Abstract
Magnesium (Mg) chelatase is a heterotrimeric enzyme complex that catalyzes a key regulatory and enzymatic reaction in chlorophyll biosynthesis, the insertion of Mg2+ into protoporphyrin IX. Studies of the enzyme complex reconstituted in vitro have shown that all three of its subunits, CHL I, CHL D, and CHL H, are required for enzymatic activity. However, a new T-DNA knockout mutant of the chlorina locus, ch42-3 (Chl I), in Arabidopsis is still able to accumulate some chlorophyll despite the absence of Chl I mRNA and protein. In barley (Hordeum vulgare), CHL I is encoded by a single gene. We have identified an open reading frame that apparently encodes a second Chl I gene, Chl I2. Chl I1 and Chl I2 mRNA accumulate to similar levels in wild type, yet CHL I2 protein is not detectable in wild type or ch42-3, although the protein is translated and stromally processed as shown by in vivo pulse labeling and in vitro chloroplast imports. It is surprising that CHL D accumulates to wild-type levels in ch42-3, which is in contrast to reports that CHL D is unstable in CHL I-deficient backgrounds of barley. Our results show that limited Mg chelatase activity and CHL D accumulation can occur without detectable CHL I, despite its obligate requirement in vitro and its proposed chaperone-like stabilization and activation of CHL D. Thus, the unusual post-translational regulation of the CHL I2 protein provides an opportunity to study the different steps involved in stabilization and activation of the heterotrimeric Mg chelatase in vivo.
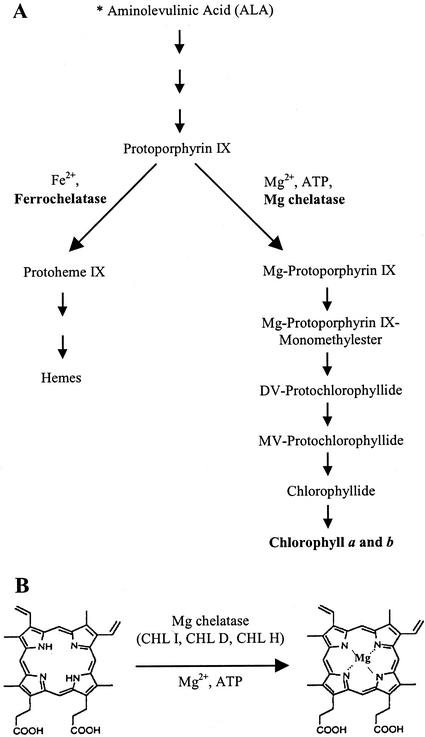
Insertion of a Mg ion into protoporphyrin IX (proto IX) by the Mg chelatase is the first committed step in chlorophyll biosynthesis and occurs at the chlorophyll and heme branch point of tetrapyrrole biosynthesis (Fig. 1A). Evidence is accumulating that this step is subject to tight regulation and that the products and substrates of the reaction may be involved in chloroplast-nuclear genome signaling and in regulation of early steps in the tetrapyrrole biosynthetic pathway (Kropat et al., 1997, 2000; Papenbrock et al., 2000a). As a consequence, the Mg chelatase enzyme complex has been extensively studied in an effort to understand the interactions among its three subunits, their respective roles in Mg-proto IX synthesis, and regulation of proto IX flux through the chlorophyll and heme biosynthetic pathways (Walker and Willows, 1997).
Figure 1.
Chlorophyll biosynthetic pathway in plants. A, Mg chelatase catalyzes the insertion of Mg2+ into proto IX at the branch point between heme and chlorophyll biosynthesis. B, The enzyme is comprised of three subunits: CHL I, CHL D, and CHL H.
The Mg chelatase is a heteromeric enzyme complex composed of three subunits, CHL I, CHL D, and CHL H, which catalyze chelation of Mg by proto IX in an ATP-dependent manner (Fig. 1B). In contrast, the heme branch of tetrapyrrole biosynthesis is initiated by ferrochelatase, which is a homodimeric enzyme that does not require ATP to catalyze the chelation of iron by proto IX (Dailey, 1997; Fig. 1A). In vitro reconstitution of Mg chelatase, first undertaken in Rhodobacter sphaeroides and confirmed in other plant and bacteria species, showed that Mg chelatase activity is obtained only when all three subunits are combined (Gibson et al., 1995; Willows et al., 1996). In vivo evidence has also clearly demonstrated the necessity for three subunits such that the xantha-f, -g, and -h mutants of barley (Hordeum vulgare), which lack CHL H, CHL D, and CHL I, respectively, are unable to synthesize chlorophyll (Jensen et al., 1996; Kannangara et al., 1997). Furthermore, alteration of subunit ratios both in vitro and in vivo can adversely effect Mg chelatase activity (Gibson et al., 1999; Papenbrock et al., 2000b). It was demonstrated recently that either a reduction or excess accumulation of the CHL I subunit in transgenic tobacco (Nicotiana tabacum) plants resulted in a significant loss of total chlorophyll (Papenbrock et al., 2000b).
Proposed mechanisms for metalation of proto IX by the Mg chelatase have postulated a two-step process: 1) an activation step in which CHL I and CHL D interact in an ATP- and Mg2+-dependent manner and 2) an ATP hydrolysis-dependent chelation step involving the interaction of the CHL I/CHL D complex with CHL H, which binds proto IX (Walker and Willows, 1997; Gräfe et al., 1999). Formation of a 200-kD “activation complex” composed of CHL I (BCH I) and CHL D (BCH D) was shown to be dependent on the presence of ATP and Mg2+ in R. sphaeroides and Synechocystis sp. PCC6803 (Gibson et al., 1999; Jensen et al., 1999). An additional role for the CHL I/CHL D complex may be to facilitate folding and stability of the CHL D subunit. In barley mutants that completely lack the CHL I subunit, CHL D is also absent (Hansson et al., 1999; Petersen et al., 1999b). It is interesting that CHL I was shown by sequence homology searches to be a member of the AAA-ATPase family of proteins, which have functions including chaperone activity and protein remodeling (Neuwald et al., 1999). Determination of the three-dimensional structure of BCH I from Rhodobacter capsulatus also revealed a structural similarity to several members of the AAA-ATPase family (Fodje et al., 2001). Furthermore, CHL I from Synechocystis sp. PCC6803 exhibits ATPase activity in vitro (Jensen et al., 1999; Petersen et al., 1999a). Therefore, it seems plausible that CHL I may have a dual function: to assist in folding or stabilization of CHL D and to hydrolyze ATP, which may drive the chelation of Mg2+ by proto IX via protein remodeling. Much progress has been made toward an understanding of the mechanism of the Mg-chelatase enzyme complex. However, the specific role of individual subunits and the stoichiometry required for optimal activity in vivo remain unclear.
