Abstract
The plasma membrane of eukaryotic cells differs in lipid composition from most of the internal organelles, presumably reflecting differences in many of its functions. In particular, the plasma membrane is rich in sphingolipids and sterols, one property of which is to decrease the permeability and increase the thickness of lipid bilayers. In this paper, we examine the length of transmembrane domains throughout the yeast secretory pathway. Although the transmembrane domains of cis and medial Golgi residents are similar to those of endoplasmic reticulum proteins, these domains lengthen substantially beyond the medial Golgi, suggesting a thickening of the bilayer. Yeast sphingolipids have particularly long acyl chains, and Aur1p, the inositol phosphorylceramide synthase that initiates yeast sphingolipid synthesis, was found to be located in the Golgi apparatus by both immunofluorescence and membrane fractionation, with its active site apparently in the Golgi lumen. Thus, it appears that sphingolipid synthesis in yeast takes place in the Golgi, separated from glycerophospholipid synthesis in the endoplasmic reticulum. A similar separation has been found in mammalian cells, and this conservation suggests that such an arrangement of enzymes within the secretory pathway could be important for the creation of bilayers of different thickness within the cell.
INTRODUCTION
The membranes of eukaryotic cells vary in their lipid composition, both between the membranes of different organelles and between the two sides of particular membranes (van Meer, 1989). This diversity presumably reflects the differing functional roles of the membranes beyond providing a basic hydrophobic bilayer. Differences in permeability, fluidity, microdomain heterogeneity, and surface charge will allow each bilayer to accommodate different biochemical processes, to provide varying ease of passage to small molecules, and to recruit specific subsets of cytosolic proteins to provide signaling and structural functions. One example of lipid heterogeneity is the high levels of sterols and sphingolipids found at the plasma membrane compared with many of the internal membranes (Patton and Lester, 1991; Hechtberger et al., 1994). These lipids are believed to restrict acyl chain mobility, increase lateral compaction of lipids, and thicken the bilayer, all of which contribute to reducing the permeability of the cell's outer membrane.
How such lipid heterogeneity within the cell is established and maintained is largely unresolved. In those eukaryotes examined so far, phospholipids are mostly synthesized in the endoplasmic reticulum (ER) and associated mitochondria, although some of the enzymes involved have not yet been localized precisely (Kent, 1995; Kohlwein et al., 1996; Vance, 1998). Likewise, sterols such as cholesterol are synthesized in the ER, with mammalian cells also having the capacity to generate cholesterol in lysosomes by hydrolysis of endocytosed cholesterol esters. However, not only do the relative levels of the different phospholipids vary between different post-ER compartments, but sterols are also found at higher levels in the plasma membrane than in their site of synthesis in the ER (Lange et al., 1989). This implies that mechanisms must exist for the selective sorting of lipids in the secretory pathway, most obviously the selective anterograde transport of sterols and certain classes of phospholipids (van Meer, 1989, 1998). The transport vesicles that connect compartments provide one opportunity for lipid transport, and it is also possible that nonvesicular mechanisms can contribute to the movement of lipids between organelles. Thus, it has been proposed that cholesterol can move from its site of synthesis in the ER to the plasma membrane in a nonvesicular manner, although this is controversial (Liscum and Underwood, 1995).
The third class of abundant lipids are the sphingolipids, which have a backbone of ceramide rather than diacylglycerol. In mammalian cells, the major sphingolipids are sphingomyelin and the glycolipids, and although these are most abundant in the plasma membrane, both sphingomyelin and glucosylceramide (the precursor of most glycolipids) are synthesized in the Golgi apparatus from ceramide that is synthesized in the ER (Futerman et al., 1990; Jeckel et al., 1990, 1992; Mandon et al., 1992; Hirschberg et al., 1993). In the yeast Saccharomyces cerevisiae, sphingolipids comprise inositol phosphorylceramide (IPC) and its mannosylated derivatives, and, as in mammalian cells, these lipids are most abundant in the plasma membrane (Patton and Lester, 1991; Hechtberger et al., 1994; Daum et al., 1998). However, in contrast to the situation in mammalian cells, in yeast IPC synthesis is believed to occur in the ER (Futerman, 1995; Daum et al., 1998; van Meer, 1998; Dickson and Lester, 1999). This is based on the fact that IPC synthesis from the ER-derived precursors ceramide and phosphatidylinositol (PI) continues even when ER-to-Golgi vesicle transport is blocked by use of temperature-sensitive sec mutations (Puoti et al., 1991).
The site of sphingolipid synthesis within the cell may have several important biological consequences. First, sterols have an apparent affinity for sphingolipids, and it has been proposed that sphingolipid synthesis could initiate the formation of domains rich in sterols and sphingolipids, which would drive anterograde cholesterol transport, or even the organization of domains within the plasma membrane (Brown, 1998). Second, there is an increasing interest in the possibility that lipid microheterogeneity could have a role in the sorting and compartmentalization of proteins (Recktenwald and McConnell, 1981; Simons and van Meer, 1988; Brown and London, 1998). Thus, it has been proposed that lipid composition could contribute to sorting between the apical and basal-lateral surfaces of polarized cells, to sorting during ER exit, and to sorting to the internal membranes of endosomes (Sutterlin et al., 1997; David et al., 1998; Kobayashi et al., 1998). In addition, we have proposed that sphingolipids and sterols could play a role in the sorting of proteins in the Golgi apparatus (Bretscher and Munro, 1993). We observed that in mammalian cells, the transmembrane domains (TMDs) of Golgi proteins are on average five residues shorter than those of plasma membrane proteins. This led to the suggestion that the shorter TMDs of Golgi enzymes could help exclude them from the carriers rich in cholesterol and sphingolipids destined for the plasma membrane.
Thus, the location of sphingolipid-synthesizing enzymes to the Golgi in mammalian cells may have an important role both in maintaining a distribution of sterols and sphingolipids that is distinct from that of phospholipids and in protein sorting. However, the apparent difference in the spatial organization of sphingolipid synthesis between yeast and mammalian cells would undermine a potential role in the fundamental sorting processes conserved between eukaryotes (Dickson, 1998). To investigate this further, we have examined in detail the location of IPC synthesis in yeast. IPC is made by the transfer of the head group of PI to ceramide, and IPC synthase, the enzyme responsible for this activity, is encoded by the AUR1 gene (Nagiec et al., 1997; Dickson and Lester, 1999). Nonetheless, the intracellular location of the protein product of the AUR1 gene has not yet been reported. We initially used TMD length to examine bilayer thickness through the secretory pathway of yeast and found that the bilayer appears to be of constant thickness until late in the Golgi apparatus, and then it is apparently more than 50% thicker in the plasma membrane, an even larger increase than that found in mammalian cells. Yeast sphingolipids are notable for having 26 carbon fatty acyl chains, which are considerably longer than those found in both yeast glycerophospholipids and many mammalian sphingolipids. We then examined the distribution of Aur1p and found that it is localized primarily in the Golgi and not in the ER. These results suggest that bilayer thickening in the secretory pathway, and the separation of phospholipid synthesis in the ER from sphingolipid synthesis in the Golgi, is a conserved feature of eukaryotic cells.
