Abstract
Feline immunodeficiency virus (FIV) provides a valuable animal model by which criteria for lentivirus control strategies can be tested. Previous studies have shown that a 20-mer synthetic peptide of the membrane-proximal ectodomain of FIV transmembrane glycoprotein, designated peptide 59, potently inhibited the growth of tissue culture-adapted FIV in feline fibroblastoid CrFK cells. In the present report we describe the potential of this peptide to inhibit the replication of primary FIV isolates in lymphoid cells. Because antiviral activity of peptide 59 was found to map to a short segment containing three conserved Trp residues, further analyses focused on a derivative of eight amino acids (770W-I777), designated C8. Peptide C8 activity was found to be dependent on conservation of the Trp motif, to be removed from solution by FIV absorbed onto substrate cells, and to be blocked by a peptide derived from the N-terminal portion of FIV transmembrane glycoprotein. Structural studies showed that peptide C8 possesses a conformational propensity highly uncommon for peptides of its size, which may account for its considerable antiviral potency in spite of small size.
Initiation of cell infection by lentiviruses is a complex process involving multiple interactions between the glycoproteins (gp) of the viral envelope (Env) and cell surface molecules, with the transmembrane (TM) gp mediating the final event of membrane fusion and entry (6, 46, 68). In recent years, improved understanding of the structural features and functions of human immunodeficiency virus type 1 (HIV-1) TM gp41 (8, 15, 36, 38, 42, 44, 45, 59) has permitted the development of antiviral peptides derived from amino-terminal (N peptides) and carboxy-terminal (C peptides) ectodomains of this molecule, which effectively inhibit both HIV-1 entry into cells and HIV-1-induced cell fusion in vitro (17, 28, 29, 31, 32, 57, 65; A. R. Neurath, K. Lin, N. Strick, and S. Jiang, Letter, AIDS Res. Hum. Retrovir. 11:189-190, 1995). Some such peptides are currently under clinical evaluation for their therapeutic and prophylactic potentials (35). For example, in recent studies T-20, a 36-amino-acid C peptide also known as DP-178, has been found to be safe and powerfully suppress viral loads in HIV-1-infected individuals (33). It may also have applications for gene therapy (26). T-20 is believed to owe its antiviral activity primarily to hindrance of the receptor-triggered conformation rearrangements of TM gp41 that mediate virion and cell membrane fusion (9, 30, 34, 36, 56, 64). Although T-20 exerts its antiviral activity specifically against HIV-1 (65), similar results have been reported with simian immunodeficiency virus-derived peptides, supporting the concept that the viral entry functions associated with the TM gp ectodomain are essentially conserved among lentiviruses (39, 59).
Feline immunodeficiency virus (FIV) is a naturally occurring pathogen that causes an AIDS-like syndrome in domestic cats (50), and it is a valuable model system by which criteria for antilentiviral vaccine and drug development can be investigated (reviewed in references 5, 20, 21, and 66). The surface and TM gp of FIV and HIV-1 exhibit a common structural framework and appear to play similar roles in cell entry (6, 11, 19, 23, 46-48, 54, 60, 67, 68). We have previously screened 20- to 23-mer peptides covering the entire Env of FIV and found especially potent in vitro antiviral activity associated with peptides derived from two regions, one located in the N-terminal domain of the surface gp and the other located membrane proximally in the ectodomain of the TM gp (37). In subsequent studies (40), we focused on peptides of the surface gp, because the most efficacious of the TM peptides, referred to as peptide 59 (Fig. 1), had not appeared promising as a therapeutic strategy for naturally occurring infections since it had potently inhibited tissue culture-adapted (TCA) FIV in fibroblastoid cells (CrFK) but not a primary isolate in feline lymphoid cells. Prompted by the HIV-1 findings discussed above, we have now reexamined the antiviral properties of peptide 59 under modified experimental conditions. Removal of unadsorbed input virus from the test cultures revealed that peptide 59 and smaller derivatives potently inhibited the replication of both TCA and fresh isolates of FIV in feline lymphoid cells. Because the activity of peptide 59 mapped to a short stretch encompassing three conserved Trp residues, further analyses were carried out with a peptide of eight amino acids containing such a Trp motif (770W-I777), designated C8. Peptide C8 exerted a powerful antiviral effect on all the FIV isolates tested, and this activity was dependent on an intact Trp motif, was removed by FIV that had interacted with substrate cells, and was blocked by a peptide derived from the N-terminal segment of the FIV TM gp. Additional investigations have shown that peptide C8 possesses a conformational propensity highly uncommon for peptides of this size, which might explain why it is potently inhibitory in spite of its reduced size.
FIG. 1.
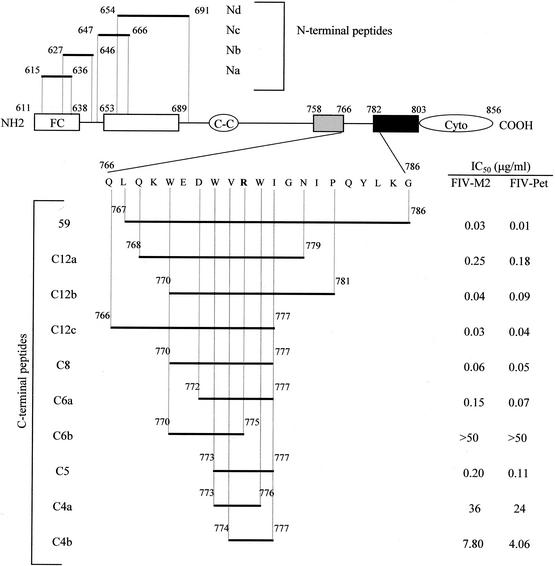
Schematic representation of the TM gp of FIV and derived synthetic peptides used in this study. Superscript numbers indicate amino acid positions starting with the first methionine of Env, according to the sequence of prototype FIV-Pet, clone 34TF10 (62). Domains are indicated as previously described (53). C peptides include peptide 59, described in a previous report (37), and nine shorter peptides. At position 775, the latter had a Gly instead of the Arg present in peptide 59, to better conform to the great majority of FIV published sequences (55). The IC50 for FIV-M2 and FIV-Pet of the C peptides found in one representative experiment out of three performed are also shown. N peptides were used only to investigate the possible site of action of peptide C8. Abbreviations and symbols: FC, fusion peptide; □, leucine zipper region; C-C, cysteine loop; ░⃞, membrane-proximal α-helix; ▪, membrane-spanning segment; Cyto, cytoplasmic region.
