Abstract
The minichromosome maintenance protein 10 (Mcm10) is an evolutionarily conserved factor that is essential for replication initiation and elongation. Mcm10 is part of the eukaryotic replication fork and interacts with a variety of proteins, including the Mcm2-7 helicase and DNA polymerase alpha/primase complexes. A motif search revealed a match to the proliferating cell nuclear antigen (PCNA)-interacting protein (PIP) box in Mcm10. Here, we demonstrate a direct interaction between Mcm10 and PCNA that is alleviated by mutations in conserved residues of the PIP box. Interestingly, only the diubiquitinated form of Mcm10 binds to PCNA. Diubiquitination of Mcm10 is cell cycle regulated; it first appears in late G1 and persists throughout S phase. During this time, diubiquitinated Mcm10 is associated with chromatin, suggesting a direct role in DNA replication. Surprisingly, a Y245A substitution in the PIP box of Mcm10 that inhibits the interaction with PCNA abolishes cell proliferation. This severe-growth phenotype, which has not been observed for analogous mutations in other PCNA-interacting proteins, is rescued by a compensatory mutation in PCNA that restores interaction with Mcm10-Y245A. Taken together, our results suggest that diubiquitinated Mcm10 interacts with PCNA to facilitate an essential step in DNA elongation.
Minichromosome maintenance protein 10 (Mcm10) is an essential replication factor that plays a central role in replication fork assembly (22, 25, 44, 48, 50, 64). Initially identified in two independent genetic screens (44, 54), the biochemical characterization of Mcm10 in Saccharomyces cerevisiae and Schizosaccharomyces pombe has proven difficult because of the insoluble nature of the protein (40, 48). Nevertheless, a number of reports have demonstrated physical and genetic interactions between Mcm10 and other replication factors. Among those interacting proteins are the replication initiator, the origin recognition complex (ORC), components of the Mcm2-7 complex, the putative replication helicase, and a cofactor of the Mcm2-7 complex, Cdc45 (9, 24, 25, 33, 50). In addition, we and others have shown that Mcm10 associates with DNA polymerase alpha/primase (pol-α/primase) in S. cerevisiae and S. pombe (19, 48). Pol-α/primase is the only enzyme in eukaryotic cells capable of initiating DNA synthesis de novo (6). The interaction between Mcm10 and pol-α is likely direct and mediated through the catalytic subunit, Cdc17/Pol1 (19). Importantly, Mcm10 is required to stabilize Cdc17/Pol1 in budding yeast. In the absence of Mcm10, the catalytic subunit of pol-α is rapidly degraded (48).
In S. cerevisiae, Mcm10 binds chromatin in a cell cycle-dependent manner (48). It associates with DNA after origin licensing (22, 48, 50, 64). Origin licensing is accomplished by loading the Mcm2-7 complex onto replication origins (2, 13, 14). This process ensures that replication origins are not activated more than once during S phase and is thus crucial for maintaining genome integrity (5). Once bound to replication origins, Mcm10 appears to coordinate multiple steps in the assembly of the replication complex. It has been implicated in the activation of the replicative helicase by regulating the Cdc7/Dbf4-dependent phosphorylation of Mcm2-7 (36) and the subsequent recruitment of Cdc45 (22, 50, 64), which is required for DNA unwinding (61). Upon DNA unwinding, replication protein A stabilizes single-stranded DNA and likely provides a molecular signal for pol-α/primase (stabilized in a complex with Mcm10) to assemble onto the DNA (57). In this process, Mcm10 presumably positions pol-α/primase next to ORC, via a direct Mcm10-ORC interaction, where replication initiates (3, 4). However, Mcm10 is not only required to facilitate the initiation of DNA replication; it is also essential for elongation, at least in budding yeast (48). This might be simply explained by the finding that pol-α/primase is displaced from chromatin in the absence of Mcm10 (48).
To ensure timely replication of the entire genome, a polymerase switch occurs from the nonprocessive pol-α/primase to pol-δ and pol-ɛ that cooperate with a trimeric sliding clamp, the processivity factor proliferation cell nuclear antigen (PCNA) (42). PCNA is the eukaryotic counterpart of the prokaryotic β clamp. While pol-ɛ associates with DNA even before recruitment of pol-α (43, 45) and likely facilitates leading strand synthesis (20, 60), pol-δ might be responsible for Okazaki fragment synthesis on the lagging strand (21). Both pol-δ and pol-ɛ interact with PCNA through a common PCNA-interacting protein (PIP) motif, the PIP box, which has the following consensus motif: QXX(M/L/I)XX(F/Y)(F/Y) (62). Strikingly, Mcm10 also has a close match to the PIP box, which is highly conserved in other species, although in humans it more closely resembles the prokaryotic β-clamp binding consensus motif (11, 38). In S. cerevisiae, an eight-amino-acid stretch, 239QHTLDVYI246, corresponds to a 7/8 match to the PIP box. In a previous study that tested all proteins containing an 8/8 match to the PIP box (51), Mcm10 was not identified as a potential PCNA-interacting protein because it did not fulfill the search criteria. The significance of this putative PIP box is unclear, however, as another study that identified a match to the prokaryotic β-clamp binding site in Mcm10/Cdc23 of S. pombe tested for binding between purified S. pombe Mcm10 and S. pombe PCNA but failed to detect a physical interaction (19).
Besides the two polymerases mentioned above, a large number of replication and repair proteins have been shown to interact with PCNA (42, 59). Two domains in PCNA, the interdomain connector loop (IDCL) and the carboxy terminus, contribute to the formation of a large hydrophobic pocket which might be induced upon interaction with the PIP box (23). This was best illustrated in two cocrystallization studies of PCNA with the cell cycle inhibitor p21 (23) and the flap endonuclease Fen1 (49), respectively, and is further supported by numerous genetic analyses (1, 8, 17). Interestingly, PIP boxes embedded in different protein complexes might facilitate distinct primary interactions with either the IDCL or the C-terminal residues of the hydrophobic pocket (16, 59).
In this study, we demonstrate that Mcm10 interacts with PCNA in S. cerevisiae. Moreover, our results indicate that PCNA recognizes only the diubiquitinated form of Mcm10 and not the unmodified protein. Posttranslational modification of Mcm10 is cell cycle regulated and occurs in G1 and S phase. Furthermore, we provide evidence that the PIP box in Mcm10 facilitates the interaction with the carboxy terminus of PCNA. Importantly, a tyrosine-to-alanine mutation in the seventh position of the PIP box abolishes binding to PCNA in a two-hybrid system. The same PIP box mutation abolishes cell growth in budding yeast, suggesting that Mcm10 binding to PCNA serves an essential function during DNA replication.
MATERIALS AND METHODS
Strains.
