Abstract
Peptic ulcer disease (PUD) occurs after a long-term Helicobacter pylori infection. However, the disease can develop earlier, and rare cases have been observed in children, suggesting that these H. pylori strains may be more virulent. We used suppressive subtractive hybridization for comparative genomics between H. pylori strains isolated from a 5-year-old child with duodenal ulcer and from a sex- and age-matched child with gastritis only. The prevalence of the 30 tester-specific subtracted sequences was determined on a collection of H. pylori strains from children (15 ulcers and 30 gastritis) and from adults (46 ulcers and 44 gastritis). Two of these sequences, jhp0562 (80.0% versus 33.3%, P = 0.008) and jhp0870 (80.0% versus 36.7%, P = 0.015), were highly associated with PUD in children and a third sequence, jhp0828, was less associated (40.0% versus 10.0%, P = 0.048). Among adult strains, none of the 30 sequences was associated with PUD. However, both jhp0562 and jhp0870 were less prevalent in adenocarcinoma strains than in PUD strains from children and adults, the difference being statistically significant for jhp0870. In conclusion, two H. pylori genes were identified as being strongly associated with PUD in children, and their putative roles as an outer membrane protein for jhp0870 and in lipopolysaccharide biosynthesis for jhp0562, suggest that they may be novel virulence factors of H. pylori.
Helicobacter pylori, a spiral-shaped gram-negative bacterium, can lead to various gastroduodenal diseases. A causal association has been established with chronic gastritis, peptic ulcer, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (10). The mechanisms for such a clinically diverse profile are not totally clear but may include host and environmental factors, as well as bacterial virulence factors (7, 20, 23, 47).
In adult patients, severe gastroduodenal diseases such as duodenal ulcer, gastric adenocarcinoma, and MALT lymphoma occur after a long-term colonization (22, 47), while the onset of H. pylori infection is essentially during childhood (26). Indeed, the majority of H. pylori-infected children remain asymptomatic, except for a very small group which will develop peptic ulcer. Other factors such as nonsteroidal anti-inflammatory drug use, physiologic stress, and vascular insufficiency also play an important role beside H. pylori infection in the development of peptic ulcer in children (28). Moreover, colonization with H. pylori strains that are considered virulent in adults does not always correlate with the severity of endoscopic and histologic findings in children (24). These conflicting results may be due to the short-term infection, the association becoming possibly stronger as the duration of infection increases (53). Ulcers in children, in contrast to adults, appear shortly after H. pylori colonization, suggesting that differences exist between the ulcerogenic strains which infect children and adults and that strains associated with peptic ulcer in children may be more aggressive against the gastroduodenal epithelium. According to this hypothesis, we chose to use molecular methods to identify new genes involved in the virulence of H. pylori by comparing the genome of clinical isolates, using suppressive subtractive hybridization (SSH) (1, 3). Thus, the aim of the present study was to look for new markers that could be potential virulence candidates by studying H. pylori DNA fragments from an ulcer strain and a gastritis strain isolated from two young children and obtained by genetic subtraction. The new bacterial genotypic markers found were then tested on a larger population, including adult patients with peptic ulcer disease (PUD), gastric adenocarcinoma, and gastric MALT lymphoma, to measure the strength of the association.
MATERIALS AND METHODS
H. pylori strains.
Two H. pylori strains were compared: the tester strain (68/00), isolated from a 5-year-old male Caucasian child a presenting a duodenal ulcer, and the driver strain (139/00), also isolated from a young age- and sex-matched Caucasian child, presenting with recent abdominal pain and gastritis alone. Both children were subjected to upper gastrointestinal endoscopy because of vomiting and abdominal pain. Several episodes of bleeding were reported for the patient presenting with the duodenal ulcer. The isolated strains were previously typed for some H. pylori genes and presented the same genotype. Both strains were negative for the entire cag pathogenicity island (PAI), namely, cagA (54), cagE, cagG, and cagM (56), and the “cag empty site” was positive (2). They were vacAs1m2 (8), babA2+, (21), iceA1 (55), hopQI (13), and both had a functional oipA gene (56) and nonfunctional sabA (40) and hopZ (46) genes. Another characteristic shared by the strains is that they both carry plasmidic DNA, which was estimated to have approximately 10.0 and 6.0 kb for the tester and driver strains, respectively.
The distribution of the ulcer-specific sequences obtained was evaluated in a panel of H. pylori strains isolated from pediatric and adult patients, presenting either peptic ulcer or non-ulcer dyspepsia. The pediatric group was comprised of 45 patients: 15 with PUD, of whom 2 had gastric ulcers (GU), and 13 with duodenal ulcers (DU) (64% male; mean age, 11.7 ± 3.8 years), and another 30 presenting with recurrent abdominal pain and gastritis (57% male; mean age, 9.1 ± 3.6 years). All of these pediatric patients were members of different families, with no parental relationship. In the adult population, strains from 90 patients were included; 46 of these patients presented with peptic ulcer, 12 presented with GU, and 34 presented with DU (52% male; mean age, 49.5 ± 16.2 years), and 44 presented with non-ulcer dyspepsia (44.7% male; mean age, 51.3 ± 14.6 years). A group of 15 strains isolated from patients with gastric adenocarcinoma (43% male; mean age, 62.3 ± 10.8 years) were also included in order to study the prevalence of specific-subtracted sequences associated with PUD in this particular group. All of these strains were isolated from Portuguese patients and are part of the collection of Centro Bacteriologia, from Instituto Nacional Saúde Dr Ricardo Jorge. All of the strains used in the present study were cultured from antral biopsies, preserved in brucella broth supplemented with 20% glycerol, and maintained at −80°C until used.
In addition, a French collection of 43 H. pylori strains obtained from gastric MALT lymphoma patients (62.8% male; mean age, 48.2 ± 13.4 years) described previously (39) was also included.
The two H. pylori strains with completely sequenced genomes, 26695 (ATCC 700392) and J99 (ATCC 700824), were used as reference strains (6, 52).
Total DNA from all strains was extracted from a 48-hour-old culture grown in 10% horse blood agar, using a QIAamp DNA minikit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer's instructions.
SSH.
The genomic content of the two H. pylori strains, the tester (DU) strain and the driver (gastritis) strain, was compared by PCR-SSH, using the PCR-Select bacterial genome subtraction kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions, with some minor modifications. The restriction enzyme used was AluI (New England Biolabs, Ipswich, MA) according to the method of Akopyants et al. (3). The treatment of both tester and driver DNAs with the AluI restriction enzyme generated fragments with sizes between 1,250 and 178 bp, as previously reported (3, 39). The analysis of the subtraction efficacy was performed by real-time PCR with the SYBR-Green format in a LightCycler thermocycler (Roche Diagnostics GmbH, Mannheim, Germany), using the H. pylori 16S rRNA gene as the target gene. The 10-μl PCR mixture contained 1 μl of LightCycler DNA Master SYBR Green I mixture (Roche Diagnostics), 3 mM MgCl2, 0.4 μM concentrations of forward MON1 (5′-GGATGAAGGTTTTAGGATTGT-3′) and reverse MON2 (5′-CATCCATCGTTTAGGGCGT-3′) primers and 1 μl of template DNA. After an initial denaturation for 10 min at 95°C, amplification steps (95°C for 0 s, 54°C for 10 s, and 72°C for 17 s) were repeated 40 times with a temperature transition rate of 20°C/s. Fluorescence was measured at 300 nm after each cycle and plotted versus the cycle number to determine the threshold cycle at the end of the PCR.