MATERIALS AND METHODS
Strains, Plasmids, and Antibodies
Yeast strains are listed in Table 1. Aur1p (YKL004w) was tagged at its COOH terminus with either three copies of the hemagglutinin (HA) epitope tag or protein A with the use of the PCR knock-in approach (Wach et al., 1997). Plasmid p3xHA-HIS5 was modified by the insertion of the ADH1 terminator downstream of the HA tags to create p3xHAt-HIS5 (Jungmann et al., 1999). Protein A fusions, comprising two copies of a Z domain separated from the reading frame by a cleavage site for the tobacco etch virus (TEV) protease, were created with the use of plasmid pZZ-HIS5 (Rayner and Munro, 1998). SEC7 was tagged with green fluorescent protein (GFP) at its COOH terminus with the use of integration plasmid pUSE-URA3 (Seron et al., 1998). Mnt1p tagged at the COOH terminus with three copies of the myc epitope was expressed from its own promoter in CEN plasmid pM3 M-416. Myc-tagged Emp47p was expressed from its own promoter with the use of either Myc-EMP47, a LEU2 integration plasmid, or pEmpM-416, a CEN URA plasmid (Schröder et al., 1995; Lewis and Pelham, 1996). The HA epitope tag was detected with either rabbit anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA), rat mAb 3F10 (Roche, Basel, Switzerland), or mouse mAb 12CA5, and the myc tag was detected with mouse mAb 9E10. Other antibodies were mouse mAbs against Vma1p (Molecular Probes, Eugene, OR) and BiP (2E7; Napier et al., 1992) and rabbit antisera against Anp1p (Jungmann and Munro, 1998), Vam3p and Tlg1p (Holthuis et al., 1998), Bet1p (M.J. Lewis, MRC-LMB, Cambridge UK), and Sec61p (C.J. Stirling, University of Manchester, United Kingdom). Protein blots were probed with peroxidase-coupled secondary antibodies (Bio-Rad, Richmond, CA), except for protein A–tagged proteins, which were detected with the use of peroxidase-antiperoxidase complexes (DAKO, Carpenteria, CA); immunofluorescence was with Alexa 488 or Alexa 568 conjugates (Molecular Probes).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα ura3-52 his3-Δ200 leu2-3,112 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 |
| SEY6211 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ901 ade2-101 suc2-Δ9 | Robinson et al., 1988 |
| AUR1-HA | SEY6210 aur1::AUR1 3xHA HIS5Sp | This work |
| AUR1-ZZ | SEY6210 aur1::AUR1 ZZ HIS5Sp | This work |
| VAN1-ZZ | SEY6210 van1::VAN1 ZZ HIS5Sp | Jungmann et al., 1999 |
| TLY138 | SEY6210 aur1::AUR1 3xHA HIS5Sp and pEmpM-416 | This work |
| TLY376 | SEY6210 aur1::AUR1 3xHA, HIS5Sp sec7::SEC7 GFP URA3 | This work |
| JWY47 | SEY6211 sec6-4 | J. Whyte |
| RSY282 | MATa sec23-1 ura3-52 leu2-3,112 his4-619 | R. Schekman |
| TLY137 | RSY282 leu2::EMP47-myc LEU2, aur1::AUR1 3xHA URA3Kl | This work |
Fractionation of Membranes by Centrifugation
Yeast organelles were separated on sucrose velocity gradients essentially as described by Antebi and Fink (1992), except that spheroplasted cells were lysed by freeze-thaw and passage through a syringe needle (Baker et al., 1988; Antebi and Fink, 1992). Thus, 1 l of log-phase cells [OD = 1 (600 nm)] was harvested by centrifugation (1000 × g for 2.5 min), washed in 100 ml of water, resuspended in 50 ml of 100 mM Tris, pH 9.4, 10 mM DTT, and incubated at 30°C for 10 min. Cells were pelleted (1000 × g for 10 min) and resuspended in 50 ml of spheroplasting buffer (0.6 M sorbitol, 50 mM Tris, pH 7.4, 10 mM DTT), 0.35 mg of oxalyticase (Enzogenetics, Corvallis, OR) was added, and the cells were incubated at 30°C until <20% were lysis resistant (typically 20 min). The spheroplasts were harvested (1000 × g for 5 min), washed twice with 30 ml of ice-cold freezing buffer (0.4 M sorbitol, 20 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8, 150 mM potassium acetate, 2 mM magnesium acetate), resuspended in the same at 130 OD units (600 nm)/ml, and frozen in 200-μl aliquots above liquid nitrogen, and the tubes were stored at −70°C until required.
For fractionation, 6 ml of spheroplasts was thawed, combined in three tubes, and pelleted in a microfuge (10,000 × g, 10 s), and the pellet was washed twice in 1 ml of permeabilizing buffer (20 mM HEPES, pH 6.8, 150 mM potassium acetate, 50 mM sorbitol, 5 mM magnesium acetate) and then lysed in lysis buffer (0.8 M sorbitol, 10 mM triethanolamine acetate, pH 7.6, 1 mM EDTA) by passage four times through a 26-gauge needle, followed by incubation on ice for 5 min. Unlysed cells were pelleted (10,000 × g, 5 min), and the supernatant was removed and saved. This lysis step was repeated twice more. All of the supernatants were pooled (S1) and protease inhibitors were added (1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mM PMSF) and then centrifuged (10,000 × g, 10 min), yielding a final supernatant (S2) and a pellet containing large debris. Typically, 60–70% of the Aur1p and IPC synthase activity was left after this initial centrifugation. Similar yields were seen for other Golgi markers such as Anp1p and Tlg1p.
One milliliter of cell supernatant (S2) was loaded onto sucrose gradients of 1-ml steps of 22, 26, 30, 34, 38, 42, 46, 50, 54, and 60% (wt/vol) sucrose and then 1 ml of 60% (wt/wt) sucrose, all in 10 mM HEPES, pH 7.5, 1 mM MgCl2. Gradients were spun for 2.5 h at 37,100 rpm in a SW40Ti rotor at 4°C, and 20 fractions (0.66 ml) were removed by pipette from top to bottom and stored at −20°C after addition of protease inhibitors. For analysis by protein blotting, membranes were pelleted either by centrifugation (100,000 × g, 30 min) or methanol/chloroform precipitation and resuspended in SDS sample buffer by sonication and vortexing. For SDS-PAGE, samples were warmed only to 37°C to prevent precipitation of polytopic membrane proteins.
Immunofluorescence Microscopy
Yeast growing in log phase was fixed in 3.7% formaldehyde for 45 min (except Sec7p-GFP strains, which were fixed for 15 min). Growth media were supplemented with 5 mM d-myo-inositol to maximize expression of Aur1p (Ko et al., 1994). Fixed cells were spheroplasted with Glusulase (1000 U/ml; New England Nuclear, Boston, MA) and Zymolyase 20T (100 μg/ml; ICN Biomedical, Costa Mesa, CA) at 30°C for 90 min, applied to poly-l-lysine–coated slides, fixed with methanol and then acetone (−20°C; 300 and 30 s, respectively), and probed with antibodies as described previously (Holthuis et al., 1998).
Assay of IPC Synthase Activity In Vitro
To assay for modification of C6-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) ceramide (C6-NBD-ceramide), 60 μl of gradient fractions was incubated with an equal volume of reaction mix (10 μM C6-NBD-ceramide [Molecular Probes; 4 mM stock solution in DMSO], 10 mg/ml defatted BSA, 1 mM PI, 2 mM 3-([3-chloramidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate, 2 mM manganese chloride, 2 mM magnesium chloride, 250 mM sucrose, 10 mM HEPES, pH 7.2, 1 mM EDTA) for 4 h at 30°C. If present, aureobasidin A (Takara Biomedicals, Shiga, Japan) was added from a 5 mg/ml stock in ethanol. The lipids were then extracted by sequential additions of 400 μl of a 4:10:1 mixture of chloroform:methanol:1 M HCl, 50 μl of 1 M HCl, and finally 100 μl of chloroform to achieve phase separation. The chloroform layer was collected after centrifugation for 5 min at 13,000 × g, dried down, resuspended in 15 μl of chloroform, and run out on a Silica Gel thin-layer chromatography (TLC) plate (Whatman, Clifton, NJ) in an 11:9:2 mixture of chloroform:methanol:30 mM potassium chloride. Fluorescent bands on the TLC plates were quantified with the use of an ARTHUR multiple wavelength fluorescence imager (Wallac, Perkin Elmer, Gaithersburg, MD) with excitation at 480 nm and emission at 530 nm.
C6-NBD-Ceramide Labeling of Live Yeast
Yeast in log phase was pelleted and resuspended at 5 OD (600 nm)/ml in minimal complete medium and preincubated with or without aureobasidin A (AbA) for 10 min at 30°C. Then, defatted BSA (5 mg/ml) and C6-NBD-ceramide (20 μM) were added and the cells were incubated at 30°C for another 20 min, followed by pelleting the cells, washing in ice-cold medium, and back-extracting for 1 h at 4°C in medium containing 5 mg/ml defatted BSA. NBD-labeled lipids were extracted from cell medium as for the in vitro assay (see above) and from whole cells by resuspending pelleted cells directly in 500 μl of a 5:12:4 mixture of chloroform:methanol:1 M HCl and vortexing for 5 min with glass beads (425–600 μm). After centrifugation (10,000 × g, 5 min), supernatants were transferred to fresh tubes, 125 μl of chloroform was added to achieve phase separation, and lipids in the chloroform layer were treated as described above. For fluorescence microscopy, cells were mounted in medium under a coverslip and photographed on a Zeiss (Thornwood, NY) Axioskop microscope with the use of conventional FITC filters and a Princeton Instruments (Trenton, NJ) CCD-1300 camera.