YK10MYC has been previously described (25). ABy025 contains MCM10 with nine Myc tags. ABy021 is a W303 strain in which the BAR1 gene has been disrupted. ABy078 and ABy104 were constructed by transforming plasmid YEp105 (Myc-tagged synthetic ubiquitin gene) (18) into a triple-hemagglutinin (HA)-tagged MCM10 strain (ABy078) or nontagged MCM10 strain (ABy104). RDKY3552 (ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.POL30) and RDKY3556 (ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104) were a gift from Richard Kolodner. A triple-HA-epitope gene fusion construct, a mcm10-PIP box (L242A) mutant, was integrated at the endogenous loci in RDKY3552 (ABy142) and RDKY3556 (ABy143). A triple-HA-tagged mcm10-PIP box (Y245A) mutant was integrated at the endogenous locus in RDKY3556 (ABy144). The ABy145 strain was constructed by transforming pRS316-Mcm10 plasmid (MCM10 URA3) into RDKY3552. ABy146 and ABy147 were constructed by integrating triple-HA-tagged mcm10-PIP box (L242A and Y245A) mutants into the endogenous locus in ABy145. A nine-Myc-tagged gene fusion of Mcm10 was integrated at the endogenous loci in RDKY3552 (ABy210) and RDKY3556 (ABy199). A nine-Myc-tagged mcm10-PIP box (Y245A) mutant was integrated at the endogenous loci in RDKY3556 (ABy200) and ABy145 (ABy201). DK1 (MATa ade 2-1 ura3-1 his3-11,15 trp1-1 can1-100 ERG6::LEU2) was a gift from Deanna Koepp. ABy198 was generated by integrating Myc-tagged MCM10 at the endogenous locus in DK1. ABy212 was generated by transforming pGal-HA-Clb2 (GAL-HA-CLB2) plasmid (gift from Deanna Koepp) into DK1. ABy211 was constructed by transforming pRS423gal-Mcm10-3HA plasmid (GAL-MCM10-3HA) and YEp105 (Myc-tagged ubiquitin gene) into ABy021.
Coimmunoprecipitations.
Cells were arrested in S phase with 200 mM hydroxyurea (HU), and whole-cell extracts (WCEs) were prepared as described previously (10) except that N-ethylmaleimide (NEM) was added to a final concentration of 5 mM. For samples treated with DNase I, 10 mM MgCl2 was added with 150 kU DNase I for 5 min on ice. Immunoprecipitations were performed with antibodies to Myc (9E11), HA (12CA5), PCNA (PC10), or immunoglobulin G (IgG) (Oncogene Research Products) for 2 h at 4°C. Antibodies used for Western blotting were against Mcm10, Myc (9E11 used for coimmunoprecipitations [co-IPs] and immunoblots if not indicated otherwise and 9E10 used for immunoblots as indicated), HA (16B12), PCNA (Neomarkers), and ubiquitin (P4G7-H11).
Histone association assay.
Cells were cross-linked with formaldehyde and sonicated to fragment the chromatin, and then histone H3 was immunoprecipitated as described previously (47, 48). The cross-links were reversed by boiling, and histone-associated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (48). Antibodies used for detection were anti-Myc (9E11) and anti-histone H3 (Abcam).
WCE preparation using lyticase.
Asynchronous yeast cultures (W303 and ABy021) were grown overnight at 30°C. Cells were converted to spheroplasts as reported previously (15). Spheroplasts were lysed in SNH buffer (0.4 M sorbitol [pH 7.5], 150 mM NaCl, and 50 mM HEPES-KOH [pH 7.5], including standard protease inhibitors, such as 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mM benzamidine, and 5 mM NEM as indicated) by addition of Triton X-100 to 0.2%. WCEs were fractionated into supernatant and pellet after centrifugation at 15,000 × g for 15 min at 4°C. The insoluble pellet was resuspended in 500 μl of SNH buffer. To 100 μl of either soluble supernatant or insoluble pellet, 100 μl of 2× Laemmli buffer was added, boiled, and fractionated on SDS-PAGE for Western blotting.
Flow cytometry.
Flow cytometry was performed as described previously (48) using a Becton Dickinson FACSCalibur.
Yeast two-hybrid assays.
The L40 [MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ GAL4] yeast strain was cotransformed with a LexA fusion construct (TRP1) and a Gal4 fusion prey construct (LEU2) using the polyethylene glycol-lithium acetate method. Transformants were selected on synthetic complete (SC) medium plates lacking leucine and tryptophan grown for 2 to 4 days at 30°C and then transferred to triple dropout plates (SC-Leu-Trp-His) at 30°C. Finally, lacZ reporter gene expression was studied using SC-Leu-Trp-His plates containing 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 30°C for 2 to 3 days. The two-hybrid plasmids used in this study, pBL240 (Gal4 activation domain, POL30) and pBTM.M10 (LexA binding domain, MCM10), have been previously described (11, 30). Amino acid substitutions in the putative PIP box of Mcm10 (L242A, L242Y, and Y245A) and in the C terminus and interdomain connector loop of PCNA (A251V, Y133A, and Y133L) were introduced by site-directed mutagenesis using a QuikChange kit (Stratagene).
Verification of bait and prey expression by Western blot analysis.
Ten-milliliter cultures of the yeast strain L40 carrying prey and bait constructs were grown overnight in liquid SC-Leu-Trp-His medium to an optical density of 0.6 at 600 nm. Trichloroacetic acid (TCA)-precipitated proteins were separated by SDS-PAGE and Western blotting using anti-LexA (Invitrogen) and anti-PCNA (Neomarkers) antibodies, which confirmed the expression of the wild-type and mutant Mcm10 and PCNA proteins, respectively.
Preparation of WCEs from yeast two-hybrid strains for quantification of β-galactosidase activity.
A published method (51) was modified for preparation of WCEs. A single colony from the yeast two-hybrid strain of interest was inoculated into 10 ml of liquid SC-Leu-Trp-His medium and grown overnight at 30°C to an optical density of 1.2 to 1.5 at 600 nm. Cells were pelleted and washed once with 25 ml of ice-cold, distilled water and once with 1 ml of ice-cold P buffer (50 mM sodium phosphate [pH 7.7], 300 mM sodium acetate, 10% glycerol, 1 mM 2-mercaptoethanol, 500 nM dithiothreitol, 1 μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 1 mM benzamidine). The cells were resuspended in 100 μl of P buffer and lysed in a beater in the presence of an equal amount of acid-washed glass beads. The extract was centrifuged, and the resulting supernatant was transferred to a fresh tube. The supernatant was further cleared by centrifugation for 1 h at 14,000 rpm at 4°C. β-Galactosidase activity in cleared WCEs was measured using a β-galactosidase assay kit from Invitrogen. β-Galactosidase activity and total protein (Bradford assay) were determined in duplicates from at least three transformants for each two-hybrid strain. The interaction between Pol32 and PCNA served as a positive control (30).
RESULTS
Mcm10 exists in two forms, one interacting with pol-α and the other with PCNA.
In a previous report, we showed that budding-yeast Mcm10 exists in a complex with pol-α (48). In the course of these studies, we observed that Mcm10 is present in two different molecular weight forms in asynchronously growing cells, as an Mcm10-specific antibody detected two proteins in both the soluble and the insoluble fractions of a WCE with sizes of 66 and 82 kDa, respectively (Fig. 1a). No higher-molecular-weight forms of Mcm10 were observed (Fig. 1a). We detected the 82-kDa band only when we added a large excess of a cocktail of freshly prepared protease inhibitors that contained NEM, an inhibitor of cysteine proteases that is often used to block deubiquitinating enzymes (18). When we conducted co-IP experiments with WCEs from S-phase-arrested cells expressing an HA-tagged gene for the catalytic subunit of pol-α, CDC17/POL1, and Myc-tagged MCM10, we pulled down only the unmodified form of Mcm10 (Fig. 1b). Since Mcm10 contains a close match to the canonical PIP box, we performed similar experiments utilizing PCNA-specific antibodies. Surprisingly, in these assays, we exclusively coimmunoprecipitated the modified form of Mcm10 (Fig. 2a). The interaction between Mcm10 and PCNA was confirmed by performing the reverse co-IP experiment (Fig. 2b). Although PCNA is known to be posttranslationally modified (32), we observed only unmodified PCNA interacting with Mcm10. Moreover, association of Mcm10 and PCNA was detectable in extracts that had been treated with DNase I, suggesting that the interaction was not mediated by DNA (Fig. 2c).
FIG. 1.
