Abstract
Apyrase activity is present in the saliva of haematophagous arthropods. It is related to blood-feeding because of the apyrase ability to hydrolyse ADP, a key component of platelet aggregation. Five apyrases with apparent molecular masses of 88, 82, 79, 68 and 67 kDa were identified in the saliva of the vector of Chagas disease, Triatoma infestans. The large size observed during purification of these enzymes suggested oligomerization. In the present study, we confirmed, using gel-filtration and analytical ultracentrifugation, the presence of apyrase oligomers with molecular masses of 200 kDa in the saliva. Electrophoretic analyses showed that disulphide bonds were involved in homo-oligomerization. In addition, heterogeneity in disulphide bonds and in pI was detected, with the pI ranging from 4.9 to 5.4. The present study gives the first insights into the quaternary structure of soluble apyrases.
Keywords: anti-platelet, apyrase, Chagas disease, haematophagy, insect saliva, Triatoma infestans
Abbreviations: DTSSP, 3,3′-dithiobis(sulphosuccinimidyl propionate); DTT, dithiothreitol; IEF, isoelectric focusing; NEM, N-ethylmaleimide; PAF, platelet-activating factor
INTRODUCTION
The adaptation of arthropods to haematophagy required the development of molecular tools able to counteract the host haemostatic response, which relies mainly on vasoconstriction, blood coagulation and platelet aggregation. Many such antihaemostatic compounds have been described, providing some perspectives on biomedical applications [1–4]. Platelet aggregation is one of the first and most important haemostatic responses following injury of small blood vessels and capillaries. Platelets can be activated by many factors, such as thrombin, collagen, adrenaline and ADP, and subsequently aggregate through a mechanism involving fibrinogen receptors. It is therefore expected that haematophagous arthropod saliva would contain compounds degrading or inhibiting those host factors.
The identification of triabin, which was characterized as a ligand of the thrombin anion-binding exosite in the saliva of the haematophagous insect Triatoma pallidipennis [5], led to the determination of the triabin–thrombin complex structure [6]. Furthermore, it was shown that this triatomine possesses pallidipin, which is an inhibitor of the collagen-induced blood-clotting pathway [7,8]. In addition, nitrophorins present in Rhodnius prolixus display vasodilatory, as well as anticoagulant, activities; the nitric oxide they carry inhibits platelet aggregation [9,10]. More recently, a 19 kDa protein, RPAI-1 (Rhodnius platelet aggregation inhibitor 1), was characterized in this triatomine [11]. This new anti-platelet factor belongs to the lipocalin family and acts by sequestration of free ADP. The saliva of the tick Ornithodoros moubata contains disagregin, an inhibitor of platelet fibrinogen receptors [12,13]. Similar inhibitors were identified in another tick species [14,15] and in deerfly [16]. Platelet aggregation can also be triggered by the lipid PAF (platelet-activating factor) that is also involved in inflammatory and allergic reactions. A phospholipase C that specifically degrades PAF has been identified in the saliva of the mosquito Culex quinquefasciatus [17]. In addition, a PAF acetylesterase activity was also reported in cat flea [18].
The saliva of haematophagous insects therefore present a wide range of anti-platelet activities, which inhibit the factors that are released in response to a vascular injury. Among them, anti-ADP activities are attractive therapeutic targets. Indeed, activated platelets release ADP that recruits and activates more platelets, inducing their aggregation. Thus ADP represents a final mediator and amplifier of different activation pathways of platelet aggregation [19]. The inhibition of collagen- and thrombin-induced platelet reactivity by apyrase, an ADP-degrading enzyme [20,21] reveals the central role of ADP in this process. Confirming the importance of ADP, the human CD39 protein has been identified as an apyrase present at the vascular endothelial surface [22]. This protein is thought to play a major role in the prevention of thrombus formation, by controlling the extracellular level of ADP. Although some 5′-nucleotidases and vertebrate ecto-apyrases are assembled as oligomeric structures [23,24], the structural characterization of soluble apyrases from the invertebrate salivary glands is lacking.
Interestingly, the importance of salivary apyrases for obtaining a blood meal is suggested by its ubiquitous presence in the saliva of haematophagous insects. The apyrases of such insects were shown to belong to two genetic families. The so-called Cimex family is found in the Cimicidae (bedbugs) (Cimex lectularius) and in the Phlebotominae (sandflies) (Phlebotomus papatasi) [25–27]. Expressed sequence tags presenting significant similarities with this family of apyrases have been also identified in rat and human with no clear associated physiological role [28,29]. The other apyrase family appeared to have originated from the intracellular 5′-nucleotidases and has been described in the Culicidae Aedes aegypti, Anopheles gambiae and Anopheles stephensi [30–32]. The information available in the literature for each species ranges from reports of the DNA sequence of the apyrase genes and detection of enzyme activity to a few extensive biochemical studies.
Previously, we showed that the Triatoma infestans saliva contains five apyrases from the 5′-nucleotidase family displaying differences in primary sequences and originating from a least three genetic loci. These five proteins are likely to be active enzymes as they bind an ATP analogue [33]. During purification, we noticed that these proteins displayed a high native molecular mass. In the present paper, we report the biochemical and biophysical studies of the oligomeric nature of these salivary proteins.
EXPERIMENTAL
Triatomines, saliva and apyrase activity
T. infestans were reared in an insectary maintained at 28±2 °C, 70% ±5 relative humidity, with photoperiods of 12 h. Saliva (0.5–1 μl per adult triatomine) was collected in pipette tips placed on the triatomines' mouthparts. All experiments were performed with pooled saliva obtained from insects at 20 days following a blood meal. The saliva used in the reducing experiments was collected using a pipette tip containing an equal volume of 2 mM NEM (N-ethylmaleimide). The saliva was filtered through a 0.22-μm-pore-size membrane and stored at −80 °C until use. T. infestans salivary apyrase activity was determined as described previously [33].
IEF (isoelectric focusing) in agarose gels
Agarose gels (0.5 mm thick) were cast in a vertical mould, according to the manufacturer's recommendation (Amersham Biosciences). The gels were made with 2 g of sorbitol, 19 ml of water, 0.16 g of agarose IEF and 1.3 ml of ampholine, at pH ranging from 3.5 to 10. The distance between electrodes was 9 cm. Focusing was achieved with 1500 V, 5 mA/cm and 0.25 W/cm for 1000 V/h. The sample (5 μl) was applied on to the gel surface. In-gel apyrase activity was revealed by the formation of a calcium phosphate precipitate following incubation of the gel in a solution containing 0.1 M Tris/HCl, pH 8.0, 20 mM NaCl, 20 mM CaCl2, 1 mM MgCl2 and 5 mM ATP, ADP or AMP, as described by Mathews et al. [34] with washing steps performed in the presence of substrate.
