Abstract
Mutations in the EDA gene cause anhidrotic/hypohidrotic ectodermal dysplasia, a disorder characterized by defective formation of hair, sweat glands, and teeth in humans and in a mouse model, “Tabby” (Ta). The gene encodes ectodysplasin, a TNF ligand family member that activates the NF-κB-signaling pathway, but downstream targets and the mechanism of skin appendage formation have been only partially analyzed. Comparative transcription profiling of embryonic skin during hair follicle development in WT and Ta mice identified critical anhidrotic/hypohidrotic ectodermal dysplasia (EDA) effectors in four pathways, three already implicated in follicle formation. They included Shh and its effectors, as well as antagonists for the Wnt (Dkk4) and BMP (Sostdc1) pathways. The fourth pathway was unexpected, a variant NF-κB-signaling cascade based on lymphotoxin-β (LTβ)/RelB. Previously known to participate only in lymphoid organogenesis, LTβ was enriched in developing hair follicles of WT but not in Ta mice. Furthermore, in mice lacking LTβ, all three types of mouse hair were still formed, but all were structurally abnormal. Guard hairs became wavy and irregular, zigzag/auchen hairs lost their kinks, and in a phenocopy of features of Ta animals, the awl hairs doubled in number and were characteristically distorted and pinched. LTβ-null mice that received WT bone marrow transplants maintained mutant hair phenotypes, consistent with autonomous LTβ action in skin independent of its expression in lymphoid cells. Thus, as an EDA target, LTβ regulates the form of hair in developing hair follicles; and when EDA is defective, failure of LTβ activation can account for part of the Ta phenotype.
Keywords: collagen, ectodermal dysplasia, hair type, NF-κB, skin appendages
Ectodermal dysplasias comprise >175 genetic disorders that cause aberrant formation of two or more skin appendages. Anhidrotic/hypohidrotic ectodermal dysplasia (EDA) is the most frequent ectodermal dysplasia. Affected boys and model (Ta) mice have mutations in the EDA gene, resulting in defective hair, missing sweat glands, and rudimentary teeth (1–3). EDA dependence is more pronounced in EDA patients (Online Mendelian Inheritance in Man accession no. 305100), who lack essentially all hair, than in Tabby (Ta) mice. In Ta mice, two of three hair types (guard and zigzag) are absent, but the third, straight “awl” hair, is still made, although in an abnormal form (4).
A transgene (4, 5) or injected ligand (6) of the A1 isoform of Eda restores sweat glands and guard hair but not zigzag hair to Ta mice. Our findings are consistent with primary EDA action to regulate the formation of hair follicle subtypes rather than triggering follicle induction (7).
Patient mutations and animal models have established that EDA acts through the canonical NF-κB-signaling pathway (8–10). As a TNF superfamily member (11), EDA binds to its receptor EDAR (12) and a receptor adaptor protein, EDARADD (13). With further involvement of TRAF6 (14, 15), the NEMO–IkBα–NF-κB (p65/p50)-signaling cascade is activated (8–10, 16). Thus, overexpression of EDAR leads to p65 activation (8), and ablation of p65 results in loss of the EDA-dependent guard hair in mice deficient in another NF-κB subunit (c-Rel; see ref. 17). However, the downstream effectors of EDA–NF-κB are poorly understood (18, 19).
To characterize EDA action, we profiled RNA from embryonic WT and Ta mouse skin with genome-wide cDNA probes. A small group of genes were affected at embryonic day 13.5 (E13.5), just before guard hair formation. They included components of the Shh- (20), Wnt- (21), and bone morphogenic protein (BMP)-signaling pathways (22), and in addition, lymphotoxin-β (LTβ), another TNF superfamily member (23). LTβ, like other detected targets, was highly expressed in hair follicles in WT mice but was selectively low in Ta mice. Furthermore, we found that mice lacking LTβ have characteristically abnormal hair, including large numbers of Ta-like hair. Thus, LTβ functions as a critical EDA target during hair follicle development.
Results
Dkk4, Shh, and LTβ as Candidate EDA Targets at E13.5.
As noted in ref. 24, the first hair follicles formed, for guard hair, were not yet seen in E13.5 embryo back skin of WT, but follicle germs were apparent at E14.5 and were growing massively by E16.5 and thereafter (see Fig. 5, which is published as supporting information on the PNAS web site). Ta lack the guard hair wave, but at E16.5, secondary hair follicles started in both WT and Ta mice. Thus, genes differentially expressed in WT and Ta skin at E13.5 should include early EDA targets for hair follicle development.
Microarray and real-time PCR analysis with total RNAs from back skin samples at E13.5 found a small, distinct group of genes significantly more expressed in WT than in Ta mice (13 gene probes of 44,000; Table 1 and see Table 2, which is published as supporting information on the PNAS web site). They included critical components of several signaling pathways.
Table 1.