We have identified a lethal T-DNA-tagged mutant (ch42-3) in Arabidopsis in which the T-DNA insertion occurs 6 bp downstream from the start Met of the Chl I1 gene. Although Chl I mRNA and protein are undetectable by northern- and western-blot analyses, ch42-3 is able to accumulate 17% (w/w) wild-type chlorophyll. Southern-blot analyses indicate that Chl I is a single copy gene in tobacco as well as barley (Kruse et al., 1997; Petersen et al., 1999b). Although tobacco, which is an amphidiploid, contains two homeologous copies of Chl I (Kjemtrup et al., 1998). Searches of sequence databases revealed a second Chl I gene (Chl I2) in Arabidopsis that shows 82% similarity to the Chl I1 gene. Expression of Chl I2 in ch42-3, however, is insufficient to support viable levels of chlorophyll biosynthesis. Because of novel traits of the ch42-3 mutant and the CHL I2 protein in Arabidopsis, we have a unique system for examining the role of the CHL I subunit in Mg-proto IX biosynthesis.
RESULTS
Genetic Characterization of Chlorina Mutant ch42-3
Screening of the Feldman T-DNA populations (Forsthoefel et al., 1992) for lethal chlorotic lines resulted in identification of the T-DNA-tagged mutant, ch42-3. Complementation tests by reciprocal crosses (data not shown) with the chlorina alleles ch1, ch3, ch5, and ch42-1 revealed that ch42-3 is a recessive mutation and is allelic to ch42. Segregation of kanamycin resistance in CH42-3/ch42-3 reciprocal crosses (data not shown) indicated that there was only one T-DNA insertion, and it was linked to ch42-3. A genomic library of ch42-3 was probed with the left and right borders of the T-DNA and five genomic clones containing the T-DNA and flanking genomic DNA were isolated. Linkage of Chl I1 to the T-DNA insertion was confirmed by Southern-blot analysis of genomic DNA probed with Chl I1 and T-DNA left border fragments (data not shown).
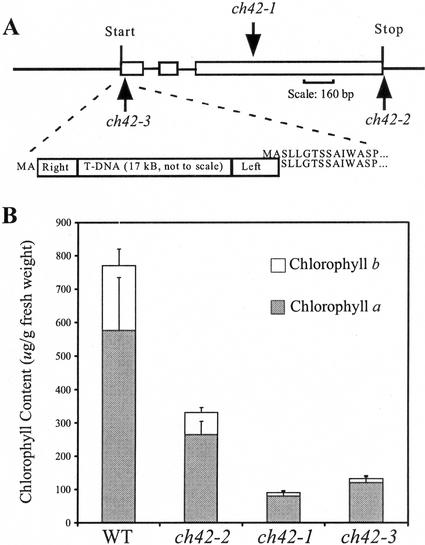
Comparison of sequence flanking the right border of the T-DNA to the Chl I1 sequence showed that the T-DNA insertion occurred 6 bp downstream of the start Met (Fig. 2A). There are two other reported ch42 alleles, the inviable, chlorotic ch42-1, an x-ray-induced mutation that was defined as a lesion in the Chl I1 gene (Fischerova, 1975), and the viable pale green ch42-2, a T-DNA-tagged mutant where the insertion was near the 3′ end of the gene resulting in a C-terminal fusion of 11 amino acids (Koncz et al., 1990).
Figure 2.
Chlorophyll accumulation is reduced in the three ch42 alleles. A, Map of chl I1 gene showing site of T-DNA insertions ch42-3 and ch42-2 (Koncz et al., 1990). ch42-1 has an x-ray-induced eight-nucleotide deletion in exon 3 (Fischerova, 1975). B, ch42-3 chlorophyll a/b (chl a/b) content of leaves is 17% (w/w) wild-type chlorophyll (μg g−1 fresh weight). Identification of chl a was verified by spectrophotometry and mass spectrometry.
ch42-3 Is Capable of Accumulating Chlorophyll Despite the Absence of Chl I1 mRNA
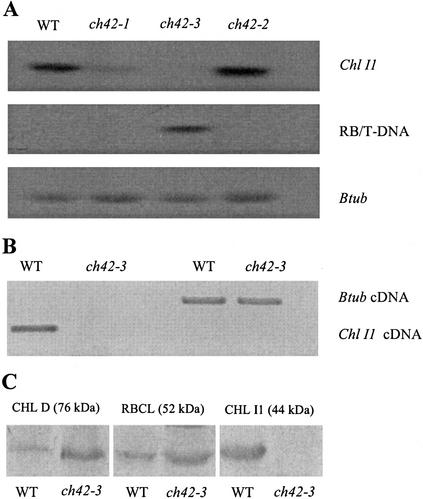
Insertion of the T-DNA 6 bp downstream of the start Met in ch42-3 should disrupt transcription of Chl I1 and prevent synthesis of a functional CHL I protein. Northern-blot analysis of total RNA from wild type, ch42-1 (Fischerova, 1975), ch42-2 (Koncz et al., 1990), and ch42-3 probed with a Chl I1 fragment show that the Chl I1 mRNA was undetectable in ch42-3 and reduced in ch42-1 and ch42-2 (Fig. 3A). The absence of Chl I1 mRNA in ch42-3 was confirmed by RT-PCR using gene-specific primers (Fig. 3B).
Figure 3.