Mcm10 exists in two forms, and unmodified Mcm10 interacts with pol-α. (a) WCEs were prepared from asynchronous W303 cells (MCM10). Spheroplasts were lysed with 0.2% Triton X-100, and lysates were fractionated into a soluble supernatant and an insoluble pellet. Two forms of Mcm10 were detected in soluble and pellet fractions, using an anti-Mcm10 antibody. (b) ABy003 cells (MCM10-9MYC, CDC17-3HA) were synchronized in S phase with HU. WCEs were immunoprecipitated with an anti-HA antibody in the presence (+) or absence (−) of DNase I. Mcm10-9Myc was detected by an anti-Myc antibody. Protein A Sepharose beads were mock treated as a control (Mock). The asterisk indicates a nonspecific band.
FIG. 2.
Modified Mcm10 interacts with PCNA. (a) ABy025 cells (MCM10-9MYC) were synchronized in S phase with HU. WCEs were immunoprecipitated with an anti-PCNA antibody or IgG as indicated. Mcm10-9Myc (unmodified and modified) was detected with an anti-Myc antibody by Western blotting. The asterisk indicates a nonspecific band. (b) ABy025 cells (MCM10-9MYC) were synchronized in S phase with HU. WCEs were immunoprecipitated with an anti-Myc antibody or IgG. PCNA was detected by Western blotting using an anti-PCNA antibody. (c) ABy025 (MCM10-9MYC) cells were synchronized in S phase with HU. WCEs were immunoprecipitated with anti-PCNA or anti-Myc antibodies in the presence (+) or absence (−) of DNase I as indicated. PCNA was detected using an anti-PCNA antibody.
The experiments described above utilized WCE from S-phase-arrested cells. To investigate whether the modification of Mcm10 was S phase specific, cells expressing Myc-tagged Mcm10 were either synchronized with α-factor in G1, synchronized with HU in S phase, synchronized with nocodazole in G2/M, or left untreated. Total protein was TCA precipitated, and Mcm10 was detected with Myc-specific antibodies. This analysis revealed that the high-molecular-weight form of Mcm10 was present in G1 and S phase of the cell cycle but not at the G2/M transition (Fig. 3). Taken together, these results suggested that Mcm10 is modified in a cell cycle-dependent manner and that this modification directs Mcm10 towards a different interaction partner, namely, PCNA.
FIG. 3.
Modified Mcm10 is cell cycle regulated. ABy025 (MCM10-9MYC) cells were synchronized in G1, S, and G2/M phases of the cell cycle using α-factor, HU, and nocodazole, respectively. Myc-tagged Mcm10 was detected in TCA-precipitated proteins using an anti-Myc antibody. The asterisks indicate nonspecific bands. Another nonspecific band served as a loading control.
Mcm10 is diubiquitinated.
While posttranslationally modified forms of Mcm10 in humans have been described (28), modifications of Mcm10 in other species have not been reported. Human Mcm10 is phosphorylated in mitosis, and two molecular-weight forms of Mcm10 that are consistent with mono- and diubiquitination can be detected in late G1 and throughout S phase (28). Whether human Mcm10 is indeed ubiquitinated has not been rigorously demonstrated, and it has remained unclear whether this modification of human Mcm10 serves any biological function. Because NEM is commonly used to inhibit deubiquitinating enzymes (18) and the size difference of 16 kDa was consistent with budding-yeast Mcm10 being diubiquitinated, we immunoprecipitated Myc-tagged Mcm10 from WCEs of S-phase-arrested cells and immunoblotted the precipitate with either Myc- or ubiquitin-specific antibodies on samples that were fractionated side by side on the same gel. WCEs from nontagged cells were analyzed in parallel. While the Myc-specific antibody detected two bands that differed by 16 kDa in their molecular masses, the ubiquitin-specific antibody decorated only the higher-molecular-weight form of Mcm10 (Fig. 4a). No bands were detected in samples prepared from the nontagged strain. We obtained the same result when we analyzed cells that carried an HA epitope tag at the endogenous MCM10 locus (data not shown). To verify that budding-yeast Mcm10 was indeed ubiquitinated, we performed an independent assay. Cells expressing non- or HA-tagged Mcm10 were transformed with a Myc-tagged, copper-inducible synthetic ubiquitin gene (18). Ubiquitin expression was induced in HU-arrested cells with copper. WCEs were incubated with HA-specific antibodies, and precipitates were subjected to immunoblotting with either HA- or Myc-specific antibodies. We detected two forms of HA-tagged Mcm10 (Fig. 4b, left). The slow-migrating form of Mcm10 was also detected by the Myc-specific antibody, whereas the fast-migrating form was not (Fig. 4b, right). We did not detect any bands in nontagged cells. To show that the modification of Mcm10 is truly a diubiquitination, we prepared spheroplasts by addition of lyticase and lysed them with Triton X-100 on ice in the absence of NEM. We reasoned that we might be able to detect mono- and diubiquitinated Mcm10 if we did not inhibit deubiquitin enzymes. Under these conditions, we observed three forms of Mcm10-9Myc (Fig. 4c) (please note that we used the antibody 9E10, which did not cross-react with an ∼82-kDa protein in WCEs from nontagged cells). Immunoprecipitation with Myc-specific antibodies and subsequent immunoblotting with ubiquitin-specific antibodies revealed that the two higher-molecular-weight forms corresponded to mono- and diubiquitinated Mcm10 (Fig. 4c). As mentioned earlier, when we lysed cells with glass beads (which process is not as gentle as spheroplast lysis) and omitted NEM, we observed only unmodified Mcm10 (data not shown). We then attempted to cooverexpress Mcm10-3HA and Myc-tagged ubiquitin in an effort to up-regulate the production of diubiquitinated Mcm10. Under these conditions, we detected two higher-molecular-weight forms of Mcm10 in TCA precipitates (Fig. 4d); however, we were unable to immunoprecipitate significant amounts of mono- or diubiquitinated Mcm10 (data not shown), suggesting that these modified forms of Mcm10 are highly unstable when Mcm10 is overexpressed. Based on the data shown in Fig. 4d, we inferred that the ratio of diubiquitinated to unmodified Mcm10 was approximately the same regardless of whether Mcm10 was overexpressed or expressed from its endogenous promoter (compare Fig. 4b and d). Since we detected only diubiquitinated Mcm10 but not monoubiquitinated Mcm10 in wild-type cells (Fig. 1a), except when we omitted NEM in the preparation of WCEs (Fig. 4c), we concluded that diubiquitination is the predominant modification of Mcm10. Together, these results demonstrate that a fraction of the cellular Mcm10 pool is diubiquitinated and that the amount of diubiquitinated Mcm10 is tightly regulated.
FIG. 4.
S. cerevisiae Mcm10 is diubiquitinated. (a) ABy025 (MCM10-9MYC) and ABy021 (nontagged) cells were synchronized in S phase. WCEs were immunoprecipitated with an anti-Myc antibody or IgG. Ubiquitinated Mcm10 was detected by an ubiquitin-specific antibody. Unmodified Mcm10 (Mcm10-9Myc) and diubiquitinated Mcm10 [Mcm10-9Myc-(Ub)2] were also detected by an anti-Myc antibody. The asterisk indicates a nonspecific band. (b) Expression of a Myc-tagged synthetic ubiquitin gene was induced in Aby078 (MCM10-3HA) and ABy104 (nontagged) cells with 100 μM copper sulfate for 4 h. Cells were allowed to grow to a cell density of 0.6 at 600 nm and then arrested in S phase by HU. WCEs were immunoprecipitated with anti-HA antibody. Mcm10-3HA (unmodified and diubiquitinated) was detected by Western blotting with an anti-HA antibody (left). Diubiquitinated Mcm10 was also detected with an anti-Myc antibody (right). (c) WCEs were prepared from asynchronous ABy003 (MCM10-9MYC, CDC17-3HA) and W303 (MCM10) cells in the presence of protease inhibitors except for N-ethylmaleimide. Mono- and diubiquitinated Mcm10 were detected in Myc-specific immunoprecipitates by a ubiquitin-specific antibody (right). Western blot analysis with the anti-Myc (9E10) antibody revealed the unmodified, mono-, and diubiquitinated forms of Mcm10 (left). All samples were fractionated on the same gel. (d) Expression of Mcm10 was analyzed in asynchronous ABy211 (GAL-MCM10-3HA CUP1-MYC-Ub), ABy035 (MCM10-3HA), and ABy021 (MCM10) cells grown overnight in 2% raffinose at room temperature. Expression of Myc-Ub was induced by addition of 100 μM copper sulfate for 4 h at a cell density of 0.4 at 600 nm. At a cell density of 0.6 at 600 nm, overexpression of Mcm10 was either induced in 2% galactose (Gal.) or repressed in 2% glucose (Gluc.) for 2 h. Unmodified, mono-, and diubiquitinated Mcm10 were detected in TCA protein precipitates using an anti-HA antibody. A nonspecific band served as a loading control. Gels in panels c and d were run much longer than those shown in panel b for higher resolution.
