Abstract
The titan (ttn) mutants of Arabidopsis exhibit dramatic alterations in mitosis and cell cycle control during seed development. Endosperm development in these mutants is characterized by the formation of giant polyploid nuclei with enlarged nucleoli. Embryo development is accompanied by significant cell enlargement in some mutants (ttn1 and ttn5) but not others (ttn2 and ttn3). We describe here the molecular cloning of TTN5 using a T-DNA–tagged allele. A second allele with a similar phenotype contains a nonsense mutation in the same coding region. The predicted protein is related to ADP ribosylation factors (ARFs), members of the RAS family of small GTP binding proteins that regulate various cellular functions in eukaryotes. TTN5 is most closely related in sequence to the ARL2 class of ARF-like proteins isolated from humans, rats, and mice. Although the cellular functions of ARL proteins remain unclear, the ttn5 phenotype is consistent with the known roles of ARFs in the regulation of intracellular vesicle transport.
INTRODUCTION
Embryo-defective mutants of Arabidopsis have been used for many years to identify genes with essential functions during plant embryogenesis (Goldberg et al., 1994; Jurgens et al., 1994; Meinke, 1995). Recent advances in gene isolation have made it possible to recover increasing numbers of these genes and study their role in plant growth and development (Li and Thomas, 1998; Lotan et al., 1998; Patton et al., 1998; Uwer et al., 1998; Albert et al., 1999). Development of the female gametophyte and early endosperm in Arabidopsis has also been subjected to extensive genetic analysis (Drews et al., 1998; Berger, 1999). Particular attention has been given to mutants with defects in gene imprinting and mutants in which endosperm development begins in the absence of fertilization (reviewed in Preuss, 1999). Molecular cloning of the medea (med) and fertilization-independent endosperm and seed (fie and fis, respectively) genes (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Ohad et al., 1999) has renewed interest in the biochemical signals associated with fertilization and the coordinated development of the gametophyte, embryo, and endosperm tissue in plants.
The titan (ttn) mutants of Arabidopsis represent another interesting class of mutants with defects in embryo and endosperm development (Liu and Meinke, 1998). The most striking feature of these mutants is the formation of giant endosperm nuclei and nucleoli during early stages of seed development. Embryo development is arrested shortly after fertilization in most ttn mutants and in some cases is accompanied by dramatic cell enlargement. Three nonallelic ttn mutants (ttn1, ttn2, and ttn3) with related but distinct phenotypes were first described by Liu and Meinke (1998). The ttn1 phenotype includes extraordinary enlargement of nuclei in the embryo and endosperm, similar enlargement of cells in the arrested embryo, and a disruption of endosperm nuclear migration to the chalazal end of the seed. Defects in ttn2 are limited to early embryonic lethality and enlargement of endosperm nuclei. Embryo development in ttn3 is surprisingly normal but is accompanied by the formation of giant endosperm nuclei early in development and the appearance of aberrant mitotic figures with numerous condensed chromosomes. Another unique feature of the ttn3 phenotype is cellularization of the mutant endosperm late in development. The ttn4 mutant identified recently resembles ttn2 in phenotype (Wu, 1999). The ttn5 mutant described here is most similar to ttn1 except that migration of endosperm nuclei is not disrupted.
Several additional mutants of the ttn1 class were recently isolated in the Jurgens laboratory and named pilz mutants because of the mushroom shape of arrested embryos (Mayer et al., 1999). On the basis of mutant phenotypes and map locations, it appears that champignon (cho) corresponds to ttn1 and hallimasch (hal) is allelic to ttn5. Additional ttn mutants with more variable embryo and endosperm phenotypes have been identified during the course of our T-DNA insertional mutagenesis project. These mutants are currently at different stages of phenotypic and molecular characterization.
Several models have been proposed for TTN gene function (Liu and Meinke, 1998). Nuclear enlargement in ttn mutants may result from a disruption of either cell cycle control or the structural mechanics of mitosis. Aberrant migration of endosperm nuclei in ttn1 appears most consistent with a disruption of cytoskeletal organization (Liu and Meinke, 1998). The observation that microtubule organization is altered in pilz seeds (Mayer et al., 1999) provides further evidence that cytoskeletal defects may be responsible for some of the abnormalities seen in the ttn1 (pilz) class of mutants. The recently cloned TTN3 gene has been shown to encode a protein related to the SMC2 family of condensins (C.M. Liu and D. Meinke, unpublished results). These proteins, components of the nuclear scaffold required for chromosome condensation in yeast and animal systems (Hirano, 1999), appear to have evolved more specialized functions in plants. The ttn3 phenotype therefore results from disruption of a protein required for normal chromosome mechanics and nuclear behavior at mitosis.
We describe here the molecular cloning of TTN5 using a T-DNA–tagged allele. The predicted gene product is related to the ADP ribosylation factor (ARF) family of small GTP binding proteins known to regulate a wide range of cellular processes in eukaryotes. Based on sequence analysis, TTN5 is most closely related to the ARL2 subfamily of ARF-like (ARL) proteins. Much less is known about these ARL proteins than other ARF proteins, which appear to play an important role in vesicle transport and phosphoinositide-mediated signal transduction pathways (Martin, 1998; Chavrier and Goud, 1999; Godi et al., 1999). The ttn5 phenotype is consistent with a general disruption of intracellular transport processes during seed development and may provide valuable insights into ARL function throughout eukaryotic organisms.
RESULTS
Identification of Mutant Alleles
The ttn5-1 mutant was identified in a large-scale screen for T-DNA insertional mutants of Arabidopsis defective in embryo development. Glufosinate ammonium (BASTA)–resistant plants that developed from seeds produced after Agrobacterium-mediated transformation were scored for seed phenotypes, and the presence of embryo-defective mutations was confirmed in the following generation. Putative ttn mutants were identified by the presence of watery seeds with enlarged endosperm nuclei. Plants heterozygous for the ttn5-1 mutation produced siliques with ∼25% mutant seeds, as expected for a recessive lethal mutation. Mapping with visible markers localized the mutation to chromosome 2, ∼15 cM from erecta. Seeds from the parental line produced a high ratio (99:1) of BASTA-resistant:BASTA-sensitive plants, indicative of multiple sites of T-DNA insertion. Two subfamilies with a low ratio (<3:1) of resistant:sensitive plants, indicative of a single insertion site, were identified when progeny seeds from 29 resistant plants were plated on media containing BASTA. No wild-type plants were identified among 108 resistant plants derived from these low-ratio subfamilies. This result is consistent with the presence of a tagged mutant allele.
A second mutant allele (ttn5-2) was identified by W. Lukowitz (Carnegie Institution of Washington, Stanford, CA) as part of a large-scale ethyl methanesulfonate mutagenesis project. Mapping this allele demonstrated close linkage between the mutant phenotype and cleaved amplified polymorphic sequence markers on chromosome 2. One recombinant F2 plant was found with PHYB and seven with THY1 after scoring of 186 chromosomes (W. Lukowitz, B. Fang, and C. Somerville, personal communication). These results are consistent with those for ttn5-1 and a location above erecta on chromosome 2. Complementation tests confirmed allelism when crosses between ttn5-1 and ttn5-2 heterozygotes produced siliques with ∼25% mutant seeds.
The ttn5 Phenotype
The ttn5 seed phenotype closely resembles that of ttn1 described by Liu and Meinke (1998). Embryo development in mutant seeds is limited to the formation of a few giant cells. In some cases, these embryo cells can exceed 150 μm in diameter. This is larger than a normal heart-stage embryo composed of several hundred cells. Giant nuclei and nucleoli are found in both the embryo and endosperm tissue. The number of endosperm nuclei is reduced, often by 10- to 20-fold, and endosperm cellularization is blocked. Heterozygous plants appear normal except for the production of 25% defective seeds. Terminal phenotypes of ttn5-1 and ttn5-2 mutant seeds are illustrated in Figure 1 and summarized in Table 1. Both alleles exhibit similar defects in seed development. The ttn5-2 allele may be slightly weaker, given its somewhat greater extent of embryo development.
