, the coding DNA sequence of a T-Cc like gene was also isolated from cow cDNA (GenBank accession no. AAI02715.1). These observations strongly imply that, no matter whether functional or not, the T-Cc gene is not only present in rodent mammals but is very prevalent in mammals in general.
Cc serves as an electron carrier in bacteria (27). As evolution progressed, new roles appeared to be added to this antique protein in different organisms. There are two main possibilities by which this can be obtained, either by conserving the essential domains while mutating others, such as in the case of the apoptotic function of mammalian somatic Cc, or by gene duplication followed by divergent mutations to obtain novel functions, such as the Cyt-cd and Cyt-cp cytochromes c of Drosophila (24, 25). In Drosophila, the Cyt-cd form is required for the final stage of spermatic terminal differentiation through an apoptosis-like mechanism (39), whereas the Cyt-cp isoform is the usual form of Cc used by all tissues of the fly. The mouse T-Cc gene, apparently a duplicated form of the S-Cc gene with independent evolution for >100 million years, protects sperm from ROS with its enhanced anti-H2O2 capability on the one hand and might be adapted to eliminate damaged male germ cells by its high apoptotic activity on the other hand. Thus, it is very likely that T-Cc has evolved to ensure the fidelity and efficiency of the DNA transmission by sperm.
Materials and Methods
Construction and Expression of Plasmids.
The cDNA of T-Cc was synthesized with eight single-strand primers, each ≈80 bp in length, in a single PCR. The Escherichia coli pBTR expression system for yeast iso-1 Cc (40) was converted to express a variety of cytochromes c. The cDNA of T-Cc, S-Cc, and yeast iso-2 Cc was cloned into pBTR through two BamHI restriction sites (see Supporting Text). Because of its poor yield in the E. coli system, S-Cc cDNA was further cloned into YEp13 plasmid with a CYC1 promoter, which was well expressed in the GM-3C-2 (ΔCtmp) yeast strain whose Cc methyltransferase was knocked out by an efficient site-specific homologous recombination method to avoid the trimethylation of Lys-72 (41). The directions and sequences of all constructs were confirmed by DNA sequencing. cytochromes c, except S-Cc, were overexpressed under the conditions of 37°C, lack of oxygen, pH 8.0, and in the DH5α E. coli strain. hh-Cc was purchased from Sigma. For S-Cc, all of the procedures of expression and pre-HPLC purification were performed according to an established protocol (42).
Purification of Cc.
The procedures of purification were based on the method described by Patel et al. (40). The purity and sequence of T-Cc protein were also confirmed by peptide mass fingerprint (43). The concentrations of the various cytochromes c were determined at 550 nm from the difference in the absorption of the ferrous and ferric forms of the protein employing the Hitachi 2800 spectrophotometer used throughout this study. The ΔεmM550(reduced − oxidized) of yeast Cc is 18.5, and that of the mammalian Cc is 19.6. The concentrations of the mammalian ferriCc’s were further confirmed at 410 nm (ΔεmM410 = 106.1).
Reactions of Cc with H2O2, Ascorbate, and Beef Heart Cc Oxidase.
The kinetics of oxidation of ferroCc by H2O2 was studied spectrophotometrically at 550 nm in 0.2 M Tris·Cl (pH 7.0), 170 μM H2O2, and 1–30 μM ferroCc. The degradation of ferriCc heme was estimated from the dissipation of the Soret band absorption at 408 nm in 50 mM phosphate buffer (pH 6.1) containing 3 mM H2O2. The degradation constant (Kd) was calculated from a first-order equation A = Ao·e−Kdt obtained from the residual absorption versus time.
For the assay of reduction by ascorbate, different concentrations of ferriCc ranging from 0.1 to 50 μM were added to 50 mM phosphate Na+, 25 mM Na2SO4 (pH 7.0) containing 0.2 mM ascorbate in a cuvette, which was then sealed to block the access of air. The initial rate of reduction was measured at 550 nm, and the different rates obtained were plotted as a function of the Cc concentration.
The electron transfer activity of Cc to CcO was measured spectrophotometrically. The membrane-bound form of the enzyme, namely a Keilin–Hartree particle preparation, was prepared according to the procedure of Ferguson-Miller et al. (44). Initial rates of oxidation of reduced Cc at two pH values were followed at 550 nm in 25 mM acetate/Tris, pH 7.8 (the standard ionic conditions for polarographic assays), and in 50 mM phosphate/Tris, pH 6.5 (a more acidic buffer reported to be optimal for the spectral assays). The concentration of beef CcO was calculated from the difference between the oxidized and dithionite-reduced spectra using a ΔεmM of 24 at 605 nm. Reproducibility of all of the biochemical assays was confirmed by taking each measurement at least three times and showed a fluctuation <8%. Samples with all components except Cc served as negative controls.