Inhibition of the 26S proteasome does not affect Mcm10 turnover.
Because Mcm10 was diubiquitinated in a cell cycle-dependent manner, we wondered whether diubiquitinated Mcm10 was polyubiquitinated and subsequently degraded by the 26S proteasome. To experimentally address this question, we monitored Mcm10 levels in erg6Δ mutants, which are sensitive to the proteasome inhibitor MG132 (34). Deletion of ERG6 did not significantly affect S-phase progression (data not shown). Regardless of whether cells were grown in the presence or absence of MG132, we did not detect any differences in the amounts of unmodified and diubiquitinated Mcm10, nor did we detect any higher-molecular-weight bands that would have been consistent with polyubiquitination (Fig. 5a). In contrast, the B-type cyclin Clb2, a known target of the 26S proteasome (66), was stabilized in the presence of MG132 (Fig. 5b). In an independent experiment, we examined Mcm10-9Myc in cells, which overexpressed mutant ubiquitin that carried arginine substitutions in either K29, K48, K63, or all seven lysine residues and thus inhibited the formation of ubiquitin chains (26). We did not detect an accumulation of unmodified or diubiquitinated Mcm10 (Fig. 5c). Moreover, we did not observe any conversion of diubiquitinated to monoubiquitinated Mcm10, suggesting that Mcm10 is monoubiquitinated at two distinct residues (Fig. 5c). Together, these results argue that Mcm10 is not turned over by polyubiquitination and subsequent 26S proteasome-mediated degradation.
FIG. 5.
Mcm10 is not turned over by polyubiquitination and subsequent 26S proteasome-mediated degradation. (a) Asynchronous ABy198 (erg6Δ MCM10-9MYC) and DK1 (erg6Δ MCM10) cells were treated with either dimethyl sulfoxide (DMSO) or 100 μM MG132 (MG132) for 2 h. Unmodified and diubiquitinated Mcm10 were detected in TCA-precipitated proteins using an anti-Myc antibody. On the right, we show a darker exposure of the same gel. The number sign indicates a nonspecific band that is not identical to the one detected in W303 cells (Fig. 1b). The triangle depicts a second nonspecific band. We failed to detect any high-molecular-weight forms of Mcm10, indicative of polyubiquitination, in the top portion of the gel. Ponceau S staining served as a loading control. (b) Asynchronous ABy212 (ergΔ GAL-CLB2-HA) and DK1 (erg6Δ CLB2) cells were grown in 2% raffinose overnight at room temperature. Overexpression of Clb2 was induced by addition of 2% galactose. Samples were taken at 0 and 30 min after induction. Thirty minutes after induction, DMSO or 100 μM MG132 was added for 2 h. Clb2 was detected in TCA-precipitated proteins using an anti-HA antibody. Ponceau S staining served as a loading control. (c) Expression of untagged ubiquitin was induced with 100 μM copper sulfate in asynchronous ABy025 cells (MCM10-9MYC) transformed with plasmid coding for either wild-type ubiquitin (WT ub) (ABy177) or mutant ubiquitin ABy172 (ub-K48R), ABy174 (ub-K29R), ABy176 (ub-K63R), and ABy178 (ub-KO indicates K6R, K11R, K27R, K29R, K33R, K48R, and K63R). WCEs were immunoprecipitated with an anti-Myc antibody. Mcm10 (unmodified and diubiquitinated) was analyzed by Western blotting with an anti-Myc antibody.
Unmodified and diubiquitinated Mcm10 cycle on and off chromatin.
To better understand whether diubiquitinated Mcm10 served a function directly related to DNA replication, we asked whether this form of Mcm10 was bound to chromatin during S phase. In a previous study, we investigated the cell cycle-dependent chromatin association of unmodified Mcm10 (48). Because Mcm10 is partially insoluble in budding (48) and fission (40) yeasts, commonly used chromatin fractionation procedures that rely on the solubility of unbound proteins could not be employed for these experiments. Instead, we applied the histone association assay to monitor DNA binding of Mcm10 in vivo (47, 48). Cells expressing Myc-tagged Mcm10 were synchronized in G1 with α-factor and then released at a low temperature to slow down cell cycle progression. Samples were taken in 15-min intervals and split in half, and cell cycle progression was analyzed by fluorescence-activated cell sorting (Fig. 6a). In parallel, cells were cross-linked with formaldehyde and sonicated. The soluble fraction of this WCE was used for immunoprecipitation with histone H3-specific antibodies to pull down bulk chromatin. Precipitates were boiled and subsequently analyzed by Western blotting. Both forms of Mcm10 were bound to chromatin in G1 and S phase (Fig. 6b). Upon S-phase completion, however, both forms were displaced from DNA and reloaded only when cells had gone through mitosis and entered the subsequent G1 phase. Dissociation from chromatin was unaffected by addition of MG132 (Fig. 6c and d), arguing that it was not regulated by proteasome-mediated degradation. In fact, it seems unlikely that any of the steps required for S-phase completion are dependent on the 26S proteasome, as the presence of MG132 did not impede the ability to release from a HU block and progress through S phase (Fig. 6c and data not shown). With regard to the function of Mcm10, we conclude from these results that both unmodified and diubiquitinated Mcm10 play a role on chromatin during S phase.
FIG. 6.
Unmodified and diubiquitinated Mcm10 cycle on and off chromatin. (a) YK10MYC cells (MCM10-9MYC) were synchronized in G1 and released at 22°C. Samples were taken at the times indicated. DNA content was monitored using flow cytometry. (b) The histone association assay was performed on cells released from α-factor as shown in panel a. Immunoprecipitated proteins (Histone H3 IP) were analyzed by immunoblotting. Mcm10-9Myc, Mcm10-9Myc-(Ub)2, and histone H3 were detected. The asterisk indicates a nonspecific band. The blot for Mcm10-9Myc was overexposed to visualize diubiquitinated Mcm10. The majority of Mcm10 remained in the supernatant (not shown). Mcm10 was also detected in the cross-linked extracts (Input; 1/10 was loaded onto the gel). We could not detect diubiquitinated Mcm10 in these extracts, likely due to the cross-linking procedure. Total protein was stained with Ponceau S (Input). (c) Cultures of ABy198 (erg6Δ MCM10-9MYC) were blocked in S phase with the addition of HU. Once arrested, cells were released into yeast-peptone-dextrose containing 20 μg/ml of nocodazole at 30°C and were then treated with either 100 μM MG132 or DMSO as a control. After 90 min, cell cycle progression in the presence of the inhibitor (+MG132) or in its absence (+DMSO) was monitored using flow cytometry. (d) Samples shown in panel c were analyzed by the histone association assay for chromatin binding of Mcm10. Cross-linked extract from HU-arrested cells was mock treated as a control (Mock). Histone H3 immunoprecipitations (Histone H3 IP) and 1/10 of the input samples (Input) were analyzed by immunoblotting as described for panel b. Total protein was stained with Ponceau S (Input). Importantly, inhibition of the 26S proteasome by treatment with MG132 did not alter the capability of Mcm10 to dissociate from chromatin.