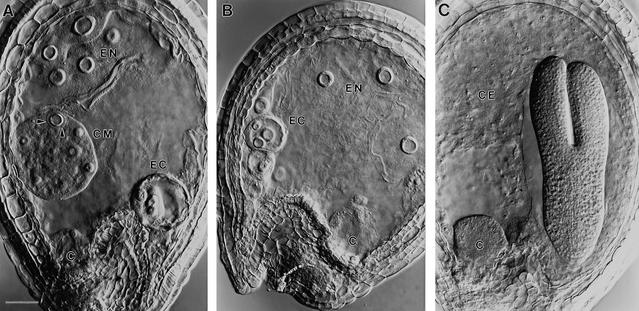
Figure 1.
Phenotype of Mutant Seeds Examined with Nomarski Optics.
Giant endosperm nucleoli (EN) and embryo cells (EC) are visible in cleared mutant seeds from heterozygous siliques at the torpedo stage of normal embryo development.
(A) ttn5-1 mutant seed with a large cytoplasmic mass (CM) near the chalazal (C) end. Arrowheads mark the edge of an endosperm nucleus.
(B) ttn5-2 mutant seed with enlarged nuclei in both the embryo and endosperm.
(C) Wild-type seed with an embryo at the torpedo stage, cellular endosperm tissue (CE), and prominent chalazal endosperm (C).
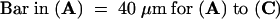 .
.
Table 1.
Phenotypic Characterization of Immature Mutant Seedsa
| Phenotypic Characterb | ttn5-1 | ttn5-2 |
|---|---|---|
| No. of seeds examined | 85 | 11 |
| No. of endosperm nucleoli | 27 (8–71) | 27 (12–53) |
| Largest endosperm nucleolus (μm) | 24 (12–41) | 21 (9–32) |
| No. of embryo cells | 2 (1–4) | 3 (2–5) |
| Largest embryo cell (μm) | 100 (48–166) | 71 (51–99) |
Cleared seeds at the cotyledon stage of normal development were examined with Nomarski optics. Numbers represent averages followed by range observed.
Wild-type seeds have ∼150 to 300 endosperm nuclei at the heart stage of development and endosperm nucleoli that average 4 μm in diameter (Wu, 1999). Normal embryo cell diameter is 8 μm. Embryo cell number includes the suspensor.
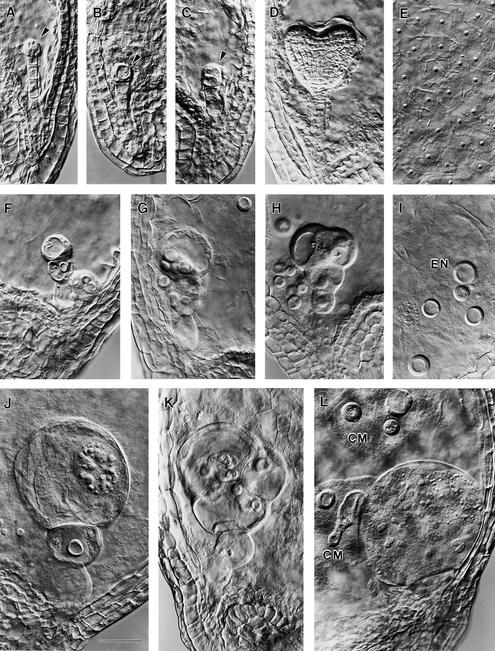
Nomarski images (see Methods) of cleared seeds at three stages of development are shown in Figure 2. Mutant seeds are first distinguished from wild type at the proembryo stage by abnormalities in both the embryo proper and the suspensor (Figures 2A to 2C). The mutant suspensor at this stage is short, has altered morphology, and contains fewer cells than normal. The embryo proper is delayed in division and has become vacuolated. The number of endosperm nuclei is also reduced. Unusual cell enlargement in the mutant embryo is apparent at the equivalent of the globular to heart stages of normal development (Figures 2F to 2H) and continues through subsequent cotyledon stages (Figures 2J to 2L). Giant embryo cells often appear to be multinucleate. Endosperm nuclei and nucleoli (Figures 2E and 2I) continue to enlarge throughout development.
Figure 2.
Developmental Changes in the Morphology of Mutant Seeds.
(A) to (C) Wild-type (A), ttn5-1 (B), and ttn5-2 (C) embryos at the proembryo stage of development. Arrowheads mark the embryo proper.
(D) and (E) Wild-type embryo (D) and endosperm (E) at the heart stage. Small dots in (E) correspond to endosperm nucleoli.
(F) to (H) ttn5-2 mutant embryos from siliques at the globular to heart stages of normal development.
(I) Endosperm tissue with prominent nucleoli (EN) present in the mutant seed shown in (H).
(J) to (L) ttn5-2 ([J] and [K]) and ttn5-1 (L) mutant embryos from siliques at a mature cotyledon stage of normal development. Two cytoplasmic masses (CM) with enlarged endosperm nucleoli are visible in (L) adjacent to the giant embryo cell.
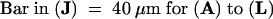 .
.
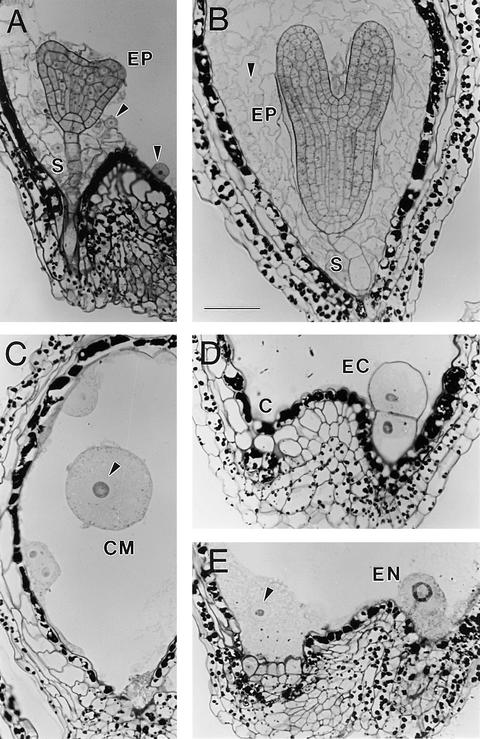
Light micrographs of sections through mutant seeds confirmed the phenotypes observed with Nomarski optics, including embryo cell enlargement and vacuolization, increase in nuclear volume, decrease in number of endosperm nuclei, and elimination of endosperm cellularization. Examples of such abnormalities are shown in Figure 3. One consistent difference between ttn1 and ttn5 phenotypes is the presence of chalazal endosperm in ttn5. This specialized mass of cytoplasm and endosperm nuclei is found at the chalazal pole of wild-type seeds and becomes a prominent feature at the globular to torpedo stages of development (Figure 1C). The chalazal endosperm is present but less prominent than normal in ttn5 mutant seeds (Figures 1A, 1B, and 3E) and is missing altogether in ttn1 mutant seeds (Liu and Meinke, 1998).
Figure 3.
Light Microscopy of Mutant and Wild-Type Seeds.
(A) and (B) Wild-type seeds at the heart (A) and torpedo (B) stages. The suspensor (S) attaches the embryo proper (EP) to the seed coat. Endosperm nucleoli are visible as dark dots surrounded by a pale nucleus (arrowheads) and variable amounts of cytoplasm.
(C) Part of a ttn5-1 mutant seed from a silique at the cotyledon stage of normal development. A giant endosperm nucleolus (dark inner circle) and nucleus (arrowhead) are visible within a large cytoplasmic mass (CM).
(D) and (E) Basal portion of a ttn5-1 mutant seed from a silique at the cotyledon stage of normal development. Vacuolated embryo cells (EC) and a large endosperm nucleus (EN) are visible at the micropylar end of the seed. A chalazal endosperm nucleus (arrowhead) and surrounding cytoplasm are found at the chalazal (C) pole.
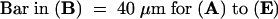 .
.