RESULTS
TMD Length Along the Yeast Secretory Pathway
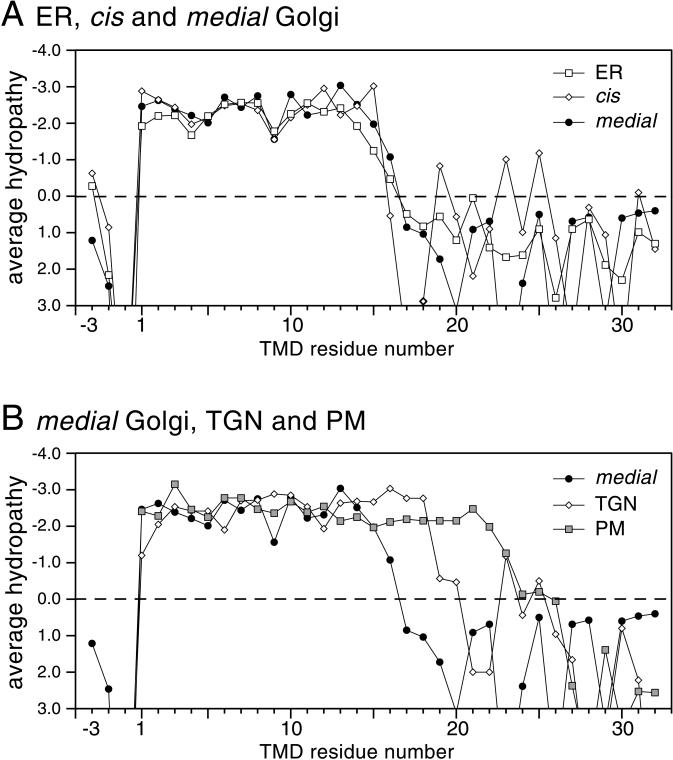
To compare the TMDs of proteins from different parts of the yeast secretory pathway, data on yeast proteins with established locations at different points along the secretory pathway were collected from the databases (Table 2). As with our previous analyses of mammalian proteins, only those with a single TMD were selected in an attempt to minimize interference from nonhydrophobic residues involved in helix-packing interactions. In addition, for single TMDs, the cytoplasmic tails usually end at the bilayer with a strongly positively charged sequence, allowing the beginning of the TMD to be clearly defined (von Heijne and Gavel, 1988; Landolt-Marticorena et al., 1993). The proteins were thus aligned at the start of their TMDs, and average hydropathies were then calculated for each position in the sequence and displayed graphically. Figure 1A shows a comparison of the TMDs of proteins from the ER and from the cis and medial Golgi. In all three cases, the plots show a similar overall shape, with the TMDs showing a hydrophobic plateau of 15 residues before hydrophilic resides start to predominate. This 15-residue stretch is of the same length as that seen with mammalian Golgi proteins (Bretscher and Munro, 1993; Munro, 1995). In contrast, the plots for the latter parts of the pathway showed a very different result, with the profiles of the proteins of the trans-Golgi network (TGN) and the plasma membrane having longer hydrophobic plateaus of 18 and 23 residues, respectively (Figure 1B). This 8-residue difference between Golgi and plasma membrane proteins is even greater than the 5-residue difference observed in mammalian cells.
Table 2.
Yeast genes encoding organelle-specific proteins with a single TMD
| Organelle | Corresponding genes |
|---|---|
| ER | ALG5, CNE1, CPR4, CUE1, CWH41, DPM1, FKB2, GPI12, GPI3, GPI8, HRD3, IRE1, MNS1, OST1, OST4, PBN1, SBH1, SCS2, SEC12, SEC20, SEC66, SED4, SPC3, SRP102, SSS1, TSC10, UBC6, UFE1, WBP1 |
| Golgi, cis | ANP1, GDA1, HOC1, MNN5, MNN2, MNN9, MNN10, MNN11, OCHI, VAN1, SED5 |
| Golgi, medial | MNN1, MNN4, MNN6, MNTI, YGL257c, YIL014w, YOR320c, YLR361c |
| TGN | KEX1, KEX2, STE13, TLG1, TLG2, VPS10 |
| Plasma membrane | AXL2, FET3, FUS1, HKRI, MID2, MSB2, MTL1, PMP1, SLG1, SSO1, WSC2, WSC3 |
Locations are as described in YPD (Hodges et al., 1999), except YOR320c and YLR361c (C.A.R. Wiggins and S. Munro, unpublished). For some enzymes in the medial Golgi, location was inferred because the protein is required for a biochemical modification that competes with, or is immediately preceded by, that of a known medial enzyme. Such proteins are likely to be later than the cis Golgi, but it is possible that some are also found to some extent in the TGN, as has been suggested for Mnn1p itself (Graham et al., 1994).
Figure 1.
Hydropathy plots of the bilayer-spanning regions of single TMD proteins from the organelles of the yeast secretory pathway. (A) The known single TMD proteins of the ER and of the cis and medial Golgi (Table 2) were aligned by the beginning of the TMDs on the cytoplasmic side (residue 1 on the plot, defined as being the residue after the last strongly hydrophilic residue of the cytoplasmic tail). For each position in each aligned set, the average side chain hydropathy was calculated with the use of the Goldman, Engelman, Steitz scale (kcal/mol; Engelman et al., 1986). The statistical significance of differences was evaluated with the use of a two-sample t test (Satterthwaite's method to allow for unequal variances). (B) As in A, except that the proteins were from the medial Golgi, TGN, and plasma membrane (PM).
The significance of these differences was examined with the use of a two-sample t test. For the TGN proteins, the amino acids at the three positions beyond the Golgi hydrophobic plateau (residues 16–18) are each more hydrophobic than the corresponding residues of the medial Golgi plot (p < 0.01). For the plasma membrane proteins, residues 16–22 are different from those of the medial Golgi (p < 0.02), as is residue 23 (p < 0.05). Together, these analyses strongly suggest that the hydrophobic bilayer spanning sections of single-span plasma membrane proteins are substantially longer than those of the ER and Golgi, with those of the TGN having an intermediate length.
Aur1p Is Localized in the Golgi Apparatus
The increase in TMD length between the medial Golgi and the plasma membrane seems likely to reflect the hydrophobic portion of the lipid bilayer being thicker. As in mammalian cells, the yeast plasma membrane is enriched in sphingolipids and sterols, both of which can thicken bilayers (Zinser et al., 1993; Hechtberger and Daum, 1995). Moreover, in yeast, the sphingolipids contain a longer acyl chain than that found in most mammalian sphingolipids (C26 versus C18), which could account for the fact that the plasma membrane protein hydrophobic span is three residues longer in yeast than in mammals. Therefore, it might be expected that the bilayer would thicken as these long-chain ceramides are incorporated into sphingolipid. However, because blocking of ER-to-Golgi transport does not stop the synthesis of IPC from ceramide and PI, it has generally been assumed that that IPC is made in the ER (Puoti et al., 1991; Dickson and Lester, 1999). An ER location for IPC synthesis would make it difficult to explain why the bilayer along the secretory pathway does not apparently start to thicken until late in the Golgi. IPC synthase is a polytopic membrane protein encoded by the AUR1 gene, (Nagiec et al., 1997), so we next investigated in which of the compartments of the yeast secretory pathway Aur1p is actually localized.
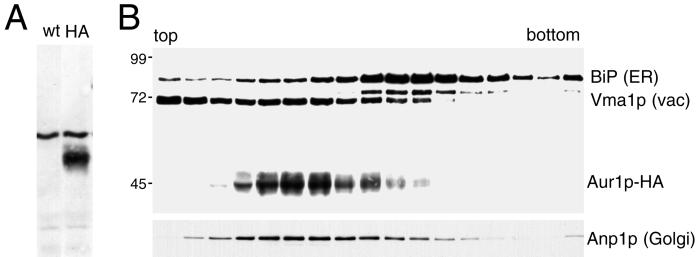
To localize the Aur1p IPC synthase, the endogenous copy of the gene was tagged by homologous recombination to insert three copies of the HA epitope tag at the COOH terminus of the ORF. The addition of the epitope tag did not affect the growth of the cells but resulted in the appearance of a 45-kDa band in protein blots of total yeast proteins probed with an anti-HA antibody (Figure 2A; our unpublished results). Because AUR1 is an essential gene, the viability of haploid cells in which the only copy of the gene is tagged indicates that attachment of the COOH-terminal epitope tag does not inactivate the enzyme. The intracellular location of the Aur1p-HA was initially investigated by fractionating organelles from the tagged strain on a velocity gradient and blotting the gradient fractions with antibodies against the HA tag. Figure 2B shows that upon such fractionation, Aur1p-HA is clearly separated from BiP, a marker for the ER, and Vma1p, a marker for the vacuole. In contrast, Aur1p-HA shows cofractionation with Anp1p, a subunit of the Golgi-localized mannan-polymerase II. This result shows that Aur1p is not localized primarily in the ER and suggests that instead it may be in the Golgi apparatus.
Figure 2.
Epitope-tagged Aur1p cofractionates with the Golgi apparatus. (A) Immunoblot of total proteins from strain SEY6210 either with (HA) or without (wt) three copies of the HA epitope tag inserted at the COOH terminus of the AUR1 gene. The background band is characteristic of the 12CA5 anti-HA mAb. (B) Immunoblot of membranes from a strain expressing Aur1p-HA separated by velocity sedimentation and probed with 12CA5 and endogenous markers for ER, vacuole (vac), and Golgi, followed by peroxidase-conjugated secondary antibodies for visualization by chemiluminescence. Mobilities of size markers are indicated (kDa), and the faint band below BiP is protein disulfide isomerase, which also reacts with the anti-HDEL mAb 2E7.