The PIP box in Mcm10 interacts with the carboxy terminus of PCNA.
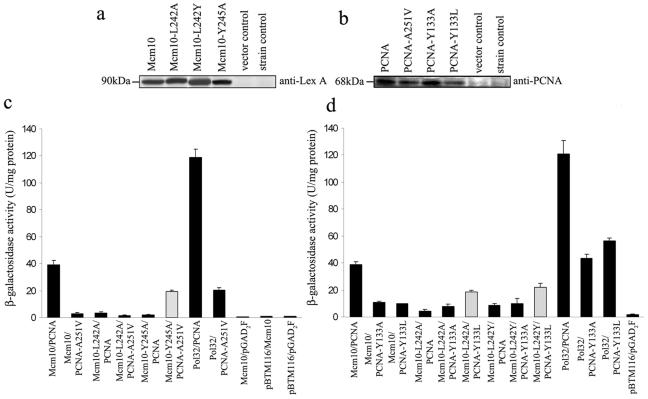
To gain further insight into how Mcm10 might affect PCNA function, we wished to delineate how Mcm10 and PCNA interacted at the molecular level. Both proteins have candidate interaction motifs that we subsequently analyzed by two-hybrid assays. We constructed PIP box mutants in Mcm10 that carried alanine substitutions at positions 242 (L242A) and 245 (Y245A). For PCNA, we initially tested a carboxy-terminal A251V mutant, which had been analyzed in a previous study (51). Expression of wild-type Mcm10 and PCNA proteins as well as mutant proteins was confirmed by Western blot analysis (Fig. 7a and b). Moreover, we found that a fraction of the LexA-Mcm10 fusion protein was indeed diubiquitinated (data not shown), suggesting that interactions between Mcm10 and PCNA measured by two-hybrid analysis accurately reflected the interaction of the endogenous proteins. Plasmids that harbored the wild-type genes or respective mutant alleles were transformed into L40 cells. For each interaction, three colonies were selected and β-galactosidase activity was measured in a liquid assay. Empty vectors served as negative controls. As a positive control, we included constructs expressing PCNA and Pol32, the PCNA-interacting subunit of the pol-δ complex (30). Mcm10 and PCNA showed a moderately strong interaction that was significantly higher than that of the negative controls but not as strong as the interaction between PCNA and Pol32 (Fig. 7c). We obtained the same results when we tested Mcm10 and PCNA in the opposite vectors (data not shown). Importantly, this interaction was decreased to 6.75% of the initial binding activity when we paired Mcm10 with the carboxy-terminal A251V mutant of PCNA. We observed a similar decrease in binding activity for wild-type PCNA and the two mcm10-PIP box (L242A and Y245A) mutants (Fig. 7c). Interestingly, when we combined the mcm10-PIP box Y245A mutant, which was mutated at the seventh position of the motif, with the carboxy-terminal PCNA A251V mutant, binding was partially restored to 50% of the wild-type interaction, indicating that these residues might interact directly with each other (Fig. 7c). This was not the case for the mcm10-PIP box L242A mutant, which carried a substitution at position 4. These results argue that the residues at positions 4 and 7 of the PIP box in Mcm10 are important for the interaction with PCNA. Moreover, the data document that the carboxy terminus in PCNA significantly contributes to the interaction with Mcm10.
FIG. 7.
Mcm10 interacts with the C terminus of PCNA through its PIP box in yeast two-hybrid assays. Immunoblotting of TCA-precipitated proteins with anti-LexA (a) and anti-PCNA (b) antibodies confirmed the expression of bait (Mcm10, Mcm10-L242A, Mcm10-L242Y, Mcm10-Y245A) and prey (PCNA, PCNA-A251V, PCNA-Y133A, PCNA-Y133L) proteins, respectively. Empty vectors (pBTM116 and pGAD2F) and the yeast strain (L40) alone served as controls. (c) β-Galactosidase activity was measured in yeast two-hybrid strains expressing PCNA and Mcm10 wild-type and mutant alleles. The Pol32 and PCNA interaction served as a positive control. pBTM116 and pGAD2F were empty-vector controls. For each combination, three transformants were tested in duplicate, and thus the average for six measurements is shown. The error bars indicate standard deviations. The light shaded bar indicates restoration of activity in the corresponding double mutant. (d) β-Galactosidase activity was measured in yeast two-hybrid strains expressing either wild-type or IDCL mutants of PCNA and wild-type or PIP box mutants of Mcm10. The Pol32 and PCNA interaction served as a positive control. pBTM116 and pGAD2F were empty-vector controls. For each combination, three transformants were tested in duplicate, so the average for six measurements is shown. The error bars indicate standard deviations. The light shaded bars indicate restoration of activity in the corresponding double mutants.
We then asked whether the IDCL of PCNA also plays a role for Mcm10 binding. To that end, we constructed two IDCL (Y133A and Y133L) mutants. We chose Y133A because it is conserved between budding yeast and human PCNA (23). Furthermore, in human PCNA this side chain interacts with leucine at position 4 of the PIP box in p21 (23). Thus, we were curious to test the effect of the Y133L substitution in PCNA in combination with the reciprocal mutation in L242 of Mcm10 (L242Y, mutated at the fourth position of the PIP box in Mcm10). When we combined wild-type Mcm10 with the two IDCL (Y133A and Y133L) mutants, we observed a reduction to about 25% of the initial binding activity (Fig. 7d). While this constitutes a significant decrease, the activity was still clearly above background levels and still higher than that determined for the carboxy-terminal A251V mutant (Fig. 7c). This indicated to us that Mcm10 binds primarily to the carboxy terminus in PCNA, although the IDCL clearly contributes to the interaction. As expected, both PIP box mutants, L242A and L242Y, also significantly decreased binding to PCNA in this system. However, binding was partially restored to 47% and 56% of the initial activity, respectively, when these mutants were combined with Y133A and Y133L mutants in PCNA (Fig. 7d). Therefore, Y133 in PCNA and L242 in Mcm10 may interact directly.
Mcm10 binding to PCNA serves an essential function in vivo.
To evaluate whether mutations in MCM10, which interfere with PCNA binding in yeast two-hybrid assays, showed any phenotype in vivo, we integrated the mcm10-L242A and mcm10-Y245A alleles into the endogenous MCM10 loci of wild-type cells and pol30-A251V mutants, respectively. Surprisingly, we were unable to isolate viable mcm10-Y245A mutants. This is somewhat unusual, because most PIP box mutations, even those in essential genes such as DNA ligase I, are viable (46). Consistent with our two-hybrid results, we obtained viable mcm10-Y245A pol30-A251V double mutants, arguing that it is indeed the inability to interact with PCNA that causes the growth defect. To confirm the phenotype of the mcm10-Y245A allele, we introduced a plasmid-borne MCM10 gene into cells before we integrated mcm10-Y245A at the endogenous MCM10 locus. We then triggered the loss of the MCM10 transgene by plating cells on fluoroorotic acid (FOA)-containing medium. While wild-type and mcm10-L242A cells retained viability, mcm10-Y245A cells did not grow on FOA, consistent with the notion that this specific allele of MCM10 is defective for cell proliferation (Fig. 8a).
FIG. 8.