RNA and protein blot of wild type and ch42. A, Total RNA was isolated from 14-d-old leaves grown at 80 μmol photons m−2 s−1 in tissue culture of wild type and three mutant alleles of the ch42 locus. Twenty micrograms of RNA was probed with a 1.2-kb genomic fragment of Chl I1, right border of the T-DNA (RB/T-DNA), and β-tubulin. Right-border transcription in ch42-3 is presumed to be because of its proximity to Chl I1 promoter. B, Absence of Chl I1 mRNA in ch42-3 was verified by reverse transcription (RT)-PCR using RNA extracted from wild-type and ch42-3 leaves from plants grown under same conditions used for northern-blot analysis. β-Tubulin primers were used as a control. Chl I1 gene-specific primers resulted in cDNA from wild-type leaves only. C, Proteins from 14-d-old leaves were extracted in SDS sample buffer, separated by SDS-PAGE, and visualized by immunodetection with anti-CHL I (from Arabidopsis), Rubisco, large subunit (rbcl), or CHL D (from pea [Pisum sativum]) polyclonal serum.
In barley, null xantha-h (Chl I) mutants are incapable of synthesizing chlorophyll; yet in Arabidopsis the null ch42-3 allele accumulated 17% of wild-type chlorophyll levels. The other two alleles, ch42-1 and ch42-2, accumulated 11% and 43%, respectively (Fig. 2B). It is interesting that less chlorophyll is synthesized in the ch42-1 allele that has detectable mRNA than the null ch42-3. Forty percent of wild-type chlorophyll levels in ch42-2 is sufficient for viability and reflects the insertion of the T-DNA near the 3′ end of the Chl I1 gene (Fig. 2A; Koncz et al., 1990).
chl a/b ratios were elevated relative to the wild-type chl a/b ratio of 3.0, ranging from 4.0 to 10.4 in the ch42 mutants examined, which is typical of chlorophyll biosynthetic mutants and is usually accompanied by a reduction in light-harvesting apoproteins. Lhcb 1 (the major light harvesting complex of photosystem II) levels were not altered in ch42-3, whereas Lhcb 2 and Lhcb 3 apoproteins were absent (data not shown).
Because ch42-3 was the only null allele, it was used for further studies. Western-blot analyses of total proteins verified that CHL I protein does not accumulate in ch42-3 (Fig. 3C), which would be expected given the location of the T-DNA insertion and the absence of detectable message (Fig. 3, A and B). It is interesting that the CHL D subunit accumulated to levels that were slightly higher than that observed in wild type (Fig. 3C). The presence of stable CHL D protein and the ability of ch42-3 to synthesize some chlorophyll in the absence of CHL I protein was in contrast to what was previously observed in null Chl I mutants in barley. These results prompted us to examine the possibility of a CHL I multigene family in Arabidopsis.
Chl I2 Is Encoded by a Multigene Family in Arabidopsis
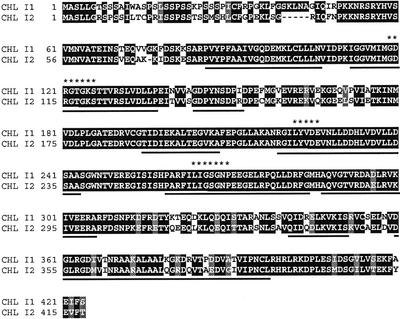
Searches of expressed sequence tag (EST) and genome sequences resulted in the identification of a second expressed Chl I gene, Chl I2, located between 91 and 101 cM on chromosome V (Transformation-competent artificial chromosome [TAC] clone K15122; GenBank accession no. AB016870 and the partial-length cDNA from EST clone E4F4T7). The amino acid sequence was obtained from the annotated K15122 clone information at the Arabidopsis thaliana Database (Arabidopsis Genome Initiative) and comparisons to the Chl I1 amino acid sequence showed an 82% identity between CHL I1 and CHL I2 (Fig. 4). The sequence of spliced Chl I2 was verified by amplification of the full-length cDNA using RT-PCR, and no premature stop codons or mis-splicings of introns were detected. The predicted cleavage site for the transit peptides of CHL I1 and CHL I2 is between Ser-60 and Val-61 and Ser-55 and Val-56, respectively (Emanuelsson et al., 1999). One notable difference between the leader sequences, however, is a small deletion in the CHL I2 transit peptide that is just upstream of the predicted cleavage site (Fig. 4).
Figure 4.
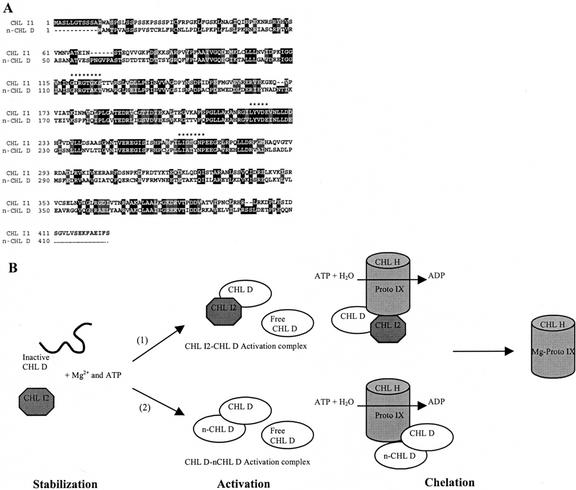
Alignment of ChlI1 and ChlI2 derived amino acid sequences. Chl I1 (GenBank accession no. X91411; chromosome IV; position, 39.4 cM) and Chl I2 (GenBank accession no. AB016870; bacteria artificial chromosome (BAC) sequence; chromosome V; position, 91–101 cM) are 82% identical at the amino acid level. The CHL I2 sequence contains all the conserved domains observed in an alignment of all CHL I sequences and contains the three conserved Mg-ATPase motifs (*******). Motifs present in members of the AAA-ATPase family are underlined.