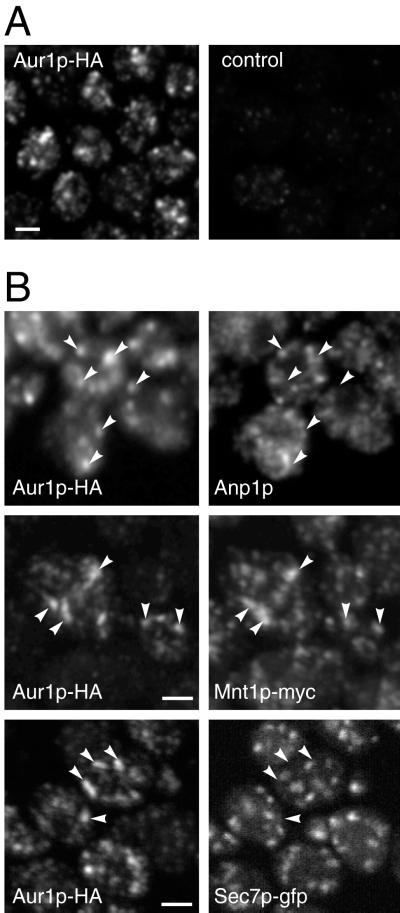
The different subcompartments of the yeast Golgi are not well resolved on velocity gradients, so immunofluorescence was used to compare the localization of Aur1p-HA with various Golgi membrane proteins, the Golgi compartments of yeast not being arranged in a stack but rather scattered unstacked throughout the cytoplasm. Figure 3 shows that anti-HA staining of Aur1p-HA–expressing cells produced a punctate pattern characteristic of the yeast Golgi apparatus and completely distinct from the nuclear envelope and subplasma membrane pattern typically seen with ER markers. These Aur1p-HA–containing spots did not show substantial colocalization with the cis Golgi marker Anp1p or with Sec7p and Tlg1p (our unpublished results), markers for the late Golgi or TGN (Franzusoff et al., 1991; Jungmann and Munro, 1998; Rossanese et al., 1999). However, the Aur1p-HA–positive structures did show considerably more colocalization with the mannosyltransferase Mnt1p, a marker for the medial Golgi marker (Lussier et al., 1995).
Figure 3.
Localization of epitope-tagged Aur1p by immunofluorescence. (A) Immunofluorescence confocal micrographs of yeast cells in which the AUR1 gene is either tagged with HA or not (control) and probed with the rat anti-HA mAb 3F10. (B) Double-label immunofluorescence confocal micrographs comparing the localization of Aur1p-HA with that of endogenous Anp1p (cis Golgi), myc-tagged Mnt1p (medial Golgi), or GFP-tagged Sec7p (TGN). Anti-HA staining was with a rabbit antiserum or, for colocalization with Anp1p, rat mAb 3F10. Most of the Aur1p-positive structures were also positive for Mnt1p, but a few showed colocalization with Anp1p (arrowheads pointing right). Bars, 2 μm.
IPC Synthase Activity Is Localized in the Golgi Apparatus
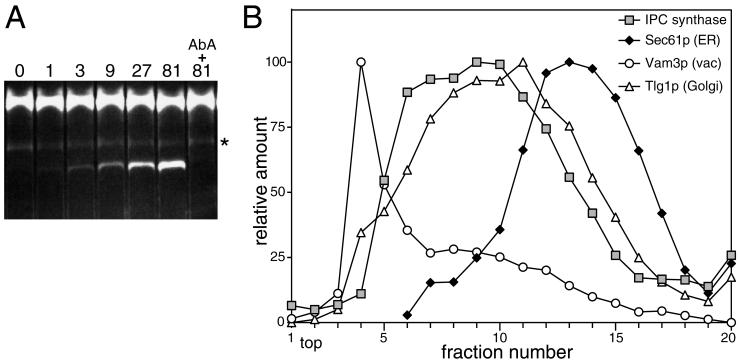
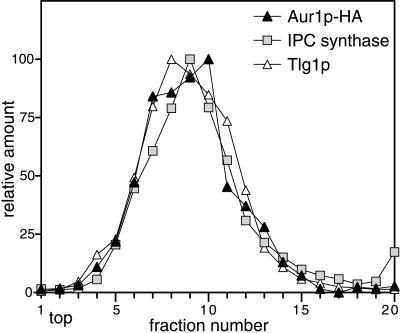
These results strongly suggest that Aur1p is a resident protein of the Golgi apparatus of yeast, primarily in the medial compartment. However, the localization relies on the use of an epitope-tagged version of the protein, so to ensure that the attachment of the epitope tag did not change the intracellular distribution by masking a localization signal, such as an ER retention signal, we also assayed membrane fractions from wild-type cells for the presence of IPC synthase activity. For these assays, we used the fluorescent substrate C6-NBD-ceramide, which has been used extensively to assay mammalian sphingolipid-synthesizing enzymes (Pagano et al., 1989). When yeast membranes were incubated with this substrate and the products separated by TLC, a more slowly migrating band appeared, and quantitation of the TLC revealed that the amount of product increased linearly with the amount of membrane present. The formation of the band was blocked if AbA, a specific inhibitor of Aur1p (Nagiec et al., 1997), was included in the reaction (Figure 4A). These results suggest that this band represents C6-NBD-ceramide modified by Aur1p (presumably C6-NBD-IPC), and indeed, the sole product formed when C6-NBD-ceramide is incubated with yeast microsomes was recently confirmed by mass spectrometry to be C6-NBD-IPC (Zhong et al., 1999). The localization of this IPC synthesis activity was then compared with the positions of specific organelle markers on a velocity gradient of intracellular membranes. Figure 4B shows that the IPC synthase activity in a strain containing the wild-type Aur1p did not fractionate with the ER but rather cofractionated with a Golgi marker. To confirm that the IPC synthase activity cofractionates with Aur1p itself, membranes from cells expressing Aur1p-HA were fractionated on a velocity gradient, and the fractions were divided and assayed for both Aur1p-HA and IPC synthase activity. As shown in Figure 5, there is a very close coincidence of the Aur1p-HA and IPC synthase peaks, indicating that there is not a small pool of the enzyme with a different specific activity.
Figure 4.
Modification of C6-NBD-ceramide by Aur1p. (A) TLC separation of the reaction products produced when C6-NBD-ceramide was incubated with gradient fractions containing Golgi membranes, as described in MATERIALS AND METHODS. The samples were diluted threefold serially from the most active membrane fractions in the gradient shown in Figure 5 (81; 25 μg/ml membranes), and the Aur1p inhibitor AbA was added to a sample containing the highest concentration of membranes (10 μg/ml AbA). The upper band corresponds to unmodified C6-NBD-ceramide, and the faint AbA-insensitive band (*) comigrates with C6-NBD fatty acid and is apparently released from C6-NBD-ceramide during the acid extraction of lipids before TLC. (B) IPC synthase activity cofractionates with Golgi and not ER membranes. Fractions from a velocity gradient separation of membranes from SEY6210 were assayed for the indicated proteins by quantitative immunoblotting with the use of 125I-protein A (Amersham, Arlington Heights, IL) and a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Fractions were also incubated with C6-NBD-ceramide, the products were separated by TLC, and the modified band was quantified with the use of a fluorescence imager. The IPC synthase activity is the mean of results from four gradients, and the peak is slightly broadened as a result.
Figure 5.
Aur1p-HA cofractionates with IPC synthase activity. Membranes prepared from strain SEY6210 with endogenous Aur1p tagged with HA were fractionated on a velocity gradient and assayed for IPC synthase activity or for the indicated proteins by quantitative immunoblotting, as in Figure 4B.
Recycling of Aur1p
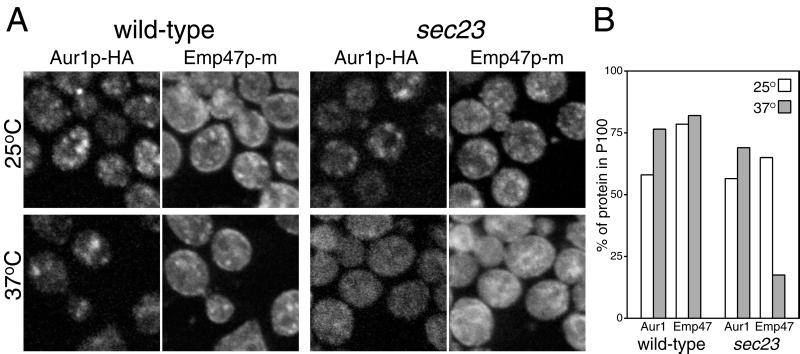
Together, these results show that Aur1p is primarily localized to the Golgi apparatus. However, it is possible that the enzyme cycles continuously through the ER, as has been observed for some Golgi proteins such as Emp47p and Sed5p (Schröder et al., 1995; Wooding and Pelham, 1998). To examine this possibility, the localization of Aur1p-HA was examined in a strain carrying sec23-1, a temperature-sensitive allele of SEC23. Sec23p is required for ER exit, and when it is inactivated in a sec23-1 strain by increasing the temperature to 37°C, recycling proteins are still able to return to the ER but cannot leave and so shift from the Golgi to the ER (Lewis and Pelham, 1996; Wooding and Pelham, 1998). However, when the localization of Aur1p-HA was examined under these circumstances, the pattern went from spots to a cytoplasmic fuzz, whereas in the same cells, Emp47p had redistributed from spots to the ER (Figure 6A). The diffuse distribution of Aur1p-HA is similar to results obtained previously with the medial Golgi proteins Mnn1p and Sft2p and is believed to reflect an incorporation into retrograde vesicles that would normally fuse with ER-derived vesicles or with a cis Golgi derived from these but that can no longer fuse in the absence of ER budding (Wooding and Pelham, 1998). The distribution of Aur1p-HA and Emp47p was also examined with the use of protein blotting of membrane fractions sedimenting at 13,000 × g (P13) and 100,000 × g (P100) (Lewis and Pelham, 1996). Figure 6B shows that, as expected, the Emp47p moves from the Golgi-enriched P100 to the ER-enriched P13 when the sec23-1 strain is shifted to the nonpermissive temperature. In contrast, Aur1p-HA does not shift, similar to the behavior observed with the Golgi proteins Mnn1p and Sft2p (Wooding and Pelham, 1998). This suggests that if Aur1p-HA is recycling, it is behaving like medial proteins and going back to earlier Golgi compartments, and hence not behaving like those early Golgi proteins that recycle through the ER.