Native PAGE and in-gel activity
Native PAGE was performed using the method of Notarnicola et al. [35], with modification. The electrophoresis buffer contained 25 mM Tris/HCl, 190 mM glycine and 10 mM sodium acetate at pH 8.3. Linear gradient gels (3–15%) were cast with the same buffer. Electrophoresis was carried out using the Multiphor II system (Amersham Biosciences) at 7.1 V/cm for 30 min and 14.3 V/cm for 2 h. In-gel activity was detected using the procedure described for IEF; however, the washing steps were performed in the absence of substrate.
Analytical gel-filtration chromatography and analytical ultracentrifugation
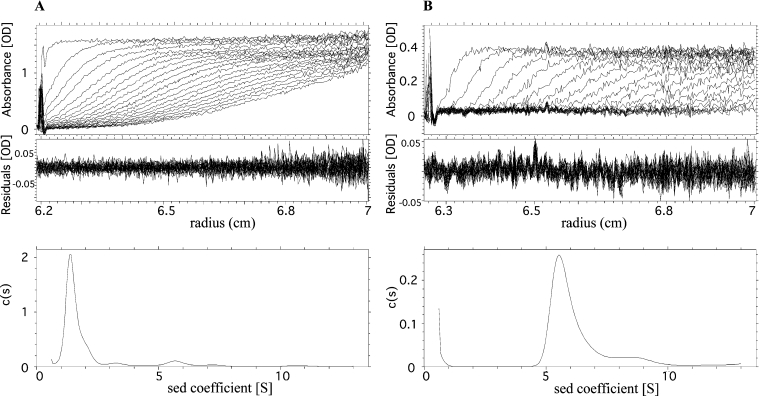
Analytical gel-filtration chromatography was conducted on a Superdex 200 column (Amersham Biosciences) in 25 mM Tris/HCl, pH 7.5, and 500 mM NaCl at a flow rate of 0.4 ml/min. The absorbance at 280 nm was recorded, and 0.4 ml fractions were tested for enzyme activity. The column was calibrated using the known hydrodynamic radii (Rh) of thyroglobulin, ferritin, aldolase and BSA [36]. Sedimentation velocity experiments were performed using a Beckman XL-I analytical ultracentrifuge and an An-60 Ti rotor. Experiments were carried out at 10 °C in 25 mM Tris/HCl, pH 7.5, and 500 mM NaCl. Samples of 100 μl of total saliva at protein concentrations of 2.2, 4.4 and 10 mg/ml and purified apyrase at 0.3 mg/ml were loaded into 3 mm sapphire cells and centrifuged at 42000 rev./min. Scans were recorded at 278 nm for total saliva or 228 nm for purified apyrase, using a 0.003 cm radial spacing. We used the continuous distribution c(s) analysis of P. Schuck (Sedfit: http://www.analyticalultracentrifugation.com) to treat the data [37,38] (see [39] for a review on the recent progresses in analytical ultracentrifugation). The c(s) method considers the sample as being composed of a number of non-interacting species. For all species, a common relationship is assumed that links the molar mass and the hydrodynamic radius of the macromolecule, and thus its sedimentation and diffusion coefficients, through the solvent density and viscosity and reasonable values of the partial specific volume and frictional ratio of the macromolecule. It allows the calculation of theoretical sedimentation profiles for any species. The c(s) distribution corresponds to the best linear combination of species that describes the experimental set of sedimentation profiles. Sedfit also takes advantage of a radial and time-independent noise-subtraction procedure. Because the sedimentation profiles are mainly determined from the values of the sedimentation coefficients, and in a minor way from the values of the diffusion coefficients, the c(s) analysis is robust, even if the details of the distribution can be sensitive to the hypothesis used. The c(s) curves were obtained considering 200 particles with sedimentation coefficients between 0.3 and 13 S, with a regularization procedure leading to a smooth curve (F-ratio=0.68). We considered partial specific volumes, v, of 0.74 ml/g for the polypeptide and 0.609 ml/g for the glycosylation contribution, and tabulated values for the solvent density (ρ=1.021 g/ml) and viscosity (η=1.377 cp), for the c(s) analysis. From the inferred sedimentation coefficient, s, and the hydrodynamic radius, Rh, from calibrated gel-filtration, the molar mass, M, of apyrase was calculated using the Svedberg equation:
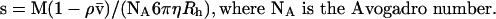 |
Non-reducing/reducing two-dimensional SDS/PAGE
Free cysteine residues from purified apyrase were blocked by incubation for 10 min at room temperature (25 °C) with 10 mM iodoacetamide to avoid formation of non-specific disulphide bonds during sample preparation. Non-reducing Laemmli SDS/PAGE buffer was added, and samples were boiled for 5 min and submitted to non-reducing SDS/6% PAGE. Half of the gel lane was then cut out longitudinally and disulphide bonds reduced by incubation in reducing Laemmli sample buffer for 10 min at room temperature. The remaining part of the gel containing non-reduced apyrase and molecular-mass markers was silver-stained. The in-gel reduced lane was horizontally applied on top of a 6% gel and run into a second dimension. Molecular-mass markers and reduced apyrase samples were simultaneously run only in this second reducing dimension as references. Gels were silver-stained.
Disulphide bond reduction and apyrase activity
Purified apyrases (2 μg) were incubated with different concentrations of DTT (dithiothreitol) for 30 min at room temperature. Thereafter, free cysteine residues were blocked with a 10-fold excess of iodoacetamide. Aliquots were diluted 10-fold and were used to measure apyrase activity. Changes in band patterns were detected by SDS/PAGE under non-reducing conditions. Controls contained the same quantity of saliva and iodoacetamide in the absence of DTT.
DTSSP [3,3′-dithiobis(sulphosuccinimidyl propionate)] cross-linking experiments
DTSSP (Pierce) cross-linking was performed essentially as described by the manufacturer. In short, purified apyrases (0.1 mg/ml) were incubated for 30 min with 0–25 μM DTSSP in 50 mM Hepes, pH 7.5, and 500 mM NaCl to a final volume of 15 μl. After quenching by the addition of 0.5 μl of 1 M Tris/HCl, pH 6.8, the reaction products were resolved by SDS/6% PAGE under non-reducing conditions.