Expression of Chl I1 and Chl I2 in Arabidopsis
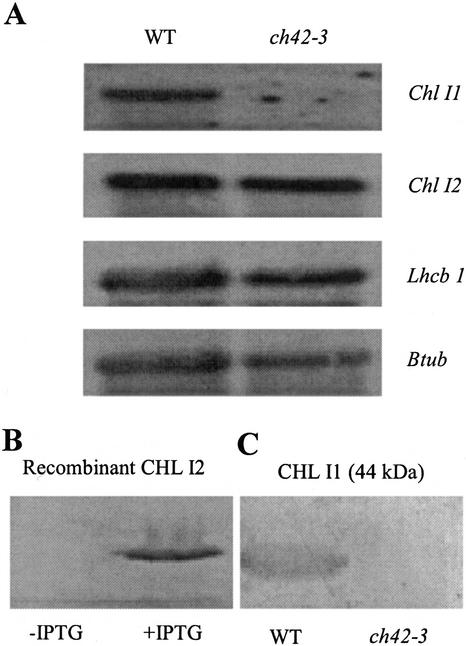
Because of the high identity between Chl I1 and Chl I2, gene-specific probes were designed by sequence analysis and tested by demonstrating an absence of cross-hybridization to dot blots of the respective cDNA clones. Northern-blot analysis of total RNA from wild type showed that Chl I1 and Chl I2 mRNA accumulated to similar levels in wild type (Fig. 5A). In addition, Lhcb 1 mRNA accumulates to wild-type levels in ch42-3 (Fig. 5A). Abundance of mRNA for Chl I2 was similar in ch42-3 and wild-type leaves (Fig. 5A), suggesting that the suboptimal levels of chlorophyll biosynthesis in ch42-3 was not because of a lack of Chl I2 mRNA.
Figure 5.
RNA and protein blots of CHL I1 and CHL I2. A, Total RNA was isolated from 14-d-old leaves grown at 80 μmol photons m−2 s−1 in tissue culture of wild type and ch42-3. Twenty micrograms of RNA was probed with LhcbII, Chl I1, Chl I2, Lhcb 1, and β-tubulin. B, Polyclonal CHL I antisera (from Synechocystis sp. PCC6803) reacts to recombinant CHL I2 overexpressed in Escherichia coli. C, Chloroplasts were isolated from 14-d-old leaves of wild type and ch42-3. Proteins were extracted in SDS sample buffer, separated by SDS-PAGE, and visualized by immunodetection with anti-CHL I serum (from Synechocystis sp. PCC6803).
CHL I2 Accumulation Is Regulated Post-Translationally
Despite the abundance of Chl I2 mRNA, the CHL I2 protein is not detected in ch42-3 by two different polyclonal CHL I antisera (from Arabidopsis and Synechocystis sp. PCC6803) or by an anti-CHLD serum that reacts to CHL I. The CHL I antisera from Synechocystis sp. PCC6803 reacts with recombinant CHL I2 expressed in E. coli indicating that the polyclonal antisera can detect the CHL I2 isoform (Fig. 5B). Proteins from isolated chloroplasts were also analyzed for presence of the CHL I2 protein in ch42-3. Although CHL I was very abundant in chloroplast protein samples from wild type, no CHL I2 was detected in ch42-3 (Fig. 5C). Titration and serial dilutions demonstrated that there is at least a 1,000-fold reduction in CHL I2 in ch42-3 compared with CHL I1 levels in wild type (data not shown).
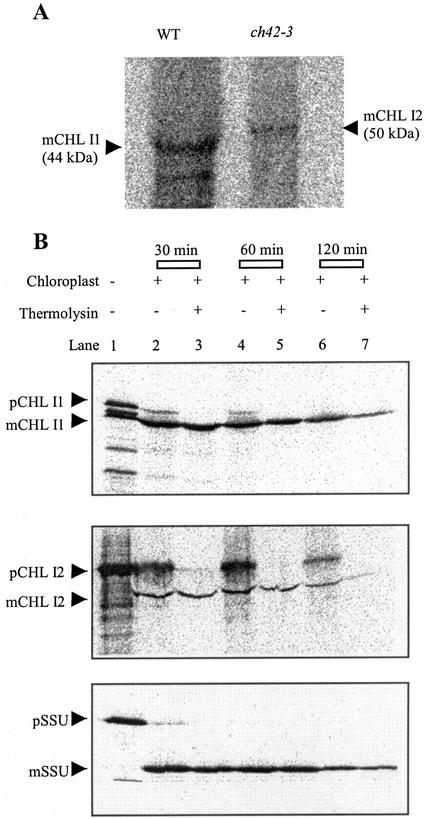
Pulse labeling of ch42-3 leaves with a [35S]Met/Cys mixture and subsequent immunoprecipitation of labeled CHL I1 and CHL I2 protein using the polyclonal CHL I antisera was performed to determine whether or not the abundant Chl I2 mRNA was translated into protein. CHL I1 protein was detectable by pulse labeling in wild type, with an apparent molecular mass of approximately 44 kD, which is slightly larger than the predicted mass of the stromally processed CHL I1 (39.9 kD; Fig. 6A). In ch42-3, which only accumulates Chl I2 mRNA, CHL I2 protein was detected with an apparent molecular mass of approximately 50 kD. This is more consistent with the predicted molecular mass of precursor CHL I2 (46 kD), than the mature form of CHL I2 (39 kD). In addition, the immunoprecipitation of CHL I2 in ch42-3 further confirms that the polyclonal antisera can detect the CHL I2 isoform.
Figure 6.
CHL I2 is imported into chloroplasts and processed to the mature form. A, CHL I2 is detected by pulse labeling of ch42-3 leaves. Leaves from 14-d-old wild type and ch42-3 were labeled with 125 μCi of a [35S]Met/Cys mixture. After immunoprecipitation with polyclonal CHL I antiserum (from Arabidopsis), proteins were separated by SDS-PAGE and visualized by autoradiography. B, In vitro chloroplast import assays. Precursor proteins were expressed in a coupled transcription-translation system (Promega, Madison, WI) and import assays were carried out in isolated pea chloroplasts. The small subunit of Rubisco (SSU) was used as a control. Proteins were monitored for 120 min after import, separated by SDS-PAGE, and analyzed by autoradiography. The size difference between the precursor (p) and mature (m) forms for CHL I1, CHL I2, and SSU was approximately 6 kD. mCHL I2 was 6 kD larger than mCHL I1 (a similar size difference was observed between pCHL I1 and pCHL I2).