Cell-Free Caspase Activity Assay.
The apoptotic activity of Cc in interacting with apoptotic protease-activating factor 1 (apaf-1) and activating caspase-3 was studied through the cleavage of a caspase-3 substrate, N-acetyl-Asp-Glu-Val-Asp-pNA (DEVD-pNA) from Sigma, measured spectrophotometrically in a time course at 405 nm. A 70-μl reaction mixture containing 20 μl of cell extract from Xenopus egg was incubated with varied concentrations of cytochromes c (from 0.01 to 0.5 μM) in 25 mM Hepes (pH 7.5), 50 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, and 1 mM DTT with a final dATP concentration of 1 mM. After incubation at 23°C for 30 min, DEVD-pNA was added to a concentration of 100 μM, and the increase of absorption at 405 nm was measured every 10 s for a total of 4,000 s. Using the standard spectral absorption curve of p-nitroanilide (Sigma), the initial rate of substrate cleavage was calculated from the slope of the linear portion of the curve of absorption change versus time.
Apoptosis Induction in Chicken Erythrocyte Nuclei and Assembled Nuclei.
A Xenopus egg extract obtained by ultraspeed centrifugation, termed extract S-150, and chicken erythrocyte nuclei were prepared as described by Lu et al. (45). A total of 1 × 105 chicken erythrocyte nuclei were mixed with 30 μl of S-150 and a series of Cc concentrations from 0.01 to 1 μM, and were incubated at 23°C for the times shown in Fig. 3B. After DAPI staining, the changes in the appearance of the chromatin of nuclei were monitored by using an Olympus IX71 fluorescence microscope and images were captured by a cooled charge-coupled device camera. For the DNA fragmentation assay, the reaction mixture was diluted with 10 volumes of 100 mM Tris·Cl (pH 8.0), 5 mM EDTA, 0.2 M NaCl, 0.4% SDS, and 0.2 mg/ml proteinase K and incubated at 37°C for 3 h. The breakage of the DNA, as an indication of apoptosis, was analyzed by electrophoresis on a 1.5% agarose gel.
Crystallization and X-Ray Data Collection.
Crystals of T-Cc were grown by the hanging drop method with reservoir solutions containing 27.5–22% polyethylene glycol 1000 (wt/vol), 30 mM potassium phosphate (pH 7.1) and drops prepared by mixing 1 μl of protein solution (20 mg/ml) and 1 μl of the reservoir solution. After setup at 16°C, crystals appeared several hours later and kept growing for 2–3 days. These conditions are similar to those used by Sanishvili et al. (22) for the crystallization of hh-Cc. The x-ray diffraction data for T-Cc crystals were collected at 100 K (i.e., −173°C) by using a Rigaku R-Axis IV++ image plate with a Rigaku FRE rotating CuKα anode home x-ray generator at 40 kV and 50 mA (wavelength = 1.5418 Å) and diffracted to at least 1.55-Å resolution. All of the intensity data were processed and scaled by using the hkl2000 program (46). Data collection statistics are summarized in Table 3.
Model Building and Refinement.
The structure of T-Cc was solved by the molecular replacement method with the coordinates of ferrihh-Cc (Protein Data Bank ID 1CRC) (22). Cross-rotation function and translation function searches were performed with the program cns (47), and a clear solution for a monomer was found. The residues that differ between hh-Cc and T-Cc were then replaced and geometrically adjusted by using the program o (48) under the guidance of 2Fo − Fc and Fo − Fc difference maps, followed by cycles of iterative rebuilding and refinement with the program cns, until no better resolution of the protein could be obtained.
Phylogenetic Analysis.
A phylogenetic tree was constructed with the amino acid sequences of various cytochromes c, obtained by searching the National Center for Biotechnology Information database (49) using “cytochrome c” as the key word and then applying the neighbor-joining method with “the number of differences” model by the mega3 program (50).
Supplementary Material
Acknowledgments
We thank Prof. A. Grant Mauk (University of British Columbia, Vancouver) for the generous gift of PBTR plasmid, Dr. Zhigang Lu (Peking University) for support and valuable comments, and Dr. Ruslan Sanishvili (Argonne National Laboratory, Argonne, IL) for discussions on the crystal structures. This project was funded by Key Projects of the Chinese Ministry of Education Nos. 03180 and 104232, the TransCentury Training Program Foundation for the Talents of the Ministry of Education, the Tsinghua-Yue-Yuen Medical Sciences Fund, and National Natural Science Foundation of China (NSFC) Grants 30225016 and 30221003.