MATERIALS AND METHODS
Cells and viruses.
MBM cells are a line of CD3+, CD4−, and CD8− T lymphocytes originally established from the peripheral blood mononuclear cells (PBMC) of an FIV-negative and feline leukemia virus-negative cat (10). They are routinely grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 5 μg of concanavalin A, and 20 U of recombinant human interleukin-2 per ml. The viruses used included 11 primary isolates of subtype B (FIV-M2, -M3, -M19, -M22, -M45, -M73, -M82, -M88, -M91, -M92, and -M97) described in a previous report (10), one primary isolate of subtype A (FIV-GL8; kind gift of Os Jarrett, Glasgow, United Kingdom), and one TCA strain of subtype A (FIV-Pet; kindly provided by J. K. Yamamoto, Gainesville, Fla.). The viral stocks consisted of cell-free supernatant of chronically infected FL4 cells (FIV-Pet [70]) or of acutely infected MBM cells (all other viruses). Titration of the viral stocks was carried out in MBM cells and, depending on experiment design, included or did not include washout of unadsorbed virus after 5 h at 37°C to reproduce the conditions used in the FIV inhibition assay (see below). Internal inactivation of the viruses used in peptide binding experiments was carried out with 2,2′-dithiodipyridine (also known as aldrithiol-2 [AT2]) as previously described (24). In brief, viral stocks were incubated with 300 μM AT2 at 4°C for 2 h, pelleted in the cold at 20,000 × g for 90 min, and resuspended at 1 mg/ml in phosphate-buffered saline.
Peptide synthesis and purification.
Synthetic peptides (Fig. 1) were prepared manually using a conventional solid-phase strategy using N-α-fluorenylmethoxycarbonyl-protected amino acids and a p-alkoxybenzyl alcohol resin as solid support, as previously described (37). All were N-terminally acetylated and C-terminally amidated in order to improve enzymatic stability. Crude peptides were purified to homogeneity by semipreparative reverse-phase high-pressure liquid chromatography, with purity greater than 95%, and lyophilized. Final products were characterized by analytical high-pressure liquid chromatography and electrospray mass spectrometry.
Assays for FIV inhibition.
Cell-free virus inhibition assays were performed in 96-well flat-bottom microplates against 10 50% tissue culture infectious doses (TCID50) of FIV, using MBM cells as substrate and supernatant p25 antigen quantification as an end point. In some experiments, larger doses of virus were also used. Virus was appropriately diluted and mixed with an equal volume of the test peptides diluted from 0.0005 to 50 μg/ml (final concentrations) or with diluent alone. The mixtures were then immediately inoculated (100 μl/well) into two to four wells containing 105 MBM cells in 100 μl of culture medium. After 5 h at 37°C, the inocula were rinsed out and replaced with fresh medium, while after 4 additional days 100 μl of supernatant was removed from each well and replaced with fresh medium. Deviations from this standard protocol are described below. In any case, the cultures were stopped on day 8, when p25 production by control wells inoculated with virus alone was well evident. The enzyme-linked immunosorbent assay for measuring p25 has been described previously (37). Peptide inhibition of virus growth was calculated using the formula (mean p25 concentration in wells inoculated with FIV + peptides/mean p25 concentration in wells inoculated with FIV alone) × 100. Fifty percent inhibitory concentrations (IC50) were calculated using the predicted exponential growth function in Microsoft Excel, which uses existing x-y data to estimate the corresponding peptide concentration (x) from a known value (y), which in this case was 50% infectivity. Mean IC50 ± standard deviations (SD) were calculated using all replicates. All experiments were repeated at least twice.
The assay for cell-associated FIV inhibition was performed as described above, except that the viral inoculum was 105 fresh Ficoll-separated PBMC pooled from three specific-pathogen-free cats infected with FIV-M2 6 months earlier, the wells were not rinsed after contact, and the medium added on day 4 contained appropriate concentrations of test peptides. Parallel wells showed that such inoculum titrated approximately 102 infectious units in substrate MBM cells but produced no detectable p25 antigen if incubated alone.
CD and NMR.
Circular dichroism (CD) spectra were recorded using a Jasco J-710 spectropolarimeter, with a cell of 1 mm in path length. The CD measures were performed in the near-UV CD range with 1-nm bandwidth, four accumulations, and scanning speed of 20 nm/min, at room temperature. The pH of the aqueous sample was adjusted to 6.6 by adding small amounts of phosphate buffer solution. Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker DRX-600 spectrometer. Samples were prepared by dissolving the appropriate amount of the test peptides in 0.5 ml of H2O-phosphate buffer (pH 6.6) to obtain a 1 mM solution. Lyophilized peptides were dissolved in 8 mM sodium phosphate buffer containing 80% dimethyl sulfoxide (DMSO). One-dimensional NMR spectra were recorded in the Fourier mode with quadrature detection, and the water signal was suppressed by a low-power selective irradiation in the homogated mode. Double-quantum-filtered correlated spectroscopy, total correlated spectroscopy (TOCSY), and nuclear Overhauser enhancement spectroscopy (NOESY) experiments (4, 27, 51) were run in the phase-sensitive mode using quadrature detection in 1 by time-proportional phase increase of initial pulse. Data block sizes were 2,048 addresses in t2 and 512 equidistant t1 values. Before Fourier transformation, the time domain data matrices were multiplied by shifted sin2 functions in both dimensions. NOESY experiments were run at 270 K in DMSO-water. A mixing time of 70 ms was used for the TOCSY experiments, while mixing times in the range of 150 to 300 ms were used for NOESY experiments. The qualitative and quantitative analyses of double-quantum-filtered correlated spectroscopy, TOCSY, and NOESY spectra were obtained using the interactive program package SPARKY. The nuclear Overhauser effect (NOE)-based distance restraints were obtained from NOESY spectra collected with a mixing time of 250 ms.
Structure calculation.