Construction and identification of the subtracted library.
For this purpose, the mixture of fragments resulting from the secondary suppressive PCR was cloned in the pCR2.1-TOPO plasmid, using the TOPO TA Cloning kit, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The recombinant DNAs were transformed into Escherichia coli TOP10 cells and selected by the blue/white screening on selective Luria-Bertani (LB) agar plates supplemented with 100 μg of ampicillin (Sigma-Aldrich, Madrid, Spain)/ml. White colonies were randomly picked and grown in LB liquid medium supplemented with 100 μg of ampicillin/ml at 37°C overnight with agitation. This culture was used to prepare boiled supernatants and also to preserve the clones in brucella broth supplemented with 20% glycerol and 100 μg of ampicillin/ml at −80°C.
For the identification of clone inserts, an amplification reaction was performed by PCR, in a 25-μl final volume, using 5 μl of crude DNA in boiled suspensions and the vector specific primers M13 forward and M13 reverse. The products were then purified using the Jetquick kit (Genomed, Lohne, Germany) and subjected to sequencing on both strands, with the PCR primers, by the dye termination method using the Big Dye Terminator v1.1 Sequencing standard kit (PE Applied Biosystems Chemistry, Foster City, CA) and the Automated Sequencer Genetic Analyser ABI-Prism 3130 xl (PE Applied Biosystems). Sequence analysis was performed with the DNAstar Lasergene (version 5.0) (12). Homology searches with GenBank data were done by using BLAST at the Web server at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Functional classification of genes from both sequenced H. pylori genomes was carried out according to the method of Boneca et al. (11; http://genolist.pasteur.fr/PyloriGene/genome.cgi).
Validation of the subtracted sequences.
The validation of each subtracted sequence as tester specific was performed by PCR using the DNAs of both tester and driver strains, as well as the DNAs of the J99 and 26695 strains as positive controls. The primers were designed by using the Web Primer3 software (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi) in regions of homology between the H. pylori databases and the subtracted sequences. PCRs were carried out under a final volume of 25 μl with the following composition: 1× PCR Buffer (Roche Diagnostics GmbH), 1.5 mM MgCl2 (Roche), 200 μM concentrations of each deoxynucleoside triphosphate (Roche Diagnostics GmbH), 0.5 μM concentrations of each primer, 1 U of Taq polymerase (Roche Diagnostics GmbH). The amplification reaction consisted of 1 cycle of 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55 to 60°C, and 30 s at 72°C, with a final extension cycle of 7 min at 72°C, to complete elongation.
Evaluation of the prevalence of subtracted tester-specific sequences in H. pylori strains by dot blotting and PCR.
The prevalence of subtracted tester-specific sequences was evaluated by dot blotting, except for two open reading frames (ORFs) (jhp0562 and jhp0870), whose prevalence was determined by PCR, because of the high degree of homology displayed with other H. pylori genes.
Dot blot.
All of the sequences with a PCR-positive result for the tester-strain and PCR negative for the driver-strain were analyzed by dot blot DNA hybridization according to conditions previously reported (39, 44) in the collection of H. pylori strains, in order to look for possible associations. The J99 and 26695 strains were also included as controls for hybridization. All of the probes were digoxigenin-labeled by PCR using the same primers for the PCR validation previously mentioned.
PCR amplification of the jhp0562 gene.
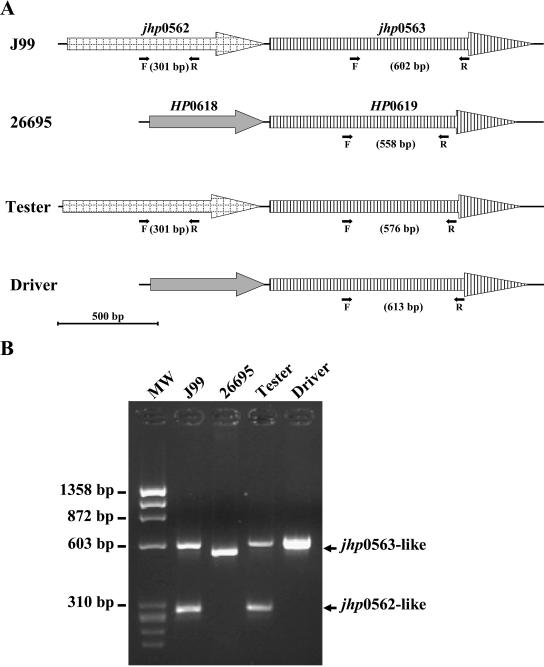
In order to screen for the presence of the jhp0562 gene, PCR primers were designed in the region of homology between subtracted and database sequences, allowing the amplification of an internal region that enables a distinction between the two 80% homologous genes, jhp0562 and jhp563 (Fig. 1A). The primers designed by using the Web Primer3 software, F1-jhp0562/jhp0563 (5′-TGAAAAGCCCTTTTGATTTTG-3′) and R1-jhp0562/jhp0563 (5′-GCTGTAGTGGCCACATACACG-3′), were used under the following PCR cycle conditions: initial denaturation for 5 min at 95°C, 35 amplification steps (95°C for 30 s, 56°C for 30 s, and 72°C for 30 s), and a final extension cycle of 7 min at 72°C. These primers generate two PCR products with 301 and 602 bp in the J99 strain and only one PCR product with 558 bp in the 26695 strain (Fig. 1).
FIG. 1.
Schematic representation of the PCR assay for the detection of the jhp0562, jhp0563(HP0619)-like genes in H. pylori strains (A) and analysis of their detection by PCR in an ethidium bromide-stained 1.5% agarose gel (B). (A) The black arrows represent the forward F1-jhp0562/jhp0563, labeled F, and the reverse R1-jhp0562/jhp0563, labeled R, conserved primers between H. pylori strains, respectively. These F and R primers hybridize both on jhp0562 and on jhp0563-like genes. Genes that belong to the same family are represented in an identical way. (B) MW, molecular weight marker ΦX174 RF DNA/HaeIII fragments (72-1353 pb; Invitrogen).
PCR amplifications of the jhp0870 gene.
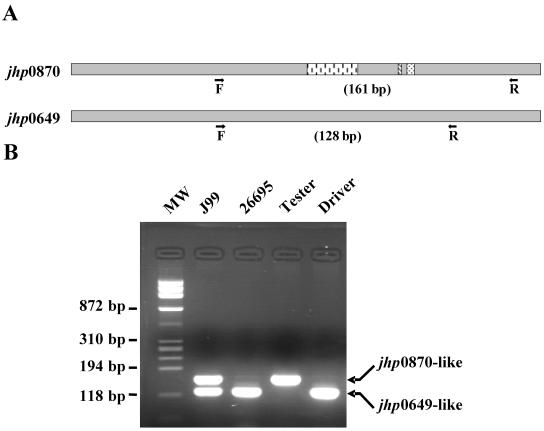
The two 90% homologous genes, jhp0870 and jhp0649 (5), were aligned by using a multiple sequence alignment with hierarchical clustering (http://prodes.toulouse.inra.fr/multalin/multalin.html) (15), and a search was carried out to identify conserved regions outside of the divergent 27-, 2-, and 4-bp additional sequences present in jhp0870 but absent in jhp0649 (Fig. 2 and 3A). Primers were designed using the Primer Express software package (PE Applied Biosystems). The primers F1-jhp0870/jhp0649 (5′-AGAGGGTGTTTGAAACGCTCAATA-3′) and R1-jhp0870/jhp0649 (5′-GGTGAATTCTTCTGCGGTTTG-3′) were designed to amplify two sequences of 128 and 161 bp in the J99 strain and only one 128-bp sequence in the 26695 strain (Fig. 3B). The PCR cycles conditions were as follows: initial denaturation for 5 min at 95°C, 35 amplification steps (95°C for 30 s, 60°C for 30 s, and 72°C for 17 s), and a final extension cycle of 7 min at 72°C.