Figure 6.
Aur1p-HA does not return to the ER when ER-to-Golgi transport is blocked. (A) Double-label immunofluorescence confocal micrographs of SEY6210 (wild type) and RSY282 (sec23-1) cells expressing both HA-tagged Aur1p and myc-tagged Emp47p and probed with rabbit anti-HA and a mouse anti-myc mAb. Cells were shifted to 37°C for 1 h or left at 25°C before fixation as indicated. (B) Quantitation of the amount of Aur1p-HA (Aur1) and Emp47-Myc in P100 membrane fractions. Cells of the indicated strains were treated as in A, with the addition of cycloheximide (67 μg/ml) from 10 min before temperature shift to prevent the accumulation of newly made proteins in the ER. Membranes were isolated and fractionated as described by Holthuis et al. (1998), and the tagged proteins were quantified as in Figure 4.
In Vivo Localization of IPC Synthase Activity
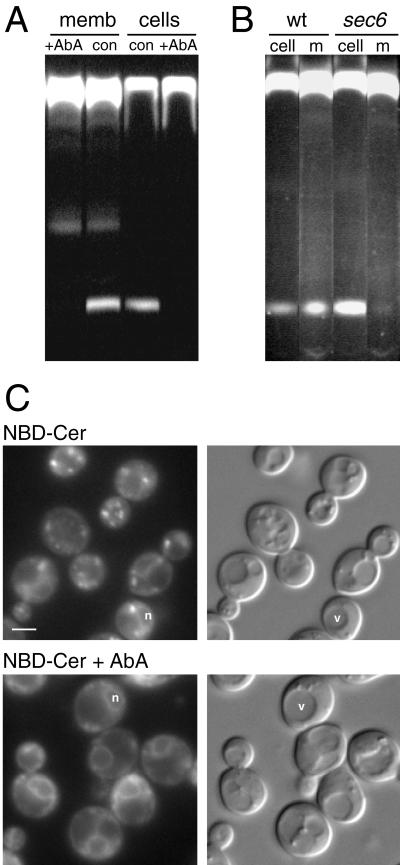
The ability of mammalian enzymes involved in the synthesis and breakdown of sphingolipids to use fluorescent substrates applied to living cells has provided many insights into sphingolipid trafficking and enzyme subcellular localization. The modification of C6-NBD-ceramide by Aur1p in vitro suggests that such an approach could be extended to yeast. Because no such studies have been reported, we initially investigated the incorporation and trafficking of the fluorescent precursor by live yeast. Yeast cells were incubated with C6-NBD-ceramide at 30°C and then back-extracted on ice to reduce unincorporated substrate. When lipids isolated from these cells were separated by TLC, a band was present of the same mobility as the C6-NBD-IPC produced by isolated membranes in vitro (Figure 7A). The appearance of this band was sensitive to the addition of AbA, with 5 min of preincubation sufficient for a complete block. The lack of any additional bands from the labeled cells compared with the microsomes suggests that the C6-NBD-IPC is not mannosylated by yeast, in contrast to endogenous IPC, some of which is mannosylated to MIPC. The reasons for this are not clear, but it may be that C6-NBD-IPC is a poor substrate for the mannosylating enzyme, because other enzymes in sphingolipid synthesis show varied recognition of short-chain substrates (Pagano et al., 1991). Indeed, in mammalian cells, C6-NBD-ceramide is efficiently converted to C6-NBD-glucosylceramide but does not proceed efficiently to higher glycosylated forms (Lipsky and Pagano, 1983).
Figure 7.
Modification of C6-NBD-ceramide by living yeast cells. (A) TLC separation of the products of lipids C6-NBD-ceramide extracted either from microsomes (memb), as in Figure 4, or from live yeast incubated with C6-NBD-ceramide, as described in MATERIALS AND METHODS. AbA treatment was at 10 μg/ml, with a 10-min preincubation for the cells. The faster migrating band is the precursor C6-NBD-ceramide. (B) As in A except that wild-type (SEY6211; wt) and sec6-4 (JWY47; sec6) cells were incubated with C6-NBD-ceramide for 10 min at 25°C and then for 5 min at 37°C. After washing and back-extraction for 60 min on ice, cells were resuspended into fresh, prewarmed medium containing 10 mg/ml defatted BSA and 10 μg/ml AbA and incubated for 10 min at 37°C, and lipids were extracted from cells and medium. (C) Live yeast labeled with C6-NBD-ceramide with or without AbA, as described in MATERIALS AND METHODS, and photographed with both fluorescence and Nomarski optics. Rings can be seen around the nucleus (n) distinct from the vacuole (v). Bar, 1.5 μm.
A feature of the metabolism of C6-NBD-ceramide in mammalian cells is that the resulting NBD sphingomyelin and glucosylceramide appear at the cell surface, from which they can be extracted into the medium (Lipsky and Pagano, 1985; van Meer et al., 1987). This was initially interpreted as movement of the labeled lipids from the Golgi to the cell surface by vesicular transport. However, it was subsequently shown that NBD-lipid export could also occur in the absence of vesicular transport if the lipids had access to the cytosolic leaflet of an internal membrane and that this export was mediated by a plasma membrane–localized ATP-binding cassette transporter (van Helvoort et al., 1996, 1997). Examination of the medium of yeast labeled with C6-NBD-ceramide revealed that the C6-NBD-IPC was accessible for extraction from the plasma membrane (Figure 7B). However, in strains carrying a temperature-sensitive allele of SEC6, a gene involved in Golgi-to-plasma membrane transport, the appearance of this product in the medium was completely blocked at the nonpermissive temperature, indicating that vesicular transport is required for delivery to the plasma membrane. In contrast, a strain lacking two major ATP-binding cassette transporters, Snq2p and Pdr5p, showed unaffected transport into the medium (our unpublished results). Overall, these results demonstrate that C6-NBD-ceramide can be modified by IPC synthase in living yeast and that the product then moves to the cell surface by vesicular transport, in agreement with the previously reported requirement for vesicular transport for movement of endogenous sphingolipids to the plasma membrane (Hechtberger and Daum, 1995).
Fluorescence microscopy of live yeast incubated with C6-NBD-ceramide revealed small, bright, cytoplasmic patches similar in appearance to the Golgi (Figure 7C). The staining was photolabile, fading under the fluorescence microscope in a few seconds, a behavior of NBD probes also seen in mammalian cells after cholesterol deprivation (Martin et al., 1993). The bright patches were absent in cells treated with AbA, indicating that their formation required the action of Aur1p, and hence that they constitute the fluorescent IPC derivative of C6-NBD-ceramide that has become resistant to back-extraction or intraorganellar partitioning, perhaps because the addition of the charged phosphorylinositol head group has trapped it on the lumenal leaflet of the Golgi membranes. Fainter ring staining was also visible that was unaffected by AbA, indicating that it corresponds to unmetabolized C6-NBD-ceramide (Figure 7C). These rings do not correspond to the vacuole visible in Nomarski optics, indicating that the C6-NBD-ceramide preferentially incorporates into the nuclear envelope (i.e., the ER), a phenomenon also seen in mammalian cells (Lipsky and Pagano, 1985).
Topology of IPC Synthesis in the Golgi
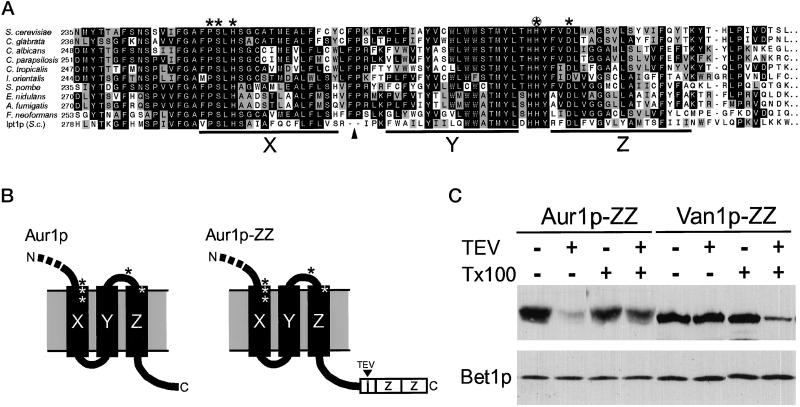
Together, these results suggest that Aur1p is localized primarily to the Golgi apparatus. However, the substrates used by Aur1p (PI and ceramide) are likely to be available on both sides of the lipid bilayer, so IPC could be made on either the cytoplasmic or the lumenal leaflet of the Golgi membranes. The sequence of Aur1p has been found to contain a motif originally identified in a superfamily of integral membrane phosphatases and soluble haloperoxidases (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997; Heidler and Radding, 2000), and the residues that match this motif are found in the most highly conserved regions of the Aur1p homologues cloned to date from different fungi and yeast (Figure 8A). In the lipid phosphate phosphohydrolases, these residues are believed to constitute the active site for the cleavage of the bond between the lipid hydroxyl group and the phosphate group (Neuwald, 1997; Brindley and Waggoner, 1998). In the case of Aur1p, this reaction would represent the first step in the transfer of inositol phosphate from PI, with the resulting phosphate intermediate presumably being subjected to nucleophilic attack by the oxygen of ceramide rather than the oxygen of the water used by the phosphatases. This sequence motif is found between the last two predicted TMDs of the protein (Figure 8A), the same position it occupies in the distantly related lipid phosphate phosphohydrolases (Brindley and Waggoner, 1998).