The NOE cross peaks were integrated with the SPARKY program and converted into upper distance bounds using the CALIBA module of DYANA software (25). Cross peaks which overlapped by more than 50% were treated as weak restraints in the DYANA calculation. An ensemble of 50 structures was generated with distance geometry-simulated annealing by program DYANA using 75 NOE-based distance constraints. No dihedral angle restraints nor hydrogen bond restraints were used. The necessary pseudoatom corrections were applied for nonstereospecifically assigned protons at prochiral centers and for the methyl group. After discarding redundant and duplicated constraints, the final list included 51 intraresidue and 113 interresidue constraints, which were used to generate an ensemble of 50 structures by the standard protocol of simulated annealing in torsion angle space implemented in DYANA (using 6,000 steps) The best 20 structures with low values of target functions (0.83 to 1.19) and small residual violations (maximum violation = 0.38 Å) were refined by in vacuo minimization in the AMBER 1991 force field, using the program SANDER of the AMBER suite (version 5.0; University of California, San Francisco [63]). To mimic the effect of solvent screening, all net charges were reduced to 20% of their real value, and moreover a distance-dependent dielectric constant (ɛ = r) was used. The cutoff for nonbonded interactions was 12 Å. The NMR-derived upper bounds were imposed as semiparabolic penalty functions, with force constants of 16 Kcal/mol Å2; the function was shifted to linear when the violation exceeded 0.5 Å. The best 10 structures after minimization had AMBER energies ranging from −441.4 to −391.1 kcal/mol. The final structures were analyzed using the Insight 98.0 program. Computations were performed on SGI Indigo II computers.
RESULTS
Inhibition of FIV replication in feline lymphoid cells by peptide 59.
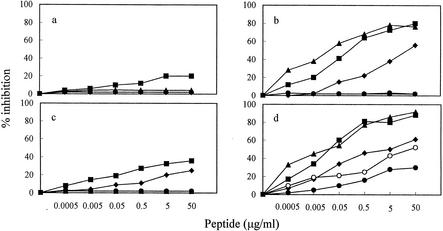
To establish the optimal conditions for the antiviral action of peptide 59 in vitro, we examined the effects of varying the conditions of virus-peptide-cell contact. As shown by Fig. 2, removing the inoculum virus-peptide mixtures after virus-cell adsorption greatly affected outcome. When the inocula were left in the MBM cell cultures throughout the incubation period, TCA FIV-Pet inhibition was less evident (Fig. 2c) than that previously observed with CrFK cells (37), and FIV-M2 inhibition was virtually absent (Fig. 2a). In contrast, removing the inocula after 5 h of contact markedly augmented FIV-Pet inhibition (Fig. 2d) and rendered FIV-M2 inhibition also very clear (Fig. 2b). Time of peptide addition proved also critical. Pretreating cells and, in particular, adding the peptide 1 h after the virus markedly reduced the effect. In contrast, mixing virus and peptide immediately before inoculation, or 1 h earlier, yielded equally optimal inhibition. In subsequent experiments virus and peptides were therefore mixed just before addition to cells and washed off 5 h later.
FIG. 2.
Effect of varying the experimental conditions on FIV inhibition by peptide 59. Test viruses were the primary isolate FIV-M2 (a and b) and the TCA strain FIV-Pet (c and d). The peptide was mixed with virus 1 h before infection (▪) or added to cells 1 h before (♦), simultaneously with (▴), or 1 h after the virus. In the latter case, virus-cell contact was carried out at 4°C (○) or 37°C (•). Inoculum virus-peptide mixtures were washed out after 5 h of contact (b and d) or left in the cultures throughout the incubation period (a and c). A 20-mer control peptide derived from a different region of the FIV TM was devoid of activity (not shown).
FIV inhibition by peptides of reduced size.
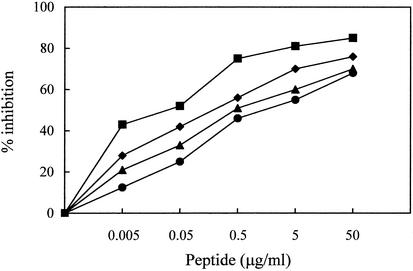
Peptide 59 contains a motif of three Trp residues separated, from each other, by 2 amino acids (Fig. 1). The fact that several Trp residues are highly conserved in the membrane-proximal ectodomain of the TM gp of lentiviruses suggests that these residues exert an essential role for viral infectivity (59). Moreover, an inhibitory C peptide of HIV-1 corresponding in location to peptide 59 was shown to lose much activity following deletion of the Trp-rich domain (64, 65). For these reasons, the derivatives of peptide 59 that were assayed for anti-FIV activity (Fig. 1) had all, or most, of the Trp residues conserved. As summarized by Fig. 1, all peptides comprising eight (peptide C8) or more amino acids inhibited both FIV-M2 and FIV-Pet, with IC50 ranging between 0.03 and 0.25 μg/ml. On the other hand, of the two hexamer peptides examined, the one with Trp-770 truncated (C6a) showed reduced activity, while the one with Trp-776 truncated (C6b) was totally ineffective. Finally, the 5-mer peptide having both Trp-773 and Trp-776 included in it still had some activity, and the two 4-mer peptides were essentially ineffective. Thus, peptide size could be reduced considerably with little or no loss of antiviral activity. Subsequent studies focused on peptide C8, i.e., on the shortest among the above peptides that was fully active and had all three Trp conserved. This peptide was tested for its ability to inhibit graded doses of FIV. The results of a representative experiment are shown in Fig. 3. Overall, in three experiments the mean IC50 ± SD were 0.07 ± 0.01, 0.31 ± 0.07, 0.92 ± 0.05, 2.02 ± 0.1 μg/ml against 10, 32, 100, and 320 TCID50, respectively. Similar to peptide 59, peptide C8 had no detectable effects on MBM cell viability and proliferation (data not shown).
FIG. 3.
Inhibition of graded doses of cell-free FIV-M2 by peptide C8. Peptide diluted from 0.005 to 50 μg/ml (final concentration) was mixed with 10 (▪), 32 (♦), 100 (▴), and 320 (•) TCID50 of virus. Except for virus dose, the procedure was as described in Materials and Methods. The experiment was repeated three times with comparable results.
Importance of the Trp motif for antiviral activity of peptide C8.
To assess the importance of the Trp residues for antiviral activity of peptide C8, we performed an Ala scan study, in which modified peptides were compared for anti-FIV activity with the wild-type peptide and a scrambled form of it (C8scr), thus obtaining information about each side chain. Test viruses were the ones used above plus FIV-GL8, a primary isolate of subtype A. As shown in Table 1, Ala substitution for Trp-770 markedly reduced effectiveness, while Ala substitution for Trp-773 or Trp-776 completely eliminated it. All other Ala substitutions had little or no effect, whereas C8scr was completely inactive. Thus, these findings suggest that an intact Trp motif is crucial for a robust antiviral activity.
TABLE 1.