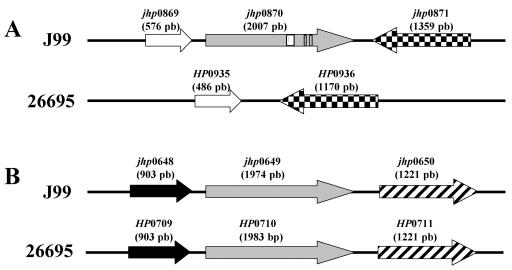
FIG. 2.
Genomic organization of the jhp0870 (A) and jhp0649/HP0710 (B) loci in J99 and 26695 H. pylori strains. The jhp0870 gene is specific for the J99 strain. No homologue of this gene exists in the 26695 strain (5). Instead, an intergenic space is observed, flanked by genes homologous to those flanking jhp0870 in the J99 strain (52). Genes which belong to the same family are represented in an identical way. The boxes represent the 27-, 2-, and 4-bp additional sequences present in jhp0870 but absent in jhp0649/HP0710.
FIG. 3.
Schematic representation of the PCR assay for the detection of the jhp0870 and jhp0649-like genes in H. pylori strains (A) and analysis of their detection by PCR in ethidium bromide-stained 2,5% agarose gel (B). (A) The black arrows represent the forward F1-jhp0870/jhp0649, labeled F, and reverse F1-jhp0870/jhp0649, labeled R, conserved primers between the jhp0870 and jhp0649-like genes in H. pylori strains, respectively. These F and R primers hybridize both on jhp0649 and jhp0870-like genes. The three internal boxes in the jhp0870 scheme represent the 27-bp, 2-bp and 4-bp additional sequences present in jhp0870 but absent in jhp0649. (B) MW, molecular weight marker ΦX174 RF DNA/HaeIII fragments (72-1353 pb; Invitrogen).
In order to determine the genomic positions of the jhp0870 and jhp0649 genes in the clinical strains, two different PCRs were developed. The flanking regions of the jhp0870 and jhp0649(HP0710) genes (Fig. 2) were aligned between the J99 and 26695 strains (15), and a search was carried out to identify conserved regions. As a result, two sets of primers were designed by using the Web Primer3 software in the jhp0869(HP0935)/jhp0871(HP0936) and jhp0648(HP0709)/jhp0650(HP0711) regions for amplification of the entire jhp0870 and jhp0649(HP0710) genes, respectively. The primer sets F1-jhp0869/HP0935 (5′-AAGAGGATTGCGTGGTGGAGTTG-3′) with R1-jhp0871/HP0936 (5′-TCCAAGCCCAAAGGCAACCC-3′) and F1-jhp0648/HP0709 (5′-TAATTTCGCGCAAAAACATC-3′) with R1-jhp0650/HP0711 (5′-ATTCCAGCGCCTAATGGAC-3′) were used, under the following amplification conditions: initial denaturation for 5 min at 95°C, 35 amplification steps (95°C for 30 s, 58°C for 30 s, and 72°C for 2 min), and a final extension cycle of 7 min at 72°C.
In cases where PCR products of different lengths were obtained, the results were confirmed by sequencing.
Statistical analysis.
For univariate analysis, the Fisher exact test and the χ2 test, with Yates' continuity correction, were used to compare differences in the prevalence of subtracted sequences between groups. P values of <0.05 were considered significant.
RESULTS
Analysis of the subtracted library.
The real-time PCR quantification analysis of the SSH between DNA from the tester and driver strains indicated a successful subtraction experiment since a difference of 17 threshold cycles between the subtracted and unsubtracted products was observed (data not shown).
Among the 138 clones analyzed by sequencing, 71 (51.4%) were H. pylori plasmid-related sequences, and 67 clones (48.6%) showed homology with H. pylori chromosomic DNA. Of the 71 plasmid-related clones, only 12 were nonredundant sequences, whereas for the 67 chromosome-related clones, 63 were unique sequences. Overall, there were 75 unique DNA sequences, indicating 45.7% redundancy, the plasmid sequences being the most represented, in this category.
The assignment of each clone as tester specific was performed by amplification of the region of homology between the cloned and the database sequences, using both tester and driver strain DNAs. For the chromosomic sequences, PCRs were validated with the DNAs of H. pylori J99 and 26695 reference strains, whereas the screening of plasmid-related clones was validated by a positive result with the tester strain. Accordingly, 30 clones were identified to be tester specific either by the absence of amplification with the driver strain DNA or by the generation of PCR products with different lengths between the tester and the driver strain DNAs, including 9 of the 12 plasmid-related sequences (75%) and 21 of the 67 chromosome-related sequences (33.3%).
With regard to the nine tester-specific plasmid-related sequences, the great majority showed full-length extent homology with H. pylori plasmids, with a degree of homology from 87 to 97% (Table 1). Six fragments were homologous to sequences within singular coding regions, while three comprised the end of one gene and the beginning of the flanking downforward gene. All of these tester-specific sequences matched with database sequences, namely, with ORFs described for H. pylori plasmid pHel4 and, to a lesser extent pHel5, two cryptic plasmids described in H. pylori, which suggests the presence of a similar plasmid in the tester strain (30). Three functional categories were distinguished, along with a group of proteins of unknown function (Table 1). Thus, three clones matched with putative mobilization genes involved in conjugative transfer and one matched with replication protein A, which is the predominant replication protein of H. pylori (29). Two clones matched two ORFs described for pHel4, orf4A and orf4B, with significant homology to two genes of the microcin mccC7 gene cluster of E. coli plasmid pMccC7, involved in the MccC7 biosynthesis and secretion (30).
TABLE 1.
Results from the screening of plasmid-subtracted sequences in H. pylori strains isolated from children with peptic ulcer or non-ulcer gastritis
| Subtracted sequence
|
Homology with H. pylori plasmids
|
H. pylori strains
|
|||||
|---|---|---|---|---|---|---|---|
| No. (%)
|
Pc | ||||||
| Clonea | Length (bp) | GenBank accession no. | ORF, plasmid | Gene product and/or functionb | Ulcers (n = 15) | Gastritis (n = 30) | |
| 5p | 438 | DQ364775 | ORF4O, pHel4 | Putative | 5 (33.3) | 9 (30.0) | 0.909 |
| 12p | 727 | DQ364770 | ORF4C, pHel4 | MobA-like protein | 5 (33.3) | 9 (30.0) | 0.909 |
| 13p* | 491 | DQ364771 | ORF4M, pHel4 | Putative | 6 (40.0) | 16 (53.3) | 0.598 |
| ORF4N, pHel4 | Putative | 6 (40.0) | 14 (46.7) | 0.916 | |||
| 24p | 353 | DQ364772 | ORF4N, pHel4 | Putative | 6 (40.0) | 14 (46.7) | 0.916 |
| 25p | 367 | DQ364779 | ORF4C, pHel4 | MobA-like protein | 5 (33.3) | 9 (30.0) | 0.909 |
| 62p | 677 | DQ364780 | repA, pHel5 | H. pylori replication protein A | 8 (53.3) | 17 (56.6) | 0.916 |
| 99p | 349 | DQ364781 | ORF4A, pHel4 | MccC-like protein | 4 (26.7) | 7 (23.3) | 0.902 |
| B1-F1p* | 969 | DQ364776 | ORF4C, pHel4 | MobA-like protein | 5 (33.3) | 9 (30.0) | 0.909 |
| ORF4F, pHel4 | MobC-like protein | 7 (46.6) | 9 (30.0) | 0.441 | |||
| B1-F16p* | 935 | DQ364778 | ORF4A, pHel4 | MccC-like protein | 4 (26.7) | 7 (23.3) | 0.902 |
| ORF4B, pHel4 | MccB-like protein | 4 (26.7) | 7 (23.3) | 0.902 | |||
*, Tested separately by dot blotting.