Despite the apparent abundance of Chl I2 mRNA and its translation in vivo, CHL I2 protein does not accumulate in ch42-3. Therefore, the apparent instability is presumably the result of post-translational processes. Initial results indicated that the CHL I2 protein may not be imported and processed because the size detected by pulse labeling was approximately 6 kD larger than CHL I1. In addition, there is a deletion in the CHL I2 transit peptide in a region conserved in all plant CHL I proteins. Deletion of a similar region in the ferredoxin transit peptide of Silene pratensis impaired processing of preferredoxin to ferredoxin in transgenic Arabidopsis plants (Rensink et al., 2000). Results from repeated in vitro import into pea chloroplasts, however, showed that both pre-CHL I1 and pre-CHL I2 are efficiently imported and processed. The higher apparent mass of mature CHL I2 was also observed in these imports, suggesting that the apparent size difference reflects different relative mobility of the CHL I2 isoform during SDS-PAGE rather than mis-processing. Furthermore, both CHL I1 and CHL I2 were stable after processing of the leader sequence. The mature form of CHL I2 was as stable as the small subunit of Rubisco after import and processing in isolated pea chloroplasts (Fig. 6B). Therefore, we conclude that CHL I2 is imported and stromally processed in ch42-3, yet CHL I2 fails to accumulate in vivo.
DISCUSSION
Differential Accumulation of Two CHL I Isoforms in Arabidopsis
We have identified and characterized a null mutant of the ch42 allele in Arabidopsis, ch42-3, that is capable of synthesizing chlorophyll despite the absence of Chl I1 mRNA and protein. In fact, all three ch42 (Chl I1) lines are still able to synthesize some chlorophyll. The capacity to synthesize chlorophyll in a knockout allele is most surprising because in vitro studies of bacterial and plant subunits and previous analyses of barley mutants demonstrated that the absence of the CHL I subunit (xantha-h) completely inactivates the Mg chelatase (Kannangara et al., 1997; Papenbrock et al., 1997; Jensen et al., 1998; Willows and Beale, 1998). To explain the ability of ch42-3 to accumulate 17% of wild-type chlorophyll levels, a mechanism in which the CHL D and CHL H subunits alone exhibit some activity may be invoked. However, the simpler possibility of a Chl I multigene family in Arabidopsis was a more plausible explanation. Even then, this is unusual because neither tobacco nor barley shows any evidence of a second gene, and only one CHL I isoform is reportedly present in photosynthetic bacteria and in most higher plants studied thus far. However, in addition to a second gene in Arabidopsis, there are two genes in soybean based on searches of EST databases. Whereas soybeans are a considerable phylogenetic distance from Arabidopsis, the two are phylogenetically closer to each other than to tobacco (Bremer et al., 1998).
The second isoform, CHL I2, is over 80% identical to CHL I1 (Fig. 4), and amino acid residues that are strictly conserved among CHL I sequences from 11 different species, including higher plants and bacteria, are also conserved in the CHL I2 sequence. Furthermore, both CHL I1 and CHL I2 show homology to the AAA family of ATPases, which is comprised of a variety of proteins including Bch I, the analogous ATPase subunit of the cobalt chelatase, and Rubisco activase (Neuwald et al., 1999). Yet, in spite of the high degree of identity between the two isoforms, expression of the Chl I2 gene is insufficient to maintain viable rates of chlorophyll biosynthesis and results in inviable chlorotic plants in the absence of Chl I1. Because the mRNA accumulation of both genes is similar in both wild-type and ch42-3 mutant leaves, transcriptional regulation of Chl I2 does not account for the severe phenotype of ch42-3.
An additional factor that may contribute to the lack of CHL I2 protein could be an intrinsic instability of the protein. Yet, in vitro chloroplast import assays showed that both CHL I1 and CHL I2 were relatively stable 2 h after import and processing compared with the Rubisco control (Fig. 6B). This is in complete contrast to what is observed in vivo, where CHL I2 is only detectable by pulse labeling (Fig. 6A), suggesting that rapid turnover of CHL I2 prevents sufficient accumulation of the protein (Fig. 5C). The nature of this instability remains unclear but is nonetheless intriguing. Why is CHL I2 unstable when the mRNA accumulates normally, translated into protein efficiently, imported into the chloroplasts, and processed to a mature form that is stable in vitro, yet targeted for rapid degradation in vivo? Specific proteolytic degradation of CHL I2 could also account for the instability of CHL I2 in vivo. A similar phenomenon is observed at another key step in the chlorophyll biosynthetic pathway, namely protochlorophyllide oxidoreductase (POR). In most plant species there are two isoforms: PORA and PORB. Both isoforms are present in etiolated plants, but PORA is rapidly degraded during the greening process (Reinbothe et al., 1995; Runge et al., 1996). However, a survey of tissues from different developmental stages and during greening did not reveal any stable CHL I2 protein (data not shown).
Implications for the Mechanism of Mg Chelatase Activity in Arabidopsis
The fact that a transient presence of CHL I2 is sufficient to support chlorophyll biosynthesis in ch42-3 raises questions regarding the mechanism for Mg chelatase activity. Several studies have suggested that CHL I may exhibit chaperone-like activities or prevent proteolytic degradation of CHL D. For example, null xantha-h56,57 mutants in barley, which lack CHL I mRNA and protein, are also deficient in the xantha-g (CHL D) protein (Hansson et al., 1999; Petersen et al., 1999b). It is surprising that in ch42-3, the CHL D subunit accumulates to wild-type levels, even though CHL I is undetectable. The CHL I2 protein has all of the conserved amino acids of a CHL I subunit and could presumably bind ATP and facilitate folding of CHL D (Figs. 4 and 7B). However, reduced levels of chlorophyll in ch42-3 indicate that the accumulation of stabilized CHL D is insufficient to support maximal Mg chelatase activity. These results support a model for Mg chelatase activity in which CHL I has two functions: to fold and stabilize CHL D and to assist CHL H in catalyzing chelation of Mg2+ by proto IX. How then, does Mg chelatase activity occur in ch42-3 given that CHL I levels are reduced by at least 1,000-fold?