Effect of alanine scan on FIV-inhibitory activity of peptide C8
| Peptide | Mean IC50 ± SD (μg/ml)a |
||
|---|---|---|---|
| FIV-M2 | FIV-GL8 | FIV-Pet | |
| C8 | 0.07 ± 0.02 | 0.46 ± 0.20 | 0.05 ± 0.01 |
| C8scr | >50 | >50 | >50 |
| C8W→A770 | 4.30 ± 0.70 | >50 | 2.10 ± 0.10 |
| C8E→A771 | 0.16 ± 0.03 | 0.22 ± 0.05 | 0.12 ± 0.01 |
| C8D→A772 | 0.10 ± 0.02 | 0.40 ± 0.22 | 0.03 ± 0.06 |
| C8W→A773 | >50 | >50 | >50 |
| C8V→A774 | 0.07 ± 0.03 | 0.54 ± 0.02 | 0.15 ± 0.12 |
| C8G→A775 | 0.09 ± 0.03 | 0.38 ± 0.04 | 0.25 ± 0.06 |
| C8W→A776 | >50 | >50 | >50 |
| C8I→A777 | 0.15 ± 0.01 | 0.64 ± 0.08 | 0.13 ± 0.04 |
Values obtained in two to three independent assays.
Peptide C8 inhibition of different primary FIV isolates.
As described above, primary isolate FIV-GL8 had appeared somewhat more resistant to C8 inhibition than primary isolate FIV-M2 (Table 1). It appeared therefore of interest to extend the study to a larger number of primary isolates. Ten additional local isolates selected at random from our collection were infectivity titrated in MBM cells and then assayed for susceptibility to C8. All were effectively inhibited, with IC50 ranging between 0.03 and 0.63 μg/ml (data not shown), thus revealing a considerable breadth of activity although with some variation depending on the FIV isolate tested.
C8 inhibition of cell-associated FIV.
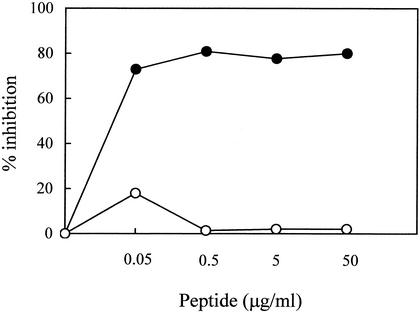
Since lentiviruses can spread directly via cell-to-cell contact (5), it was of interest to verify whether peptide C8 might block this form of infection. PBMC of FIV-M2 infected cats were cocultivated with MBM cells in the presence or absence of C8. Figure 4 shows that C8, but not C8scr, inhibited cell-associated infection with an IC50 of 0.015 μg/ml. This observation agrees with HIV-1 studies showing that inhibitory peptides block cell-associated infectivity at lower concentrations than that required for cell-free virus (65).
FIG. 4.
Inhibition of cell-associated FIV infection by peptide C8. PBMC, freshly harvested from long-term FIV-M2-infected cats and containing approximately 102 infectious units as determined in substrate MBM cells, were mixed with C8 (•) or C8scr (○). Test cultures were then processed as described for inhibition of cell-free virus except that the wells were not rinsed after contact and the medium added on day 4 contained appropriate concentrations of test peptides.
Peptide C8 binds to cell-associated FIV.
In the experiments in Fig. 2, the antiviral effects of peptide 59 were reduced when the peptide was preincubated with substrate cells prior to their infection, suggesting that it may bind to, or be taken up by, the cells, as reported for certain anti-HIV-1 peptides (61). To investigate this possibility, peptide C8 was incubated with either MBM cells alone, FIV particles alone, or both, and the residual inhibitory activity in the culture supernatant was determined after 1 h at 4°C. The FIV particles used in this study were rendered noninfectious by the internal inactivating agent AT2 (24). This agent has previously been shown to inactivate HIV and SIV without affecting Env structure and functions, including cell entry (2, 58). We also investigated the effect of conformational changes in the viral Env on the capacity to bind peptide C8. This was done by comparing the adsorption capacity of virus-cell mixtures preincubated at either 4°C for 1 h or 37°C for 3 h. The results are shown in Table 2. Preincubation with MBM cells alone had a negligible effect on the antiviral properties of peptide C8. Likewise, two primary isolates of FIV, FIV-M2 and FIV-GL8, also appeared unable to bind the peptide. However, the conformational changes which occurred in Env following binding of these primary FIV isolates to cells markedly enhanced the capacity to remove peptide C8 activity, as shown by the fact that virus-MBM cell mixtures preincubated under conditions which permitted virus adsorption to cells but not under conditions which also permitted cell entry effectively removed C8 activity from solution. On the other hand, TCA FIV-Pet removed much peptide C8 activity, even without having previously interacted with cells. Thus, these findings showed that FIV has the capacity to bind peptide C8 but, in the case of primary isolates, this occurs only after virus has adsorbed onto cells.
TABLE 2.
Effect of preincubation with FIV, substrate cells, or virus-cell mixtures on FIV-inhibitory activity of peptide C8
| Expt no. | Virus used | Mean IC50 ± SD (μg/ml) of C8 preincubated witha: |
||||
|---|---|---|---|---|---|---|
| Nothing | Virionsb alone | Cellsc alone | Virions + cellsd (no entry allowed) | Virions + cellse (entry allowed) | ||
| 1 | FIV-M2 | 0.04 ± 0.02 | 0.03 ± 0.01 | 0.08 ± 0.03 | >1 | 0.04 ± 0.03 |
| 2 | FIV-GL8 | 0.46 ± 0.10 | 0.77 ± 0.20 | 0.38 ± 0.03 | >1 | 0.46 ± 0.13 |
| 3 | FIV-Pet | 0.03 ± 0.02 | >1 | 0.06 ± 0.04 | >1 | 0.03 ± 0.01 |
Peptide C8 (0.8 ml of a 5-μg/ml solution) was preincubated with the indicated material at 4°C for 1 h, centrifuged at 600 × g for 15 min (to remove cells) followed by 20,000 × g for 90 min (to remove virions), diluted from 0.001 to 1 μg/ml, and then assayed for inhibition of the same FIV strain used in C8 preincubation, by the standard procedure. Values obtained in two to three independent assays.
FIV inactivated with AT2 (5 μg protein).
Viable MBM cells (5 × 105).
FIV inactivated with AT2 (5 μg of protein) and incubated with MBM cells (5 × 105) at 4°C for 1 h before C8 addition, to permit virus adsorption but not entry (24).