The mcm10-Y245A allele does not support cell growth. (a) Growth phenotype of RDKY3552 (MCM10 POL30) cells that express MCM10 from plasmid pRS316 and carry integrations of either mcm10-L242A (ABy146) or mcm10-Y245A (ABy147) at the endogenous MCM10 loci. RDKY3552 cells transformed with plasmid pRS316-Mcm10 (ABy145) alone served as a control. Cells were grown for 2 to 3 days at 30°C on SC-Ura and 5′-FOA plates. (b) Successive 10-fold serial dilutions of strains RDKY3552 (MCM10 POL30), ABy142 (mcm10-L242A POL30), RDKY3556 (MCM10 pol30-A251V), ABy143 (mcm10-L242A pol30-A251V), and ABy144 (mcm10-Y245A pol30-A251V) were grown for 3 to 4 days at 30°C on yeast-peptone-dextrose (YPD) plates and YPD containing 25 mM, 50 mM, and 75 mM HU. All mutant alleles were integrated into the chromosome.
To determine whether the viable mcm10-PIP box mutants exhibited any growth defect, we plated wild-type cells and mcm10-L242A POL30, mcm10-L242A pol30-A251V, and mcm10-Y245A pol30-A251V mutants on complete medium in serial dilutions. Consistent with previous reports on PIP box mutants of other PCNA-interacting proteins (16, 46), none of the viable mcm10 mutants or mcm10 pol30 double mutants showed a significant growth defect (Fig. 8b). However, when we added low amounts of the DNA synthesis inhibitor HU (25 mM) to induce replication stress, the mild growth defect observed in the mcm10-L242A and pol30-A251V single mutants became more severe in the mcm10-L242A pol30-A251V double mutant (Fig. 8b), mimicking the complete loss of interaction that we detected for these mutants by two-hybrid assay interaction (Fig. 7c). This became even more obvious at higher concentrations of HU (50 and 75 mM) (Fig. 8b). In contrast, the mcm10-Y245A pol30-A251V double mutants behaved like wild-type cells, agreeing remarkably well with our two-hybrid result that supported the notion that the two mutations are compensatory and restore the interaction between Mcm10 and PCNA (Fig. 7c). Taken together, these results suggested that the interaction between Mcm10 and PCNA serves an essential function during DNA replication.
Ubiquitination of Mcm10 occurs independently of its interaction with PCNA.
Having identified a mutation in Mcm10 that abrogates the binding to PCNA, we then asked whether the diubiquitination of Mcm10 occurred dependently or independently of the interaction with PCNA. To this end, we compared the statuses of Myc-tagged wild-type Mcm10 and Mcm10-Y245A (Fig. 9a). in cells that expressed a nontagged copy of MCM10 from a plasmid to keep the mcm10-Y245A mutants alive. In both cases, Mcm10 was diubiquitinated, arguing that ubiquitination of Mcm10 occurred independently of the interaction with PCNA (Fig. 9b). In a second set of experiments, we also tested whether the PCNA A251V mutant, which binds to Mcm10-Y245A in yeast two-hybrid assays (Fig. 7c), interacted with unmodified or ubiquitinated Mcm10. The data shown in Fig. 9c and d document that both Mcm10 and Mcm10-Y245A were diubiquitinated when bound to PCNA. From the results presented in Fig. 9, we concluded that diubiquitination of Mcm10 is a prerequisite for PCNA binding rather than a consequence of the interaction.
FIG. 9.
Ubiquitination is a prerequisite for Mcm10 binding to PCNA. (a) Schematic structure of protein motifs identified in Mcm10. Mcm10 is a 571-amino-acid protein that contains an OB fold from amino acid 201 to 297, a PIP domain (239 to 245), a zinc (Zn) finger domain (309 to 335) (11), and two C-terminal nuclear localization signals (NLS) (435 to 451 and 512 to 527) (7). The N-terminal half of the protein, including the OB fold domain, has previously been implicated in the interaction with pol-α (19). The PIP box Mcm10-Y245A mutant, defective in PCNA binding, is shown. (b) Mcm10-Y245A is diubiquitinated. ABy210 (MCM10-9MYC POL30) and ABy201 cells (mcm10-Y245A-9MYC POL30 pRS316.MCM10) were synchronized in S phase with HU. Unmodified and diubiquitinated Mcm10 were detected in TCA-precipitated proteins using an anti-Myc antibody. Ponceau S staining of the same extracts served as a loading control. The asterisk indicates a nonspecific band. (c) PCNA mutants express both unmodified and diubiquitinated Mcm10. ABy210 (MCM10-9MYC POL30), ABy199 (MCM10-9MYC pol30-A251V), ABy200 (mcm10-Y245A-9MYC pol30-A251V), and RDKY3552 (MCM10 POL30) cells were synchronized in S phase with HU. Unmodified and modified Mcm10 were detected in TCA-precipitated proteins using an anti-Myc antibody. The asterisk indicates a nonspecific band. Another nonspecific band served as a loading control. (d) ABy210 (MCM10-9MYC POL30), ABy199 (MCM10-9MYC pol30-A251V), ABy200 (mcm10-Y245A-9MYC pol30-A251V), and RDKY3552 (MCM10 POL30) cells were synchronized in S phase with HU. WCEs were immunoprecipitated with an anti-PCNA antibody. Diubiquitinated Mcm10 was detected by Western blotting with an anti-Myc antibody.
DISCUSSION
Nonproteolytic ubiquitination modulates the function of Mcm10.
Mcm10 is an essential replication factor that has been implicated in both the initiation and the elongation steps of DNA replication (22, 25, 33, 44, 48, 64). Consistent with this notion, Mcm10 has been shown to interact with ORC, Mcm2-7, Cdc7/Dbf4, and pol-α/primase (9, 24, 25, 27, 33, 36). In the course of our experiments, we discovered that Mcm10 is posttranscriptionally modified by diubiquitination and that diubiquitinated Mcm10 binds to PCNA but not to pol-α/primase. Interactions with Mcm2-7 subunits, Dbf4, and the catalytic subunit of pol-α do not require ubiquitination of Mcm10 and are likely mediated through surfaces that map to the conserved core of Mcm10 (19, 25, 27, 36). This core region (amino acids 147 to 356) includes a predicted oligonucleotide/oligosaccharide binding (OB) fold domain (spanning amino acids 201 to 297) (http://scop.mrc-lmb.cam.ac.uk/scop/), consistent with the ability to bind single-stranded DNA (19), and a zinc finger domain (309CX10CX11CX2H335), which has been implicated in the formation of higher-order Mcm10 complexes (11). The PIP box of Mcm10 (239QHTLDVYI246) lies within the OB fold directly in the center of the protein. This is unusual among PCNA-interacting proteins as the majority of them (with the exception of Pol2) contain a PIP box at either the N or the C terminus (16, 51, 59). Therefore, it is possible that ubiquitination induces a conformational change, which exposes the normally buried PIP box and allows Mcm10 to interact with PCNA (Fig. 10). This happens seemingly at the expense of rendering other interaction domains nonfunctional (e.g., diubiquitinated Mcm10 does not interact with pol-α). Because the majority of Mcm10 is required to stabilize the catalytic subunit of pol-α (48), it makes sense that only a small fraction of Mcm10 is ubiquitinated in the cell. Indeed, the amount of diubiquitinated Mcm10 appears to be tightly regulated. Even when we overexpressed Mcm10, the normal ratio of diubiquitinated to unmodified protein was not altered, although ubiquitin was overexpressed at the same time. This suggests that the cell somehow measures the total pool of Mcm10 and diubiquitinates a given fraction, regardless of how much diubiquitinated Mcm10 is necessary for the interaction with PCNA. This notion is also consistent with our finding that diubiquitination of Mcm10 occurs independently of its interaction with PCNA, as a mutant form of Mcm10, unable to bind PCNA, was still ubiquitinated. It is also interesting that we did not detect any polyubiquitinated forms of Mcm10, arguing that the cell cycle-dependent turnover of ubiquitinated Mcm10 likely requires deubiquitinating enzymes rather than 26S proteasome activity. In the same vein, we did not find any evidence that unmodified Mcm10 is turned over by proteasomal degradation, as had been reported previously for its human counterpart (28). This is also true for other replication factors, such as Cdt1, which is polyubiquitinated and degraded in humans (interestingly, upon binding to PCNA) (52) but not proteolytically regulated, by nuclear import and export, in budding yeast (56).