Genes down-regulated in embryonic Ta skin starting at E13.5
| Genes | Fold differences (WT/Ta) |
||||
|---|---|---|---|---|---|
| Back skin |
Footpads |
||||
| E13.5 | E14.5 | E16.5 | E18.5 | E18.5 | |
| Eda | 4.9 | 5.8 | 4.6 | 5.1 | 3.7 |
| Dkk4 | 5.0 | 6.0 | 4.9 | 1.6 | 3.5 |
| Dkk1 | 1.4 | 1.5 | 2.1 | 3.0 | 1.7 |
| Kremen2 | 1.7 | 1.4 | 2.1 | 1.3 | 2.6 |
| Wnt10b | 2.4 | 1.2 | 1.3 | 1.3 | 1.3 |
| Shh | 8.0 | 10.0 | 7.4 | 4.3 | 33.0 |
| Ptc | 1.5 | 2.8 | 3.3 | 1.8 | 2.0 |
| Gli1 | 3.3 | 3.0 | 2.3 | 1.5 | 1.8 |
| Sostdc1 | 1.4 | 2.3 | 2.9 | 2.0 | 1.0 |
| Bmp4 | 1.0 | 2.0 | 1.3 | 1.2 | 1.1 |
| LTβ | 2.9 | 3.0 | 1.4 | 0.8 | 1.1 |
| Relb | 2.0 | 2.5 | 1.4 | 1.4 | 1.4 |
| K17 | 1.6 | 4.1 | 11.4 | 3.2 | 3.1 |
| Fgf10 | 1.0 | 2.0 | 1.3 | 1.1 | 1.0 |
Fold differences in RNA from WT compared to Ta skin are from averages of triplicate real-time PCR assays on three independent samples.
The Wnt pathway is known to be important in hair follicle development (25–27). At E13.5 in Ta, moderate down-regulation of Wnt10b was detected (27), but it was transient at this time point (Table 1; back skin). More persistent down-regulation was seen for Wnt antagonists Dkk4 (21) and Dkk1 (27) and the Dkk receptor Kremen2 (28). In keeping with other findings (12, 19, 24), Shh and its transcription factor Gli1 were sharply down regulated in Ta. For the BMP pathway, already known to participate in hair follicle formation (29), Sostdc1, a secreted antagonist (30), was down-regulated slightly at E13.5 and more extensively thereafter.
Strikingly, a candidate pathway responsive to EDA in hair follicle development, acting through LTβ and RelB, was also significantly down-regulated in Ta skin at E13.5 (Table 1).
EDA-Responsive Genes at E14.5–E18.5.
At E14.5, the guard hair germ stage, BMP4 and Fgf10, also previously implicated in hair follicle formation (29, 31), were transiently down-regulated, but most of the E13.5 candidate EDA targets including Dkk4, Shh, Sostdc1, and LTβ continued to be >2-fold down-regulated from this stage onward (Table 1 and see Table 2). In addition, keratin 17, a component of maturing hair follicles, has an active Gli-binding site in its promoter region (32) and was also significantly down-regulated in Ta from E14.5 (Table 1). From E16.5, Dkk1 (27) was strongly down-regulated, presumably complementing the action of Dkk4, which, like LTβ, decreased sharply even during normal development (see Fig. 6 A and B, which is published as supporting information on the PNAS web site). At E18.5, down-regulation was also seen for Shh antagonist Hhip (30) and many hair keratin genes (see Table 2). At later times, WT and Ta hair follicles are morphologically quite different, and additional genes affected at these times are likely secondary rather than direct downstream targets of EDA.
Up-Regulation of EDA-Responsive Genes in Adult Transgenic Mice.
Higher EDA activity in adult EDA-A1 transgenic skin (4) was correlated with the reported activation of c-fos and Lef1 (ref. 18; see Table 3, which is published as supporting information on the PNAS web site). In line with their responses to EDA, Wnt genes and their antagonists (Wnt10b, Wnt6, Wnt5a, Wnt11, Dkk1, Kremen2, and Wif1) and Shh, Gli1, and their negative regulators Ptc and Hhip were all activated, as were LTβ, RelB, Sostdc1, and keratin 17; but Dkk4, as seen in other tissues (21), was undetectable in adult skin (see Table 3).
LTβ and RelB Are Enriched in Developing WT Mouse Hair Follicles.
The LT pathway in the immune system includes LTβ, LTα, and several receptors (33). In Ta compared with WT mouse skin, however, only LTβ was down-regulated by microarray analysis (Table 1).