Cloning the TTN5 Gene
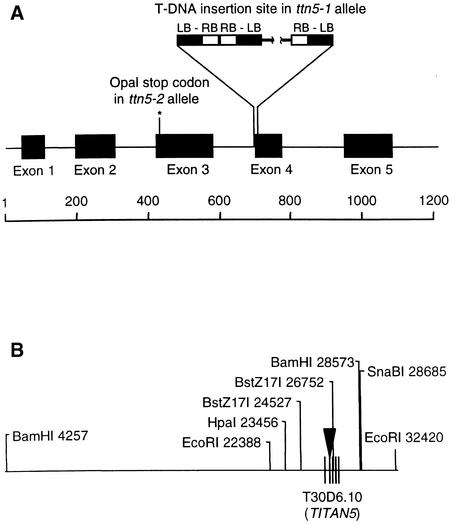
Right-border plasmid rescue was used to recover DNA sequences flanking the T-DNA insert in ttn5-1 heterozygotes. Two overlapping plasmids were recovered and found to contain GC-rich inserts with regions similar to bacterial sequences. Another fragment with similar features was obtained using an alternative cloning procedure. Two lines of evidence indicate that these inserts correspond to a portion of the Agrobacterium chromosome: (1) polymerase chain reaction (PCR) primers based on these sequences amplified a product from Agrobacterium DNA but not from wild-type plant DNA; and (2) DNA gel blots indicated the presence of these sequences in heterozygous plants but not in wild-type plants. In our current model, shown in Figure 4, ∼30 kb of Agrobacterium chromosomal DNA is present between T-DNA borders at the ttn5-1 locus.
Figure 4.
Gene Structure at the TTN5 Locus.
(A) Schematic representation of the TTN5 gene (T30D6.10) sequenced as part of the Arabidopsis Genome Initiative. Locations of exons and mutation sites in wild-type and mutant alleles are indicated. T-DNA left border (LB) and right border (RB) sequences at the ttn5-1 insertion site are shown. The complex insert contains several copies of T-DNA and ∼30 kb of Agrobacterium DNA (interrupted line). The location of the TGG-to-TGA mutation site in ttn5-2 is marked with an asterisk. Numbers on the scale bar correspond to base pairs.
(B) Map of genomic region flanking TTN5, indicating the relevant restriction sites used for DNA gel blot analysis and GenomeWalker cloning. Coordinates listed refer to numbered bases in BAC T30D6 (GenBank accession number AC006439). Some recognition sequences have been omitted for clarity. The triangle represents the T-DNA insertion site in ttn5-1. Vertical lines mark the locations of TTN5 exons. Note that TTN5 is in reverse-complement orientation relative to the BAC sequence. GenomeWalker clones were obtained from the T-DNA insertion site to the HpaI site at 23,456 bp, the BstZ17I site at 24,527 bp, and the SnaBI site at 28,685 bp.
Plant sequences flanking T-DNA left borders were then recovered using the GenomeWalker (Clontech) procedure. Two products derived from opposite ends of the T-DNA insert were found to match a portion of a bacterial artificial chromosome (BAC) chromosome 2 (T30D6) sequenced as part of the Arabidopsis Genome Initiative (www.tigr.org). Thirteen bases (positions 26,469 to 26,481) located between the two left border junction sites appear to have been deleted as a result of T-DNA insertion. Probing DNA gel blots with a PCR product amplified from this region, as illustrated in Figure 5, revealed a polymorphism between heterozygous and wild-type plants, consistent with the presence of a T-DNA insertion at the mutant locus.
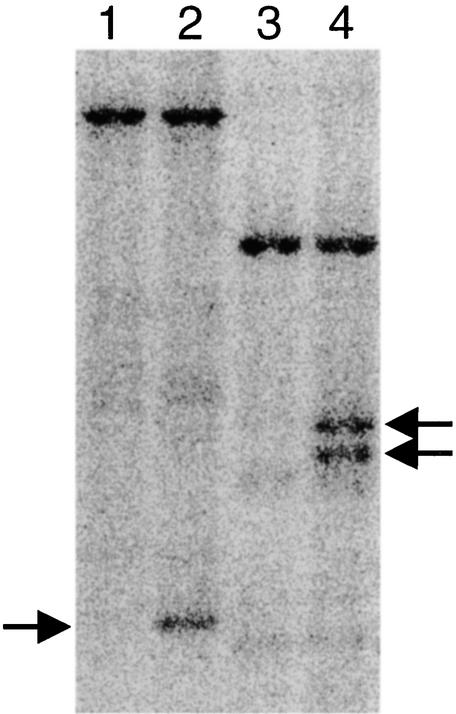
Figure 5.
DNA Gel Blot of Heterozygous and Wild-Type Plants.
Genomic DNA from wild-type plants (lanes 1 and 3) and ttn5-1 heterozygotes (lanes 2 and 4) was digested with either BamHI (lanes 1 and 2) or EcoRI (lanes 3 and 4) and probed with a 32P-labeled PCR product corresponding to the TTN5 gene (nucleotide coordinates 26,901 to 27,130 of BAC T30D6). The presence of polymorphic bands (arrows) indicates that the cloned region has been disrupted by T-DNA insertion. The lengths of hybridizing fragments are consistent with the restriction map shown in Figure 4. The predicted 22-kb polymorphic band in lane 2 cannot be resolved from the 24-kb (wild-type) band seen in lanes 1 and 2.
TTN5 Gene Organization
The TTN5 coding region was amplified from a yeast expression library (pFL61) prepared from Landsberg erecta seedlings (Minet et al., 1992) to confirm gene predictions for this region of the genome. The PCR products obtained by using primers flanking the start and stop codons were sequenced and found to match the product predicted from annotation of BAC T30D6. The TTN5 open reading frame encodes a predicted protein of 185 amino acids. The wild-type gene illustrated in Figure 4 is ∼1.1 kb long and contains five exons and four introns of similar sizes. The T-DNA insertion site in ttn5-1 is located near the 5′ end of the fourth exon.
Confirmation of Gene Identity
A second mutant allele was sequenced to confirm that disruption of the TTN5 locus was responsible for the mutant phenotype. Because tissue could not be obtained from ttn5 homozygotes, DNA was isolated from ttn5-2 heterozygotes, and the PCR products derived from mutant and wild-type alleles were sequenced together to look for polymorphisms. This approach is similar to that frequently used to detect sequence polymorphisms associated with human diseases (Barbetti et al., 1990; Bell et al., 1999). Molecular complementation was not attempted because with embryo-lethal mutations, this requires the additional step of crossing transformed wild-type plants with mutant heterozygotes and then looking for a disruption of the expected ratio of mutant seeds in the next generation. Such an extended approach will not meet the demands of high-throughput analysis of hundreds of EMB genes in the future. The direct sequencing method outlined here should therefore assist in the analysis of embryo-defective mutants and facilitate large-scale functional analysis of essential genes in Arabidopsis.
Two clear polymorphisms between the Landsberg (ttn5-2) and Columbia (ttn5-1 and BAC T30D6) sequences were found. Both were single-base changes located within introns. In addition, one ambiguous base was noted in the third exon, at nucleotide 26,738 in BAC T30D6. The uncertain (N) base call, as shown in Figure 6, resulted from the presence of peaks for both guanine and adenine. This polymorphism was found at the same location when both strands were sequenced and when PCR products from a second DNA sample were analyzed. These results are precisely what should be found if the mutation caused a G-to-A transition in this region of the TTN5 gene and there were equal amounts of wild-type (G) and mutant (A) sequences in PCR products derived from heterozygotes. The resulting G-to-A transition creates a premature stop codon near the 5′ end of the gene, at the location of amino acid 65 (tryptophan) in the protein. The presence of this nonsense mutation is consistent with the strong mutant phenotype.
Figure 6.