RESULTS
Charge heterogeneity of T. infestans apyrases
In previous work, we reported that pooled T. infestans saliva contains five apyrases of 88, 82, 79, 68 and 67 kDa in molecular sieving experiments [33]. The large size of the apyrase oligomers prevented proper IEF in polyacrylamide gels, therefore IEF was performed in an agarose gel. Apyrase activity focused as a smear in a region ranging from pH 4.9 to 5.4, as revealed by the precipitate of Pi released from ATP and ADP, but not AMP (Figure 1, lanes 1–2, 3–4 and 5–6 respectively). This activity reresents truly focused apyrase, as it migrated to the same position when sample was applied on either side of the gel (Figure 1, compare odd and even lanes for each substrate) and pI markers were finely resolved into sharp bands (results not shown). This pI heterogeneity may reflect various post-translational modifications of the apyrases, and Western-blot experiments showed that all of the five apyrases are distributed along the activity area (results not shown). When IEF experiments were performed with saliva collected from individual insects, the same pattern of multiple-activity bands was observed, revealing that the observed heterogeneities do not derive from variation within the insect population.
Figure 1. IEF of T. infestans salivary apyrases.
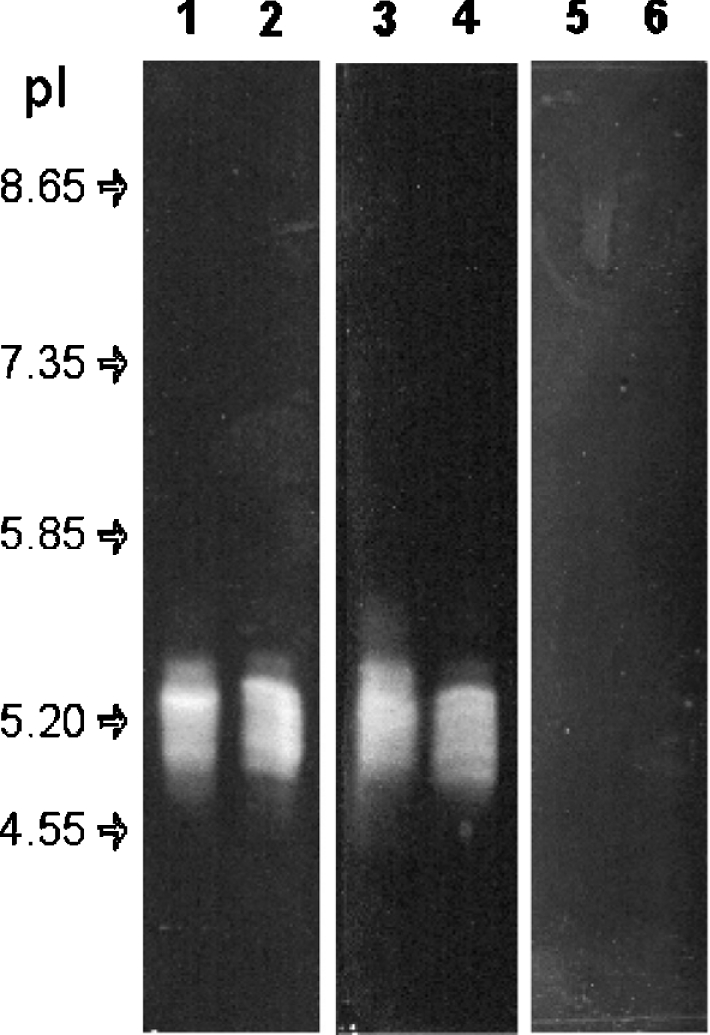
IEF (pH 3.5–10) followed by in-gel activity detected by the formation of a whitish precipitate resulting from the reaction of calcium, with Pi released from ATP (lanes 1 and 2) and ADP (lanes 3 and 4), but not AMP (lanes 5 and 6). These samples were applied 1.5 cm from the activity area on the basic (lanes 1, 3 and 5) or acidic (lanes 2, 4, and 6) sides of the gel. Arrows indicate positions of focused pI standards.
Despite these multiple charges, the apyrase activity of crude saliva upon native PAGE was rather homogeneous (Figure 2). In this separation technique based on both charge and size of the proteins, the enzymatic activity migrated as a broad band, releasing Pi from ATP and ADP, but not from AMP (Figure 2, lanes 1, 2 and 3 respectively). Pi release from ATP by the purified apyrases was observed at the same position (lane 4).
Figure 2. Native PAGE of T. infestans salivary apyrases.
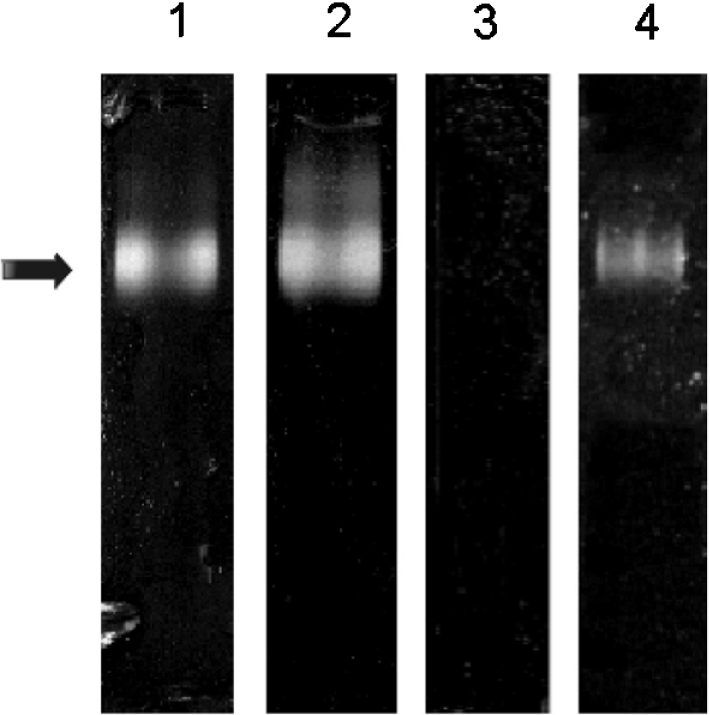
A 0.2 μl aliquot of crude saliva (lanes 1, 2 and 3) and purified apyrase (lane 4) were resolved in native PAGE, using a Tris/glycine/acetate system in a 3–15% gradient gel. In-gel activity (arrow) was detected by the formation of calcium phosphate precipitate after incubation, in the presence of ATP (lanes 1 and 4) and ADP (lane 2), but not AMP (lane 3).