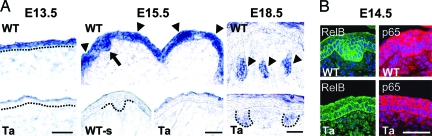
In lymphoid organ development, LTβ acts via RelB, a transcription factor of the NF-κB family that operates in a “noncanonical” pathway (34). Like LTβ, RelB expression was down-regulated in Ta mice (Table 1). To assess whether EDA up-regulates LTβ-RelB for hair follicle development, we first analyzed their expression pattern in embryonic back skin. In EDA+ mice, LTβ mRNA was broadly expressed in the cytoplasm of single-layer ectoderm at E13.5, enriched in the epidermis of nascent hair germs and pregerms at E15.5; and later (E18.5) enriched in matrix of hair follicles but not in interfollicular epidermis (Fig. 1A Upper). By contrast, Ta skin showed only very weak expression and only at late stages in awl hair pegs (Fig. 1A; E18.5). LTβ expression overlaps EDAR expression in developing follicles (19), consistent with LTβ as a downstream target of EDA. The inferred LTβ pathway component RelB was seen in the cytoplasm of basal cells in both WT and Ta skin and showed high levels in EDA-dependent guard hair germs at E14.5 (Fig. 1B). Instead, p65, working in the canonical NF-κB pathway, showed ubiquitous expression (Fig. 1B).
Fig. 1.
Expression of LTβ pathway genes in developing mouse hair follicles. (A) In situ hybridization of an LTβ-specific probe. Broad expression in epidermis at E13.5 (Upper Left), high expression in hair germ (arrow) and pregerms (arrowheads) at E15.5 (Upper Center), and in the matrix region of maturing hair follicles (arrowheads) at E18.5 (Upper Right) in WT mice. (Lower Right) Weak expression in Ta hair pegs only at E18.5. (Lower) No signal from a control sense probe (WT-s). Dotted lines demarcate epidermis and hair pegs from the dermis. (B) NF-κB expression. (Left) RelB protein localized by immunohistochemistry in cytoplasm of basal layer of epidermis both in WT and Ta and highly expressed in guard hair germs at E14.5. (Right) p65 highly expressed throughout epidermis in WT and Ta. (Scale bars: 50 μm.)
LTβ-Deficient Mice Form Characteristically Abnormal Hair.
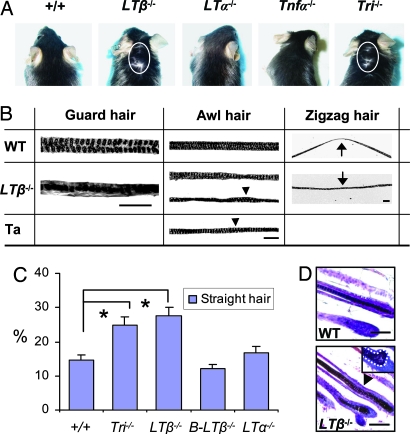
Definitive tests of LTβ action in skin appendage development were possible with LTβ−/− mice (34, 35). Unlike WT, skin could be seen through the hair around the neck of LTβ−/− mice (Fig. 2A). All three types of hair developed in LTβ−/− mice, but all mice showed striking morphological changes. Compared with WT, guard hair shafts were wavier, with irregular medullary granules in single rather than double file and a thick keratinized layer around medullary granules (Fig. 2B). Awl hairs were irregular and often thinner, with one or two medullary granule rows, similar to the abnormal awl hair formed in Ta mice (Fig. 2B). Similarly, zigzag hairs had irregular medullary granules, and many lacked the WT kinks. Consequently, they looked more like Ta awl hair (Fig. 2B). The number of awl-like hairs doubled in LTβ−/− and triple mutant LTβ−/−/LTα−/−/Tnfα−/− mice (Fig. 2C), and hair follicles themselves were also frequently distorted, producing abnormally wavy hair shafts (Fig. 2D).
Fig. 2.
Hair phenotypes in WT and LT–TNF mutant mice. (A) Gross hair phenotype. Skin visible in LTβ−/− and triple mutant LTβ−/−/LTα−/−/Tnfa−/− (Tri−/−) mice through the hair around the neck (circled) but not in other mice. (B) Abnormal hair shafts in LTβ−/− mice. Guard hair, irregular medullary granules (black dots) in a single row in LTβ−/− mice compared with WT. Awl hair, abnormal in LTβ−/− mice, resembling Ta awl (Ta) rather than WT. Arrowheads, segments with two separated medullary granule rows; zigzag hair, missing bends in LTβ−/− mice. In WT mice, the bent area is pronounced and structurally different from LTβ−/− (arrows; see Results). (C) Straight hair (%), mainly abnormal awl-like hair, significantly increased in LTβ−/− and Tri−/− mice. (D) Distorted anagen phase hair follicle in LTβ−/− mice, producing wavy hair shaft with irregular medullary granules (arrowhead in Lower) compared with WT follicles (Upper). Insert shows an abnormally asymmetric bulb region of a hair follicle in LTβ−/− mice. (Scale bars: 100 μm.)