Gene product or function is according to Hofreuter and Haas (30).
The P value was determined by using the Fisher exact test.
Concerning the 21 tester-specific chromosome-related sequences, all of them displayed homology with chromosomic and/or plasmidic H. pylori database sequences, although in one case (clone 34/1), only partial homology was observed, with 30% of the sequence displaying no homology with database sequences. In this case, the 5′ and 3′ regions of this original sequence showed homologies with nonadjacent H. pylori genes. In addition, the majority of cloned sequences presented each homology with a single coding region, whereas in four cases (clones 34/1, 13/2, 48/2, and 68/2) the homology was with more than one ORF. Different situations must, however, be distinguished: in two cases (clones 34/1 and 68/2), the 5′ and 3′ ends of the subtracted sequences showed homology with two nonadjacent H. pylori ORFs: jhp1081/HP1154 and jhp1329/HP1436 ORFs, respectively, for the clone 34/1, and jhp0742/HP0806 and jhp0142/HP0154, respectively, for the clone 68/2. In both of these cases, the two ORFs were screened separately and, for each clone, only one ORF was tester specific (Table 2).
TABLE 2.
Results from the dot blot screening of genome-subtracted sequences in H. pylori strains isolated from children and adults with peptic ulcers or non-ulcer gastritis
| Subtracted sequence
|
Homology with ORFs of H. pylori reference strains
|
Strains from children
|
Strains from adults
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%)
|
Pc | No. (%)
|
Pc | ||||||||
| Namea | Length (bp) | GenBank accession no. | J99 | 26695 | Gene product and/or functionb | Ulcers (n = 15) | Gastritis (n = 30) | Ulcers (n = 46) | Gastritis (n = 44) | ||
| 18/1 | 588 | DQ364763 | jhp0820 | None | Putative LPS biosynthesis | 6 (40.0) | 8 (27.0) | 0.569 | 19 (41.3) | 20 (45.4) | 0.854 |
| 27/1 | 218 | DQ364766 | jhp1272 | HP1354 | Putative adenine-specific DNA methyltransferase | 7 (46.7) | 13 (43.0) | 0.915 | 27 (58.7) | 22 (50.0) | 0.538 |
| 32/1 | 526 | DQ364777 | jhp0899 | HP0965 | Remnant pseudogene of an ancestral ATP/GTP-binding protein | 14 (93.3) | 26 (86.7) | 0.866 | 45 (97.8) | 42 (95.5) | 0.593 |
| 34/1* | 487 | DQ364787 | jhp1081 | HP1154 | Putative | 13 (86.7) | 25 (83.3) | 0.884 | 44 (95.7) | 40 (90.9) | 0.632 |
| 39/1 | 535 | DQ364786 | jhp1050 | HP1121 | Putative type II cytosine-specific DNA methyltransferase | 12 (80.0) | 24 (80.0) | 0.693 | 43 (93.5) | 41 (93.2) | 0.714 |
| 41/1 | 363 | DQ364785 | jhp0650 | HP0711 | Putative | 9 (60.0) | 19 (63.3) | 0.913 | 31 (67.4) | 26 (59.0) | 0.550 |
| 4/2 | 427 | DQ364782 | jhp0784 | HP0846 | Type I restriction enzyme | 13 (86.7) | 22 (73.3) | 0.526 | 39 (84.8) | 37 (84.1) | 0.841 |
| 13/2*§ | 422 | DQ364783 | jhp0562 | None | Putative LPS biosynthesis | 12 (80.0) | 10 (33.3) | 0.008 | 30 (65.2) | 30 (68.2) | 0.941 |
| 30/2 | 639 | DQ364784 | jhp0439 | HP0487 | Putative OMP | 13 (86.7) | 27 (90.0) | 0.867 | 44 (95.6) | 42 (95.4) | 0.641 |
| 47/2 | 210 | DQ364762 | jhp0816 | HP0884 | Putative | 13 (86.7) | 25 (83.3) | 0.884 | 42 (91.3) | 35 (79.5) | 0.198 |
| 48/2*§ | 1089 | DQ364761 | jhp0870 | None | Putative OMP | 12 (80.0) | 11 (36.7) | 0.015 | 28 (60.9) | 24 (54.5) | 0.694 |
| 52/2 | 598 | DQ364758 | jhp0100 | HP0108 | Putative | 7 (46.7) | 12 (40.0) | 0.915 | 27 (58.7) | 23 (52.3) | 0.689 |
| 64/2 | 603 | DQ364759 | None | HP0446 | Putative | 5 (33.3) | 4 (13.5) | 0.236 | 7 (15.2) | 5 (11.4) | 0.820 |
| 68/2* | 362 | DQ364760 | jhp0142 | HP0154 | Putative enolase | 11 (73.3) | 23 (76.7) | 0.902 | 38 (82.6) | 31 (70.5) | 0.266 |
| 82/2 | 737 | DQ364769 | jhp0600 | HP0655 | Putative OMP | 14 (93.3) | 28 (93.3) | 0.526 | 45 (97.8) | 42 (95.5) | 0.969 |
| 89/2 | 427 | DQ364768 | jhp0755 | None | Putative type II cytosine-specific DNA methyltransferase | 3 (20.0) | 2 (6.6) | 0.402 | 3 (6.5) | 5 (11.4) | 0.663 |
| 103/2# | 262 | DQ364767 | jhp0825 | None | Putative | 7 (46.7) | 8 (26.7) | 0.314 | 14 (30.4) | 14 (31.2) | 0.931 |
| 108/2# | 158 | DQ364764 | jhp0828 | None | Putative | 6 (40.0) | 3 (10.0) | 0.048 | 13 (28.3) | 8 (18.2) | 0.378 |
| B3/1 | 178 | DQ364765 | jhp0797 | HP0863 | Putative | 15 (100) | 29 (96.7) | 0.721 | 46 (100) | 43 (97.7) | 0.672 |
| B3/8 | 510 | DQ364773 | jhp0110 | HP0120 | Putative | 14 (93.3) | 24 (80.0) | 0.467 | 33 (71.7) | 36 (81.8) | 0.827 |
| B4/2 | 128 | DQ364774 | None | HP0317 | Putative OMP | 3 (20.0) | 3 (10.0) | 0.642 | 5 (10.9) | 2 (4.5) | 0.432 |
*, Clones that showed sequence homologies with more than one ORF; #, clones that showed high-level sequence homologies also with H. pylori plasmidic sequences; §, tested only by PCR and sequencing. ORFs significantly associated with PUD in children are indicated in boldface.
Gene product or function according to Boneca et al. (11; http://genolist.pasteur.fr/PyloriGene/genome.cgi).
The P value was determined by using the Fisher exact test.
In the two other cases (clones 13/2 and 48/2), the subtracted sequence recognized more than one ORF in the H. pylori J99 strain genome, probably reflecting the existence of duplicated genes or genes belonging to the same family of paralogous genes.