Figure 7.
Model for mechanism of Mg chelatase activity in ch42-3. A, Alignment of CHL I1 and the N terminus of CHL D from Arabidopsis illustrating conserved Mg-ATP binding motifs (******). B, Two proposed models for Mg-proto IX synthesis in ch42-3. CHL I2 interacts with inactive CHL D in an ATP- and Mg2+-dependent manner to facilitate folding and stabilization of CHL D. (a) In the first model, formation of CHL I2/CHL D activation complexes occurs, although most of the CHL D present in ch42-3 would be free because of the instability of CHL I2. The sub-stoichiometric levels of CHL I2/CHL D complexes could then interact with CHL H-proto IX to catalyze chelation. (b) As an alternative, after CHL D is stabilized, the N terminus of free CHL D subunits could substitute for the CHL I subunit (n-CHL D) and form a CHL D/n-CHL D activation complex that could then interact with CHL H to catalyze Mg-proto IX synthesis.
One possible explanation is that very dilute concentrations of CHL I/CHL D activation complexes are sufficient for interaction with CHL H-proto IX and subsequent chelation of Mg2+ (Fig. 7B). In vitro reconstitution studies of the R. sphaeroides Mg chelatase demonstrated that a 9-fold reduction in BCH I levels resulted in a 60% reduction in Mg chelatase activity (Gibson et al., 1999). Similar results using recombinant Mg chelatase from Synechocystis sp. PCC6803 suggest that lowering the concentration of BCH I by approximately 30-fold results in a 70% reduction in Mg chelatase activity (Jensen et al., 1998). Based on reconstitution studies, it is actually quite surprising that ch42-3 accumulates 17% of wild-type chlorophyll levels given the greater than 1,000-fold reduction in levels of the CHL I subunit of the Mg chelatase. However, it cannot be discounted that sub-stoichiometric levels of CHL I2 support the formation of activation complexes in vivo.
An alternative hypothesis to account for Mg-proto IX synthesis in the absence of a stable CHL I subunit is based on observations that the N-terminal region of CHL D has a high degree of homology to CHL I (Fig. 7A). The hypothesis that CHL D and CHL H alone can insert Mg into proto IX has been discounted by their lack of activity in vitro in the absence of CHL I (Willows et al., 1996; Papenbrock et al., 1997). However, if CHL D is unable to form an active conformation in the absence of CHL I, then in vitro assays have not actually addressed the question of whether an “activated” CHL D subunit can, in conjunction with CHL H-proto IX and ATP, catalyze the chelation of Mg2+. In fact, a CHL I-CHL D fusion protein from tobacco, where the N terminus of CHL D was replaced by the CHL I sequence, actually exhibited some activity when assayed in vitro with the CHL H subunit (Gräfe et al., 1999). The activity of the CHL I-CHL D fusion protein suggests that some Mg chelatase activity can occur in the absence of “free” CHL I and that CHL I still exhibits some activity when fused to the C terminus of CHL D. We hypothesize that synthesis of Mg-protoIX in ch42-3 may result when transient CHL I2 undertakes a chaperone function and activates CHL D, which could not occur in null xantha-g (CHL I) mutants of barley. The N terminus of some of the activated CHL D subunits may then substitute for CHL I, albeit inefficiently, to allow Mg chelatase activity (Fig. 7B).
Based on the in vivo results herein and in conjunction with published studies, we present a model for the Mg chelatase that depicts three steps leading to the synthesis of Mg-protoIX (Fig. 7B). Stabilization of the CHL D subunit occurs via the chaperone-like activity of CHL I. Accumulation of CHL D in ch42-3 suggests that transient, undetectable levels of CHL I2 are sufficient to support CHL D stabilization. Stabilization of CHL D, however, is not sufficient for Mg chelatase activity and, thus, there is a second activation step before chelation that is reported to involve the formation of a 200-kD activation complex (Gibson et al., 1999; Jensen et al., 1999). Given that CHL I2 is undetectable in ch42-3, then either the transient presence of a CHL I2/CHL D activation complex is sufficient for some Mg chelatase activity or the N-terminal domain of CHL D will support the formation of partially functional activation complexes, providing CHL D has been stabilized first by CHL I2. The activation complexes then interact with CHL H-protoIX to catalyze the chelation of Mg2+. Whether it is the non-detectable levels of CHL I2 that form trace levels of CHL I/CHL D activation complexes or whether activated CHL D and CHL H alone can facilitate residual chlorophyll biosynthesis in ch42-3 is subject to further investigation.
MATERIALS AND METHODS
Growth of Plant Material, Identification of Mutants, and Pigment Analysis
The Feldman T-DNA populations (Forsthoefel et al., 1992) were screened for chlorotic lines to identify null lesions in pigment biosynthetic pathways as described (Pogson et al., 1996). Complementation tests by reciprocal crosses with chlorina alleles from the Arabidopsis Biological Resource Center were undertaken. The segregation of the T-DNA, as determined by kanamycin resistance with the loss of chlorophyll, was established by growing seedlings on kanamycin media. The genomic DNA flanking the T-DNA was identified by the probing of a genomic library prepared from ch42-3 DNA cloned into lambda GEM-12 (Promega) with the right and left borders of the T-DNA. Five genomic clones ranging from 11 to 17 kb that hybridized predominantly to the left border were characterized in detail by restriction mapping (Koncz et al., 1990) and sequencing across the insertion junction (Sanger and Coulsen, 1977). The five clones all contained the left border of the T-DNA adjacent to Chl I1 genomic DNA. A 1.2-kb fragment containing almost exclusively Chl I1 genomic DNA was isolated and used for subsequent Southern- and northern-blot analyses.
The BAC genomic and EST sequence databases were periodically screened during the course of the project with the ChlI1 gene sequence. BACs and ESTs encoding the Chl I1 gene show that it maps to chromosome IV at 39.4 cM. A second open reading frame with very high homology to the Chl I1 gene was identified on TAC clone K15122 (GenBank accession no. AB016870), which is located on chromosome V between 91 and 101 cM. An EST of this Chl I2 gene was identified (GenBank accession no. AO42226).