Analytical ultracentrifugation and analytical gel-filtration chromatography
To determine the molecular mass of the large species displaying apyrase activity in gel-filtration, we combined the results of analytical gel-filtration and sedimentation velocity experiments. Sedimentation velocity profiles of total saliva and purified apyrase are shown on the top panels of Figures 3(A) and 3(B) respectively. These profiles correspond to successive records of the protein concentration along the centrifuge cell radius during ultracentrifugation. Each set of velocity profiles was globally analysed in terms of a continuous distribution of sedimentation coefficients, c(s) [37,38]. The sample is qualitatively described as an assembly of non-interacting particles, defined by their s values, and the correctness of the fit is estimated by the residuals, the differences between the experimental and the modelled data. Sedimentation velocity profiles of total saliva and purified apyrase (Figures 3A and 3B, top panels, respectively) were well modelled using size-distribution analysis, as indicated by the uniform residuals (Figure 3, middle panels). Comparison of the s-distribution from total saliva and purified apyrase samples revealed enrichment in the 5.7 S species, which appeared to be the main species after purification (Figure 3A and 3B, bottom panels, respectively). In total saliva, the more abundant species with sedimentation coefficients ranging from 1 to 1.5 S must correspond to the salivary proteins with a molecular mass below 45 kDa seen in SDS/PAGE [33]. For total saliva, the same distributions of sedimentation coefficients were obtained at 10, 4.4 and 2.2 mg/ml, which suggested that the proteins were not undergoing dissociation in the tested range of concentrations. The same absence of dissociation upon dilution was observed throughout the gel-filtration experiments on a Superdex 200 column. The Rh of 6.1 nm obtained from calibrated gel-filtration experiments allowed us to calculate for apyrase species a total molar mass between 220 and 190 kDa depending on the hypothesis made for the glycosylation level. The former considers the case of a non-glycosylated form, the latter assumes a glycosylation of 30%, a high value for a glycoprotein. We considered different apyrase glycosylation levels because these levels could not be inferred from SDS/PAGE experiments, since glycosylated proteins often migrate abnormally. From the values of the molecular mass and Rh, we derived a frictional ratio of 1.5 corresponding to a relatively asymmetrical molecule.
Figure 3. Sedimentation velocity analysis of total saliva and purified apyrases.
Superposition of sedimentation profiles for the total saliva at 4.4 mg/ml protein (A, top) and purified apyrase at 0.3 mg/ml (B, top), obtained at 10 °C at a rotor speed of 42000 rev./min. Scans (top panels) recorded the absorbencies at 280 and 228 nm respectively as a function of the radius (cm). The last scans shown correspond to centrifugation for 10 h (A) and 3.5 h (B). Units in (A) and (B) are absorbance units (‘OD’). The sedimentation profiles were modelled in terms of a continuous distribution c(s) of sedimentation coefficients s (S) (sed coefficient; bottom panels). The residuals (middle panels) give the differences between the experimental and modelled profiles.
Electrophoretic analysis
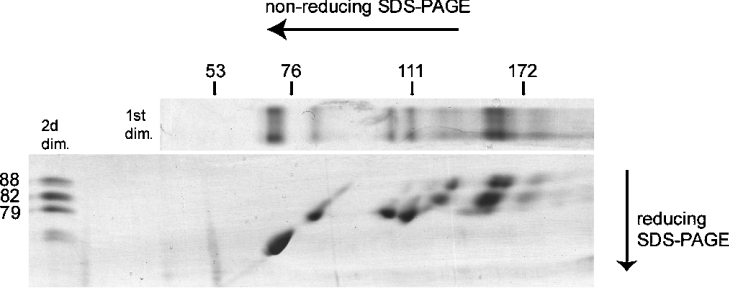
Previous analysis showed that T. infestans saliva contains three more abundant apyrases, showing apparent molecular masses of 88, 82 and 79 kDa on reducing SDS/PAGE, and two less abundant species migrating as a doublet of 68/67 kDa [33]. Figure 4 shows that protein bands of up to 180 kDa were formed under non-reducing conditions (first dimension). These bands in the first dimension (non-reducing SDS/PAGE) were resolved in a second dimension under reducing conditions to investigate the presence of interchain disulphide bonds forming these high-molecular-mass species. In this experiment, proteins were first separated under non-reducing conditions, allowing disulphide-linked proteins to co-migrate, and then in a second dimension under reducing conditions, to subsequently separate the proteins involved in disulphide-linked oligomers. Hetero-oligomer subunits linked by disulphide bonds form vertically aligned protein spots and disulphide-linked homo-oligomers form a single spot, as described by Molinari and Helenius [40]. The results shown in Figure 4 indicate that the 88, 82 and 79 kDa proteins formed disulphide-linked homo-oligomers, because they were not found vertically aligned with other molecular species. However, owing to the complexity of the band pattern in the 150 kDa region, we cannot exclude the possibility that the 88 and 82 kDa proteins also assemble as a hetero-oligomer. Nevertheless, it is likely that each of the 88, 82 and 79 kDa proteins self-associates to form the species seen under non-reducing conditions. A similar experiment, which was performed with a 15% gel for the second dimension, showed the absence of a low-molecular-mass protein linked to apyrases by disulphide bonds (results not shown).
Figure 4. SDS/PAGE analysis of T. infestans apyrases.
Purified apyrases were submitted to non-reducing SDS/6% PAGE, then the gel lane was cut out, and disulphide bonds were reduced by incubation in reducing sample buffer. The lane was then applied horizontally on top of a 6% gel and run into a second dimension. Upper and left-hand lanes represent controls run only in the first non-reducing dimension (1st dim.) and the second reducing dimension (2d dim.) respectively.
The 88 kDa apyrase forms species of 192, 155, 153 and 128 kDa apparent molecular masses under non-reducing conditions. Similarly, the 82 kDa apyrase forms bands of 180, 149, 147 and 121 kDa, whereas the 79 kDa apyrase forms species of 145, 110, 103 and 79 kDa. These multiple bands are likely to represent heterogeneity in disulphide bonds: the presence/absence of intrachain disulphide bonds or a variable number of subunits held by disulphide bonds, for example. The minor doublet of 68/67 kDa does not exhibit any disulphide bond as it migrates at the same position under non-reducing and reducing conditions. On one hand, the sedimentation experiment showed an average molecular mass of 205 kDa and the absence of monomeric species. On the other hand, disulphide bond analysis showed that the 68/67 kDa proteins and the 79 kDa protein fraction have no interchain disulphide bonds. Thus these species must form non-covalent oligomers. The molecular-mass estimations must be considered cautiously as intrachain disulphide bonds are known to modify protein migration, making the protein more compact, thus migrating faster under non-reducing conditions [41]. Therefore no conclusion on complex stoichiometry should be formulated.