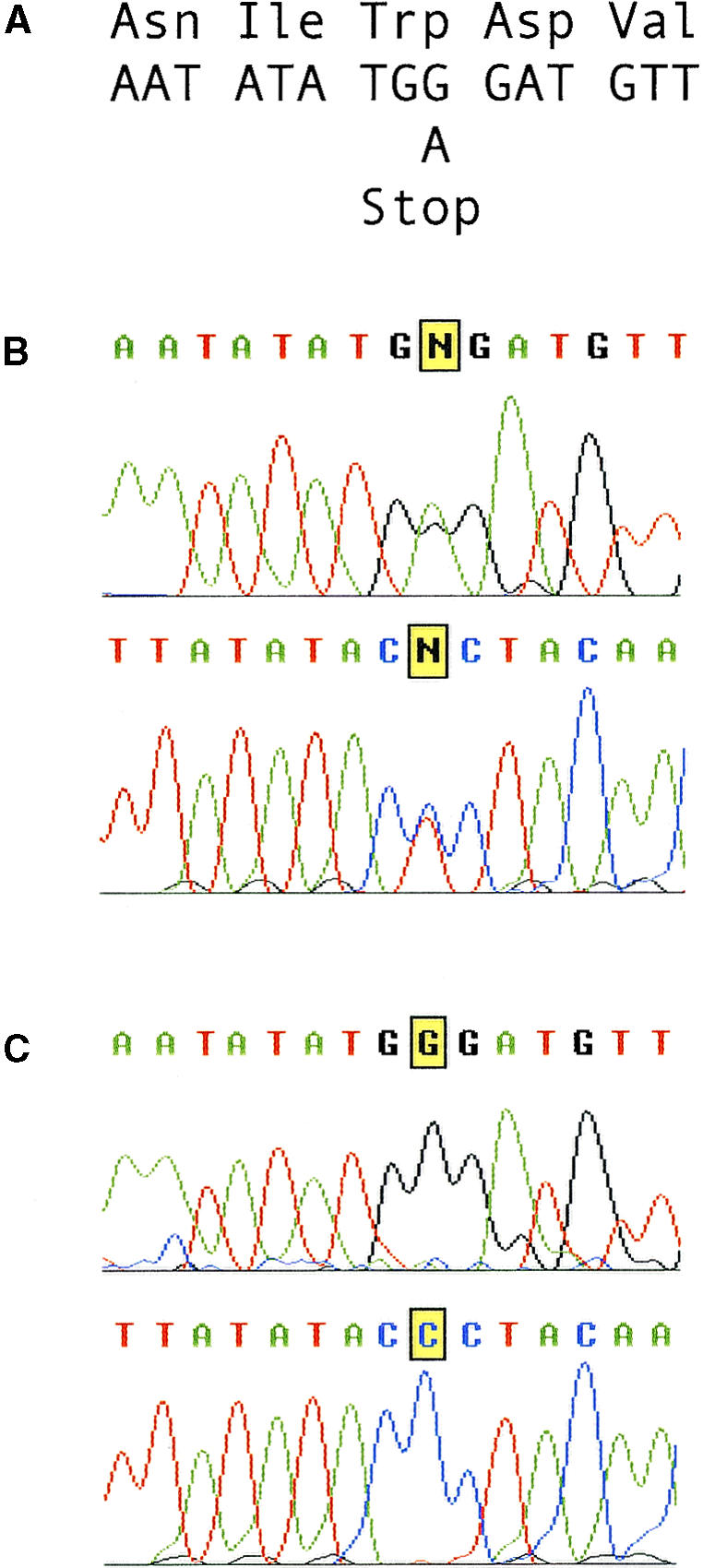
Direct Sequencing of PCR Products from Heterozygous (ttn5-2) and Wild-Type Genomic DNA.
(A) DNA and predicted amino acid sequence of the region around the mutation site in ttn5-2. The G-to-A transition in the mutant allele generates a premature opal stop codon.
(B) Sequence chromatograms from direct sequencing of PCR products derived from a ttn5-2 heterozygote. Results of sequencing both strands are presented. Note the highlighted (N) base call derived from an equal amount of wild-type and mutant products in the sequencing reaction. Small peaks for both A + G (top strand) and C + T (bottom strand) are visible at this position.
(C) Control chromatograms for the same region from wild-type sibling plants. Note the absence of any sequence ambiguities.
TTN5 Encodes an ARL Protein
RNA gel blots were first used to determine whether TTN5 expression could be detected in wild-type plants. Poly(A)+ RNA was isolated from 14-day-old seedlings and mature leaves and probed with the TTN5-derived PCR product described in Figure 5. Single hybridizing bands of the expected size (∼600 bp) were detected with both samples (data not shown). Reverse transcription (RT)–PCR with gene-specific primers was then used to confirm expression. The results shown in Figure 7 indicate that TTN5 is expressed in seedlings, leaves, roots, and inflorescences of wild-type plants. This gene therefore appears to function throughout the Arabidopsis life cycle.
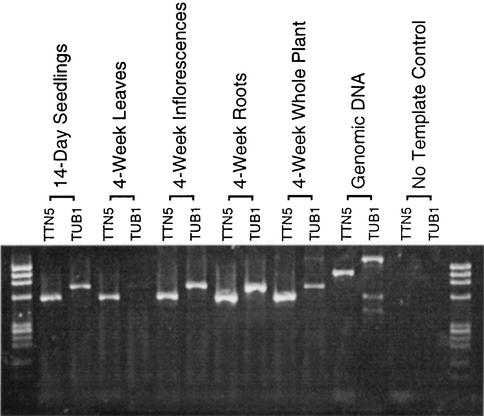
Figure 7.
DNA Gel Blot of RT-PCR Products Confirms TTN5 Expression.
The tubulin (TUB1) gene is included as a control. The expected product sizes for RT-PCR reactions are 570 (TTN5), 739 (TUB1), 1052 (TTN5 genomic DNA control), and 1654 bp (TUB1 genomic DNA control).
The original annotation for BAC T30D6 indicated that the predicted TTN5 gene product was related in sequence to ARFs. We used the TTN5 protein sequence to perform a more thorough analysis of the ARF gene family in Arabidopsis and related plants. The results of BLASTP searches against all GenBank sequences revealed that TTN5 is most closely related in sequence to the ARL2 subfamily of ARL proteins found in animals. These results are summarized in the phylogram shown in Figure 8. TTN5 has the conserved GTP binding domains expected for ARF proteins and exhibits 63% identity to human and mouse ARL2 and 62% identity to rat ARL2. The high degree of sequence conservation throughout the protein is shown in Figure 9A. The distinction between ARF and ARL proteins was originally based on functional assays in mammals and yeast but has become more blurred with the identification of additional sequences in other organisms. The top 10 BLASTP scores with TTN5 were nevertheless ARL2-related proteins, consistent with the conclusion that TTN5 represents the first example of a plant ARL2 homolog.
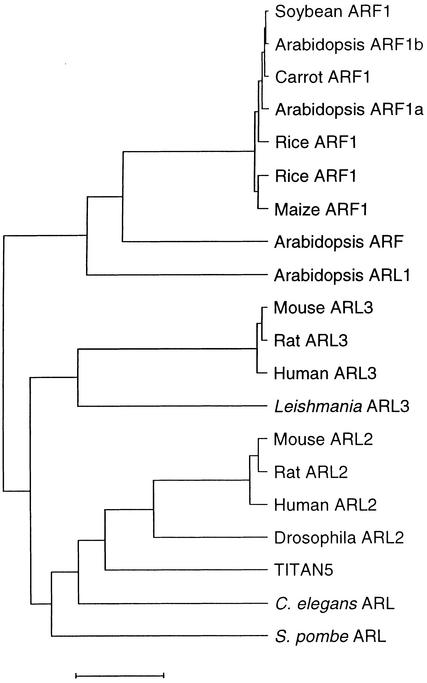
Figure 8.
Relationship between TTN5 and Selected ARF/ARL Proteins.
This GrowTree (Version 10.0; Genetics Computer Group, Madison, WI) phylogram was constructed using 10 representative protein sequences with high BLASTP scores to TTN5, four Arabidopsis sequences with high scores, and five high scores from other plants. Alignments were calculated using the blosum62 matrix, uncorrected distances, and the UPGMA tree-building method. Tree branch lengths are proportional to evolutionary distance, as indicated by the scale. Entries separated by short horizontal lines exhibit a high degree of sequence identity. Note that TTN5 falls within the ARL2 cluster of sequences, distinct from ARL3 and ARF1. Protein sequences and GenPept accession numbers used in the alignment are soybean ARF1, AAD17207; Arabidopsis ARF1b, P36397; carrot ARF1, P51822; Arabidopsis ARF1a, AAC98042; rice ARF1, P51823; rice ARF1, AAB65432; maize ARF1, P49076; Arabidopsis ARF, AAD26902; Arabidopsis ARL1, P40940; mouse ARL3, AAD33067; rat ARL3, P37996; human ARL3, NP_004302; Leishmania ARL3, CAA65780; mouse ARL2, AAD33908; rat ARL2, O08697; human ARL2, NP_001658; Drosophila ARL2, Q06849; TITAN5, AAD15498; Caenorhabditis elegans ARL, Q19705; and Schizosaccharomyces pombe ARL, Q09767.  .