The sequence of clone 13/2 showed homology with the J99 ORFs jhp0562 and jhp0563. The jhp0562 gene is a J99-specific ORF located upstream of jhp0563. No homologue of this gene exist in the 26695 strain, whereas the HP0619 in the 26695 strain is homologous to the jhp0563 (Fig. 1A). Both of these genes code for putative glycosyltransferases involved in the synthesis of the lipopolysaccharide (LPS) core (16).
There was another situation (clone 48/2) of homology of the subtracted sequence with two ORFs present in the J99 strain genome (jhp0870 and jhp0649) and one present in the 26695 strain (HP0710, a jhp0649 homologue) (see Fig. 2). The two ORFs, jhp0870 and jhp0649, are 90% identical and encode for putative outer membrane proteins (OMPs) (5) but are located in different regions of the genome from J99 strain. The jhp0649 and its homologue HP0710 are flanked by the same ORFs in both strains. On the other hand, no homologue of the jhp0870 exists in the 26695 strain. Instead, an intergenic space is observed, flanked by ORFs homologous to those which flank jhp0870 in strain J99 (Fig. 2).
Overall, within the 21 chromosome-related tester-specific sequences, 8 (38.1%) were previously identified as strain specific, 6 being specific for the J99 strain and 2 specific for the 26695 strain (6). In terms of function, three clones exhibited homology with restriction modification systems, namely, putative methyltransferases, four putative OMPs, two matched putative LPS biosynthesis genes, one restriction enzyme, one putative enolase, one remnant pseudogene of an ancestral ATP/GTP-binding protein, and nine clones exhibited homology with a predicted coding region with no assigned function (Table 2).
The prevalence of subtracted tester-specific sequences was then determined by using the collection of H. pylori strains provided from children and from adults.
PCR screening of the jhp0562 and jhp0870 subtracted sequences.
The two homologous genes, jhp0562 and jhp0563 (6), share no specific region long enough to design a dot blot probe to differentiate these genes. Thus, a PCR was developed allowing the amplification of an internal region which enable the distinction between these two genes (Fig. 1A). The results are presented on Fig. 1B. Two PCR products of 602 and 301 bp were obtained for J99 strain, and only one 558-bp amplicon was obtained for the 26695 strain. Concerning the tester strain, a profile similar to that of the J99 strain was obtained, with two PCR products with 576 and 301 bp, while for the driver strain only one 613-bp fragment was obtained. PCR products were identified by sequencing, showing the presence of both jhp0562 and jhp0563 genes in J99 and tester strains (301 bp for jhp0562 in J99 and tester strains; 576- and 602-bp products for jhp0563 in the J99 and tester strains, respectively), and the presence of only the jhp0563 homologue in both the 26695 (HP0619) and the driver strains (558- and 613-bp products, respectively). This PCR was performed on the H. pylori strain collection; PCR products from several clinical strains were also sequenced, showing the validity of this PCR (data not shown). Moreover, among the clinical strains, the sequencing results revealed a size variable sequence for the jhp0563/HP0619 gene, in contrast to the well-conserved jhp0562 gene.
An identical situation was observed for the two genes, jhp0870 and jhp0649, which are 90% homologous (5), excluding the possibility of dot blot experiments. In order to determine which gene was present in H. pylori clinical strains, a PCR was developed to detect the three additional sequences with 27, 2, and 4 bp present in jhp0870 but absent in jhp0649 (Fig. 3A). The results are presented in Fig. 3B. As expected, two PCR products of 161 and 128 bp were obtained for J99 strain, and a 128-bp amplicon was obtained for the 26695 strain. Concerning the tester and driver strains, single PCR products of 161 and 128 bp, respectively, were obtained. This PCR was performed on the H. pylori strain collection, and PCR products from several clinical strains were sequenced, confirming the presence or absence of the three additional sequences (data not shown).
The jhp0870 and jhp0649 genes are located in different positions in the genome of the J99 strain. Thus, in order to understand the genomic position of these genes in the tester and driver strains, two PCRs were developed to amplify specifically the jhp0870 and jhp0649/HP0710 regions, respectively, by using primers designed in the conserved flanking genes of these two regions [jhp0869(HP0935)/jhp0871(HP0936) and jhp0648(HP0709)/jhp0650(HP0711); see Fig. 2]. PCRs with these primers led to the following results. For the jhp0870 flanking regions a 3,500-bp fragment was obtained for the tester and J99 strains, whereas for the driver strain a 1,350-bp fragment was observed, similar to that obtained for the 26695 strain, where an intergenic space is observed since no jhp0870 homologue exists (Fig. 2A). On the other hand, for the jhp0649 flanking regions, an approximate 2,600-bp product was observed for all of the strains (not shown), suggesting that this region is always occupied. Sequencing of the PCR products showed, for the tester strain, the presence of a jhp0870-like gene at the genomic position jhp0869(HP0935)/jhp0871(HP0936) but also at the genomic position jhp0648(HP0709)/jhp0650(HP0711), these two jhp0870-like genes being 99% identical. Concerning the driver strain, the genomic position jhp0869(HP0935)/jhp0871(HP0936) is occupied by an intergenic space, and a jhp0649-like gene is observed in the other position, a situation similar to that observed for the 26695 strain (Fig. 2).
The results of the PCR screenings for jhp0562 and jhp0870 are presented in Table 2 and described below.
Evaluation of the prevalence of subtracted tester-specific sequences in H. pylori strains by dot blotting and PCR.
The distribution of the tester-specific sequences, 9 plasmid and 21 chromosome related, was evaluated by dot blot DNA hybridization or PCR only for the jhp0562 and jhp0870 genes, as described in the previous paragraph, in a panel of H. pylori strains isolated from patients presenting with either PUD (n = 61) or non-ulcer gastritis (n = 74) in order to test whether these sequences were disease specific. The results are presented separately according to the type of patients, adults or children, and the type of sequences (Tables 1 and 2).
The results of the dot blot analysis were in agreement with the first validation PCR results, i.e., the absence of hybridization with the driver strain and a positive signal with the tester strain. This suggests that the negative results obtained by PCR truly reflect the absence of the sequence and not a mispairing of the primers.
The plasmid-related sequences were only screened in the group of strains isolated from children and none was PUD associated. Their prevalence varied between 23.3 and 56.6% (Table 1).
Regarding the distribution of the 21 chromosome-related sequences in the clinical strains three of these ORFs were significantly associated with PUD, but only in the strains isolated from children: jhp0562 (80.0% versus 33.3%, respectively, P = 0.008), jhp0828 (40.0% versus 10.0%, respectively, P = 0.048), and jhp0870 (80.0% versus 36.7%, respectively, P = 0.015) (Table 2). Interestingly, all of these three genes were reported to be strain specific, being also present in the genome of the J99 strain and absent in the 26695 strain (Table 2). In strains isolated from adults, both jhp0562 and jhp0870 were present in more than 50% of the strains, either isolated from patients with PUD or isolated from subjects with gastritis, whereas jhp0828 showed a low prevalence (<30%). Also, in the children's strains, although a significant difference exists between the two diseases, the prevalence of jhp0828 was less than 50% in both cases (Table 2). The jhp0562 ORF codes for a putative LPS biosynthesis, while jhp0870 codes for a putative OMP; however, no function could be assigned for jhp0828 (6). Among the 18 other subtracted sequences in the present study, none of the five other strain-specific genes from 26695 or J99 strains were associated with PUD, nor were any of the 13 conserved ORFs in 26695 and J99 H. pylori strains (Table 2).