FIV inactivated with AT2 (5 μg of protein) and incubated with MBM cells (5 × 105) at 37°C for 3 h before C8 addition, to permit both virus adsorption and entry (24).
In a second approach, we examined whether C8 activity was affected by mixing with peptides derived from the N-terminal portion of FIV TM gp (N peptides), as suggested by previous studies on HIV-1 which indicated that inhibitory C peptides act by binding to specific N-terminal segments of the TM gp41 (9, 30, 34, 36, 56, 64). The synthetic N peptides used comprised amino acid positions 615 to 636 (Na), 627 to 646 (Nb), 647 to 666 (Nc), and 654 to 691 (Nd) (Fig. 1). Since these N peptides were also weakly inhibitory for FIV replication (reference 37 and data not shown), the mixtures contained a fixed volume of the N peptides diluted to the respective IC50. As shown in Table 3, peptide Nb produced a 25-fold reduction of the expected IC50 value of C8, whereas the other N peptides left C8 activity essentially unchanged. The antiviral effect in control cultures in which C8 was substituted with either C8W→A776 or C8scr was as with the N peptides alone. Thus, the presence of a specific N-terminal segment of the TM gp of FIV eliminated C8-mediated inhibition of FIV replication.
TABLE 3.
Effect of mixing with N peptides on FIV-inhibitory activity of peptide C8
| Test peptide | Mean IC50 ± SD (μg/ml) of mixture of test peptide and N peptidea |
||||
|---|---|---|---|---|---|
| None | Na (1.20)b | Nb (1.20) | Nc (1.20) | Nd (0.05) | |
| C8 | 0.06 ± 0.01 | 0.06 ± 0.01 | 1.5 ± 0.80 | 0.07 ± 0.03 | 0.01 ± 0.01 |
| C8W→A776 | >50 | 1.50 ± 0.10 | 1 ± 0.20 | 1 ± 0.24 | 0.06 ± 0.02 |
| C8Scr | >50 | 1.50 ± 0.15 | 1 ± 0.23 | 1 ± 0.20 | 0.06 ± 0.02 |
| None | >50 | 1.20 | 1.20 | 1.20 | 0.05 |
Test peptides diluted to contain final concentrations ranging from 0.0005 to 50 μg/ml were mixed with fixed IC50 of the indicated N peptides, incubated at 4°C for 30 min, and then assayed for cell-free FIV inhibition by the standard procedure. Values obtained in two to three independent assays.
The IC50 of the indicated N peptide is given in parentheses in micrograms per milliliter.
Conformational analysis of peptide C8.
The above findings were conducive to the idea that peptide C8 had a stable conformation in spite of its small size. This possibility was addressed by comparing the CD and NMR properties of C8 and the inactive Ala analogue C8W→A776. A preliminary screening of the conformational preferences of the two peptides as a function of the environment was performed by CD spectroscopy. CD spectra were run in water and DMSO-water (80:20, vol/vol). The spectra in water showed that both peptides, like most similarly short linear peptides, were present in extended, random-coil conformations. On the contrary CD spectra recorded in the near-UV CD range showed well-defined bands at the longer wavelength adsorption maxima of tryptophan, anticipating the NMR finding that the backbone and consequently the indole side chains are in a defined structural orientation (see below).
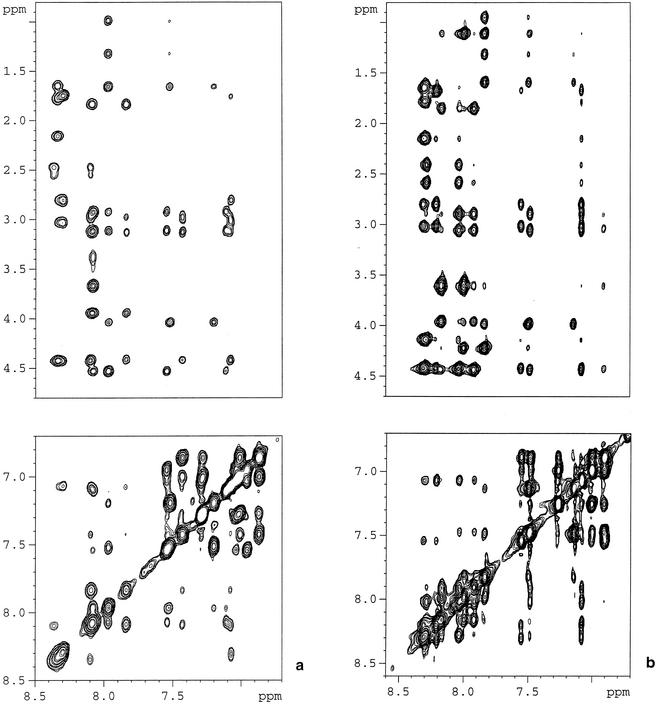
Previous findings have shown that the DMSO-water solution, due to its high viscosity, affects the equilibrium among isoenergetic conformers, thus selecting the more-ordered ones (1), and produces physicochemical conditions compatible with those of biological fluids (16). Based on these findings and the above CD data, analysis of the conformational preferences of C8 and C8W→A776 by NMR spectroscopy was performed in DMSO-water. Figure 5 shows amide and fingerprint regions of the NOESY spectra (270 K; 600 MHz; mixing time, 300 ms). The presence of a high number of well-resolved cross peaks, unusual for peptides as short as the ones under scrutiny, is evident: particularly notable are a series of sequential (NHi, NHi + 1) NOEs and of diagnostically critical (i, i + 3) effects, which are strongly indicative of the predominance of ordered conformers.
FIG. 5.
NMR spectra of peptide C8 and its inactive Ala analogue C8W→A776. Shown are the amide (top) and the fingerprint (bottom) regions of the NOESY spectra of C8 (a) and C8W→A776 (b) collected in DMSO-water at 270 K (600 MHz; mixing time, 300 ms).