.
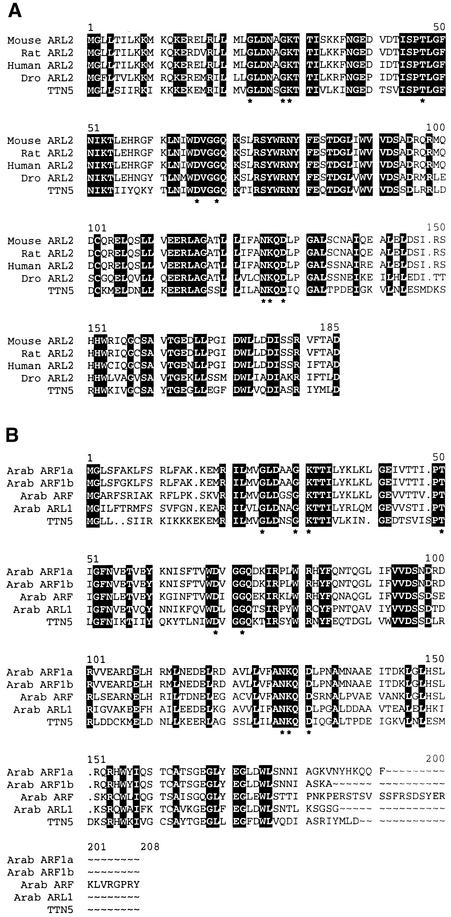
Figure 9.
Sequence Alignment of TTN5 with Related ARF and ARL Proteins.
Identical amino acids found in all proteins are highlighted with black background. Dots represent gaps introduced to maximize alignment (Genetics Computer Group Pileup Program; parameters set as in Figure 8). Asterisks mark the locations of conserved GTP binding domains (Bourne et al., 1991). Accession numbers are given in Figure 8.
(A) Alignment with ARL2 proteins identified in humans, mouse, rat, and Drosophila (Dro).
(B) Alignment with ARF and ARL proteins identified in Arabidopsis (Arab).
The ARF Family in Arabidopsis
TTN5 shares <50% sequence identity with every other plant ARF protein identified to date and, as shown in Figure 9B, clearly differs from several ARF proteins found in Arabidopsis. The ARF1 gene family exhibits a marked degree of sequence conservation in plants. Six ARF1 family members have been identified in Arabidopsis. One member (ARF1a) is located on chromosome 1 and encodes a predicted protein of 188 amino acids. Another member (ARF1b) is found on chromosome 2 and encodes a predicted protein of 181 amino acids. The sequence of ARF1b has been confirmed through isolation of a full-length cDNA (Regad et al., 1993). ARF1a appears to have a novel C-terminal extension (amino acids 179 to 188) not found in other ARF1 proteins, but only a single amino acid differentiates ARF1a and ARF1b in the remainder of the protein. This sequence conservation also extends to other plants. For example, Arabidopsis ARF1b differs from rice and carrot ARF1 proteins by only three amino acids. Four additional ARF1 family members have recently been identified through the Arabidopsis genome project. Three of these are located on chromosome 1 (BACs F20B4, F28C1, and F24J13) and one on chromosome 3 (BAC T17J13). The only differences identified among the six predicted proteins are the C-terminal extension and two amino acids located near the N terminus.
Two other ARF family members were found by searching Arabidopsis sequence databases. One (Arabidopsis ARF in Figure 8) is located on chromosome 2 (BAC F27O10) and is difficult to classify because it shares only 67% sequence identity with other plant ARF1 proteins, 44% identity with TTN5, and similar identities with a number of different ARF and ARL proteins from animal systems. Another Arabidopsis ARF sequence located on chromosome 2 (BAC F27C12) shares 47% identity with TTN5 and corresponds to a published cDNA sequence (Lebas and Axelos, 1994). This member was originally named ARF3 but appears to be most closely related (65% identity) to human and rat ARL1. We therefore believe this sequence corresponds to an Arabidopsis ARL1 protein. In addition, a putative GTPase-activating protein (GAP) that may associate with TTN5 has been identified on sequenced BAC F13M22 located on chromosome 2. Additional members of the ARF and GAP families may be uncovered when the Arabidopsis genome project is completed.
DISCUSSION
Many cellular processes in eukaryotes are regulated by small GTP binding proteins (Boguski and McCormick, 1993; Garcia-Ranea and Valencia, 1998; Hall, 1998). These monomeric GTPases alternate between active (GTP-bound) and inactive (GDP-bound) states, in part through association with GAPs and guanine exchange factors (GEFs). The active state interacts with membranes and effector molecules that function in a variety of signal transduction networks. The RAS superfamily of GTP binding proteins is divided into several families with diverse but overlapping functions: RAS (cell growth and differentiation), RHO/RAC/CDC42 (cell motility and polarity), RAN (nuclear protein import), RAB (vesicle transport), and ARF/SAR (vesicle formation). Although characterized most extensively in yeast and mammalian systems, RAS-like proteins have been identified in a wide range of organisms, and their cellular functions appear in general to be conserved.
GTP binding proteins have also been isolated and characterized in various plants (Ma, 1994; Bischoff et al., 1999). Recent studies have examined the role of RAC proteins in pollen tube growth (Kost et al., 1999), RAB proteins in desiccation tolerance (O'Mahony and Oliver, 1999) and intracellular protein trafficking (Sanderfoot and Raikhel, 1999), RAN proteins in nuclear protein import (Smith and Raikhel, 1999), and SAR proteins in vesicle formation (d'Enfert et al., 1992; Takeuchi et al., 1998). Although mutants defective in such proteins have been studied for many years in yeast, similar mutants have been difficult to identify in plants and have not contributed in a major way to our understanding of plant processes. The TTN5 gene described here illustrates how an Arabidopsis mutant with a novel phenotype can be used to examine not only the regulation of mitosis and cytokinesis during seed development but also the function of ARF-like proteins in plants and other eukaryotes.
The ARF proteins are conserved GTP binding proteins that function in vesicle formation and intracellular vesicle trafficking in yeast and mammalian systems (Chavrier and Goud, 1999). ARF proteins also stimulate phospholipase D activity (Brown et al., 1993; Martin, 1998) and serve as cofactors for ADP ribosylation of heterotrimeric G proteins by cholera toxin (Kahn and Gilman, 1984). The ARF family has been divided into ARF and ARL proteins on the basis of structural and functional characteristics. ARL proteins are unable to function as effective activators of cholera toxin or to rescue the lethal phenotype of yeast arf1 arf2 double mutants. Several ARF and ARL proteins have been examined in detail in yeast (Lee et al., 1997). Humans encode at least six ARFs and perhaps even more ARLs (Hong et al., 1998; Jacobs et al., 1999). ARF1 is localized to the Golgi complex and plays a well-documented role in coat assembly and vesicle budding (Schekman and Orci, 1996; Chavrier and Goud, 1999). In contrast, the functions of ARL proteins have remained elusive despite their identification in a wide range of organisms (Tamkun et al., 1991; Clark et al., 1993; Cavenagh et al., 1994; Lee et al., 1997; Jacobs et al., 1999).
Several examples of plant ARFs have been described in the literature (Bischoff et al., 1999) and uncovered through the Arabidopsis genome project (Meinke et al., 1998). These include ARF1 homologs from rice (Higo et al., 1994), carrot (Kiyosue and Shinozaki, 1995), maize (Verwoert et al., 1995), and Arabidopsis (Regad et al., 1993). The TTN5 gene product is most closely related in sequence to the ARL2 class of proteins. This class was originally identified in humans and Drosophila using degenerate primers based on conserved domains (Lee et al., 1997). Disruption of the ARL1 locus in Drosophila results in zygotic lethality (Tamkun et al., 1991). Mutants defective in ARL2 genes have not been described in any organism, and no plant ARL2 gene sequences other than TTN5 have been submitted to GenBank.