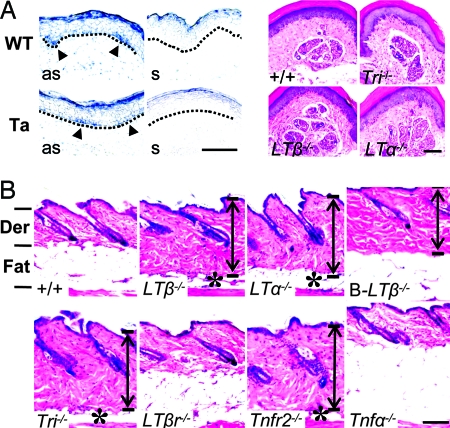
LTβ plays a pivotal role in immunity (33), but that role apparently does not underlie its action in skin. In B cell-specific LTβ−/− mice (36), skin showed more collagen, but the fatty layer and hair type composition and structure were completely normal (Fig. 2C and Fig. 3B Lower). Also, LTβ−/− mice that received WT bone marrow transplants (see Materials and Methods) maintained the exposed skin on the neck, thick collagen and thin fatty layers in skin dermis, and characteristic abnormal hair shaft structure (see Fig. 6C). Thus, the effects of LTβ are “tissue autonomous,” consistent with its local expression in skin keratinocytes.
Fig. 3.
Normal sweat glands, but abnormal skin in LT pathway-deficient mice. (A Left) LTβ expression in sweat gland germs in WT mice and basal layer of epidermis in Ta mice (arrowheads) at E18.5, by in situ hybridization with antisense (as) but not control sense probes (s). (A Right) Normal sweat glands in adult LTβ−/−, LTα−/−, and Tri−/− mice by hematoxylin/eosin staining. (B) Telogen phase LTβ−/−, LTα−/−, Tri−/−, and Tnfr2−/− mice showed a thick dermis (double-headed arrows) and thin fatty layer (asterisks). B cell-specific conditional LTβ−/− mice showed a normal fatty layer but somewhat thickened dermis; Tnfa−/− mice showed only a thick fatty layer. Der, dermis; Fat, fatty layer. (Scale bars: 100 μm.)
LTβ Is Not Required for Sweat Gland Formation or Function.
Sweat glands begin at E18.5 from germs that are histologically similar to hair follicle germs, and mutations in EDA also abolish sweat gland germ formation (4, 6). Because all sweat glands are localized in the hairless mouse footpads, expression profiling of nascent WT and Ta sweat glands (at E18.5) can provide independent information about EDA targets. Expression profiling showed many of the same small number of genes significantly altered in Ta (see Footpads in Table 1). However, Sostdc1 and LTβ expression were the same in Ta and WT footpads; comparably low LTβ expression was seen by in situ assays in Ta and WT (Fig. 3A Left). Furthermore, the sweat glands in LTβ−/− mice were morphologically and functionally normal [see Fig. 3A Right and by sweat tests (data not shown)]. Thus, LTβ is activated by EDA in hair follicle formation, with no comparable action in developing sweat glands.
Partners of LTβ in Hair Follicle Development.
LTβ and LTα form a heterotrimer for lymphoid organ development (23). LTα−/− mice also showed a thickened collagen layer in dermis and a thin fatty layer in the subcutis (Fig. 3B); but LTα−/− (and Tnfα−/−) mice showed normal hair, and triple mutant LTβ−/−/LTα−/−/Tnfa−/− mice showed only the LTβ−/−-related defects in hair, including the doubling of awl-like hair (Fig. 2C).
In lymphoid organs, the predominant LTβ2LTα1 heterotrimer binds to the LTβ receptor (LTβR) (33); but skin and hair were normal in LTβr−/− mice (Fig. 2C and 3B), and LTβR target gene expression in Ta mice showed no strict correlation; for example, the secondary lymphoid-tissue chemokine was not affected and cxcl13 was up-regulated (see Table 2; E14.5). Alternative heterotrimers of LTβ1LTα2 bind to TNFR1 and TNFR2 in the immune system (33). Tnfr2−/− mice (although not Tnfr1−/−) showed thickened collagen, stubbier awl hair, and thin fatty layers (Fig. 3B), but lacked the characteristic abnormalities of LTβ−/− hair.
Discussion
Skin appendage formation requires reciprocal signaling of mesenchyme and ectoderm. EDA plays a regulatory role in the ectoderm (1, 4, 19, 24). Our study suggests that an alternative NF-κB/LTβ pathway is integrated with Shh, Wnt, and BMP pathways downstream of EDA.
Other Pathways Interacting with EDA Regulation in Hair Follicle Development.
Shh, already noted as down-regulated in Ta mice (12, 19), was severely affected throughout the development of the various types of hair follicles and sweat glands, and its pathway was also up-regulated in the presence of an additional EDA-A1 transgene (Table 1 and see Table 3), suggesting that it is a direct target of EDA action. However, Shh is also further required for the development of all types of hair follicles including EDA-independent awl hair (20).
Overexpression of one inhibitor of the BMP pathway, Noggin, increased the number of hair follicles and reverted sweat glands to hair follicles in footpads (37), and Noggin-null mice showed decreased numbers of hair follicles (29). There is no evidence for EDA control of Noggin because its levels were unchanged in Ta. However, down-regulation of the BMP antagonist Sostdc1 was seen, and consistent with an EDA target, it is highly expressed in developing hair follicles (22, 30) and teeth (38).