To avoid formation of artefactual disulphide bonds, saliva was collected directly in NEM solution to block free cysteine residues. Analyses of NEM-treated samples by electrophoresis and gel-filtration chromatography confirmed the above findings (results not shown). Taken together, these data suggest that the 88, 82 and 79 kDa polypeptides assemble into homo-oligomeric structures, most probably homodimers, with intermolecular disulphide bonds.
Intermolecular disulphide bonds stabilize apyrase structure and activity
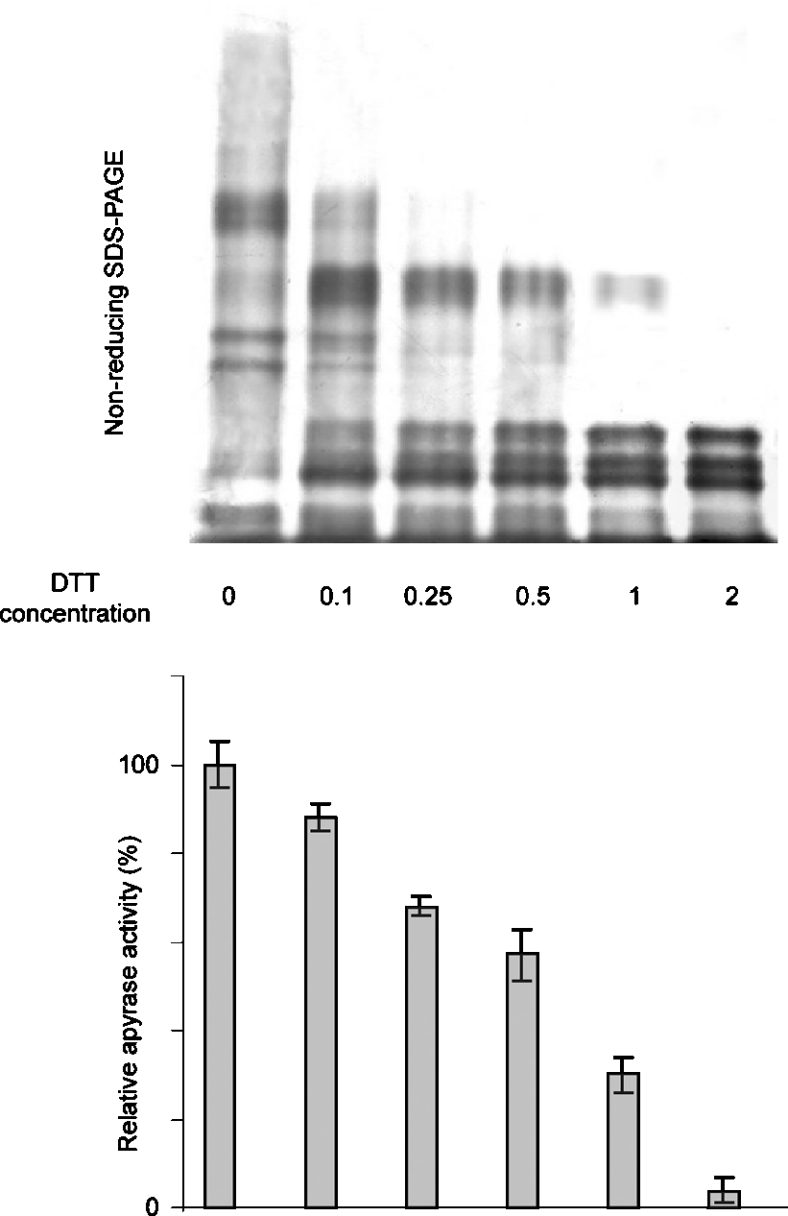
The relationship between the presence of inter-molecular disulphide bonds and the activity of the T. infestans apyrases was analysed by measuring the activity and monitoring the band pattern using non-reducing SDS/PAGE (Figure 5). The enzymatic activity and band pattern of purified apyrases were determined after incubation with increasing concentrations of DTT and subsequent addition of iodoacetamide to block the free cysteine residues and remove the unreacted DTT. The extent of reduction of the intermolecular disulphide bonds correlates with the loss of apyrase activity. A complete reduction was observed upon incubation with 2 mM DTT, concomitant with a 96% reduction of enzymatic activity.
Figure 5. Effects of DTT on intermolecular disulphide bonds and apyrase activity.
Purified apyrases were incubated with increasing concentrations of DTT (0, 0.1, 0.25, 0.5, 1 and 2 mM) for 30 min. After quenching with iodoacetamide, part of the sample was analysed by non-reducing SDS/PAGE, and the other part was used to determine the relative enzymatic activity in relation to the mock sample.
Chemical cross-linking
To investigate the involvement of the 88, 82, 79, 68 and 67 kDa species in oligomers, purified apyrase aliquots were incubated with increasing concentrations of DTSSP and were analysed by SDS/PAGE under non-reducing conditions. Figure 6 shows that incubation with an increasing concentration of DTSSP resulted in the gradual disappearance of the bands observed under non-reducing conditions along with the appearance of bands in the 200 kDa region. As expected, electrophoresis in a second dimension under reducing conditions showed that the 200 kDa bands formed upon incubation with 25 μM DTSSP are composed of the five apyrases with the same apparent stoichiometry as in the saliva (results not shown). These findings are consistent with the molecular mass determined by analytical ultracentrifugation and confirm that, in addition to disulphide bonds, other types of interactions are responsible for the quaternary structure of apyrases.
Figure 6. DTSSP cross-linking of T. infestans apyrases.