Abbreviations
- ROS
reactive oxygen species
- Cc
cytochrome c
- T-Cc
testis-specific Cc
- S-Cc
somatic Cc
- CcO
Cc oxidase
- hh-Cc
horse heart Cc.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2AIU).
References
- 1.Sharlip I. D., Jarow J. P., Belker A. M., Lipshultz L. I., Sigman M., Thomas A. J., Schlegel P. N., Howards S. S., Nehra A., Damewood M. D., et al. Fertil. Steril. 2002;77:873–882. doi: 10.1016/s0015-0282(02)03105-9. [DOI] [PubMed] [Google Scholar]
- 2.Aitken R. J., Gordon E., Harkiss D., Twigg J. P., Milne P., Jennings Z., Irvine D. S. Biol. Reprod. 1998;59:1037–1046. doi: 10.1095/biolreprod59.5.1037. [DOI] [PubMed] [Google Scholar]
- 3.Sharma R. K., Agarwal A. Urology. 1996;48:835–850. doi: 10.1016/s0090-4295(96)00313-5. [DOI] [PubMed] [Google Scholar]
- 4.Jones R., Mann T., Sherins R. J. Fertil. Steril. 1979;31:531–537. doi: 10.1016/s0015-0282(16)43999-3. [DOI] [PubMed] [Google Scholar]
- 5.Turrens J. F., Alexandre A., Lehninger A. L. Arch. Biochem. Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 6.Gavella M., Lipovac V. Arch. Androl. 1992;28:135–141. doi: 10.3109/01485019208987691. [DOI] [PubMed] [Google Scholar]
- 7.Cadenas E., Davies K. J. Free Radical Biol. Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 8.Baker M. A., Krutskikh A., Curry B. J., Hetherington L., Aitken R. J. Biol. Reprod. 2005;73:334–342. doi: 10.1095/biolreprod.104.037960. [DOI] [PubMed] [Google Scholar]
- 9.Cross A. R., Jones O. T. G. Biochim. Biophys. Acta. 1991;1057:281–298. doi: 10.1016/s0005-2728(05)80140-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y., Wang Z. B., Xu J. X. J. Biol. Chem. 2003;278:2356–2360. doi: 10.1074/jbc.M209681200. [DOI] [PubMed] [Google Scholar]
- 11.Bielski B. J. H., Arudi R. L., Sutherland M. W. J. Biol. Chem. 1983;258:4759–4761. [PubMed] [Google Scholar]
- 12.Adinarayana M., Bothe E., Schulte-Frohlinde D. Int. J. Radiat. Biol. 1988;54:723–737. doi: 10.1080/09553008814552171. [DOI] [PubMed] [Google Scholar]
- 13.Morehouse L. A., Tien M., Bucher J. R., Aust S. D. Biochem. Pharmacol. 1983;32:123–127. doi: 10.1016/0006-2952(83)90663-9. [DOI] [PubMed] [Google Scholar]
- 14.Mathai J. C., Sitaramam V. J. Biol. Chem. 1994;269:17784–17793. [PubMed] [Google Scholar]
- 15.Zhao Y., Xu J. X. Biochem. Biophys. Res. Commun. 2004;317:980–987. doi: 10.1016/j.bbrc.2004.03.144. [DOI] [PubMed] [Google Scholar]
- 16.Barros M. H., Netto L. E., Kowaltowski A. J. Free Radical Biol. Med. 2003;35:179–188. doi: 10.1016/s0891-5849(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z. B., Li M., Zhao Y., Xu J. X. Protein Pept. Lett. 2003;10:247–253. doi: 10.2174/0929866033479013. [DOI] [PubMed] [Google Scholar]
- 18.Sharma R. K., Pasqualotto F. F, Nelson D. R., Thomas A. J., Jr, Agarwal A. Hum. Reprod. 1999;14:2801–2807. doi: 10.1093/humrep/14.11.2801. [DOI] [PubMed] [Google Scholar]
- 19.Hake L. E., Hecht N. B. J. Biol. Chem. 1993;268:4788–4797. [PubMed] [Google Scholar]
- 20.Narisawa S., Hecht N. B., Goldberg E., Boatright K. M., Reed J. C., Millan J. L. Mol. Cell. Biol. 2002;22:5554–5562. doi: 10.1128/MCB.22.15.5554-5562.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess R. A., Miller L. A., Kirby J. D., Margoliash E., Goldberg E. Biol. Reprod. 1993;48:1299–1308. doi: 10.1095/biolreprod48.6.1299. [DOI] [PubMed] [Google Scholar]