For the Wnt pathway, overexpression of Dkk1 under a keratin 14 promoter blocked all hair follicle formation (27), but possible regulation of Wnt by EDA through a second member of the Dkk family, Dkk4, during guard hair formation is a previously uncharacterized finding.
Overall, activation of both effectors and antagonists was observed for Wnt, Shh, and BMP pathways (Table 1 and see Table 3). The time course of relative activation during development is consistent with a refined pattern of feedback action. For example, the activation of Shh precedes the marked activation of its inhibitors, Ptc and Hhip, and after EDA is up-regulated by Wnt/Lef1 (19, 39), there may be a feedback interaction of EDA and Wnt through the balance of Lef1 and stage-specific action of Dkk4/Dkk1.
Toward a Mechanism of Action for LTβ in Hair-Type Determination.
LTβ has an expression pattern like the EDA receptor EDAR (Fig. 1A; see ref. 19) and was down-regulated in embryonic Ta skin (Table 1). It likely functions in skin through RelB, its mediator in lymphoid organ development, which showed the same EDA activation and was also similarly highly expressed in guard hair germs. Interestingly, in the report of Relb−/− mice (40), skin appendages were not examined in detail, but the authors commented that mice showed “disheveled” hair, perhaps reflecting a block similar to that seen in LTβ-null mice.
Both LTβ and RelB have active NF-κB-binding sites in their promoter regions, and both were activated by the p65/p50 heterodimer (41), suggesting that the noncanonical LTβ/RelB pathway is activated as a downstream target of the canonical EDA–NF-κB (p65/p50)-signaling during hair follicle development (8, 24). In accord with sequential activation, when the EDA pathway was initiated in cells transfected with the EDAR receptor, we confirmed direct activation of p65 (8, 24) but not RelB (see Fig. 3D).
The signaling proteins in an LTβ pathway in skin are apparently somewhat different from those in the immune system (23, 34). There, LTβ and LTα form heterotrimers that activate p100/RelB-signaling through LTβR (23). However, the changes in mouse hair were specific for LTβ; LTα−/− (and Tnfα−/−) mice showed normal hair and hair-type composition. Furthermore, B cell-specific LTβ−/− mice showed completely normal hair-type composition and hair structure, and LTβ−/− mice fortified with WT bone marrow retained mutant hair phenotypes. Interestingly, LTβ, LTα, and TNFα are clustered in the MHC region, but the six known ectodermal dysplasia genes all map to other chromosomes (1, 8, 9, 12, 13, 33, 42). We conclude that LTβ apparently acts in hair follicle development independently of its immune system involvement. In addition, mouse models show that LTβR and all other known LT receptors, which recognize various LTβ–LTα heterotrimers, are dispensable for LTβ action in hair follicles. We found no evidence for LTβ interaction with receptors Troy, XEDAR, or TNFR2 in cotransfection or immunoprecipitation experiments (unpublished data). Further studies are needed to see whether LTβ binds to other TNF receptor family members, including EDAR and HVEM (33) or whether an as yet uncharacterized LTβR mediates its function in hair follicle development.
Notably, like Shh and NF-κBs (19, 24), LTβ expression was detectable, although very weakly, in Ta hair follicles at E18.5, suggesting that additional upstream regulators may exist for LTβ in skin at late stages. Also, LTβ was equally expressed in WT and Ta footpads (sweat gland germs). Thus, the full range of regulatory mechanisms of LTβ expression in skin remains to be elucidated.
Relative of Roles of LTβ and Other Signaling Pathways in Skin Appendage Formation.
Whereas the Wnt, BMP, and Shh pathways act in both hair follicle and sweat gland formation, the LTβ pathway affects only hair follicles. Also, Wnt (26) and BMP (37) pathways determine the number of hair follicles (and possibly the level of EDA), and Shh is required for the formation of all follicles (ref. 20; Fig. 4); although LTβ was expressed at the induction stage for hair follicles, all types of hair were formed in its absence. The basis for time- and tissue-specific access to LTβ and its targets are unknown; however, we infer that like its activator EDA, LTβ contributes primarily to modulate the form of hair produced during differentiation (Fig. 4). Hair shaft production and keratinization are most markedly affected in its absence, producing hair with “Ta-like” features. Thus, failure of LTβ activation could contribute to the increased numbers of abnormal awl-like hairs in EDA-null Ta mice.
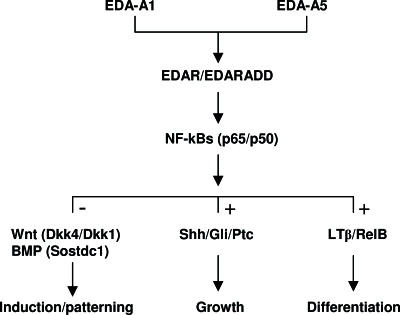
Fig. 4.