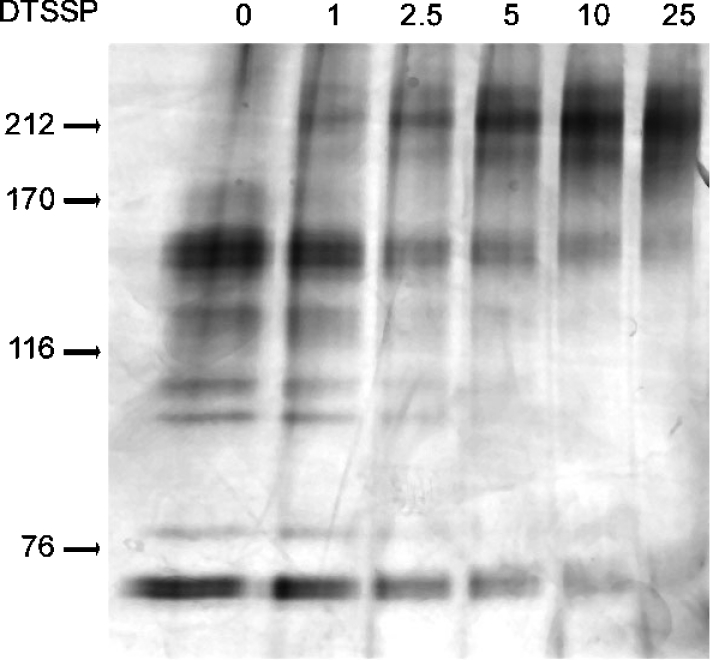
Purified apyrases were incubated with increasing concentrations of DTSSP (0, 1, 2.5, 5, 10 and 25 μM) and submitted to non-reducing SDS/6% PAGE. The position of molecular-mass standards is indicated by arrows: myosin, 212 kDa; α-macroglobulin, 170 kDa; β-galactosidase, 116 kDa and transferrin, 76 kDa.
DISCUSSION
Triatomine vectors of Chagas disease were shown to overcome host platelet aggregation by secreting apyrase in their saliva during feeding [42], and we established further that T. infestans presents five apyrases from the 5′-nucleotidase family [33]. In the present report, we provide evidence that these proteins are assembled into oligomers.
Comparison of T. infestans apyrase molecular masses determined by SDS/PAGE (ranging from 67 to 88 kDa) and by analytical centrifugation (205±15 kDa) shows that these proteins form oligomers, probably dimers or trimers. However, the exact stoichiometry could not be solved because their apparent masses obtained from SDS/PAGE are not appropriate for this purpose. To our knowledge, soluble oligomeric apyrases have not been reported previously, although this feature is present in membrane-associated apyrases [43–45].
The 5′-nucleotidases that were shown to form oligomers have their subunits linked sometimes by disulphide bonds [24], and this information is consistent with our observation that the T. infestans apyrases belong to this enzyme family [33]. Our electrophoresis findings indicated that disulphide bonds could assemble apyrases into homo-oligomers. The possibility of the hetero-oligomers involving the 88 kDa and 82 kDa species is consistent with our previous demonstration that these two molecular species correspond to different alleles [33]. We hypothesize that the heterogeneity in disulphide bonds of some species, and their absence for the 68/67 kDa apyrases and the 79 kDa fraction, are probably due to the disulphide bonds being an alternative oligomerization mechanism for the apyrases. In favour of this reasoning is the existing heterogeneous population of oligomers, part of which being assembled by disulphide bonds and another part or subpopulation being held by non-covalent interactions. In this respect, co-existing covalent and non-covalent oligomers have been described for alternative oxidases of plants [46]. The advantage of a multimeric over a monomeric structure remains unclear, but it is possible that the quaternary structure allows both hydrophobic regions to be buried within the protein assemblage and reduction of the molecule surface with the medium, thus limiting the amount of water required to stabilize these proteins [47].
Cross-linking with DTSSP analysed by SDS/PAGE resulted in the formation of high-molecular-mass bands with apparent molecular mass close to 200 kDa, showing that interactions other than disulphide bonds participate to form the quaternary structure and that the different apyrase oligomers have similar mobility when cross-linked. This is consistent with the repeated observation that apyrase activity of T. infestans saliva is related to five proteins that could not be distinguished under native conditions, i.e. gel-filtration, native PAGE and ultracentrifugation and during the various purification methods tested. Despite heterogeneity in disulphide bonds, the overall biophysical properties of the oligomers seem to be very similar. We hypothesize that an existing heterogeneity in inter- and/or intra-chain disulphide bonds may result from domain swapping. Three-dimensional domain swapping is an oligomerization mechanism whereby a domain from one protein subunit replaces the identical domain of a neighbouring subunit and can involve rearrangement of disulphide bonds, passing from intrachain in the monomers to interchain in dimers [48]. In addition, both swapped and unswapped conformations could coexist [49]. This feature might be present in T. infestans apyrases, resulting in the formation of different conformers bearing different inter- and intra-chain disulphide bonds. This hypothesis remains to be tested by crystallographic studies.
T. infestans apyrases display pI values between 4.9 and 5.4, in agreement with the broad elution pattern over the salt gradient on an ion-exchange column and in chromatofocusing experiments (results not shown). In this regard, apyrases can be classified into two groups based on their pI: apyrases from mosquitoes and sandflies with basic pI [25,26,50,51] and apyrases from T. infestans and C. lectularius [52] with acidic pI. This classification is independent of the apyrase genetic families, as mosquitoes and T. infestans apyrases belong to the 5′-nucleotidase family, and sandflies and C. lectularius apyrases to the Cimex apyrase family. Thus one may speculate on the physiological reasons related to these pI differences. Neither should they be related to feeding mechanism because sandflies are capillary-feeding insects, whereas mosquitoes, T. infestans and C. lectularius are vessel-feeding. However, we noted a correlation between the pI of the enzymes and blood meal duration, which is more important for T. infestans and C. lectularius than for mosquitoes or sandflies. Consistently, an apyrase from the 5′-nucleotidase family has been detected in another arthropod with long feeding behaviour, the tick Ixodes scapularis, with a predicted acidic pI [53]. However, the biological mechanism, if any, underlying this pI segregation remains to be determined.
We have shown that the T. infestans saliva in-gel activity after IEF staining presents various active forms of apyrase with distinct pI values, which supports further the originality of these apyrases, as previous studies of pI values from apyrases in other species revealed a single active form. However, the Anopheles stephensi is an exception; its saliva apyrase displays three activities at pI values of 6.9, 7.2 and 7.6 [34]. We believe the diversity of pI values in T. infestans might result from different post-translational modifications of the corresponding proteins and is related, at least partially, to the disulphide bond heterogeneity. Indeed, the contribution of genetic polymorphism to the molecular diversity described in the present study is limited as it merely accounts for the presence or absence of the 88 and 82 kDa apyrases [33]. Additional experiments will be conducted with recombinant proteins in order to uncover the exact organization of the oligomeric structures and the role of post-translational modifications, as a prerequisite to further pharmaceutical studies.