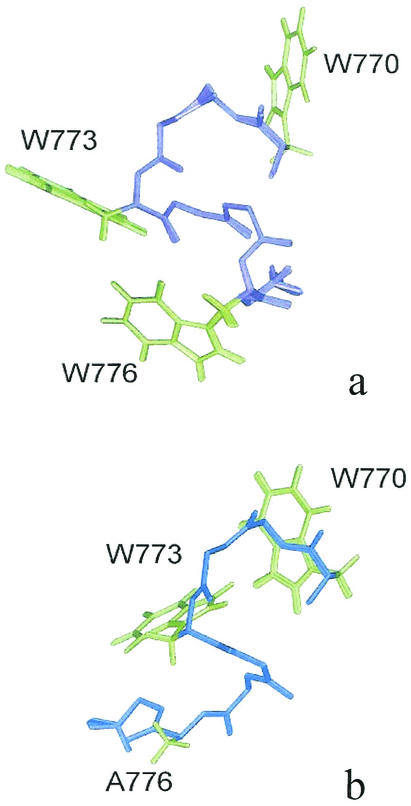
Three-dimensional structures were calculated from NMR data by simulated annealing in torsion angle space using the DYANA software package and refined by a minimization procedure using the SANDER module of AMBER software (version 5.0). Figure 6 shows the overlap of the best 20 structures of peptides C8 and C8W→A776. The fit was calculated on the backbone heavy atoms of all residues 770 to 777. Peptide C8 structure is characterized by a well-defined α-turn structure in the 773 to 776 segment, stabilized by the presence of H bonds between the C=O of Asp-772 and the NH of Gly-775, as well as between the C=O of Trp-773 and the NH of Trp-776 (Fig. 6a). Also, peptide C8 conformation is characterized by a well-defined orientation of the side chains, including the Trp rings. Peptide C8W→A776 is characterized by a folded, ordered conformation in the four N-terminal residues, whereas it shows a prevailing extended-coil structure in the remaining C-terminal residues (Fig. 6b). Consistently, overlapping of the NMR structures of C8 and C8W→A776 leads to a good backbone atoms fitting in the four N-terminal residues and shows divergent conformations in the C-terminal portion. Inspection of the side chain arrangement of peptide C8 shows that the aromatic rings of Trp-773 and Trp-776 are located on the same side of the structure, whereas in the inactive analogue C8W→A776 the region corresponding to Trp-776 in C8 is occupied by the extended N-terminal backbone.
FIG. 6.
Three-dimensional structure of peptide C8 and its inactive Ala analogue C8W→A776. Shown are the superpositions of the best 20 structures of peptides C8 (a) and C8W→A776 (b) obtained from DYANA structure calculations and following minimization using the SANDER module of AMBER software (version 5.0). The backbones are displayed in violet for C8 and in blue for C8W→A776. The side chains of Trp residues are shown (green) as well as the side chains of the Ala-776 of C8W→A776.
DISCUSSION
Peptide 59, a 20-mer C peptide of FIV that had previously proven inhibitory for TCA FIV in vitro (37), was found to effectively inhibit a primary FIV isolate as well, provided that test conditions were appropriately set. Prerequisite for inhibition—absolute for the fresh isolate but well evident also for the TCA virus—was that the inoculum virus-peptide mixtures were removed from the test cultures after adsorption, which suggests that peptide activity is short-lived, possibly due to rapid uptake and/or degradation by cells. Analysis of smaller derivatives of this peptide mapped the inhibitory activity to a small segment containing three Trp residues, separated from one another by two amino acids. Peptide C8, i.e., the shortest derivative of peptide 59 that had this Trp motif wholly conserved and was still fully active, was further investigated for inhibitory potency and breadth, critical residues, and site of action.
Peptide C8 inhibited all the primary isolates tested as cell-free virus across two FIV clades with moderate variations in IC50 and was also active against cell-associated virus. In particular, cell-free FIV-M2 was inhibited also when used at concentrations as high as 320 TCID50 per well. Furthermore, its activity proved markedly dependent on an intact Trp motif, as revealed by testing a series of analogues in which each individual amino acid Ala was substituted. In addition, peptide C8 activity was removed by preincubation with two primary FIV isolates adsorbed onto substrate cells but not by preincubation with the same viruses alone, cells alone, or cells which had permitted virus entry, demonstrating that its target is viral in nature but becomes significantly accessible or functional only after FIV has undergone postadsorption changes. That TCA FIV-Pet bound substantial C8 peptide even without prior interaction with substrate cells might be a consequence of the differences evolved by TCA relative to freshly isolated lentiviruses, including a looser Env organization (3, 43), and possibly explains the great sensitivity of this virus to peptide inhibition. Finally, peptide C8 was found to be blocked in its antiviral action by a peptide corresponding to a conserved segment at the N terminus of FIV TM gp, near the fusion domain, suggesting that this might be the site of its action. Proving unequivocally the latter aspect will, however, require a finer dissection of the phenomenon.
Collectively, the above findings are markedly reminiscent of observations with HIV-1-inhibitory C peptides (28, 29, 31, 32, 65; Neurath et al., letter). In particular, HIV-1-inhibitory C peptides act on gp41 that has undergone receptor-activated conformational changes and not on native, resting gp41 (6, 8, 15, 35, 42, 65). Also, the already-mentioned T-20, as well as C34, T649, and other peptides, seems to act by binding the N-terminal domain of gp41, as shown by peptide mixing experiments similar to the ones reported here and other evidence (9, 30, 34, 36, 56, 64). In turn, such binding is believed to inhibit the otherwise-ensuing association between structures at the N terminus of gp41, including a hydrophobic cavity and the C terminus of the same molecule, thus ultimately preventing fusion and entry. Note, however, that recent findings that T-20 activity can be modulated by HIV-1 coreceptor specificity, which is known to be defined by V3 sequences in the surface gp, suggest the existence of additional elements regulating T-20 target exposure (12, 13).
A peculiar aspect of the present results is the small size of some FIV-inhibitory C peptides studied here. As a rule, oligopeptides of such small size are devoid of consistent activities due to absent or poor conformation stability. Indeed, all consistently effective HIV fusion-inhibitory C peptides described to date are composed of 30 or more residues (31, 64). Our structural studies of peptide C8, however, suggest that at least this peptide might be unusually conformationally stable. In fact, NMR analysis of peptide C8 dissolved in DMSO-water to recreate physicochemical conditions compatible with biological environments (1, 16) revealed a remarkable degree of conformational propensity, highly uncommon in similarly short peptides. Moreover, the presence in C8 of three equally distanced Trp residues was found to determine a folded compact structure which appeared to correlate with antiviral activity, since substitution of an Ala for Trp-776 greatly affected conformational features and eliminated antiviral activity as well. The orientation of the aromatic side chains of Trp-773 and Trp-776 on the same side of the C8 structure, which was observed in the main conformer, is suggestive of a possible direct involvement of these residues in hydrophobic interaction with the C8 target site. As determined by mutagenesis and crystal structure analyses, the target cavity of HIV-1 gp41 where C peptide inhibitors are believed to exert their action is susceptible to filling by small-molecule compounds. Nevertheless, small compounds capable of filling such a cavity—identified by combinatorial chemistry and other techniques—have exhibited no anti-HIV-1 activity, and this has been attributed to conformational instability (7, 9, 18, 22). Attempts to increase the conformational stability of small peptides have included chemical cross-linking, introduction of helix-capping motifs, and substitutions with amino acids of high helical propensity with variable results (31, 32). In any case, a stable helical structure appears to be important for both binding affinity to the gp41 target and antiviral activity (22).