Schematic representation of EDA signaling in hair follicle development. EDA-A1 or EDA-A5 (ref. 43) activate the canonical NF-κB pathway via EDAR and p65/p50, which then signal to Wnt and BMP pathways and their antagonists for hair follicle induction, to Shh and its inhibitors for hair follicle growth, and to the alternative NF-κB pathway, involving LTβ and RelB, for hair-type differentiation. +, positive up-regulation; −, negative regulation (suppression).
Materials and Methods
Timed Pregnancies and Genotyping of Embryos.
To obtain sibling WT and Ta male embryos, timed pregnancies were setup with C57BL/6J male and C57BL/6J-AW-J-Ta6J (Ta) female mice (The Jackson Laboratory). The morning after mating was designated as E0.5. Embryos were harvested at E13.5, E14.5, E15.5, E16.5, and E18.5. Back skin samples or footpads and livers were taken under dissection microscopy, frozen on dry ice, and stored at −80°C until use.
Genomic DNAs were isolated from each embryo liver using a DNase Tissue Kit (Qiagen, Valencia, CA) for sex and EDA mutation status by PCR-based methods. Male-specific primers were: SryF, 5′-CTGCAGTTGCCTCAACAAAA-3′; and SryR, 5′-TTGGAGTACAGGTGTGCAGC-3′. PCR analysis was carried out with cycling conditions of denaturation at 94°C for 5 min, 35 cycles at 94°C for 30 s, 58°C for 45 s, and 72°C for 1 min. EDA mutation detection was done on genomic DNA from male embryos. A primer pair spanning the mutation site was designed. The PCR fragment derived from WT has a DdeI site that is missing in Ta, permitting unequivocal identification of WT and Ta by enzyme digestion. Primer sequences were: Ta-mu-F, 5′-GGCAGCCGTCCTTTCAACA; and Ta-mu-R, 5′-GCGTACTAGCGTACCACGTGTCGACTCACCTGGTGCCGGTCCTGGGACTC. PCR conditions were: denaturation at 95°C for 5 min, 35 cycles at 95°C for 45 s, 57°C for 45 s, and 72°C for 1 min. After DdeI digestion, the PCR fragment from WT mice yielded two species of ≈50 bp, whereas DNA from Ta mice showed a single band at 106 bp.
RNA Isolation, Gene Expression Profiling, and Real-Time PCR.
Back skin and footpad samples from male embryos at each developmental stage (24 WT and 23 Ta at E13.5, 15 WT and 14 Ta at E14.5, 10 WT and 8 Ta at E16.5, 9 WT and 7 Ta at E18.5, and 3 WT and 3 WT bearing an EDA-A1 transgene at 2 months). They were divided into three pools for biological replicates and RNA was isolated (3), and cyanine-3-labeled cRNA was hybridized to the 44,000-feature 60-mer-oligo microarray analysis (44). Triplicate data were analyzed, FDR was set to ≤0.1, and genes with fold difference <1.5 were excluded from significant gene lists. All genes detected (Table 1) were confirmed by real-time PCR with TaqMan “Assays on-Demand” probe/primers (Applied Biosystems).
In Situ and Immunohistochemistry.
Frozen skin sections (14 μm thick) were fixed in 4% paraformaldehyde and hybridized with a LTβ-specific cRNA probe (42) overnight at 60°C. After washing with 2× SSC (0.3 M NaCl/0.03 M sodium citrate, pH 7.0) and 0.1× SSC at 65°C, sections were incubated with anti-digoxigenin antibody (Roche; 1:2,000 dilution) overnight at 4°C. Signals were visualized with NBT/BCIP (Roche). Anti-RelB and anti-p65 antibodies (Santa Cruz Biotechnology) and Alexa Fluor 488 (for RelB) and 594 (for p65) secondary antibody (Invitrogen) were used for immunofluorescence staining.
Skin Phenotypes of LT-Tnf Knockout Mice and Bone Marrow Transplanted Derivatives.
Four of each of LTβ/LTα/Tnfa-null mice and LTβ, B-LTβ, LTα, LTβr, Tnfr2, Tnfr1, and Tnfa-null mice in the C57BL6 background (34, 35, 38) were studied. At least 400 hairs from back skin of each mouse were studied (4).
For bone marrow chimeras, 2-month-old mice were irradiated (1,000 cGy) and reconstituted with 5 × 106 donor bone marrow cells supplied i.v. within 2 h. Efficiency of transplantation was confirmed using Ly 5.1/Ly 5.2 markers of congenic mice. Six chimeric mice were studied 3 months later.
For histological analyses, skin samples from back skin or footpads were fixed in 4% paraformaldehyde and embedded in paraffin, and 8-μm sections were stained with hematoxylin/eosin.