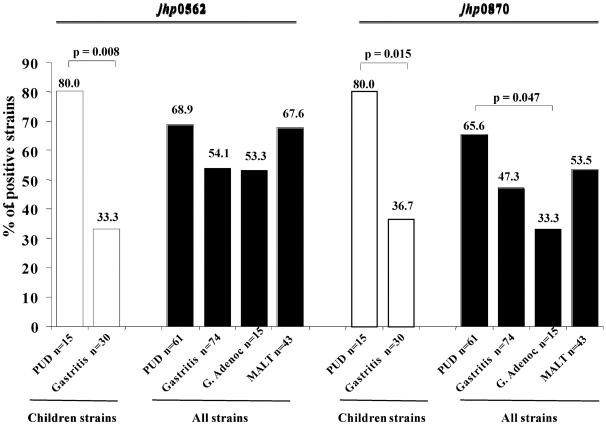
In order to verify the specificity of the PUD markers found, their distribution was also evaluated in a collection of H. pylori strains isolated from patients presenting with gastric adenocarcinoma (n = 15) and gastric MALT lymphoma (n = 43) (Fig. 4). Only the sequences that showed a high prevalence within PUD strains (jhp0562 and jhp0870) were studied. The frequency of these ORFs was lower in strains isolated from patients with adenocarcinoma than in PUD strains, with children and adults combined (53.3% versus 68.9%, P = 0.406, for jhp0562; 33.3% versus 65.6%, P = 0.047 for jhp0870) (Fig. 4). Considering only the adult strains, a difference was also observed between the prevalence of these ORFs in the two diseases, a difference that was more pronounced for jhp0870, although it was not statistically significant due to the small number of cancer strains studied (53.3% versus 65.2%, P = 0.604 for jhp0562; 33.3% versus 60.9%, P = 0.119, for jhp0870) (Table 2 and Fig. 4). On the other hand, the prevalence of jhp0562 was similar in adenocarcinoma and in gastritis strains isolated from both children and adults together (Fig. 4) but was lower than in adult strains only (53.3 and 68.2%, respectively). As for jhp0870, its prevalence in the cancer group was lower than that observed for gastritis strains in either of the two populations (Fig. 4) or in the adults only (33.3 and 54.5%, respectively). With regard to the MALT strains, the prevalence of jhp0562 was similar to that of PUD strains, either for all of the strains together (Fig. 4) or for the adult strains only (67.6 and 65.2%, respectively), whereas the jhp0870 was less frequent in both groups, although the difference was not statistically significant (Fig. 4). Comparing MALT and gastritis strains, the prevalence of both jhp0562 and jhp0870 genes was similar in the strains from adults (67.6 and 68.2%, respectively, for jhp0562; 53.5 and 54.5%, respectively, for jhp0870) but slightly higher when both populations are considered together, with no significant associations being found (Fig. 4).
FIG. 4.
Distribution of jhp0562 and jhp0870 genes among H. pylori strains isolated from subjects with PUD, gastritis, gastric adenocarcinoma, and gastric MALT lymphoma, including both children only and children and adults together (all strains). P values, determined by the Fisher exact test, are shown for the significant associations only.
Interestingly, a correlation was observed between the three PUD markers. In fact, among all strains from PUD and gastritis patients, 30 of the strains were positive for jhp0828, 16 of them were also jhp0562 positive (53.3%, P = 0.015), and 19 of them were also jhp0870 positive (63.3%, P = 0.014). Regarding the association between jhp0562 and jhp0870, 56 of the 82 strains positive for jhp0562 were also positive for jhp0870 (68.3%, P = 0.009). The presence of the three genes was observed in 12 strains. Moreover, when the association of all of the combinations of these three genotypes was evaluated in strains from PUD and gastritis, a stronger association of both jhp0562-positive and jhp0870-positive genotypes was found with PUD in strains isolated from children (69.2% versus 14.6%; P < 0.001; odds ratio [OR] = 13.18; 95% confidence interval [CI] = 3.65 to 50.58) compared to each genotype alone (80% versus 33.3%; P = 0.008; OR = 8.00; 95% CI = 1.55 to 46.78 for jhp0562; and 80% versus 36.7%; P = 0.015; OR = 6.91; 95% CI = 1.35 to 39.84 for jhp0870) (Table 2).
DISCUSSION
H. pylori is one of the most diverse bacterial species, and this variability can explain its involvement in various gastroduodenal diseases and in the establishment of a long-term infection (37). Many pathogenic factors of H. pylori have already been identified, with markedly different distributions worldwide (34, 57). However, it can be hypothesized that other genes contributing to the virulence of the bacterium still remain unidentified, and the study of strains isolated in particular clinical situations can be a useful contribution to this purpose.
There are several techniques to compare the genomic content of bacterial strains, namely, SSH, representational difference analysis (RDA), and DNA microarray (1, 18, 41). Many examples of the potency of SSH to compare bacterial genomes of related species can be found in the literature. Species- or strain-specific virulence genes (9, 38), virulence plasmid-related sequences, prophage sequences (19), and PAI (49) have been identified in several bacterial species through the use of this technique. In H. pylori, the cag PAI was discovered by using the RDA approach (14), which is also based on the principle of subtractive hybridization coupled with the use of PCR. By comparing H. pylori 26695 and J99 strains, SSH identified ca. 95% of the unique ORF in each strain, demonstrating an effective approach (1). This technique proved to be useful also in the identification of H. pylori strain-specific genes such as putative restriction modification enzymes, metabolic enzymes, a completely new ORF with an unknown function (3), transposable elements such as IS607 and ISHp608 (33, 35), and new type IV secretion systems such as tfs3 (36). Concerning the search for disease-specific genes, SSH allowed the identification of potential markers for gastric ulcer (27), intestinal metaplasia (17), and Malt lymphoma (40). In addition, SSH proved to be of great interest for comparing the gene expression of H. pylori in two different physiological states since it allowed the identification of numerous genes with enhanced expression at low pH such as the wbcJ acid-induced LPS-associated gene, thus providing new insight into mechanisms used for gastric survival (42, 43).
In the present study, SSH was used to compare the total DNA content of two H. pylori clinical strains, one isolated from a young child with DU (tester strain) and another from a child presenting with recurrent abdominal pain (driver strain), in order to isolate DNA sequences encoding determinants specific for the H. pylori ulcerogenic strain. The choice of these clinical isolates is based on the fact that, in contrast to adults, ulcer formation in children occurs shortly after H. pylori colonization, suggesting that strains associated with PUD in children may be more virulent. Moreover, we believe that in young children this disease is more “pure,” i.e., not as dependent on environmental factors, such as smoking and other toxic substances, as in adults, and thus bacterial factors can play a more important role. To overcome the differences between children and adults and, more specifically, to minimize the influence that host characteristics may have in the selection of the type of infecting strain, we compared two strains isolated from two young children who were age and sex matched.
Overall, two types of subtracted sequences were identified: chromosomic and plasmid related. A high degree of redundancy was observed, mostly due to the fact that the plasmid sequences were the most efficiently subtracted and thus represented. This suggests that (i) several copies of the plasmid exists in the bacterial cell, creating an imbalance between the number of genomic and plasmid sequences, and (ii) a high degree of divergence exists between the tester subtracted plasmidic sequences and the sequences present in the driver strain, either genomic or plasmidic, thus facilitating their subtraction. Moreover, half of the subtracted sequences were plasmid related, and thus it may be hypothesized that the presence of the plasmid decreases the overall efficiency of this subtractive experiment. However, many plasmids carry genetic determinants, also found in genomic strains without a plasmid, which may contribute to the virulence of the bacterial strains, namely, secretion systems and genes encoding antibiotic resistance (29). In addition, both isolates carry this extrachromosomal element, which could lead to the elimination of similar plasmidic sequences. A possibility would have been to determine the plasmid sequence and to test its ORF by dot bloting in the driver strain but, as demonstrated in the present study, the SSH is a powerful method that allowed the identification of very short tester-specific sequences (such as the presence of the 27-, 2-, and 4-bp additional sequences in the jhp0870). Such short sequences, if present in the plasmid, would have not been detected by dot blot hybridization. For these reasons, we decided to compare the total DNA of each strain.