FIG. 10.
Diubiquitinated Mcm10 interacts with PCNA and may facilitate its recruitment to the replication fork. Unmodified Mcm10 interacts with pol-α/primase (19, 48) and stimulates the formation of a small RNA primer (rectangle) that is extended by 10 to 20 nucleotides of DNA (black line), both of which are synthesized by pol-α/primase (Pol-α). In a previous report, we estimated that approximately four Mcm10 molecules are located at a single replication fork (48). We hypothesize that ubiquitination (stars) of Mcm10 induces a conformational change, thereby exposing its normally buried PIP box and thus allowing Mcm10 to interact with PCNA (dark circle). This may facilitate the recruitment of PCNA to the primed DNA template onto which it is subsequently loaded in an RFC-dependent manner (42, 58, 63).
A number of reports have described how mono- and diubiquitination modulate protein function. One of the best-studied cases is PCNA, which becomes monoubiquitinated following DNA damage (32). Modification of PCNA has been proposed to induce a polymerase switch from pol-δ/ɛ to pol-η, which is specialized in translesion synthesis (32, 37). Other examples are the mono- and diubiquitination of mammalian Orc1 (39), which help to recycle the protein; the nonproteolytic, ubiquitin-mediated inactivation of geminin, a component of the replication-licensing system in metazoa (38, 39); and the diubiquitination of the death effector domain containing DNA binding protein that occurs in a tissue-specific manner and regulates its interaction with other proteins (35). In the particular case of Mcm10, we postulate that the ubiquitin-driven interaction with PCNA may be evolutionarily conserved. First, human Mcm10 is modified at the G1/S transition, and the molecular-weight forms are consistent with mono- and diubiquitination (28). Similar to what we have shown for budding yeast, in humans both ubiquitinated and unmodified Mcm10 associate with chromatin at the G1/S transition and throughout S phase (28, 29). The only difference is that in humans, the mono- but not the diubiquitinated form of Mcm10 binds DNA (28). Second, the PIP box in Mcm10 appears to be conserved throughout evolution, making it likely that this motif mediates binding to PCNA in other species as well. In humans, the motif more closely resembles the consensus motif for prokaryotic β-clamp-interacting proteins, QLXLF (12, 41). An earlier report on S. pombe Mcm10 was the first to identify the putative PCNA interaction motif (19). Subsequent studies, however, with purified Mcm10 and purified PCNA failed to detect binding between these two proteins (19). We believe that this result is easily explained by the fact that Mcm10 was expressed in Escherichia coli and thus was not posttranscriptionally modified. Clearly, future studies are needed to prove that the interaction between Mcm10 and PCNA is evolutionarily conserved.
A functional PIP box in Mcm10 is essential for cell growth.
While a match to the canonical PIP box (QXXM/I/LXXF/YF/Y) is a good diagnostic marker for potential PCNA binding proteins, many factors that carry even a complete match to the consensus motif fail to interact with PCNA (31, 53). On the other hand, there are several examples of proteins that do not have a PIP box and interact with PCNA through a so-called KA motif (65). Thus, it was important to demonstrate that the interaction between Mcm10 and PCNA was truly mediated through Mcm10's PIP box. Our two-hybrid analysis suggested that tyrosine 245 in Mcm10 interacts directly with alanine 251 in the C terminus of PCNA. While the Y245A substitution in Mcm10 and the A251V substitution in PCNA are not reciprocal, they exchange a long hydrophobic side chain for a shorter one and vice versa, which explains the partial rescue of the interaction. In addition, we have shown that this interaction is crucial for cell growth, since the mcm10-Y245A allele fails to support cell proliferation. This result was entirely unexpected because corresponding PIP box mutations in other essential replication factors, such as DNA ligase I or Pol2, show no interference with cell growth at all (16, 46). Therefore, our data argue that Mcm10 regulates an essential function of PCNA during DNA replication.
Possible function of the Mcm10-PCNA interaction during DNA replication.
Because we have previously shown that Mcm10 is required for the stability of the catalytic subunit of pol-α (48), we considered the possibility that Mcm10 might also be necessary to stabilize PCNA. However, our preliminary studies suggest that this is not the case (S. Das-Bradoo and A.-K. Bielinsky, unpublished results). Thus, we believe that Mcm10 regulates PCNA on a different level. It is known that PCNA targets many different proteins and has a dynamic change in binding partners during lagging strand synthesis (42, 59, 63). Therefore, one possible role of Mcm10 might be to position PCNA in such a way that it is accessible to the correct protein at the right time. A second possible role for Mcm10 might be in loading PCNA onto DNA in vivo. These processes are facilitated by the clamp loader replication factor C (RFC) (58). While no interaction between RFC subunits and Mcm10 has been reported, both function in close proximity on the lagging strand template. RFC targets primed DNA on which it subsequently loads PCNA (58). Pol-α/primase, which is associated with Mcm10 at the replication fork (48), is in fact the only activity in the cell capable of producing primed DNA templates that are recognized by RFC. Since the interaction between diubiquitinated Mcm10 and PCNA is independent of DNA, it is conceivable that Mcm10 might assist in the recruitment of PCNA to the primed lagging strand template (Fig. 10) before RFC loads PCNA onto DNA. In support of this hypothesis, a previous study has shown that the S. pombe homolog of MCM10, cdc23+, interacts genetically with cdc24+, an essential protein required for DNA replication that interacts with RFC and PCNA (55). Because cdc24+ lacks a functional homolog in S. cerevisiae, it will be interesting to explore whether Mcm10 contributes to DNA replication by providing an analogous cdc24+ function in budding yeast.
Acknowledgments
We thank J. Berman, P. Burgers, J. Campbell, R. Harris, M. Hochstrasser, D. Koepp, R. Kolodner, and B. Tye for strains and plasmids. We also thank B. Tye and M. Lei for providing the Mcm10 antibody and C. Wieland for technical assistance. We are grateful to D. Livingston and E. A. Hendrickson for critical reading of the manuscript. We also acknowledge the assistance of the Flow Cytometry Core Facility at the University of Minnesota.
This work was supported by grants from the NIH (GM074917) and ACS (RSG0216601) to A.-K.B.