- 22.Sanishvili R., Volz K. W., Westbrook E. M., Margoliash E. Structure (London) 1995;3:707–716. doi: 10.1016/s0969-2126(01)00205-2. [DOI] [PubMed] [Google Scholar]
- 23.Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 24.Limbach K. J., Wu R. Nucleic Acids Res. 1985;13:631–644. doi: 10.1093/nar/13.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Gerstein M. Gene. 2003;312:61–72. doi: 10.1016/s0378-1119(03)00579-1. [DOI] [PubMed] [Google Scholar]
- 26.Margoliash E. Proc. Natl. Acad. Sci. USA. 1963;50:672–679. doi: 10.1073/pnas.50.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore G. R., Pettigrew G. W. Cytochromes c: Evolutionary, Structural & Physicochemical Aspects. Heidelberg: Springer; 1990. [Google Scholar]
- 28.Anderson M. J., Dixson A. F. Nature. 2002;416:496. doi: 10.1038/416496a. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse N. J., Goldstein J. C., Von A. O., Schuler M., Newmeyer D. D., Green D. R. J. Cell Biol. 2001;153:319–328. doi: 10.1083/jcb.153.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertini I., Cavallaro G., Rosato A. J. Biol. Inorg. Chem. 2005;10:613–624. doi: 10.1007/s00775-005-0011-7. [DOI] [PubMed] [Google Scholar]
- 31.Vera Y., Diaz-Romero M., Rodriguez S., Lue Y., Wang C., Swerdloff R. S., Hikim A. P. S. Biol. Reprod. 2004;70:1534–1540. doi: 10.1095/biolreprod.103.024661. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Sharma R. K., Sikka S. C., Thomas A. J., Jr, Falcone T., Agarwal A. Fertil. Steril. 2003;80:531–535. doi: 10.1016/s0015-0282(03)00756-8. [DOI] [PubMed] [Google Scholar]
- 33.Lo T. P., Komar-Panicucci S., Sherman F., McLendon G., Brayer G. D. Biochemistry. 1995;34:5259–5268. doi: 10.1021/bi00015a041. [DOI] [PubMed] [Google Scholar]
- 34.Davies A. M., Guillemette J. G., Smith M., Greenwood C., Thurgood A. G., Mauk A. G., Moore G. R. Biochemistry. 1993;32:5431–5435. doi: 10.1021/bi00071a019. [DOI] [PubMed] [Google Scholar]
- 35.Myer Y. P., Kumar S. J. Biol. Chem. 1984;259:8144–8150. [PubMed] [Google Scholar]
- 36.Pande J., Myer Y. P. J. Biol. Chem. 1980;255:11094–11097. [PubMed] [Google Scholar]
- 37.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 38.Mills G. C. J. Theor. Biol. 1991;152:177–190. doi: 10.1016/s0022-5193(05)80451-5. [DOI] [PubMed] [Google Scholar]
- 39.Arama E., Agapite J., Steller H. Dev. Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 40.Patel C. N., Lind M. C., Pielak G. J. Protein Expression Purif. 2001;22:220–224. doi: 10.1006/prep.2001.1438. [DOI] [PubMed] [Google Scholar]
- 41.Polevoda B., Martzen M. R., Das B., Phizicky E. M., Sherman F. J. Biol. Chem. 2000;275:20508–20513. doi: 10.1074/jbc.M001891200. [DOI] [PubMed] [Google Scholar]
- 42.Sherman F., Stewart J. W., Parker J. H., Inhaber E., Shipman N. A., Putterman G. J., Gardisky R. L., Margoliash E. J. Biol. Chem. 1968;243:5446–5456. [PubMed] [Google Scholar]
- 43.Mann M., Hendrickson R. C., Pandey A. Annu. Rev. Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 44.Ferguson-Miller S., Brautigan D. L., Margoliash E. J. Biol. Chem. 1978;253:149–159. [PubMed] [Google Scholar]
- 45.Lu Z., Zhang C., Zhai Z. Proc. Natl. Acad. Sci. USA. 2005;102:2778–2783. doi: 10.1073/pnas.0405374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z., Minor Z. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., et al. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S., Tamura K., Nei M. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.