Acknowledgments
We are grateful to André Zapun for helpful discussions and critical reading of the manuscript before submission. This work was partially supported by the World Bank/Financiadora de Estudos e Projetos-FINEP, and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Brazil, and Région Rhône-Alpes, Fondation pour la Recherche Médicale and Groupe d'Etude sur l'Hemostase et la Thrombose, France.
References
- 1.Law J. H., Ribeiro J. M., Wells M. A. Biochemical insights derived from insect diversity. Annu. Rev. Biochem. 1992;61:87–111. doi: 10.1146/annurev.bi.61.070192.000511. [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro J. M. C. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect. Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- 3.Arocha-Pinango C. L., Marchi R., Guerrero B. Inventory of exogenous hemostatic factors derived form arthropods. Registry of Exogenous Hemostatic Factors of the Scientific and Standardization Subcommittee of the International Society on Thrombosis and Haemostasis. Thromb. Haemostasis. 1999;81:647–656. [PubMed] [Google Scholar]
- 4.Ribeiro J. M., Francischetti I. M. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu. Rev. Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 5.Noeske-Jungblut C., Haendler B., Donner P., Alagon A., Possani L., Schleuning W. D. Triabin, a highly potent exosite inhibitor of thrombin. J. Biol. Chem. 1995;270:28629–28634. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- 6.Fuentes-Prior P., Noeske-Jungblut C., Donner P., Schleuning W. D., Huber R., Bode W. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11845–11850. doi: 10.1073/pnas.94.22.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haendler B., Becker A., Noeske-Jungblut C., Kratzschmar J., Donner P., Schleuning W. D. Expression of active recombinant pallidipin, a novel platelet aggregation inhibitor, in the periplasm of Escherichia coli. Biochem. J. 1995;307:465–470. doi: 10.1042/bj3070465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noeske-Jungblut C., Kratzschmar J., Haendler B., Alagon A., Possani L., Verhallen P., Donner P., Schleuning W. D. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J. Biol. Chem. 1994;269:5050–5053. [PubMed] [Google Scholar]
- 9.Moncada S., Palmer R. M., Higgs E. A. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 10.Ribeiro J. M., Hazzard J. M., Nussenzveig R. H., Champagne D. E., Walker F. A. Reversible binding of nitric oxide by a salivary heme protein from a bloodsucking insect. Science. 1993;260:539–541. doi: 10.1126/science.8386393. [DOI] [PubMed] [Google Scholar]
- 11.Francischetti I. M., Ribeiro J. M., Champagne D., Andersen J. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood-sucking bug, Rhodnius prolixus. J. Biol. Chem. 2000;275:12639–12650. doi: 10.1074/jbc.275.17.12639. [DOI] [PubMed] [Google Scholar]
- 12.Karczewski J., Connolly T. M. The interaction of disagregin with the platelet fibrinogen receptor, glycoprotein IIb-IIIa. Biochem. Biophys. Res. Commun. 1997;241:744–748. doi: 10.1006/bbrc.1997.7881. [DOI] [PubMed] [Google Scholar]
- 13.Karczewski J., Endris R., Connolly T. M. Disagregin is a fibrinogen receptor antagonist lacking the Arg-Gly-Asp sequence from the tick, Ornithodoros moubata. J. Biol. Chem. 1994;269:6702–6708. [PubMed] [Google Scholar]
- 14.Mans B. J., Louw A. I., Neitz A. W. Savignygrin, a platelet aggregation inhibitor from the soft tick Ornithodoros savignyi, presents the RGD integrin recognition motif on the Kunitz-BPTI fold. J. Biol. Chem. 2002;277:21371–21378. doi: 10.1074/jbc.M112060200. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Coons L. B., Taylor D. B., Stevens S. E., Jr, Gartner T. K. Variabilin, a novel RGD-containing antagonist of glycoprotein IIb-IIIa and platelet aggregation inhibitor from the hard tick Dermacentor variabilis. J. Biol. Chem. 1996;271:17785–17790. doi: 10.1074/jbc.271.30.17785. [DOI] [PubMed] [Google Scholar]
- 16.Reddy V. B., Kounga K., Mariano F., Lerner E. A. Chrysoptin is a potent glycoprotein IIb/IIIa fibrinogen receptor antagonist present in salivary gland extracts of the deerfly. J. Biol. Chem. 2000;275:15861–15867. doi: 10.1074/jbc.275.21.15861. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro J. M., Charlab R., Valenzuela J. G. The salivary adenosine deaminase activity of the mosquitoes Culex quinquefasciatus and Aedes aegypti. J. Exp. Biol. 2001;204:2001–2010. doi: 10.1242/jeb.204.11.2001. [DOI] [PubMed] [Google Scholar]
- 18.Cheeseman M. T., Bates P. A., Crampton J. M. Preliminary characterisation of esterase and platelet-activating factor (PAF)-acetylhydrolase activities from cat flea (Ctenocephalides felis) salivary glands. Insect Biochem. Mol. Biol. 2001;31:157–164. doi: 10.1016/s0965-1748(00)00113-2. [DOI] [PubMed] [Google Scholar]
- 19.Marcus A. J. Platelet and their disorders. In: Ratnoff O. D., Forbes C. D., editors. Disorders of Hemostasis. Philadelphia: W.B. Saunders; 1996. pp. 79–137. [Google Scholar]
- 20.Gayle R. B., 3rd, Maliszewski C. R., Gimpel S. D., Schoenborn M. A., Caspary R. G., Richards C., Brasel K., Price V., Drosopoulos J. H., Islam N., et al. Inhibition of platelet function by recombinant soluble ecto-ADPase/CD39. J. Clin. Invest. 1998;101:1851–1859. doi: 10.1172/JCI1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczmarek E., Koziak K., Sevigny J., Siegel J. B., Anrather J., Beaudoin A. R., Bach F. H., Robson S. C. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J. Biol. Chem. 1996;271:33116–33122. doi: 10.1074/jbc.271.51.33116. [DOI] [PubMed] [Google Scholar]