REFERENCES
- 1.Amin, N. S., and C. Holm. 1996. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics 144:479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 3.Bielinsky, A. K., and S. A. Gerbi. 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3:477-486. [DOI] [PubMed] [Google Scholar]
- 4.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 5.Blow, J. J., and A. Dutta. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6:476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgers, P. M. 1998. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107:218-227. [DOI] [PubMed] [Google Scholar]
- 7.Burich, R., and M. Lei. 2003. Two bipartite NLSs mediate constitutive nuclear localization of Mcm10. Curr. Genet. 44:195-201. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., B. J. Merrill, P. J. Lau, C. Holm, and R. D. Kolodner. 1999. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19:7801-7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, T. W., and B. K. Tye. 2003. Drosophila Mcm10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, C. R., G. Kung, F. C. Peterson, B. F. Volkman, and M. Lei. 2003. A novel zinc finger is required for Mcm10 homocomplex assembly. J. Biol. Chem. 278:36051-36058. [DOI] [PubMed] [Google Scholar]
- 12.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diffley, J. F. X., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 14.Diffley, J. F. X., and K. Labib. 2002. The chromosome replication cycle. J. Cell Sci. 115:869-872. [DOI] [PubMed] [Google Scholar]
- 15.Donovan, S., J. Harwood, L. S. Drury, and J. F. X. Diffley. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 94:5611-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dua, R., D. L. Levy, C. M. Li, P. M. Snow, and J. L. Campbell. 2002. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase ɛ in insect cells. Purification and characterization. J. Biol. Chem. 277:7889-7896. [DOI] [PubMed] [Google Scholar]
- 17.Eissenberg, J. C., R. Ayyagari, X. V. Gomes, and P. M. Burgers. 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison, M. J., and M. Hochstrasser. 1991. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 266:21150-21157. [PubMed] [Google Scholar]
- 19.Fien, K., Y. S. Cho, J. K. Lee, S. Raychaudhuri, I. Tappin, and J. Hurwitz. 2004. Primer utilization by DNA polymerase α-primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 279:16144-16153. [DOI] [PubMed] [Google Scholar]
- 20.Fukui, T., K. Yamauchi, T. Muroya, M. Akiyama, H. Maki, A. Sugino, and S. Waga. 2004. Distinct roles of DNA polymerases δ and ɛ at the replication fork in Xenopus egg extracts. Genes Cells 9:179-191. [DOI] [PubMed] [Google Scholar]
- 21.Garg, P., C. M. Stith, N. Sabouri, E. Johansson, and P. M. Burgers. 2004. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 18:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregan, J., K. Lindner, L. Brimage, R. Franklin, M. Namdar, E. A. Hart, S. J. Aves, and S. E. Kearsey. 2003. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell 14:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 24.Hart, E. A., J. A. Bryant, K. Moore, and S. J. Aves. 2002. Fission yeast Cdc23 interactions with DNA replication initiation proteins. Curr. Genet. 41:342-348. [DOI] [PubMed] [Google Scholar]
- 25.Homesley, L., M. Lei, Y. Kawasaki, S. Sawyer, T. Christensen, and B. K. Tye. 2000. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 14:913-926. [PMC free article] [PubMed] [Google Scholar]
- 26.Horak, J., and D. H. Wolf. 2001. Glucose-induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J. Bacteriol. 183:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi, M., K. Yanagi, T. Mizuno, M. Yokoi, Y. Kawasaki, K. Y. Moon, J. Hurwitz, F. Yatagai, and F. Hanaoka. 2000. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G2 phase. Nucleic Acids Res. 28:4769-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi, M., F. Yatagai, and F. Hanaoka. 2001. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 276:48526-48531. [DOI] [PubMed] [Google Scholar]
- 29.Izumi, M., F. Yatagai, and F. Hanaoka. 2004. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 279:32569-32577. [DOI] [PubMed] [Google Scholar]
- 30.Johansson, E., P. Garg, and P. M. Burgers. 2004. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279:1907-1915. [DOI] [PubMed] [Google Scholar]
- 31.Jonsson, Z. O., R. Hindges, and U. Hubscher. 1998. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 17:2412-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannouche, P. L., J. Wing, and A. R. Lehmann. 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14:491-500. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki, Y., S. Hiraga, and A. Sugino. 2000. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5:975-989. [DOI] [PubMed] [Google Scholar]
- 34.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280-27284. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. C., O. Schickling, A. H. Stegh, R. G. Oshima, D. Dinsdale, G. M. Cohen, and M. E. Peter. 2002. DEDD regulates degradation of intermediate filaments during apoptosis. J. Cell Biol. 158:1051-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, J. K., Y. S. Seo, and J. Hurwitz. 2003. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl. Acad. Sci. USA 100:2334-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann, A. R. 2005. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 579:873-876. [DOI] [PubMed] [Google Scholar]
- 38.Li, A., and J. J. Blow. 2004. Non-proteolytic inactivation of geminin requires CDK-dependent ubiquitination. Nat. Cell Biol. 6:260-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, C. J., and M. L. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang, D. T., and S. L. Forsburg. 2001. Characterization of Schizosaccharomyces pombe mcm7+ and cdc23+ (MCM10) and interactions with replication checkpoints. Genetics 159:471-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez de Saro, F. J., R. E. Georgescu, and M. O'Donnell. 2003. A peptide switch regulates DNA polymerase processivity. Proc. Natl. Acad. Sci. USA 100:14689-14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maga, G., and U. Hübscher. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116:3051-3060. [DOI] [PubMed] [Google Scholar]
- 43.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerases α and ɛ and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merchant, A. M., Y. Kawasaki, Y. Chen, M. Lei, and B. K. Tye. 1997. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimura, S., T. Masuda, T. Matsui, and H. Takisawa. 2000. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5:439-452. [DOI] [PubMed] [Google Scholar]
- 46.Refsland, E. W., and D. M. Livingston. 2005. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts. Genetics 171:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ricke, R., and A. K. Bielinsky. 2005. Easy detection of chromatin binding proteins by the histone association assay. Biol. Proced. Online 7:60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricke, R. M., and A. K. Bielinsky. 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell 16:173-185. [DOI] [PubMed] [Google Scholar]
- 49.Sakurai, S., K. Kitano, H. Yamaguchi, K. Hamada, K. Okada, K. Fukuda, M. Uchida, E. Ohtsuka, H. Morioka, and T. Hakoshima. 2005. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawyer, S. L., I. H. Cheng, W. Chai, and B. K. Tye. 2004. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J. Mol. Biol. 340:195-202. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt, K. H., K. L. Derry, and R. D. Kolodner. 2002. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 277:45331-45337. [DOI] [PubMed] [Google Scholar]
- 52.Senga, T., U. Sivaprasad, W. Zhu, J. H. Park, E. E. Arias, J. C. Walter, and A. Dutta. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281:6246-6252. [DOI] [PubMed] [Google Scholar]
- 53.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 54.Solomon, N. A., M. B. Wright, S. Chang, A. M. Buckley, L. B. Dumas, and R. F. Gaber. 1992. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 8:273-289. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, H., K. Tanaka, H. Murakami, and H. Okayama. 1999. Fission yeast Cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol. 19:1038-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka, S., and J. F. X. Diffley. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4:198-207. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka, T., and K. Nasmyth. 1998. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsurimoto, T. 1998. PCNA, a multifunctional ring on DNA. Biochim. Biophys. Acta 1443:23-39. [DOI] [PubMed] [Google Scholar]
- 59.Vivona, J. B., and Z. Kelman. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167-172. [DOI] [PubMed] [Google Scholar]
- 60.Waga, S., T. Masuda, H. Takisawa, and A. Sugino. 2001. DNA polymerase ɛ is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 98:4978-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 62.Warbrick, E. 1998. PCNA binding through a conserved motif. Bioessays 20:195-199. [DOI] [PubMed] [Google Scholar]
- 63.Warbrick, E. 2000. The puzzle of PCNA's many partners. Bioessays 22:997-1006. [DOI] [PubMed] [Google Scholar]
- 64.Wohlschlegel, J. A., S. K. Dhar, T. A. Prokhorova, A. Dutta, and J. C. Walter. 2002. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell 9:233-240. [DOI] [PubMed] [Google Scholar]
- 65.Xu, H., P. Zhang, L. Liu, and M. Y. Lee. 2001. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40:4512-4520. [DOI] [PubMed] [Google Scholar]
- 66.Zachariae, W., and K. Nasmyth. 1996. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol. Biol. Cell. 7:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]