Arabidopsis cv Columbia and Wassilewskija plants were grown on Suc-supplemented media at 80 μmol photons m−2 s−1 for 24 h per day, or in soil at 150 μmol photons m−2 s−1 for 16 h per day (Norris et al., 1995; Pogson et al., 1998). The carotenoids and chlorophylls were extracted, fractionated, and quantified by HPLC and spectrophotometry (Pogson et al., 1996, 1998).
Northern Blots and RT-PCR
Wild type, and the three ch42 alleles were grown on 2% (w/v) Suc supplemented media for 3 weeks. Leaves were harvested and RNA was extracted using either the TRIzol reagent (Gibco-BRL, Gaithersburg, MD) or by the RNeasy kit (Qiagen, Santa Clarita, CA) by manufacturers' protocols. Twenty micrograms of total RNA was fractionated and blotted as described (Pogson et al., 1995). The northern blots were probed by either the Chl I1 genomic fragment and EST or Chl I1 and Chl I2 gene-specific fragments. The gene-specific fragments were PCR-amplified fragments across a 300-bp region that showed the least homology between the two genes. The absence of cross-hybridization to dot blots of the Chl I1 genomic fragment and the Chl I2 EST was tested.
RT-PCR was performed using gene-specific primers for Chl I1 (forward primer, 5′-GCCAATGAGAAGCTGAG-3′; reverse primer, 5′-AGCTGCAAATGGATAAACCG-3′) and Chl I2 (forward primer, 5′-CGAAGAGAAAGACACTGAAATG-3′; reverse primer, 5′-AGC AGCAAACGGATAAACAG-3′) with 500 ng of total RNA. RT and PCR were performed in 50 μL using the Access RT-PCR kit as described by the manufacturer (Promega; 1 μm forward primer, 1 μm reverse primer, 1.5 mm MgS04, 0.1 units μL−1 Tfl DNA polymerase, 0.1 units μL−1 avian myeloblastosis virus reverse transcriptase, and 0.2 mm dNTP). RT was performed for 75 min at 48°C followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C. Products were analyzed on agarose gels and visualized with ethidium bromide staining.
Overexpression of Chl I2 in Escherichia coli
A partial-length Chl I2 cDNA was cloned using RT-PCR as described above (forward primer, 5′-CGAAGAGAAAGACACTGAAATCG-3′; reverse primer, 5′-GAAAACCTC CATAGAACTTC TCGGT-3′). The resulting cDNA was cloned into the pet30c+ vector (Novagen, Madison, WI) to form the plasmid pBP266. The BL21 strain of E. coli was transformed with pBP266 and cells were grown in 4 mL of Luria-Bertani medium for 4 h. Isopropylthio-β-galactoside (IPTG) was added to a final concentration of 1 mm, and cells were grown for an additional 5 h. After centrifugation, the cell pellets from both the induced (+IPTG) and noninduced (−IPTG) cultures were resuspended in 2 mL of SDS-PAGE sample buffer (125 mm Tris-Cl, pH 6.8, 4% [w/v] SDS, 2% [w/v] 2-mercaptoethanol, 1 mm amino caproic acid, 5 mm benzamidine, 0.001% [w/v] bromphenol blue, and 20% [w/v] glycerol) and boiled for 10 min before SDS-PAGE and western-blot analysis.
Western Blots, In Vivo Pulse Labeling, and Immunoprecipitations
Total proteins for western-blot analyses were extracted in SDS-PAGE sample as described above. Chloroplasts were isolated from 3 g of leaf tissue by grinding in buffer (2 mm EDTA, pH 8.0, 1 mm MgCl2, 1 mm MnCl2, 50 mm 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES]-KOH, pH 7.5, and 330 mm sorbitol) and separation on a 40%/80% (v/v) Percoll gradient. Proteins from isolated chloroplasts were extracted in SDS-PAGE sample buffer. Chloroplast proteins, total plant proteins, and recombinant CHL I2 were separated by SDS-PAGE and transferred to Immobilon-P membrane (Millipore, Bedford, MA) as previously described (Sambrook et al., 1989). Immunostaining was performed with either antiserum to CHL I at a 1:1,000 dilution, antiserum to CHL D at a 1:1,000 dilution, or antiserum to Rubisco (large subunit) at a 1:2,000 dilution followed by secondary staining with alkaline phosphatase-conjugated goat-anti-rabbit serum (Bio-Rad, Hercules, CA). Colorimetric development with 5-bromo-4-chloro-3-indoyl phosphate and nitroblue tetrazolium was utilized to visualize protein bands (Sambrook et al., 1989).
Attached leaves from wild-type and ch42-3 plants were labeled for 5 h with 125 μCi of a [35S]Met/Cys mixture as previously described (Barkan, 1998). Leaves were homogenized, and labeled CHL I1 and CHL I2 proteins were immunoprecipitated with polyclonal CHL I antiserum as previously described (Barkan, 1998) and isolated using protein-A CL-4B Sepharose (Sigma-Aldrich, St. Louis) as described previously (Sambrook et al., 1989). Proteins were separated by SDS-PAGE as described above and visualized by autoradiography.
Chloroplast Import Assays
Precursor proteins were expressed in a coupled transcription-translation system (Promega), and import assays were carried out in isolated pea chloroplasts as previously described (Bruce et al., 1994; Waegemann and Soll, 1995).
ACKNOWLEDGMENTS
We thank Drs. R. Willows (Macquarie University, Sydney, Australia), and J. Anderson (Australian National University), for many helpful discussions.
Footnotes
This work was supported by a National Science Foundation graduate research training grant at Arizona State University (to H.M.R.) and the Endowment for Excellence Award at the Australian National University (to H.M.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010625.