Figure 8.
The COOH terminus of Aur1p is cytoplasmic. (A) Sequences of the last three TMDs of Aur1p and its homologues (black bars X, Y, and Z, as predicted by PHDhtm [Rost et al., 1996]). Residues identical (black) or related (gray) in 4 or more of the 11 sequences are shaded. A 124-residue hydrophilic loop found only in Ipt1p (residues 313–436) is omitted (arrowhead). Asterisks indicate the conserved motifs originally found in integral membrane phosphatases and soluble haloperoxidases (Neuwald, 1997), and the circled asterisk indicates histidine 294, which was mutated to alanine. (B) Arrangement of the motifs and the COOH terminus with respect to the last three TMDs for Aur1p and the ZZ-tagged version. (C) Immunoblot of membranes from yeast expressing Aur1p or Van1p with a TEV-cleavable ZZ domain at the COOH terminus. Membranes were treated for 1 h at 15°C with 20 U of TEV protease (Life Technologies, Grand Island, NY) and 0.4% Triton X-100 (Tx100), as indicated. Each lane contains membranes prepared from 40 OD (600 nm) units of log-phase cells, as described previously (Holthuis et al., 1998). Membrane protein Bet1p was examined to demonstrate constant protein recovery. The apparent reduction in Aur1p-ZZ cleavage in the presence of Triton X-100 was seen reproducibly and may reflect partial masking of the cleavage site in detergent micelles.
To investigate the importance of this motif, diploid yeast strains were constructed in which one allele was tagged at the COOH terminus with myc and the remainder of the gene was either wild type or had histidine 294 mutated to alanine (Figure 8A). Protein blotting showed that the levels of myc-Aur1p were the same in the two strains, but when sporulated, only the wild type produced viable haploids. This indicates that mutation of the motif does not affect the stability of the protein but reduces the enzyme activity below that required to support growth. This is consistent with it being part of the active site of the enzyme, and because there is only one TMD COOH terminal to this sequence, the putative active site of Aur1p must be located on the opposite side of the Golgi membrane from its COOH terminus (Figure 8B). Thus, we investigated on which side of the membrane the COOH terminus of Aur1p is located by means of protein A fusion. In this chimera, a cleavage site for the sequence-specific TEV protease is located between the COOH terminus of Aur1p and the protein A domains (Figure 8B). Figure 8C shows that when membranes from cells expressing this chimera were treated with TEV protease, removal of the protein A portion was observed in the absence of detergent, whereas removal of a lumenal, COOH-terminal protein A fusion to the type II Golgi enzyme Van1p was observed only after detergent treatment. This indicates that the topology of Aur1p is such that the COOH terminus is cytosolic and hence the putative active site residues are in the lumen. This agrees with the topology of Aur1p predicted by the programs PHDhtm and TMHMM (Rost et al., 1996; Sonnhammer et al., 1998) and is the same orientation observed with the lipid phosphate phosphohydrolases (Barila et al., 1996; Brindley and Waggoner, 1998; Waggoner et al., 1999). Transfer of the PI head group to ceramide in the Golgi lumen is consistent with the C6-NBD-ceramide product remaining within the Golgi in live cells rather than being accessible to the cytosol for transfer between organelles.
DISCUSSION
The sphingolipids of S. cerevisiae consist of IPC and its derivatives, and in this paper we have examined the localization of the IPC synthase, the product of the AUR1 gene. We found that both a tagged version of Aur1p and IPC synthase activity are located primarily in the Golgi apparatus. This differs from the prevailing view that IPC is synthesized in the yeast ER (Futerman, 1995; Daum et al., 1998; van Meer, 1998; Dickson and Lester, 1999). IPC synthase activity was first detected in “crude yeast membranes” that are likely to have contained both Golgi and ER membranes (Becker and Lester, 1980). However, the most direct evidence for an ER location of Aur1p is the observation that IPC can still be synthesized from radiolabeled inositol or fatty acids, even when ER-to-Golgi vesicular transport is blocked (Puoti et al., 1991). In contrast, our results indicate that Aur1p is located mostly in the Golgi apparatus. These observations can be reconciled in two ways. First, it is possible that there is a low level of Aur1p in the ER and that this is sufficient for at least some IPC synthesis when ER-to-Golgi transport is blocked. However, it has been reported that incorporation of [3H]inositol into IPC in unaffected by a 2-h pretreatment with cycloheximide (Hechtberger and Daum, 1995). Under such conditions, there is unlikely to be any newly made Aur1p remaining in the ER. Moreover, the results presented here suggest that the Golgi pool of Aur1p does not cycle through the ER. Overall, these observations suggest that it is unlikely that Aur1p's primary site of action is in the ER with an apparent Golgi distribution at steady state.
A second possibility is that Aur1p acts in the Golgi with the use of PI and ceramide that are made in the ER and then delivered to Golgi membranes in a way that is not mediated solely by vesicular transport. Two recent observations in mammalian cells are consistent with the possibility of nonvesicular transport of ceramide from the ER to the Golgi. First, sphingolipid synthesis continues when ER-to-Golgi transport of proteins is blocked by incubation at 15°C (Kok et al., 1998). Second, a mutant Chinese hamster ovary cell line has been found with an apparent defect in ATP-dependent nonvesicular transport of ceramide to the site of sphingomyelin synthesis in the Golgi (Fukasawa et al., 1999). If such a vesicle-independent ceramide transport mechanism also exists in yeast, it would explain the delivery of ceramide to the Golgi even when ER-to-Golgi vesicular transport is blocked with conditional sec mutations. It was also observed that when such a sec block is applied to yeast, although IPC is still made it does not proceed on to its mannosylated derivative MIPC (Puoti et al., 1991). Mannosylation of IPC requires the Sur1p protein, and if this protein were in a later Golgi compartment than Aur1p, then when the Golgi vesiculated in the absence of anterograde transport the Sur1p mannosyltransferase would not be in the same vesicles as Aur1p, and no further modification could occur (Beeler et al., 1997).
The localization of IPC synthesis to the Golgi apparatus of yeast thus parallels the situation in mammalian cells, in which the major sphingolipids are also made in the Golgi (Futerman et al., 1990; Jeckel et al., 1990, 1992). This raises the question of why these widely divergent eukaryotes both appear to separate glycerophospholipid synthesis in the ER and mitochondria from sphingolipid synthesis in the Golgi. A possible reason for this, and perhaps also for why sphingolipid precursors might need to be able to bypass ER-to-Golgi transport vesicles, comes from considering the function of sphingolipids. This class of lipids has an important role as a precursor of second messengers for signal transduction in both mammalian cells and yeast (Dickson, 1998). However, sphingolipids are also abundant components of the plasma membrane of eukaryotic cells, accounting for 30% of the total phospholipids in the yeast plasma membrane (Patton and Lester, 1991; Hechtberger et al., 1994). Moreover, they differ from glycerophospholipids in that they usually have longer and more saturated acyl chains, contain hydroxylated acyl chains, and show a greater affinity for sterols (Patton, 1970; Bittman et al., 1994; Ramstedt and Slotte, 1999). This suggests that a major function of sphingolipids is to alter the overall physical properties of the plasma membrane. The properties of the acyl chains, the association with sterols, and the capacity for hydrogen bonding between hydroxyls and amide carbonyls lead to sphingolipids promoting a more compact, thicker, and less permeant bilayer. In yeast, the ceramides and sphingolipids have acyl chains that are even longer than those of mammalian cells, and they also have a greater degree of acyl chain hydroxylation and hence an increased capacity for lipid–lipid hydrogen bonding (Dickson, 1998). Yeast are exposed to harsher environments than most mammalian cells, and it may also be that because sterol synthesis becomes limiting in conditions of oxygen deprivation, they may be more reliant on sphingolipids than sterols for plasma membrane robustness. Such a bilayer-thickening role for sphingolipids is supported by the remarkable discovery of a suppresser mutant that allows yeast to grow without any sphingolipids, although only under nonstressed conditions (Lester et al., 1993; Nagiec et al., 1993). This mutation is an alteration of a fatty acyl transferase that allows the incorporation of the C26:0 long-chain fatty acids into PI, which normally has only C16 or C18 chains. This suggests that the essential function that is served by sphingolipids in yeast is their provision of long acyl chains to the plasma membrane bilayer.