In HIV-1, the C-terminal membrane-proximal ectodomain of gp41 not only has served for designing antiviral peptides but also is viewed as an extremely interesting target for broad range antibody-mediated neutralization due to its critical role in viral entry and wide conservation (49, 69, 71). Unfortunately, antibodies reactive with this region are very infrequent in HIV-1 patients and have never been elicited with candidate vaccines, so far (69). In our hands, the corresponding domain of FIV is also poorly immunogenic, as determined in infected cats and in mice repeatedly injected with derived peptides (41). However, groups using other strains of FIV have shown that the same domain is widely recognized by infected cat sera and also immunogenic when presented to cats as a fusion protein (14, 52). In addition, cat immunization with a peptide based on such a domain was seen to delay infection following FIV challenge (53). Thus, the C-terminal membrane-proximal ectodomain of the TM gp should be further evaluated also for the design of FIV immunoprophylactic agents. The small size and structural features of peptide C8 make it an ideal lead compound for the development of novel peptide-mimetic anti-FIV drugs and vaccines.
Acknowledgments
This work was supported by grants from the Ministero dell'Istruzione, dell'Università e della Ricerca, Rome, Italy.
REFERENCES
- 1.Amodeo, P., A. Motta, D. Picone, G. Saviano, T. Tancredi, and P. A. Temussi. 1991. Viscosity as a conformational sieve. NOE of linear peptides in cryoprotective mixtures. J. Magn. Reson. 95:201-207. [Google Scholar]
- 2.Arthur, L. O., J. W. Bess, Jr., E. N. Chertova, J. L. Rossio, M. T. Esser, R. E. Benveniste, L. E. Henderson, and J. D. Lifson. 1998. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc finger: a candidate SIV vaccine. AIDS Res. Hum. Retrovir. 14:S311-S319. [PubMed] [Google Scholar]
- 3.Baldinotti, F., D. Matteucci, P. Mazzetti, C. Giannelli, P. Bandecchi, F. Tozzini, and M. Bendinelli. 1994. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J. Virol. 68:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bax, A., and D. G. Davis. 1985. Mlev-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65:355-360. [Google Scholar]
- 5.Bendinelli, M., M. Pistello, S. Lombardi, A. Poli, C. Garzelli, D. Matteucci, L. Ceccherini-Nelli, G. Malvaldi, and F. Tozzini. 1995. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin. Microbiol. Rev. 8:87-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C.-H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Mauro, D., D. Matteucci, S. Giannecchini, F. Maggi, M. Pistello, and M. Bendinelli. 1998. Autologous and heterologous neutralization analyses of primary feline immunodeficiency virus isolates. J. Virol. 72:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Parseval, A., and J. H. Elder. 2001. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J. Virol. 75:4528-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Ronde, A., J. G. Stam, P. Boers, H. Langedijk, R. Meloen, W. Hesselink, L. C. E. J. M. Keldermans, A. van Vliet, E. J. Verschoor, M. C. Horzinek, and H. F. Egberink. 1993. Antibody response in cats to the envelope proteins of feline immunodeficiency virus: identification of an immunodominant neutralization domain. Virology 198:257-264. [DOI] [PubMed] [Google Scholar]
- 15.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douzou, P., and G. A. Petsko. 1984. Proteins at work: “stop-action” pictures at subzero temperatures. Adv. Protein Chem. 36:245-361. [PubMed] [Google Scholar]
- 17.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 19.Egberink, H. F., E. De Clercq, A. L. van Vliet, J. Balzarini, G. J. Bridger, G. Henson, M. C. Horzinek, and D. Schols. 1999. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J. Virol. 73:6346-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elder, J. H., G. A. Dean, E. A. Hoover, J. A. Hoxie, M. H. Malim, L. Mathes, J. C. Neil, T. W. North, E. Sparger, M. B. Tompkins, W. A. F. Tompkins, J. Yamamoto, N. Yuhki, N. C. Pedersen, and R. H. Miller. 1998. Lesson from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:797-801. [DOI] [PubMed] [Google Scholar]
- 21.Elder, J. H., and T. R. Phillips. 1995. Feline immunodeficiency virus as a model for development of molecular approaches to intervention strategies against lentivirus infections. Adv. Virus Res. 45:225-247. [DOI] [PubMed] [Google Scholar]
- 22.Ferrer, M., T. M. Kapoor, T. Strassmaier, W. Weissenhorn, J. J. Skehel, D. Oprian, S. L. Schreiber, D. C. Wiley, and S. C. Harrison. 1999. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat. Struct. Biol. 6:953-960. [DOI] [PubMed] [Google Scholar]
- 23.Frey, S. C. S., E. A. Hoover, and J. I. Mullins. 2001. Feline immunodeficiency virus cell entry. J. Virol. 75:5433-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannecchini, S., P. Isola, O. Sichi, D. Matteucci, M. Pistello, L. Zaccaro, D. Del Mauro, and M. Bendinelli. 2002. AIDS vaccination studies using an ex vivo feline immunodeficiency virus model: failure to protect and possible enhancement of challenge infection by four cell-based vaccines prepared with autologous lymphoblasts. J. Virol. 76:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guntert, P., C. Mumenthaler, and K. Wüthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 26.Hildinger, M., M. T. Dittmar, P. Schult-Dietrich, B. Fehse, B. S. Schnierle, S. Thaler, G. Stiegler, R. Welker, and D. von Laer. 2001. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J. Virol. 75:3038-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeener, J., B. H. Meyer, P. Bachman, and R. R. Ernst. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71:4546-4553. [Google Scholar]
- 28.Ji, H., W. Shu, F. T. Burling, S. Jiang, and M. Lu. 1999. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J. Virol. 73:8578-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, S., K. Lin, L. Zhang, and A. K. Debnath. 1999. A screening assay for antiviral compounds targeted to the HIV-1 gp41 core structure using a conformation-specific monoclonal antibody. J. Virol. Methods 80:85-96. [DOI] [PubMed] [Google Scholar]