Among the plasmid tester-specific sequences, none constitute a PUD marker, since their distribution in clinical isolates from children with PUD or gastritis was similar, ranging from 23.3 to 56.6%, which is in agreement with the expected prevalence of H. pylori strains carrying plasmidic DNA (48, 51).
Regarding the chromosomic tester-specific sequences, among the strain-specific genes the great majority are present in the genome of J99 but absent in the genome of 26695, which is not surprising since both the tester strain and the J99 strain were isolated from DU patients. In terms of functional categories, the most abundant class of strain-specific genes is unique to H. pylori with unknown function, as was reported in other studies (39, 50). The homologies of the other classes of strain-specific genes imply functions that may aid the bacterium under certain circumstances or in certain hosts, such as OMP and LPS genes, and also restriction modification systems that help promote the genetic diversity and thus contribute to the adaptation of the bacterium. Salama et al. (50) observed, by DNA microarray, the same functional categories among H. pylori genes presenting different patterns of distribution within clinical strains and among them the two LPS genes (jhp0820 and jhp0562) and one of the OMP (HP0317) tester-specific ORFs referred to in the present study. However, none of these ORFs belong to the group of genes showing coinheritance with the cag PAI, as observed in that same study. Other studies on functional genomics using DNA microarrays have also investigated the genetic diversity of H. pylori clinical isolates in different situations, leading to the identification of strain-specific genes important for the adaptation to a particular niche and/or to host-changing conditions, and in the induction of distinct pathological outcomes (31, 32).
The present study is in agreement with these findings, and all of them emphasize the importance of the existence of “flexible” genes that can be easily acquired or lost and allow H. pylori to adapt to different hosts, to evolve in a single host, and to modulate the host response, thereby influencing the clinical outcome.
Since at least 6 to 7% of the genes are unique features of a particular H. pylori genome, when the genomes of the two strains J99 and 26695 (6) are compared, other strain-specific genes not present in the 26695 and J99 sequences were expected; however, no original sequences were obtained in this SSH experiment. Only one sequence showed a region with no homology to database sequences, whereas in other similar studies original sequences were identified in H. pylori (3, 39). There are two possible explanations for this result: either no original sequence exists in the strain tested in the present study or such sequences exist and were missed, indicating that a more exhaustive screening of the E. coli colonies should have been performed. Both hypotheses are possible. Indeed, the plasmid copies could have masked some chromosomal sequences, and the high level of redundancy in the subtracted plasmid sequences compared to the low level of redundancy in the subtracted chromosomic sequences is an argument in favor of the latter. However, as explained above, the SSH is a powerful method that allowed the identification of very short sequences, so the probability not to have subtracted original sequences is low, since short differences between the tester and driver strains were detected.
All of the genomic subtracted sequences clearly represent genes with great potential in the virulence role of the bacterium and, when screened in a panel of H. pylori clinical strains, three sequences were found to be associated with PUD, although only in the children's isolates. Interestingly, all of them are also J99-specific genes. Two of the ORFs, jhp0562 and jhp0870, are unique members of the H. pylori J99 LPS glycosyltransferases and OMP families, respectively, and thus have sequence homology with the other members of these families. However, the fact that they were subtracted by SSH, indicates that sequence divergence does exist between these homologous genes and different numbers of copies at different genomic localizations between strains. This was verified for the strains studied, resulting in poor hybridization, and thus leading to the subtraction of one of the members. This result is consistent with a previous study where 94% of the J99-specific ORFs were identified, including jhp0562 and jhp0870, when this strain was used as the tester against 26695 as the driver strain, using the same restriction enzyme used in the present study (1). Another aspect deserving attention in the present study is the validation of the subtracted sequences as tester specific, as determined by PCR and sequencing instead of dot blotting, to avoid the elimination of tester-specific sequences containing stretches of sequences also matching with the driver strain, as was the case for jhp0562 and jhp0870, which otherwise would have been lost in subsequent analyses. Other studies have emphasized the importance of performing this screening (1, 3).
The presence of these two ORFs in clinical isolates was determined by PCR. Compared to the dot blot technique, PCR can give false-negative results due to the lack of primer annealing. For this reason, in these two cases the PCR primers were designed in a region with cross-homology with other homologous genes in the genome, and a positive PCR result was obtained for all of the strains studied, demonstrating the accuracy of the amplification reaction. Gene identification was based on the length of the amplified products, and sequencing of the PCR products has confirmed the validity of the PCR.
The three PUD-associated genes showed different prevalence profiles, with the jhp0562 and jhp0870 genes being present in almost all of the children's ulcer strains compared to the gastritis only strains. However, for adult strains these genes showed a homogeneous distribution within ulcer and gastritis strains. These results are in agreement with other studies that highlighted important differences in strains infecting the mucosa of adults and children (4, 25, 45).
Concerning the jhp0828 gene, although a different prevalence exists between PUD and gastritis strains, there is an overall weak distribution in clinical isolates and thus less probability that this particular gene may be a true virulence candidate marker.
The specificity of jhp0562 and jhp0870 as PUD markers was verified by the evaluation of their prevalence in strains isolated from gastric adenocarcinoma and MALT lymphoma. Both of these genes were less prevalent in cancer strains than in PUD strains isolated from children and adults, the difference being statistically significant for jhp0870. With regard to MALT lymphoma strains, no important difference was observed compared to other pathologies.
In conclusion, the present study has contributed to the identification of putative virulence factors in the pathogenic H. pylori species. Two genes were identified as being strongly associated with PUD in strains isolated from children, and their putative roles as OMP and in LPS biosynthesis, i.e., surface exposed and likely to have a role in the interface with the host, strongly suggest that they could be novel virulence factors of H. pylori. Moreover, a simple and reliable PCR-based technique was established for the detection of these genes in H. pylori clinical isolates.
Future studies, including an evaluation of the association between these and other previously described H. pylori virulence factors, are needed to clarify their role in the pathogenesis of this microorganism. Moreover, the study of the distribution of these genes in other populations would also be interesting to further elucidate the associations found in the present study and the possible virulent role of these factors in H. pylori infection.
Acknowledgments
We thank the Sociedade Portuguesa de Gastrenterologia and the Programme de Cooperation Scientifique et Technique Franco-Portugais offered by the French Embassy in Portugal, for supporting the project.