Our analysis of TMD length through the secretory pathway suggests that the hydrophobic portion of the bilayer is thicker in the plasma membrane. Previous electron microscopic studies of the bilayers of yeast and plant cells have found the interleaflet space to be thicker in the plasma membrane compared with the ER and the Golgi (Grove et al., 1968; Schneiter et al., 1999). Although such observations are clearly susceptible to change during fixation, they are consistent with nuclear magnetic resonance and radiographic measurements of pure lipid bilayers, which have found that bilayer thickness increases with acyl chain length and increased sterol content, features of the plasma membrane (Lewis and Engelman, 1983b; Nezil and Bloom, 1992). Although it is possible that the longer TMD length at the plasma membrane reflects the proteins in this bilayer adopting a different conformation from proteins elsewhere, we feel that these observations make it much more likely that it is the bilayer thickness that is different. If the primary function of the sphingolipids with their C26:0 acyl chains is to thicken the plasma membrane, this might provide an explanation for the conservation of a Golgi location for sphingolipid synthesis in both yeast and mammalian cells, even though the ceramide and phospholipid precursors are synthesized in the ER. The bilayer of the ER may need to be especially permeant and disordered to allow synthesis, insertion, and assembly of hydrophobic lipids and proteins. High concentrations of ceramide with long acyl chains would be expected to thicken and organize such a bilayer, and indeed, the level of ceramide in the ER is less than the 30% level that sphingolipids attain in the plasma membrane (Schneiter et al., 1999). The bilayer could be remodeled in the Golgi apparatus by the conversion of ER-derived ceramide into sphingolipids. This remodeling would require the supply of a large amount of ceramide to the Golgi. If ER-to-Golgi transport vesicles were the only source of ceramide, many rounds of vesicle recycling might be required to attain a plasma membrane level of sphingolipids. Nonvesicular transfer of ceramide could be a means to provide a delivery route to the Golgi that would reduce the requirement for either vesicle recycling or high levels of long-chain ceramide in ER membranes. Because the active site of Aur1p is apparently lumenal, ceramide delivered to the cytoplasmic leaflet would be consumed by conversion to IPC in the inner leaflet, and hence IPC would accumulate as more and more ceramide was delivered from the ER and metabolically trapped. As sphingolipids are synthesized, they may also attract ER-synthesized sterols to contribute further to the bilayer thickening and perhaps promote domain formation (Brown, 1998). This remodeling of the bilayer composition toward that of the plasma membrane also could be promoted by sterol- and sphingolipid-rich membrane being excluded from retrograde COPI vesicles. Indeed, when examined by electron microscopy, the interleaflet space of COPI vesicles appears thinner than the membranes the vesicles are budding from (Orci et al., 1996). It is tempting to speculate that the multicisternal nature of the Golgi reflects the need for a period of lipid synthesis, and multiple rounds of lipid sorting, to increase the concentration of sphingolipids and sterols until the entire bilayer is thickened. We have suggested previously that such a change in bilayer thickness could prevent Golgi residents moving toward the plasma membrane, and indeed, the short TMDs of Aur1p itself provide a possible means to link completion of bilayer thickening during cisternal maturation to retrograde transport of the enzyme to earlier cisternae.
Finally, if the difference between the thickness of the plasma membrane and the ER is as great as the TMD lengths suggest, it could raise mechanistic problems in trafficking of yeast cell surface proteins. The difference we observed between ER and plasma membrane TMDs (15 versus 23 residues) suggests a >50% increase in the bilayer thickness along the secretory pathway. ER proteins presumably have TMD lengths optimal for the thickness of the ER bilayer, and because 8 extra residues corresponds to 12 Å of α-helix, this suggests that newly made plasma membrane proteins will be in a bilayer that is 12 Å too thin. Mismatch between TMD length and bilayer thickness has been shown to induce protein aggregation in model systems (Lewis and Engelman, 1983a; Ryba and Marsh, 1992; Killian, 1998). TMD tilting could help single TMD proteins, but it would be of less help to larger polytopic proteins, although mismatch could have the benefit of ensuring that they are not active in the ER (Cornea and Thomas, 1994; Webb et al., 1998; Dumas et al., 1999). This may suggest a possible function for the several yeast proteins that have been identified recently as being required for the exit from the ER of particular, usually polytopic, plasma membrane proteins (Ljungdahl et al., 1992; Powers and Barlowe, 1998; Sherwood and Carlson, 1999; Trilla et al., 1999). Because Aur1p defines the site of sphingolipid synthesis, our description of its location and topology of action, combined with the ease of manipulation of yeast, could open the way not just to determining whether such ER proteins are indeed “mismatch chaperones” but also to examining the general features of how the secretory pathway is able to maintain bilayers with very different physical properties.
ACKNOWLEDGMENTS
We thank Carole Spibey for access to the ARTHUR fluorescence imager and advice on its use. We are indebted to Ben Glick, Mike Lewis, Karl Kuchler, Randy Schekman, and Colin Stirling for strains and antibodies and to James Whyte and members of the Pelham and Arkowitz laboratories for much useful advice and helpful discussions. T.P.L. was supported by a Research Fellowship from the British Heart Foundation.
Abbreviations used:
- AbA
aureobasidin A
- GFP
green fluorescent protein
- ER
endoplasmic reticulum
- HA
hemagglutinin
- IPC
inositol phosphorylceramide
- PI
phosphatidylinositol
- TEV
tobacco etch virus
- TGN
trans-Golgi network
- TLC
thin-layer chromatography
- TMD
transmembrane domain
REFERENCES
- Antebi A, Fink GR. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Barila D, Plateroti M, Nobili F, Muda AO, Xie Y, Morimoto T, Perozzi G. The Dri 42 gene, whose expression is up-regulated during epithelial differentiation, encodes a novel endoplasmic reticulum resident transmembrane protein. J Biol Chem. 1996;271:29928–29936. doi: 10.1074/jbc.271.47.29928. [DOI] [PubMed] [Google Scholar]
- Becker GW, Lester RL. Biosynthesis of phosphoinositol-containing sphingolipids from phosphatidylinositol by a membrane preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37°C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- Bittman R, Kasireddy CR, Mattjus P, Slotte JP. Interaction of cholesterol with sphingomyelin in monolayers and vesicles. Biochemistry. 1994;33:11776–11781. doi: 10.1021/bi00205a013. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brindley DN, Waggoner DW. Mammalian lipid phosphate phosphohydrolases. J Biol Chem. 1998;273:24281–24284. doi: 10.1074/jbc.273.38.24281. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Brown RE. Sphingolipid organization in biomembranes: what physical studies of model membranes reveal. J Cell Sci. 1998;111:1–9. doi: 10.1242/jcs.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornea RL, Thomas DD. Effects of membrane thickness on the molecular dynamics and enzymatic activity of reconstituted Ca-ATPase. Biochemistry. 1994;33:2912–2920. doi: 10.1021/bi00176a022. [DOI] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- David D, Sundarababu S, Gerst JE. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- Dumas F, Lebrun MC, Tocanne JF. Is the protein/lipid hydrophobic mismatching principle relevant to membrane organization and functions? FEBS Lett. 1999;458:271–277. doi: 10.1016/s0014-5793(99)01148-5. [DOI] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Nishijima M, Hanada K. Genetic evidence for ATP-dependent endoplasmic reticulum-to-Golgi apparatus trafficking of ceramide for sphingomyelin synthesis in Chinese hamster ovary cells. J Cell Biol. 1999;144:673–685. doi: 10.1083/jcb.144.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH. Inhibition of sphingolipid synthesis: effects on glycosphingolipid-GPI–anchored protein microdomains. Trends Cell Biol. 1995;5:377–380. doi: 10.1016/s0962-8924(00)89078-9. [DOI] [PubMed] [Google Scholar]
- Futerman AH, Stieger B, Hubbard AL, Pagano RE. Sphingomyelin synthesis in rat liver occurs predominantly at the cis and medial cisternae of the Golgi apparatus. J Biol Chem. 1990;265:8650–8657. [PubMed] [Google Scholar]
- Graham TR, Seeger M, Payne GS, Mackay VL, Emr SD. Clathrin-dependent localization of α 1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove SN, Bracker CE, Morre DJ. Cytomembrane differentiation in the endoplasmic reticulum-Golgi apparatus-vesicle complex. Science. 1968;161:171–173. doi: 10.1126/science.161.3837.171. [DOI] [PubMed] [Google Scholar]
- Hechtberger P, Daum G. Intracellular transport of inositol-containing sphingolipids in the yeast, Saccharomyces cerevisiae. FEBS Lett. 1995;367:201–204. doi: 10.1016/0014-5793(95)00567-s. [DOI] [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- Heidler SA, Radding JA. Inositol phosphoryl transferases from human pathogenic fungi. Biochim Biophys Acta. 2000;1500:147–152. doi: 10.1016/s0925-4439(99)00097-6. [DOI] [PubMed] [Google Scholar]