The role of ARF1 in vesicular transport in animal systems has been studied most extensively in the Golgi complex and endoplasmic reticulum. Vesicle formation appears to require conversion of ARF-GDP to ARF-GTP through interaction with the Sec7 domain of membrane-bound GEFs (Chavrier and Goud, 1999). The resulting change in ARF conformation facilitates recruitment of coatomer protein, leading to membrane deformation, budding, and vesicle release (Schekman and Orci, 1996). The inactive, GDP-bound form of ARF1 is later reconstituted through the action of GAPs. All known ARF GEFs contain Sec7 domains (Meacci et al., 1997; Togawa et al., 1999). Differences have been found in protein size and sensitivity to brefeldin A, a fungal toxin that disrupts Golgi structure and function. Other members of the ARF family appear to perform related cellular functions, although they differ somewhat in membrane binding specificity and associated protein cofactors. ARL proteins have been particularly difficult to analyze from a functional perspective (Hong et al., 1998; Sturm et al., 1998). No specific function has been assigned to ARLs from Drosophila, humans, rat, or mouse. Yeast ARL1 is not required for protein transport from the endoplasmic reticulum to the Golgi complex and, unlike Drosophila ARL1, is not required for cell viability (Lee et al., 1997). Despite their sequence diversity, ARLs probably function in some aspect of intracellular membrane trafficking.
Several features of the ttn5 embryo phenotype are consistent with the known functions of ARF proteins in other systems. Disruption of vesicle transport is known to interfere with cell plate formation and cytokinesis in plants, resulting in a failure to complete cell division. If accompanied by uncoupling of the cell cycle, this defect could account for the marked cell and nuclear enlargement observed in ttn5 mutant embryos. Two other embryo mutants of Arabidopsis (knolle and keule) appear to disrupt different steps in membrane trafficking (Assaad et al., 1996; Lukowitz et al., 1996). The identity of KEULE has not been published, but KNOLLE encodes a syntaxin required for vesicle transport during cytokinesis (Lauber et al., 1997). The embryo cell enlargement observed in knolle keule double mutants is consistent with the conclusion that a severe disruption of membrane transport mediated by ARL proteins could result in the formation of giant embryo cells. Alterations in the microtubule cytoskeleton observed in mutant pilz embryos (Mayer et al., 1999) may be an indirect consequence of changes in membrane transport systems and may contribute to the cell enlargement. The EMB30 (GNOM) gene, which is postulated to play an important role in embryo development, contains a Sec7 domain (Shevell et al., 1994; Busch et al., 1996) and may represent the GEF required for activation and localization of TTN5 within the membrane. The EMB30 protein has also been shown to be required for proper localization of the PIN1 auxin efflux carrier and normal pattern formation during embryo development (Steinmann et al., 1999).
The endosperm phenotype of ttn5 is somewhat more difficult to explain in light of known functions of ARF and ARL proteins. At least three different models appear to be consistent with the mutant phenotype. According to the first model, loss of TTN5 function disrupts the distribution of Golgi-derived materials critical for normal progression through mitosis but not for repeated entry into the cell cycle. This model assumes that Arabidopsis ARL2 (TTN5) performs a function in vesicular trafficking similar to that of ARF proteins examined in mammalian systems. Nuclear enlargement and the formation of giant nucleoli could then result from defects in a number of cellular processes that depend on normal Golgi function. The failure of pilz mutants to assemble microtubule networks could also be an indirect consequence of this more general defect in vesicle formation. An alternative model is that TTN5 associates with both the Golgi complex and the nuclear membrane. According to this model, TTN5 performs a secondary function distinct from that of other ARFs but not inconsistent with the sequence divergence observed between ARF and ARL proteins. A third model is that TTN5 functions not only in vesicular transport but also in signal transduction networks that play a more direct role in cytoskeletal organization and progression through mitosis. ARF1 stimulation of phospholipase D activity (Wakelam et al., 1997) and phosphatidylinositol-4,5-bisphosphate signaling pathways (Godi et al., 1999; Lorra and Huttner, 1999) illustrates how such proteins may have secondary effects on several cell processes.
Distinguishing between these models may require the isolation of a second mutant gene with a similar phenotype. Unfortunately, molecular cloning of TTN1 has been delayed by the absence of a tagged mutant allele and by a genome location that remains to be sequenced. One possibility is that TTN1 encodes a GEF or GAP that interacts with several ARF family members. This could explain the additional defect in endosperm nuclear migration observed in ttn1 mutant seeds. Differences in embryo cell morphology between ttn1/ttn5 and ttn2/ttn4 mutants also remain to be explained. With further advances in TTN gene isolation, however, valuable details should emerge about the molecular basis for the ttn phenotype and the nature of proteins required for normal progression through mitosis and cytokinesis during seed development.
METHODS
Plant Materials and Growth Conditions
T-DNA insertional mutants were generated in the Columbia ecotype of Arabidopsis thaliana according to the vacuum infiltration procedure (Bechtold and Pelletier, 1998). Progeny seeds were planted in soil, and transgenic plants were identified by their resistance to foliar application of 1.2 mM glufosinate ammonium (BASTA; Hoechst, Frankfurt, Germany). Transplanted BASTA-resistant plants were grown to maturity and scored for the presence of 25% defective seeds after self-pollination (Meinke, 1994). Heterozygous ttn5-1 plants used for subsequent analyses were maintained as described by Liu and Meinke (1998). The ttn5-2 allele was isolated by W. Lukowitz (Carnegie Institution of Washington, Stanford, CA) in the Landsberg erecta ecotype after seed mutagenesis with ethyl methanesulfonate. Seeds were imbibed in 0.3% ethyl methanesulfonate for 8 hr, and the resulting M1 plants were scored for defective seeds.
Genetic and Phenotypic Characterization
The tagging status of ttn5-1 was resolved by the strategy of Castle et al. (1993), except that selection involved growth on 50 μM glufosinate (Crescent Chemical Co., Hauppauge, NY) instead of kanamycin. Plants were scored for resistance 10 to 14 days after plating. Resistant plants were transplanted to soil and later screened for the presence of mutant seeds. Mapping of ttn5-1 with visible markers was performed as described by Franzmann et al. (1995). Approximately 160 F2 plants produced from a cross between ttn5-1 and DP23 (ch1, er, gl1, cer2, and tt3) were scored for the ttn5 and marker phenotypes to determine linkage. Mapping of ttn5-2 was performed at the Carnegie Institution by crossing heterozygotes with wild-type Columbia plants and scoring F2 plants for the cleaved amplified polymorphic sequence markers PHYB and THY1. For phenotypic characterizations, mutant seeds were cleared with Hoyers solution (7.5 g of gum Arabic, 100 g of chloral hydrate, and 5 mL of glycerol in 30 mL of water) and examined with a microscope (model BH-2; Olympus Optical Co., Tokyo, Japan) equipped with Nomarski optics, as described by Meinke (1994). Endosperm nuclei and embryo cells were counted and measured using an ocular micrometer and grid. Sectioned materials for confirmation of mutant phenotypes were prepared and examined as described by Liu and Meinke (1998).