LITERATURE CITED
- Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Method Enzymol. 1998;297:38–57. [Google Scholar]
- Bremer K, Chase MW, Stevens PF, Anderberg AA, Backlund A, Bremer B, Briggs BG, Endress PK, Fay MF, Goldblatt P et al. An ordinal classification for the families of flowering plants. Ann Missouri Bot Gard. 1998;85:531–553. [Google Scholar]
- Bruce BD, Perry S, Froelich J, Keegstra K. Plant Mol Biol Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. In vitro import of proteins into chloroplasts; pp. 1–15. [Google Scholar]
- Dailey HA. Enzymes of heme biosynthesis. J Biol Inorg Chem. 1997;2:411–417. [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischerova H. Linkage relationships of recessive chlorophyll mutations in Arabidopsis thaliana. Biol Plant. 1975;17:182–188. [Google Scholar]
- Fodje MN, Hansson A, Hansson M, Olsen JG, Gough S, Willows RD, Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J Mol Biol. 2001;311:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Yewen W, Schulz B, Bennett MJ, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust J Plant Physiol. 1992;19:353–366. [Google Scholar]
- Gibson LCD, Jensen PE, Hunter CN. Magnesium chelatase from Rhodobacter sphaeroides: initial characterization of the enzyme using purified subunits and evidence for a BCH I-BCH D complex. Biochem J. 1999;337:243–251. [PMC free article] [PubMed] [Google Scholar]
- Gibson LCD, Willows RD, Kannangara CG, von Wettstein D, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the BCH H, BCH I, and BCH D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräfe S, Saluz HP, Grimm B, Hänel F. Mg-chelatase of tobacco: the role of the subunit CHL D in the chelation step of protoporphyrin IX. Proc Natl Acad Sci USA. 1999;96:1941–1946. doi: 10.1073/pnas.96.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Kannangara CG, von Wettstein D, Hansson M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc Natl Acad Sci USA. 1999;96:1744–1749. doi: 10.1073/pnas.96.4.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Hunter CN. Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from SynechocystisPCC6803. Biochem J. 1998;334:335–344. doi: 10.1042/bj3340335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Hunter CN. ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of SynechocystisPCC6803: evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem J. 1999;339:127–134. [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Willows RD, Petersen BL, Vothknecht UC, Stummann BM, Kannangara CG, von Wettstein D, Henningsen KW. Structural genes for Mg-chelatase subunits in barley: xantha-f, -g and -h. Mol Gen Genet. 1996;250:383–394. doi: 10.1007/BF02174026. [DOI] [PubMed] [Google Scholar]
- Kannangara CG, Vothknecht UC, Hansson M, von Wettstein D. Magnesium chelatase: association with ribosomes and mutant complementation studies identify barley subunit xantha-G as a functional counterpart of Rhodobactersubunit BCH D. Mol Gen Genet. 1997;254:85–92. doi: 10.1007/s004380050394. [DOI] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, Thompson WF, Robertson D. Gene silencing from plant DNA carried by a gemnivirus. Plant J. 1998;14:91–100. doi: 10.1046/j.1365-313X.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990;9:1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rüdiger W, Beck CF. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rüdiger W, Beck CF. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 2000;24:523–531. doi: 10.1046/j.1365-313x.2000.00898.x. [DOI] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B. Isolation and characterization of tobacco (Nicotiana tabacum) cDNA clones encoding proteins involved in magnesium chelation into protoporphyrin IX. Plant Mol Biol. 1997;35:1053–1056. doi: 10.1023/a:1005913315905. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA(+): a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinones as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Gräfe S, Kruse E, Hänel F, Grimm B. Mg-chelatase of tobacco: identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 1997;12:981. doi: 10.1046/j.1365-313x.1997.12050981.x. [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mock HP, Tanaka R, Kruse E, Grimm B. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 2000a;122:1161–1169. doi: 10.1104/pp.122.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Pfündel E, Mock HP, Grimm B. Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 2000b;22:155–164. doi: 10.1046/j.1365-313x.2000.00724.x. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Kannangara CG, Henningsen KW. Distribution of ATPase and ATP-to-ADP phosphate exchange activities in magnesium chelatase subunits of Chlorobium vibrioforme and SynechocystisPCC6803. Arch Microbiol. 1999a;171:146–150. doi: 10.1007/s002030050692. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Moller MG, Jensen PE, Henningsen KW. Identification of the Xan-g gene and expression of the Mg-chelatase encoding genes Xan-f, -g and -h in mutant and wild type barley (Hordeum vulgareL.) Hereditas. 1999b;131:165–170. [Google Scholar]
- Pogson B, McDonald K, Truong M, Britton G, DellaPenna D. Arabidopsiscarotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Downs CG, Davies KM. Differential expression of two 1-aminocyclopropane-1-carboxylic acid oxidase genes in broccoli after harvest. Plant Physiol. 1995;108:651–657. doi: 10.1104/pp.108.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Niyogi KK, Björkman O, DellaPenna D. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc Natl Acad Sci USA. 1998;95:13324–13329. doi: 10.1073/pnas.95.22.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Holtorf H, Apel K. Two NADPH:protochlorophyllide oxidoreductases in barley: evidence for the selective disappearance of PORA during the light-induced greening of etiolated seedlings. Plant Cell. 1995;7:1933–1940. doi: 10.1105/tpc.7.11.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Schnell DJ, Weisbeek PJ. The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. J Biol Chem. 2000;275:10265–10271. doi: 10.1074/jbc.275.14.10265. [DOI] [PubMed] [Google Scholar]
- Runge S, Sperling U, Frick G, Apel K, Armstrong GA. Distinct roles for light-dependent NADPH:protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J. 1996;9:513–523. doi: 10.1046/j.1365-313x.1996.09040513.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger FN, Coulsen AR. DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- Walker CJ, Willows RD. Mechanism and regulation of Mg-chelatase. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows RD, Beale SI. Heterologous expression of the Rhodobacter capsulatus BCH I, -D, and -H genes that encode magnesium chelatase subunits and characterization of the reconstituted enzyme. J Biol Chem. 1998;273:34206–34213. doi: 10.1074/jbc.273.51.34206. [DOI] [PubMed] [Google Scholar]
- Willows RD, Gibson LCD, Kanangara CG, Hunter CN, von Wettstein D. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]