We are grateful to Ana Emília Carvalho and Sandrine Dupouy for technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alarcon, T., D. Domingo, M. J. Martinez, and M. LopezBrea. 1999. cagA gene and vacA alleles in Spanish Helicobacter pylori clinical isolates from patients of different ages. FEMS Immunol. Med. Microbiol. 24:215-219. [DOI] [PubMed] [Google Scholar]
- 5.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. W. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm, R. A., L. S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. UriaNickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 7.Atherton, J. C. 1997. The clinical relevance of strain types of Helicobacter pylori. Gut 40:701-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton, J. C., P. Cao, R. M. Peek, M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 9.Bae, J. W., S. K. Rhee, Y. D. Nam, and Y. H. Park. 2005. Generation of subspecies level-specific microbial diagnostic microarrays using genes amplified from subtractive suppression hybridization as microarray probes. Nucleic Acids Res. 33:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaser, M. J. 1997. Ecology of Helicobacter pylori in the human stomach. J. Clin. Investig. 100:S67-S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boneca, I. G., H. de Reuse, J.-C. Epinat, M. Pupin, A. Labigne, and I. Moszer. 2003. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res. 31:1704-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burland, T. G. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 132:71-91. [DOI] [PubMed] [Google Scholar]
- 13.Cao, P., and T. L. Cover. 2002. Two different families of hopQ alleles in Helicobacter pylori. J. Clin. Microbiol. 40:4504-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doig, P., B. L. de Jonge, R. A. Alm, E. D. Brown, M. Uria-Nickelsen, B. Noonan, S. D. Mills, P. Tummino, G. Carmel, B. C. Guild, D. T. Moir, G. F. Vovis, and T. J. Trust. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63:675-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, Q. J., M. O’Sullivan, A. Nami, P. Dowling, G. Murphy, M. Buckley, and C. O’Morain. 2005. A genetic locus of Helicobacter pylori inversely associated with gastric intestinal metaplasia. FEMS Immunol. Med. Microbiol. 44:243-249. [DOI] [PubMed] [Google Scholar]
- 18.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 19.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox, J. G., C. A. Dangler, N. S. Taylor, A. King, T. J. Koh, and T. C. Wang. 1999. High-salt diet induces gastric epithelial hyperplasia and parietal cell loss, and enhances Helicobacter pylori colonization in C57BL/6 mice. Cancer Res. 59:4823-4828. [PubMed] [Google Scholar]
- 21.Gerhard, M., N. Lehn, N. Neumayer, T. Boren, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 96:12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillen, D., and K. E. McColl. 2005. Gastroduodenal disease, Helicobacter pylori, and genetic polymorphisms. Clin. Gastroenterol. Hepatol. 3:1180-1186. [DOI] [PubMed] [Google Scholar]
- 23.Go, M. F. 1997. What are the host factors that place an individual at risk for Helicobacter pylori-associated disease? Gastroenterology 113:S15-S20. [DOI] [PubMed] [Google Scholar]
- 24.Gold, B. D., L. J. van Doorn, J. Guarner, M. Owens, D. Pierce Smith, Q. S. Song, L. Hutwagner, P. M. Sherman, O. L. de Mola, and S. J. Czinn. 2001. Genotypic, clinical, and demographic characteristics of children infected with Helicobacter pylori. J. Clin. Microbiol. 39:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Valencia, G., J. C. Atherton, O. Muñoz, M. Dehesa, A. Madrazo de la Garza, and J. Torres. 2000. Helicobacter pylori vacA and cagA genotypes in Mexican adults and children. J. Infect. Dis. 182:1450-1454. [DOI] [PubMed] [Google Scholar]
- 26.Goodman, K. J., and P. Correa. 1995. The transmission of Helicobacter pylori: a critical review of the evidence. Int. J. Epidemiol. 24:875-887. [DOI] [PubMed] [Google Scholar]
- 27.Han, F. C., M. Gong, H. C. Ng, and B. Ho. 2003. Identification of Helicobacter pylori strain specific DNA sequences between two clinical isolates from NUD and gastric ulcer by SSH. World J. Gastroenterol. 9:1747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassall, E. 2001. Peptic ulcer disease and current approaches to Helicobacter pylori. J. Pediatr. 138:462-468. [DOI] [PubMed] [Google Scholar]
- 29.Hofler, C., W. Fischer, D. Hofreuter, and R. Haas. 2004. Cryptic plasmids in Helicobacter pylori: putative functions in conjugative transfer and microcin production. Int. J. Med. Microbiol. 294:141-148. [DOI] [PubMed] [Google Scholar]
- 30.Hofreuter, D., and R. Haas. 2002. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J. Bacteriol. 184:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H.-P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersulyte, D., A. K. Mukhopadhyay, M. Shirai, T. Nakazawa, and D. E. Berg. 2000. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J. Bacteriol. 182:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. O. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kersulyte, D., B. Velapatino, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft, C., and S. Suerbaum. 2005. Mutation and recombination in Helicobacter pylori: mechanisms and role in generating strain diversity. International J. Med. Microbiol. 295:299-305. [DOI] [PubMed] [Google Scholar]
- 38.Lai, Y. C., S. L. Yang, H. L. Peng, and H. Y. Chang. 2000. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect. Immun. 68:7149-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehours, P., S. Dupouy, B. Bergey, A. Ruskone-Foumestraux, J. C. Delchier, R. Rad, F. Richy, J. Tankovic, F. Zerbib, F. Megraud, and A. Menard. 2004. Identification of a genetic marker of Helicobacter pylori strains involved in gastric extranodal marginal zone B-cell lymphoma of the MALT-type. Gut 53:931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehours, P., A. Menard, S. Dupouy, B. Bergey, F. Richy, F. Zerbib, A. Ruskone-Fourmestraux, J. C. Delchier, and F. Megraud. 2004. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect. Immun. 72:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 42.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 43.McGowan, C. C., A. S. Necheva, M. H. Forsyth, T. L. Cover, and M. J. Blaser. 2003. Promoter analysis of Helicobacter pylori genes with enhanced expression at low pH. Mol. Microbiol. 48:1225-1239. [DOI] [PubMed] [Google Scholar]
- 44.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 68:6240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oleastro, M., M. Gerhard, A. I. Lopes, P. Ramalho, J. Cabral, A. S. Guerreiro, and L. Monteiro. 2003. Helicobacter pylori virulence genotypes in Portuguese children and adults with gastroduodenal pathology. Eur. J. Clin. Microbiol. Infect. Dis. 22:85-91. [DOI] [PubMed] [Google Scholar]
- 46.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peek, R. M., Jr., and J. E. Crabtree. 2006. Helicobacter infection and gastric neoplasia. J. Pathol. 208:233-248. [DOI] [PubMed] [Google Scholar]
- 48.Penfold, S. S., A. J. Lastovica, and B. G. Elisha. 1988. Demonstration of plasmids in Campylobacter pylori. J. Infect. Dis. 157:850-851. [DOI] [PubMed] [Google Scholar]
- 49.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simor, A. E., B. Shames, B. Drumm, P. Sherman, D. E. Low, and J. L. Penner. 1990. Typing of Campylobacter pylori by bacterial DNA restriction endonuclease analysis and determination of plasmid profile. J. Clin. Microbiol. 28:83-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. Smith, O. C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 53.Torres, J., G. PerezPerez, K. J. Goodman, J. C. Atherton, B. D. Gold, P. R. Harris, A. Madrazo de la Garza, J. Guarner, and O. Munoz. 2000. A comprehensive review of the natural history of Helicobacter pylori infection in children. Arch. Med. Res. 31:431-469. [DOI] [PubMed] [Google Scholar]
- 54.Tummuru, M. K. R., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61:1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 56.Yamaoka, Y., S. Kikuchi, H. M. T. El-Zimaity, O. Gutierrez, M. S. Osato, and D. Y. Graham. 2002. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology 123:414-424. [DOI] [PubMed] [Google Scholar]
- 57.Yamaoka, Y., Y. Kodama, O. Gutierrez, J. G. Kim, K. Kashima, and D. Y. Graham. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. 37:2274-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]