- Hemrika W, Renirie R, Dekker HL, Barnett P, Wever R. From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc Natl Acad Sci USA. 1997;94:2145–2149. doi: 10.1073/pnas.94.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg K, Rodger J, Futerman AH. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem J. 1993;290:751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges PE, McKee AH, Davis BP, Payne WE, Garrels JI. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Dhruvakumar S, Pelham HRB. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeckel D, Karrenbauer A, Birk R, Schmidt RR, Wieland F. Sphingomyelin is synthesized in the cis Golgi. FEBS Lett. 1990;261:155–157. doi: 10.1016/0014-5793(90)80659-7. [DOI] [PubMed] [Google Scholar]
- Jeckel D, Karrenbauer A, Burger KN, van Meer G, Wieland F. Glucosylceramide is synthesized at the cytosolic surface of various Golgi subfractions. J Cell Biol. 1992;117:259–267. doi: 10.1083/jcb.117.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Kent C. Eukaryotic phospholipid biosynthesis. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Killian JA. Hydrophobic mismatch between proteins and lipids in membranes. Biochim Biophys Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- Ko J, Cheah S, Fischl AS. Regulation of phosphatidylinositol:ceramide phosphoinositol transferase in Saccharomyces cerevisiae. J Bacteriol. 1994;176:5181–5183. doi: 10.1128/jb.176.16.5181-5183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Kohlwein SD, Daum G, Schneiter R, Paltauf F. Phospholipids: synthesis, sorting, subcellular traffic. The yeast approach. Trends Cell Biol. 1996;6:260–266. doi: 10.1016/0962-8924(96)10025-8. [DOI] [PubMed] [Google Scholar]
- Kok JW, Babia T, Klappe K, Egea G, Hoekstra D. Ceramide transport from endoplasmic reticulum to Golgi apparatus is not vesicle-mediated. Biochem J. 1998;333:779–786. doi: 10.1042/bj3330779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J Biol Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- Lester RL, Wells GB, Oxford G, Dickson RC. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem. 1993;268:845–856. [PubMed] [Google Scholar]
- Lewis BA, Engelman DM. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J Mol Biol. 1983a;166:203–210. doi: 10.1016/s0022-2836(83)80006-0. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983b;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HRB. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- Lipsky NG, Pagano RE. Sphingolipid metabolism in cultured fibroblasts: microscopic and biochemical studies employing a fluorescent ceramide analogue. Proc Natl Acad Sci USA. 1983;80:2608–2612. doi: 10.1073/pnas.80.9.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky NG, Pagano RE. Intracellular translocation of fluorescent sphingolipids in cultured fibroblasts: endogenously synthesized sphingomyelin and glucocerebroside analogues pass through the Golgi apparatus en route to the plasma membrane. J Cell Biol. 1985;100:27–34. doi: 10.1083/jcb.100.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L, Underwood KW. Intracellular cholesterol transport and compartmentation. J Biol Chem. 1995;270:15443–15446. doi: 10.1074/jbc.270.26.15443. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Lussier M, Sdicu AM, Ketela T, Bussey H. Localization and targeting of the Saccharomyces cerevisiae Kre2p/Mnt1p α1,2-mannosyltransferase to a medial-Golgi compartment. J Cell Biol. 1995;131:913–927. doi: 10.1083/jcb.131.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon EC, Ehses I, Rother J, van Echten G, Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J Biol Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- Martin OC, Comly ME, Blanchette-Mackie EJ, Pentchev PG, Pagano RE. Cholesterol deprivation affects the fluorescence properties of a ceramide analog at the Golgi apparatus of living cells. Proc Natl Acad Sci USA. 1993;90:2661–2665. doi: 10.1073/pnas.90.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem Soc Trans. 1995;23:527–530. doi: 10.1042/bst0230527. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs: complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- Napier RM, Fowke LC, Hawes C, Lewis M, Pelham HRB. Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J Cell Sci. 1992;102:261–271. doi: 10.1242/jcs.102.2.261. [DOI] [PubMed] [Google Scholar]
- Neuwald AF. An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci. 1997;6:1764–1767. doi: 10.1002/pro.5560060817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezil FA, Bloom M. Combined influence of cholesterol and synthetic amphiphilic peptides upon bilayer thickness in model membranes. Biophys J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Schekman R, Perrelet A. Interleaflet clear space is reduced in the membrane of COP I and COP II-coated buds/vesicles. Proc Natl Acad Sci USA. 1996;93:8968–8970. doi: 10.1073/pnas.93.17.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113:1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano RE, Sepanski MA, Martin OC. Molecular trapping of a fluorescent ceramide analogue at the Golgi apparatus of fixed cells: interaction with endogenous lipids provides a trans-Golgi marker for both light and electron microscopy. J Cell Biol. 1989;109:2067–2079. doi: 10.1083/jcb.109.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton S. Correlative relationship of cholesterol and sphingomyelin in cell membranes. J Theor Biol. 1970;29:489–491. doi: 10.1016/0022-5193(70)90111-6. [DOI] [PubMed] [Google Scholar]
- Powers J, Barlowe C. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A, Desponds C, Conzelmann A. Biosynthesis of mannosylinositolphosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J Cell Biol. 1991;113:515–525. doi: 10.1083/jcb.113.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramstedt B, Slotte JP. Interaction of cholesterol with sphingomyelins and acyl-chain-matched phosphatidylcholines: a comparative study of the effect of the chain length. Biophys J. 1999;76:908–915. doi: 10.1016/S0006-3495(99)77254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem. 1998;273:26836–26843. doi: 10.1074/jbc.273.41.26836. [DOI] [PubMed] [Google Scholar]
- Recktenwald DJ, McConnell HM. Phase equilibria in binary mixtures of phosphatidylcholine and cholesterol. Biochemistry. 1981;20:4505–4510. doi: 10.1021/bi00518a042. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba NJP, Marsh D. Protein rotational diffusion and lipid/protein interactions in recombinants of bovine rhodopsin with saturated diacylphosphatidylcholines of different chain lengths studied by conventional and saturation-transfer electron spin resonance. Biochemistry. 1992;31:7511–7518. doi: 10.1021/bi00148a011. [DOI] [PubMed] [Google Scholar]
- Schneiter R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Schimmöller F, Singer-Krüger B, Riezman H. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in α-COP. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron K, et al. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood PW, Carlson M. Efficient export of the glucose transporter Hxt1p from the endoplasmic reticulum requires Gsf2p. Proc Natl Acad Sci USA. 1999;96:7415–7420. doi: 10.1073/pnas.96.13.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Ismb. 1998;6:175–182. [PubMed] [Google Scholar]
- Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Trilla JA, Duran A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Eukaryotic lipid-biosynthetic enzymes: the same but not the same. Trends Biochem Sci. 1998;23:423–428. doi: 10.1016/s0968-0004(98)01297-3. [DOI] [PubMed] [Google Scholar]
- van Helvoort A, Giudici ML, Thielemans M, van Meer G. Transport of sphingomyelin to the cell surface is inhibited by brefeldin A and in mitosis, where C6-NBD-sphingomyelin is translocated across the plasma membrane by a multidrug transporter activity. J Cell Sci. 1997;110:75–83. doi: 10.1242/jcs.110.1.75. [DOI] [PubMed] [Google Scholar]
- van Helvoort A, Smith AJ, Sprong H, Fritzsche I, Schinkel AH, Borst P, van Meer G. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell. 1996;87:507–517. doi: 10.1016/s0092-8674(00)81370-7. [DOI] [PubMed] [Google Scholar]
- van Meer G. Lipid traffic in animal cells. Annu Rev Cell Biol. 1989;5:247–275. doi: 10.1146/annurev.cb.05.110189.001335. [DOI] [PubMed] [Google Scholar]
- van Meer G. Lipids of the Golgi membrane. Trends Cell Biol. 1998;8:29–33. doi: 10.1016/s0962-8924(97)01196-3. [DOI] [PubMed] [Google Scholar]
- van Meer G, Stelzer EH, Wijnaendts-van-Resandt RW, Simons K. Sorting of sphingolipids in epithelial (Madin-Darby canine kidney) cells. J Cell Biol. 1987;105:1623–1635. doi: 10.1083/jcb.105.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, AlbertiSegui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN. Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction. Biochim Biophys Acta. 1999;1439:299–316. doi: 10.1016/s1388-1981(99)00102-x. [DOI] [PubMed] [Google Scholar]
- Webb RJ, East JM, Sharma RP, Lee AG. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: a possible mechanism for retention in the Golgi. Biochemistry. 1998;37:673–679. doi: 10.1021/bi972441+. [DOI] [PubMed] [Google Scholar]
- Wooding S, Pelham HR. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Murphy DJ, Georgopapadakou NH. Inhibition of yeast inositol phosphorylceramide synthase by aureobasidin A measured by a fluorometric assay. FEBS Lett. 1999;463:241–244. doi: 10.1016/s0014-5793(99)01633-6. [DOI] [PubMed] [Google Scholar]
- Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]