Supplementary Material
Acknowledgments
We thank Drs. M. Ko, M. Carter, C. Ottolenghi, K. Aiba, T. Tezuka, and R. Nagaraja for helpful discussions and technical advice; Drs. R. Sen and D. Longo for critical suggestions; and A. Butler, M. Michel, D. Nines, E. Douglass, and L. Drutskaya who helped with animal housing and management. This work was supported by the Intramural Research Program of the National Institute on Aging and National Cancer Institute (National Institutes of Health). S.A.N. is International Research Scholar of the Howard Hughes Medical Institute.
Abbreviations
- EDA
anhidrotic/hypohidrotic ectodermal dysplasia
- Ta
Tabby
- En
embryonic day n
- LTβ
lymphotoxin-β
- BMP
bone morphogenic protein
- LTβR
LTβ receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kere J., Srivastava A. K., Montonen O., Zonana J., Thomas N., Ferguson B., Munoz F., Morgan D., Clarke A., Baybayan P., et al. Nat. Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava A. K., Pispa J., Hartung A. J., Du Y., Ezer S., Jenks T., Shimada T., Pekkanen M., Mikkola M. L., Ko M.S., et al. Proc. Natl. Acad. Sci. USA. 1997;94:13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui C. Y., Smith J. A., Schlessinger D., Chan C. C. Am. J. Pathol. 2005;167:89–95. doi: 10.1016/S0002-9440(10)62956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui C. Y., Durmowicz M., Ottolenghi C., Hashimoto T., Griggs B., Srivastava A. K., Schlessinger D. Hum. Mol. Genet. 2003;12:2931–2940. doi: 10.1093/hmg/ddg325. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava A. K., Durmowicz M. C., Hartung A. J., Hudson J., Ouzts L. V., Donovan D. M., Cui C. Y., Schlessinger D. Hum. Mol. Genet. 2001;10:2973–2981. doi: 10.1093/hmg/10.26.2973. [DOI] [PubMed] [Google Scholar]
- 6.Gaide O., Schneider P. Nat. Med. 2003;9:614–618. doi: 10.1038/nm861. [DOI] [PubMed] [Google Scholar]
- 7.Mustonen T., Ilmonen M., Pummila M., Kangas A. T., Laurikkala J., Jaatinen R., Pispa J., Gaide O., Schneider P., Thesleff I., et al. Development (Cambridge, U.K.) 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 8.Doffinger R., Smahi A., Bessia C., Geissmann F., Feinberg J., Durandy A., Bodemer C., Kenwrick S., Dupuis-Girod S., Blanche S., et al. Nat. Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 9.Courtois G., Smahi A., Reichenbach J., Doffinger R., Cancrini C., Bonnet M., Puel A., Chable-Bessia C., Yamaoka S., Feinberg J., et al. J. Clin. Invest. 2003;112:1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt-Ullrich R., Aebischer T., Hulsken J., Birchmeier W., Klemm U., Scheidereit C. Development (Cambridge, U.K.) 2001;128:3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- 11.Ezer S., Bayes M., Elomaa O., Schlessinger D., Kere J. Hum. Mol. Genet. 1999;8:2079–2086. doi: 10.1093/hmg/8.11.2079. [DOI] [PubMed] [Google Scholar]
- 12.Headon D. J., Overbeek P. A. Nat. Genet. 1999;22:370–374. doi: 10.1038/11943. [DOI] [PubMed] [Google Scholar]
- 13.Headon D. J., Emmal S. A., Ferguson B. M., Tucker A. S., Justice M. J., Sharpe P. T., Zonana J., Overbeek P. A. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- 14.Naito A., Yoshida H., Nishioka E., Satoh M., Azuma S., Yamamoto T., Nishikawa S., Inoue J. Proc. Natl. Acad. Sci. USA. 2002;99:8766–8771. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morlon A., Munnich A., Smahi A. Hum. Mol. Genet. 2005;14:3751–3757. doi: 10.1093/hmg/ddi405. [DOI] [PubMed] [Google Scholar]
- 16.Yan M., Wang L. C., Hymowitz S. G., Schilbach S., Lee J., Goddard A., de Vos A. M., Gao W. Q., Dixit V. M. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 17.Gugasyan R., Voss A., Varigos G., Thomas T., Grumont R. J., Kaur P., Grigoriadis G., Gerondakis S. Mol. Cell. Biol. 2004;24:5733–5745. doi: 10.1128/MCB.24.13.5733-5745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui C. Y., Durmowicz M., Tanaka T. S., Hartung A. J., Tezuka T., Hashimoto K., Ko M. S., Srivastava A. K., Schlessinger D. Hum. Mol. Genet. 2002;11:1763–1773. doi: 10.1093/hmg/11.15.1763. [DOI] [PubMed] [Google Scholar]
- 19.Laurikkala J., Pispa J., Jung H. S., Nieminen P., Mikkola M., Wang X., Saarialho-Kere U., Galceran J., Grosschedl R., Thesleff I. Development (Cambridge, U.K.) 2002;129:2541–2553. doi: 10.1242/dev.129.10.2541. [DOI] [PubMed] [Google Scholar]