- 22.Marcus A. J., Broekman M. J., Drosopoulos J. H., Islam N., Alyonycheva T. N., Safier L. B., Hajjar K. A., Posnett D. N., Schoenborn M. A., Schooley K. A., et al. The endothelial cell ecto-ADPase responsible for inhibition of platelet function is CD39. J. Clin. Invest. 1997;99:1351–1360. doi: 10.1172/JCI119294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T. F., Ou Y., Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J. Biol. Chem. 1998;273:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann H. 5′-Nucleotidase: molecular structure and functional aspects. Biochem. J. 1992;285:345–365. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlab R., Valenzuela J. G., Rowton E. D., Ribeiro J. M. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela J. G., Belkaid Y., Rowton E., Ribeiro J. M. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 2001;204:229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- 27.Valenzuela J. G., Charlab R., Galperin M. Y., Ribeiro J. M. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius: a new type of nucleotide-binding enzyme. J. Biol. Chem. 1998;273:30583–30590. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- 28.Failer B. U., Braun N., Zimmermann H. Cloning, expression, and functional characterization of a Ca2+-dependent endoplasmic reticulum nucleoside diphosphatase. J. Biol. Chem. 2002;277:36978–36986. doi: 10.1074/jbc.M201656200. [DOI] [PubMed] [Google Scholar]
- 29.Smith T., Hicks-Berger C., Kim S., Kirley T. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch. Biochem. Biophys. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- 30.Arca B., Lombardo F., de Lara Capurro M., della Torre A., Dimopoulos G., James A. A., Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champagne D. E., Smartt C. T., Ribeiro J. M., James A. A. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc. Natl. Acad. Sci. U.S.A. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenzuela J. G., Francischetti I. M., Pham V. M., Garfield M. K., Ribeiro J. M. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 33.Faudry E., Lozzi S. P., Santana J. M., D'Souza-Ault M., Kieffer S., Felix C. R., Ricart C. A., Sousa M. V., Vernet T., Teixeira A. R. Triatoma infestans apyrases belong to the 5′-nucleotidase family. J. Biol. Chem. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- 34.Mathews G. V., Sidjanski S., Vanderberg J. P. Inhibition of mosquito salivary gland apyrase activity by antibodies produced in mice immunized by bites of Anopheles stephensi mosquitoes. Am. J. Trop. Med. Hyg. 1996;55:417–423. doi: 10.4269/ajtmh.1996.55.417. [DOI] [PubMed] [Google Scholar]
- 35.Notarnicola S. M., Park K., Griffith J. D., Richardson C. C. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 1995;270:20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- 36.Le Maire M., Aggerbeck L. P., Monteilhet C., Andersen J. P., Moller J. V. The use of high-performance liquid chromatography for the determination of size and molecular weight of proteins: a caution and a list of membrane proteins suitable as standards. Anal. Biochem. 1986;154:525–535. doi: 10.1016/0003-2697(86)90025-4. [DOI] [PubMed] [Google Scholar]
- 37.Dam J., Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebel C. Analytical ultracentrifugation for the study of biological macromolecules. In: Lechner M. D., Börger L., editors. Progress in Colloid and Polymer Science. Berlin: Springer-Verlag GmbH; 2004. pp. 73–82. [Google Scholar]
- 40.Molinari M., Helenius A. Analyzing cotranslational protein folding and disulfide formation by diagonal sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 2002;348:35–42. doi: 10.1016/s0076-6879(02)48623-5. [DOI] [PubMed] [Google Scholar]
- 41.Braakman I., Hoover-Litty H., Wagner K. R., Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell. Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro J. M., Schneider M., Isaias T., Jurberg J., Galvao C., Guimaraes J. A. Role of salivary antihemostatic components in blood feeding by triatomine bugs (Heteroptera) J. Med. Entomol. 1998;35:599–610. doi: 10.1093/jmedent/35.4.599. [DOI] [PubMed] [Google Scholar]
- 43.Failer B. U., Aschrafi A., Schmalzing G., Zimmermann H. Determination of native oligomeric state and substrate specificity of rat NTPDase1 and NTPDase2 after heterologous expression in Xenopus oocytes. Eur. J. Biochem. 2003;270:1802–1809. doi: 10.1046/j.1432-1033.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 44.Murphy D. M., Kirley T. L. Asparagine 81, an invariant glycosylation site near apyrase conserved region 1, is essential for full enzymatic activity of ecto-nucleoside triphosphate diphosphohydrolase 3. Arch. Biochem. Biophys. 2003;413:107–115. doi: 10.1016/s0003-9861(03)00084-5. [DOI] [PubMed] [Google Scholar]
- 45.Smith T. M., Kirley T. L. Glycosylation is essential for functional expression of a human brain ecto-apyrase. Biochemistry. 1999;38:1509–1516. doi: 10.1021/bi9821768. [DOI] [PubMed] [Google Scholar]
- 46.Umbach A. L., Siedow J. N. Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 1993;103:845–854. doi: 10.1104/pp.103.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodsell D. S., Olson A. J. Soluble proteins: size, shape and function. Trends Biochem. Sci. 1993;18:65–68. doi: 10.1016/0968-0004(93)90153-e. [DOI] [PubMed] [Google Scholar]
- 48.Knaus K. J., Morillas M., Swietnicki W., Malone M., Surewicz W. K., Yee V. C. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 49.Piccoli R., Tamburrini M., Piccialli G., Di Donato A., Parente A., D'Alessio G. The dual-mode quaternary structure of seminal RNase. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1870–1874. doi: 10.1073/pnas.89.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombardo F., Di Cristina M., Spanos L., Louis C., Coluzzi M., Arca B. Promoter sequences of the putative Anopheles gambiae apyrase confer salivary gland expression in Drosophila melanogaster. J. Biol. Chem. 2000;275:23861–23868. doi: 10.1074/jbc.M909547199. [DOI] [PubMed] [Google Scholar]
- 51.Vachereau A., Ribeiro J. M. C. Immunoreactivity of salivary gland apyrase of Aedes aegypti with antibodies against a similar hydrolase present in the pancreas of mammals. Insect Biochem. 1989;19:527–534. [Google Scholar]
- 52.Valenzuela J. G., Chuffe O. M., Ribeiro J. M. C. Apyrase and anti-platelet activities from the salivary glands of the bed bug Cimex lectularius. Insect Biochem. Mol. Biol. 1996;21:557–562. [Google Scholar]
- 53.Valenzuela J. G., Francischetti I. M., Pham V. M., Garfield M. K., Mather T. N., Ribeiro J. M. Exploring the sialome of the tick Ixodes scapularis. J. Exp. Biol. 2002;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]