Cloning of Tagged Mutant Allele
Genomic DNA was prepared using lyophilized tissue obtained from the aerial portions of heterozygous ttn5-2 plants, as described by Reiter et al. (1992). Plasmid rescue was performed as described by Castle et al. (1993) to obtain sequences adjacent to the T-DNA right border. The GenomeWalker protocol (Clontech, Palo Alto, CA) was used to recover sequences flanking the T-DNA left border. Two modifications were made to the instructions provided: (1) the blunt-end enzymes BstZ17I, HpaI, SnaBI, and StuI (New England Biolabs, Beverly, MA) were used because their recognition sites are common in Arabidopsis genomic DNA but are missing from the T-DNA vector; and (2) for primary polymerase chain reaction (PCR) amplification, the AP1 primer suggested by Clontech was modified to GTAATACGACTCACTATAGGGCA to avoid the 3′-terminal GC in the standard AP1 primer. A left border–specific primer (JM35) GCCTTTTCAGAA-ATGGATAAATAGCCTTGCTTCC was used in combination with the modified AP1 primer. Amplification was performed with Advantage Genomic Polymerase Mix (Clontech), cycling conditions described in the GenomeWalker protocol, and a thermocycler (model 9700; Perkin-Elmer) set at 9600 ramp rate. Secondary PCR was performed with a nested primer (JM34) specific to the left border (GCTTCCTATTATATCTTCCCAAATTACCAATACA) and the AP2 primer from Clontech. Products of this reaction were cloned using the TOPO T/A Cloning Kit (Invitrogen, Carlsbad, CA). DNA for four colonies per reaction was prepared using the Qiaprep Spin Miniprep kit (Qiagen, Valencia, CA), analyzed after EcoRI digestion and gel separation, and sequenced with an ABI Prism 377 using Big Dye terminator chemistry (PE Biosystems, Foster City, CA) and standard M13 forward and reverse primers.
Direct Sequencing of Second Mutant Allele
Vegetative tissue from ttn5-2 heterozygotes and wild-type siblings was frozen at −80°C and lyophilized. Genomic DNA was extracted from 10 mg of ground tissue using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) without the optional RNase step. This DNA was then used as a target for amplification with the Clontech Advantage 2 polymerase mix, which includes a proofreading polymerase to reduce the misincorporation rate, a Perkin-Elmer 9700 thermocycler set at maximum ramp rate, and the following primer pairs: DP390 (GACCATGGGACTGTTAAGCATAATC) and DP391 (GAGAGCTCTTAGTCAAGCATGTAAATCCTGGA), and DP390 in combination with JM93 (AGGCATAATGTACACTGATGAAGT-CGTCA). Cycling conditions were 95°C for 1 min followed by 30 cycles of 95°C for 15 sec, 55°C for 15 sec, and 68°C for 1.5 min, and ending with a 2-min dwell at 68°C and a 4°C hold. For each successful reaction, 1 μL of exonuclease I (10 units/μL; USB Corporation, Cleveland, OH) and 1 μL of shrimp alkaline phosphatase (1 unit/μL; USB Corporation) were added to 45 μL of reaction mix and incubated at 37°C for 60 min and then stopped by heating at 70°C for 10 min. Five microliters of this mixture was used as a template for direct sequencing according to standard ABI Big Dye terminator protocols. The following primers were used for sequencing: DP390, DP391, JM44 (TGCGTATTCTTATGGTGTATGTCA), JM64 (CGATAACCC-CTATGTCCAAATC), and JM66 (TCCGGTTATTTTCAGAACAATC). Sequences obtained for the amplified region were compared with those for bacterial artificial chromosome (BAC) T30D6 using Sequencher version 4.0 for PC (GeneCodes, Ann Arbor, MI). For additional details on the use of direct sequencing of PCR products in human genotyping using ABI automated sequencers, refer to http://www.pebio.com/ab/md/highres.html.
Gel Blot Hybridization
DNA gel blots used to analyze sequences adjacent to right-border and left-border T-DNA sequences and to confirm the presence of a sequence polymorphism at the TTN5 locus between heterozygous and wild-type plants were performed as described by Reiter et al. (1992). The PCR product used to probe the blot shown in Figure 5 was amplified from Columbia DNA using AmpliTaq (Perkin-Elmer) and standard cycling conditions with primers JM44 (TGCGTATTC-TTATGGTGTATGTCA) and JM45 (CATGTAAATCCTGGAGGCAATGTC), which flank the insertion site. RNA gel blots were performed as described by Patton et al. (1998). Poly(A)+ RNA was isolated from 100 μg of total RNA using the Micro FastTrack kit (Invitrogen).
Reverse Transcription–PCR Protocol
Approximately 0.5 g of plant tissue was ground in liquid nitrogen, and the total RNA was extracted with Trizol Reagent (Lifetech, Gaithersburg, MD) according to the manufacturer's protocol. Total RNA concentration was determined by UV absorbance at 260 nm. For each sample, 5 μg of total RNA was reverse-transcribed with an oligo dT18 primer (Ambion, Austin, TX) and Superscript II (Lifetech), as described in the Lifetech instructions. An additional incubation of 30 min at 50°C was added after the 1 hr at 42°C specified by the manufacturer. Five microliters of reaction products from each tissue was used as template in PCR reactions with the primer pairs DP390 and DP391 or TUB1F (CCACCGGACGTTACAAC) and TUB1R (CCA-CGGGAAGTGAGAGG). In control reactions, either 5 μL of water or 10 ng of genomic DNA in 5 μL of water was used as template. The reaction conditions were 0.05 unit/μL AmpliTaq, 200 nM of each dNTP, 1.5 mM MgCl2, PCR Buffer II (Perkin-Elmer), and 0.5 μM of each oligonucleotide in a 50-μL reaction volume. Cycling conditions were 95°C for 2 min followed by 25 cycles of 95°C for 15 sec, 50°C for 30 sec, 72°C for 2 min, and ending with a 5-min hold at 72°C. DNA gel analysis used 5 μL of each PCR reaction loaded on a 2% gel with Tris–borate–EDTA buffer.
Acknowledgments
We thank Wolfgang Lukowitz (Carnegie Institution of Washington, Stanford, CA) for providing seeds and mapping information for ttn5-2; Kelsey Smith, Justin Rineer, and Becky King for assistance with plant maintenance and genetic analysis at Oklahoma State University; An Hu (Novartis) for performing sequencing reactions; Greg Budzeszeweski and Joshua Levin (Novartis) for providing the RNA gel blot and TUB1 primers; and Iris Tzafrir (Oklahoma State University) for updated analysis of ARF sequences. Research on titan mutants in the Meinke laboratory is supported by a grant from the National Science Foundation, Developmental Mechanisms Program.