- 20.St-Jacques B., Dassule H. R., Karavanova I., Botchkarev V. A, Li J., Danielian P. S., McMahon J. A., Lewis P. M., Paus R., McMahon A. P. Curr. Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 21.Krupnik V. E., Sharp J. D., Jiang C., Robison K., Chickering T. W., Amaravadi L., Brown D. E., Guyot D., Mays G., Leiby K., et al. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 22.Laurikkala J., Kassai Y., Pakkasjarvi L., Thesleff I., Itoh N. Dev. Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Browning J. L., Ngam-ek A, Lawton P., DeMarinis J., Tizard R., Chow E. P., Hession C., O’Brine-Greco B., Foley S. F., Ware C. F. Cell. 1993;72:847–856. doi: 10.1016/0092-8674(93)90574-a. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt-Ullrich R., Tobin D. J., Lenhard D., Schneider P., Paus R., Scheidereit C. Development (Cambridge, U.K.) 2006;133:1045–1057. doi: 10.1242/dev.02278. [DOI] [PubMed] [Google Scholar]
- 25.van Genderen C., Okamura R. M., Farinas I., Quo R. G., Parslow T. G., Bruhn L., Grosschedl R. Genes. Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 26.Gat U., DasGupta R., Degenstein L., Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 27.Andl T., Reddy S. T., Gaddapara T., Millar S. E. Dev. Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 28.Mao B., Wu W., Davidson G., Marhold J., Li M., Mechler B. M., Delius H., Hoppe D., Stannek P., Walter C., et al. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 29.Botchkarev V. A., Botchkareva N. V., Roth W., Nakamura M., Chen L. H., Herzog W., Lindner G., McMahon J. A., Peters C., Lauster R., et al. Nat. Cell. Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 30.Rendl M., Lewis L., Fuchs E. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petiot A., Conti F. J., Grose R., Revest J. M., Hodivala-Dilke K. M., Dickson C. Development (Cambridge, U.K.) 2003;130:5493–5501. doi: 10.1242/dev.00788. [DOI] [PubMed] [Google Scholar]
- 32.Bianchi N., Depianto D., McGowan K., Gu C., Coulombe P. A. Mol. Cell. Biol. 2005;25:7249–7259. doi: 10.1128/MCB.25.16.7249-7259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ware C. F. Annu. Rev. Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- 34.Alimzhanov M. B., Kuprash D. V., Kosco-Vilbois M. H., Luz A., Turetskaya R. L., Tarakhovsky A., Rajewsky K., Nedospasov S. A., Pfeffer K. Proc. Natl. Acad. Sci. USA. 1997;94:9302–9307. doi: 10.1073/pnas.94.17.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuprash D. V., Alimzhanov M. B., Tumanov A. V., Grivennikov S. I., Shakhov A. N., Drutskaya L. N., Marino M. W., Turetskaya R. L., Anderson A. O., Rajewsky K., et al. Mol. Cell. Biol. 2002;22:8626–8634. doi: 10.1128/MCB.22.24.8626-8634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tumanov A. V., Grivennikov S. I., Shakhov A. N., Rybtsov S. A., Koroleva E. P., Takeda J., Nedospasov S. A., Kuprash D. V. Immunol. Rev. 2003;195:106–116. doi: 10.1034/j.1600-065x.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 37.Plikus M., Wang W. P., Liu J., Wang X., Jiang T. X., Chuong C. M. Am. J. Pathol. 2004;164:1099–1114. doi: 10.1016/S0002-9440(10)63197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassai Y., Munne P., Hotta Y., Penttila E., Kavanagh K., Ohbayashi N., Takada S., Thesleff I., Jernvall J., Itoh N. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- 39.Durmowicz M. C., Cui C. Y., Schlessinger D. Gene. 2002;285:203–211. doi: 10.1016/s0378-1119(02)00407-9. [DOI] [PubMed] [Google Scholar]
- 40.Weih F., Carrasco D., Durham S. K., Barton D. S., Rizzo C. A., Ryseck R. P., Lira S. A., Bravo R. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 41.Kuprash D. V., Osipovich O. A., Pokholok D. K., Alimzhanov M. B., Biragyn A., Turetskaya R. L., Nedospasov S. A. J. Immunol. 1996;156:2465–2472. [PubMed] [Google Scholar]
- 42.Pokholok D. K., Maroulakou I. G., Kuprash D. V., Alimzhanov M. B., Kozlov S. V., Novobrantseva T. I., Turetskaya R. L., Green J. E., Nedospasov S. A. Proc. Natl. Acad. Sci. USA. 1995;92:674–678. doi: 10.1073/pnas.92.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashimoto T., Cui C. Y., Schlessinger D. Gene. 2006;371:42–51. doi: 10.1016/j.gene.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Carter M. G., Hamatani T., Sharov A. A., Carmack C. E., Qian Y., Aiba K., Ko N. T., Dudekula D. B., Brzoska P. M., Hwang S. S., et al. Genome Res. 2003;13:1011–1021. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.