- 31.Jin, B.-S., J.-R. Ryu, K. Ahn, and Y. G. Yu. 2000. Design of a peptide inhibitor that blocks the cell fusion mediated by glycoprotein 41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:1797-1804. [DOI] [PubMed] [Google Scholar]
- 32.Judice, J. K., J. Y. K. Tom, W. Huang, T. Wrin, J. Vennari, C. J. Petropoulos, and R. S. McDowell. 1997. Inhibition of HIV type 1 infectivity by constrained α-helical peptides: implications for the viral fusion mechanism. Proc. Natl. Acad. Sci. USA 94:13426-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 34.Kliger, Y., S. A. Gallo, S. G. Peisajovich, I. Muñoz-Barroso, S. Avkin, R. Blumenthal, and Y. Shai. 2000. Mode of action of an antiviral peptide from HIV-1: inhibition at a post lipid-mixing stage. J. Biol. Chem. 276:1391-1397. [DOI] [PubMed] [Google Scholar]
- 35.LaBranche, C. C., G. Galasso, J. P. Moore, D. P. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antivir. Res. 50:95-115. [DOI] [PubMed] [Google Scholar]
- 36.Lawless, M. K., S. Barney, K. I. Guthrie, T. B. Bucy, S. R. Petteway, Jr., and G. Merutka. 1996. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry 35:13697-13708. [DOI] [PubMed] [Google Scholar]
- 37.Lombardi, S., C. Massi, E. Indino, C. La Rosa, P. Mazzetti, M. L. Falcone, P. Rovero, A. Fissi, O. Pieroni, P. Bandecchi, F. Esposito, F. Tozzini, M. Bendinelli, and C. Garzelli. 1996. Inhibition of feline immunodeficiency virus infection in vitro by envelope glycoprotein synthetic peptides. Virology 220:274-284. [DOI] [PubMed] [Google Scholar]
- 38.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Numberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massi, C., E. Indino, C. Lami, A. Fissi, O. Pieroni, C. La Rosa, F. Esposito, C. Galoppini, P. Rovero, P. Bandecchi, M. Bendinelli, and C. Garzelli. 1998. The antiviral activity of a synthetic peptide derived from the envelope SU glycoprotein of feline immunodeficiency virus maps in correspondence of an amphipathic helical segment. Biochem. Biophys. Res. Commun. 246:160-165. [DOI] [PubMed] [Google Scholar]
- 41.Massi, C., S. Lombardi, E. Indino, D. Matteucci, C. La Rosa, F. Esposito, C. Garzelli, and M. Bendinelli. 1997. Most potential linear B cell epitopes of Env glycoproteins of feline immunodeficiency virus are immunogenically silent in infected cats. AIDS Res. Hum. Retrovir. 13:1121-1129. [DOI] [PubMed] [Google Scholar]
- 42.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. P., P. W. H. I. Parren, and D. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muñoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pancino, G., L. Camoin, and P. Sonigo. 1995. Structural analysis of the principal immunodominant domain of the feline immunodeficiency virus transmembrane glycoprotein. J. Virol. 69:2110-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pancino, G., I. Fossati, C. Chappey, S. Castelot, B. Hurtrel, A. Moraillon, D. Klatzmann, and P. Sonigo. 1993. Structure and variations of feline immunodeficiency virus envelope glycoproteins. Virology 192:659-662. [DOI] [PubMed] [Google Scholar]
- 49.Parker, C. E., L. J. Deterding, C. Hager-braun, J. M. Binley, N. Schülke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 51.Piantini, U., O. W. Sørensen, and R. R. Ernst. 1982. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104:6800-6801. [Google Scholar]
- 52.Richardson, J., I. Fossati, A. Moraillon, S. Castelot, P. Sonigo, and G. Pancino. 1996. Neutralization sensitivity and accessibility of continuous B cell epitopes of the feline immunodeficiency virus envelope. J. Gen. Virol. 77:759-777. [DOI] [PubMed] [Google Scholar]
- 53.Richardson, J., A. Moraillon, F. Crespeau, S. Baud, P. Sonigo, and G. Pancino. 1998. Delayed infection after immunization with a peptide from the transmembrane glycoprotein of the feline immunodeficiency virus. J. Virol. 72:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson, J., G. Pancino, R. Merat, T. Leste-Lasserre, A. Moraillon, J. Schneider-Mergener, M. Alizon, P. Sonigo, and N. Heveker. 1999. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J. Virol. 73:3661-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rigby, M. A., E. C. Holmes, M. Pistello, A. Mackay, A. J. L. Brown, and J. C. Neil. 1993. Evolution of structural proteins of feline immunodeficiency virus: molecular epidemiology and evidence of selection for change. J. Gen. Virol. 74:425-436. [DOI] [PubMed] [Google Scholar]
- 56.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 58.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serres, P. F. 2000. Molecular mimicry between the trimeric ectodomain of the transmembrane protein of immunosuppressive lentiviruses (HIV-SIV-FIV) and interleukin 2. C. R. Acad. Sci. Ser. III 323:1019-1029. [DOI] [PubMed] [Google Scholar]
- 61.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiner, S. J., P. A. Kollman, D. A. Case, U. C. Singh, C. Ghio, G. Alagona, S. Profeta, and P. J. Weiner. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106:765-784. [Google Scholar]
- 64.Wild, C., T. Greenwell, D. Shugars, L. Rimsky-Clarke, and T. J. Matthews. 1995. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res. Hum. Retrovir. 11:323-325. [DOI] [PubMed] [Google Scholar]
- 65.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willett, B. J., J. N. Flynn, and M. J. Hosie. 1997. FIV infection of the domestic cat: an animal model for AIDS. Immunol. Today 18:182-189. [DOI] [PubMed] [Google Scholar]
- 67.Willett, B. J., L. Picard, M. J. Hosie, J. D. Turner, K. Adema, and P. R. Clapham. 1997. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J. Virol. 71:6407-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 69.Xiao, Y., Y. Lu, and Y.-H. Chen. 2001. Epitope-vaccine as a new strategy against HIV-1 mutation. Immunol. Lett. 77:3-6. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto, J. K., C. D. Ackley, H. Zochlinski, H. Louie, E. Pembroke, M. Torten, H. Hansen, R. Munn, and T. Okuda. 1991. Development of IL-2-independent feline lymphoid cell lines chronically infected with feline immunodeficiency virus: importance for diagnostic reagents and vaccines. Intervirology 32:361-375. [DOI] [PubMed] [Google Scholar]
- 71.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binlay, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]