References
- Albert, S., Despres, B., Guilleminot, J., Bechtold, N., Pelletier, G., Delseny, M., and Devic, M. (1999). The EMB506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J. 17, 169–179. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Mayer, U., Wanner, G., and Jurgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253, 267–277. [DOI] [PubMed] [Google Scholar]
- Barbetti, F., Raben, N., Kadowaki, T., Cama, A., Accili, D., Gabbay, K.H., Merenich, J.A., Taylor, S.I., and Roth, J. (1990). Two unrelated patients with familial hyperproinsulinemia due to a mutation substituting histidine for arginine at position 65 in the proinsulin molecule: Identification of the mutation by direct sequencing of genomic deoxyribonucleic acid amplified by polymerase chain reaction. J. Clin. Endocrinol. Metab. 71, 164–169. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bell, D.W., et al. (1999). Heterozygous germ line hCHK2 mutations in Li–Fraumeni syndrome. Science 286, 2528–2531. [DOI] [PubMed] [Google Scholar]
- Berger, F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2, 28–32. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Molendijk, A., Rajendrakumar, C.S., and Palme, K. (1999). GTP-binding proteins in plants. Cell Mol. Life Sci. 55, 233–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., and McCormick, F. (1993). Proteins regulating Ras and its relatives. Nature 366, 643–654. [DOI] [PubMed] [Google Scholar]
- Bourne, H.R., Sanders, D.A., and McCormick, F. (1991). The GTPase superfamily: Conserved structure and molecular mechanism. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brown, H.A., Gutowski, S., Moomaw, C.R., Slaughter, C., and Sternweis, P.C. (1993). ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell 75, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Busch, M., Mayer, U., and Jurgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation gene GNOM: Gene structure and intragenic complementation. Mol. Gen. Genet. 250, 681–691. [DOI] [PubMed] [Google Scholar]
- Castle, L.A., Errampalli, D., Atherton, T.L., Franzmann, L.H., Yoon, E.S., and Meinke, D.W. (1993). Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 241, 504–514. [DOI] [PubMed] [Google Scholar]
- Cavenagh, M.M., Breiner, M., Schurmann, A., Rosenwald, A.G., Terui, T., Zhang, C.J., Randazzo, P.A., Adams, M., Joost, H.G., and Kahn, R.A. (1994). ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J. Biol. Chem. 269, 18937–18942. [PubMed] [Google Scholar]
- Chavrier, P., and Goud, B. (1999). The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466–475. [DOI] [PubMed] [Google Scholar]
- Clark, J., Moore, L., Krasinskas, A., Way, J., Battey, J., Tamkun, J., and Kahn, R.A. (1993). Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: Cloning of additional human and Drosophila ARF-like genes. Proc. Natl. Acad. Sci. USA 90, 8952–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert, C., Gensse, M., and Gaillardin, C. (1992). Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. EMBO J. 11, 4205–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann, L.H., Yoon, E.S., and Meinke, D.W. (1995). Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 7, 341–350. [Google Scholar]
- Garcia-Ranea, J.A., and Valencia, A. (1998). Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 434, 219–225. [DOI] [PubMed] [Google Scholar]
- Godi, A., Pertile, P., Meyers, R., Marra, P., Di Tullio, G., Iurisci, C., Luini, A., Corda, D., and De Matteis, M.A. (1999). ARF mediates recruitment of PtdIns-4-OH kinase-B and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1, 280–287. [DOI] [PubMed] [Google Scholar]
- Goldberg, R.B., de Paiva, G., and Yadegari, R. (1994). Plant embryogenesis: Zygote to seed. Science 266, 605–614. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509–514. [DOI] [PubMed] [Google Scholar]
- Higo, H., Kishimoto, N., Saito, A., and Higo, K.I. (1994). Molecular cloning and characterization of a cDNA encoding a small GTP-binding protein related to mammalian ADP-ribosylation factor from rice. Plant Sci. 100, 41–49. [Google Scholar]
- Hirano, T. (1999). SMC-mediated chromosome mechanics: A conserved scheme from bacteria to vertebrates? Genes Dev. 13, 11–19. [DOI] [PubMed] [Google Scholar]
- Hong, J.X., Lee, F.J.S., Patton, W.A., Lin, C.Y., Moss, J., and Vaughan, M. (1998). Phospholipid- and GTP-dependent activation of cholera toxin and phospholipase D by human ADP-ribosylation factor-like protein 1 (HARL1). J. Biol. Chem. 273, 15872–15876. [DOI] [PubMed] [Google Scholar]
- Jacobs, S., Schilf, C., Fliegert, F., Koling, S., Weber, Y., Schurmann, A., and Joost, H.G. (1999). ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterized by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett. 456, 384–388. [DOI] [PubMed] [Google Scholar]
- Jurgens, G., Torres Ruiz, R.A., and Berleth, T. (1994). Embryonic pattern formation in flowering plants. Annu. Rev. Genet. 28, 351–371. [DOI] [PubMed] [Google Scholar]
- Kahn, R.A., and Gilman, A.G. (1984). Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J. Biol. Chem. 259, 6228–6234. [PubMed] [Google Scholar]
- Kiyosue, T., and Shinozaki, K. (1995). Cloning of a carrot cDNA for a member of the family of ADP-ribosylation factors (ARFs) and characterization of the binding of nucleotides by its product after expression in E. coli. Plant Cell Physiol. 36, 849–856. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jurgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebas, M., and Axelos, M. (1994). A cDNA encoding a new GTP-binding protein of the ADP-ribosylation factor family from Arabidopsis. Plant Physiol. 106, 809–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, F.J.S., Huang, C.F., Yu, W.L., Buu, L.M., Lin, C.Y., Huang, M.C., Moss, J., and Vaughan, M. (1997). Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J. Biol. Chem. 272, 30998–31005. [DOI] [PubMed] [Google Scholar]
- Li, Z., and Thomas, T.L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10, 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16, 21–31. [DOI] [PubMed] [Google Scholar]
- Lorra, C., and Huttner, W.B. (1999). The mesh hypothesis of Golgi dynamics. Nat. Cell Biol. 1, E113–E115. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jurgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. (1994). GTP-binding proteins in plants: New members of an old family. Plant Mol. Biol. 26, 1611–1636. [DOI] [PubMed] [Google Scholar]
- Martin, T.F.J.. (1998). Phosphoinositide lipids as signaling molecules: Common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14, 231–264. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inze, D., and Jurgens, G. (1999). Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78, 100–108. [DOI] [PubMed] [Google Scholar]
- Meacci, E., Tsai, S.C., Adamik, R., Moss, J., and Vaughan, M. (1997). Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc. Natl. Acad. Sci. USA 94, 1745–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke, D.W. (1994). Seed development in Arabidopsis thaliana. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 253–295.
- Meinke, D.W. (1995). Molecular genetics of plant embryogenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 369–394. [Google Scholar]
- Meinke, D.W., Cherry, J.M., Dean, C., Rounsley, S.D., and Koornneef, M. (1998). Arabidopsis thaliana: A model plant for genome analysis. Science 282, 662–682. [DOI] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony, P.J., and Oliver, M.J. (1999). Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation-tolerant grass Sporobolus stapfianus. Plant Mol. Biol. 39, 809–821. [DOI] [PubMed] [Google Scholar]
- Patton, D.A., Schetter, A.L., Franzmann, L.H., Nelson, K., Ward, E.R., and Meinke, D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D. (1999). Chromatin silencing and Arabidopsis development: A role for polycomb proteins. Plant Cell 11, 765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad, F., Bardet, C., Tremousaygue, D., Moisan, A., Lescure, B., and Axelos, M. (1993). cDNA cloning and expression of an Arabidopsis GTP-binding protein of the ARF family. FEBS Lett. 316, 133–136. [DOI] [PubMed] [Google Scholar]
- Reiter, R.S., Young, R.M., and Scolnik, P.A. (1992). Genetic linkage of the Arabidopsis genome: Methods for mapping with recombinant inbreds and random amplified polymorphic DNAs (RAPDs). In Methods in Arabidopsis Research, C. Koncz, N.H. Chua, and J. Schell, eds (Singapore: World Scientific Publishing), pp. 170–190.
- Sanderfoot, A.A., and Raikhel, N.V.. (1999). The specificity of vesicle trafficking: Coat proteins and SNAREs. Plant Cell 11, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman, R., and Orci, L. (1996). Coat proteins and vesicle budding. Science 271, 1526–1533. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.M., Gilmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H.. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Smith, H.M.S., and Raikhel, N.V. (1999). Protein targeting to the nuclear pore. What can we learn from plants? Plant Physiol. 119, 1157–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Sturm, N.R., Van Valkenburgh, H., Kahn, R.A., and Campbell, D.A. (1998). Characterization of a GTP-binding protein in the ADP-ribosylation factor subfamily from Leishmania tarentolae. Biochim. Biophys. Acta 1442, 347–352. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M., Tada, M., Saito, C., Yashiroda, H., and Nakano, A. (1998). Isolation of a tobacco cDNA encoding Sar1 GTPase and analysis of its dominant mutations in vesicular traffic using a yeast complementation system. Plant Cell Physiol. 39, 590–599. [DOI] [PubMed] [Google Scholar]
- Tamkun, J.W., Kahn, R.A., Kissinger, M., Brizuela, B.J., Rulka, C., Scott, M.P., and Kennison, J.A. (1991). The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA 88, 3120–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa, A., Morinaga, N., Ogasawara, M., Moss, J., and Vaughan, M. (1999). Purification and cloning of a brefeldin A–inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J. Biol. Chem. 274, 12308–12315. [DOI] [PubMed] [Google Scholar]
- Uwer, U., Willmitzer, L., and Altmann, T. (1998). Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoert, I.I.G.S., Brown, A., Slabas, A.R., and Stuitje, A.R. (1995). A Zea mays GTP-binding protein of the ARF family complements an Escherichia coli mutant with a temperature-sensitive malonyl-coenzyme A:acyl carrier protein transacylase. Plant Mol. Biol. 27, 629–633. [DOI] [PubMed] [Google Scholar]
- Wakelam, M.J., Hodgkin, M.N., Martin, A., and Saqib, K. (1997). Phospholipase D. Semin. Cell Dev. Biol. 8, 305–310. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q. (1999). Mutant Analysis of Endosperm Development. M.S. Dissertation (Stillwater, OK: Oklahoma State University).