Abstract
During latency, herpes simplex virus expresses a unique set of latency-associated transcripts (LATs). As the 2.0-kb LAT intron is complementary to, and overlaps, the 3′ end of the ICP0 transcript, it has been suggested that the stable LAT intron might function as an antisense repressor of ICP0 expression. We tested this hypothesis in cell culture by dissociating cis- and trans-acting effects of the 2.0-kb LAT, using a series of complementary strategies. Initially, we constructed 293T cell lines that stably express the nuclear 2.0-kb LAT intron to determine whether LAT accumulation in trans affects ICP0 expression. ICP0 mRNA and protein expression profiles were studied (i) following infections with a viral mutant containing wild-type LAT and ICP0 sequences but having deletions of other immediate-early (IE) genes, thus preventing the progression of viral early gene expression, (ii) at early time points after infection with wild-type virus, before viral LAT expression, and (iii) by plasmid transfections. Northern and Western blot analysis showed that trans expression of the 2.0-kb LAT intron does not affect ICP0 mRNA expression, stability, accumulation, splicing, or translation. In addition, suppression of viral replication by overexpression of the 2.0-kb LAT, which has been detected previously in neuronal cell lines, was not found in these nonneuronal cell lines. However, deletion of the latency-active promoter (LAP) region of the virus resulted in overexpression of IE genes, which occurred soon after infection, before viral LAT expression had commenced. This was not complemented by the expression of LAT in trans, suggesting that the LAP deletion affected transcriptional regulation of the IE genes in cis. We conclude that the function of the highly conserved LAT intron is unlikely to involve a direct-acting anti-ICP0 antisense mechanism but that the LAT region could affect ICP0 mRNA expression from the viral genome.
Herpes simplex virus (HSV) is a common human pathogen that is responsible for oral and genital infection, severe encephalitis, and blindness. Following primary mucosal or cutaneous infection, the viral nucleocapsid is transported to the sensory ganglia associated with the afferent nerve supply of the affected region. Within the nuclei of ganglionic sensory neurons, the virus is able to establish a latent infection; the viral genome may persist in this state for the lifetime of the host. Periodic reactivation triggers the formation of infectious virions that are transported by anterograde axonal transport to the periphery, where they are shed to allow infection of a new host (37).
The processes regulating the establishment and maintenance of latency are incompletely understood. During latent infection, only the latency-associated transcript (LAT) gene is expressed abundantly from the viral genome (11, 35, 42, 46). A primary 8.3-kb LAT of low steady-state abundance (56) undergoes posttranscriptional processing to form other LAT species. The 2.0-kb LAT is a stable lariat intron (36, 54) that is spliced from the primary 8.3-kb transcript (2, 17, 55), located within the nuclei of infected cells, and can be readily detected during either lytic or latent infection (13, 46). The 1.5-kb LAT is also a stable lariat intron (54) formed by the removal of a second intron from within the 2.0-kb LAT sequence (2, 44, 52). However, the 1.5-kb LAT intron has been detected only during viral latency (43, 44, 51), implying that the relevant splicing event is specific to both neurons and latent infection. The unusual stability of the LATs has been ascribed to peculiarities in the lariat structure adjacent to the branch point and splice acceptor region (25, 53, 55). The presumed 6.3-kb spliced exon product of LAT is very difficult to detect and appears to be highly unstable (12).
Despite the extensive characterization of the origin and nature of the LATs, relatively little is known about their function. Difficulty is encountered in producing viruses that are pure LAT null mutants; the genome of HSV is extremely compact, and the immediate-early (IE) gene encoding ICP0 is situated on the complementary strand of the viral genome, with the 3′ ends of ICP0 and LAT overlapping (Fig. 1). Thus, only the LAT promoters and the 5′ region of LAT may be deleted without affecting the ICP0 gene. The remainder of the LAT region has been inactivated by other means, including the use of synthetic transcriptional stop signals which do not disrupt the ICP0 reading frame on the opposite strand (4). The most striking conclusion from studies on LAT-null strains is that LAT deletion does not completely prevent the establishment of, or reactivation from, latency (22, 24, 41, 45). Various more subtle deficits have been reported, however. First, there are reports that LAT-null mutants are less efficient at establishing latent infection than wild-type or rescuant virus (40, 50). Second, latent LAT-null mutants show different reactivation kinetics than controls (3, 15, 21, 31, 32, 45), although this may be attributable in some instances to the reduced efficiency with which latent infection is established in the first place. Finally, LAT-null mutants appear to be more toxic to neurons than LAT-expressing viruses (33, 49), resulting in more cell death following infection, which may be apoptotic (33). Correspondingly, LAT expression seems to enhance cellular resistance to toxic stimuli in tissue culture (1, 33). Enhanced cytotoxicity may account for the reduced efficiency with which LAT-null viruses establish latency.
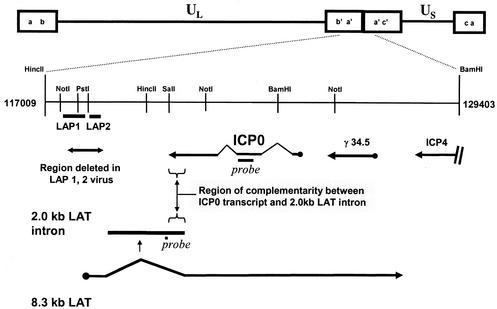
FIG. 1.
Schematic depiction of the HSV-1 genome, with the repeat region flanking the unique long (UL) region of the genome expanded to illustrate the positions of probes used in this study, the positions of LAP1 and LAP2 promoters deleted in the ΔLAP1-2 virus, and the region of antisense overlap between the 2.0-kb LAT intron and the ICP0 transcript.
The molecular events underlying these phenotypes are poorly characterized. LAT-null mutants show alterations in the regulation of other viral genes. Thus, the steady-state levels of the IE gene products ICP0, ICP4, and ICP27 are increased following infection with LAT-null mutants (7, 30). Consequently, the early gene encoding thymidine kinase is also overexpressed in the absence of LAT (7). The higher levels of IE gene products might account for the greater cell toxicity and diminished ability to establish latent infection exhibited by LAT-null mutants. However, viral deletion experiments do not distinguish between effects of LAT locus deletions occurring in cis and effects of the products of the LAT gene occurring in trans. Thus, the mechanisms responsible for dysregulation of gene expression in LAT-null mutants remain unclear.
ICP0 has profound effects on viral gene expression levels during the HSV infectious cycle (6). The ICP0 gene lies in close proximity to the LAT gene, and various mechanisms could be envisaged whereby LAT expression could affect ICP0 expression. Thus, at a cis-acting level, the LAT locus might contain regulatory elements that affect transcription of ICP0, or promoter interference might occur through two transcriptionally active genes being in opposite orientation and in close proximity. At a trans-acting level, LAT RNA might interact with the viral genome or trigger secondary effects on the chromatin structure and accessibility of viral gene promoters to transcription factors, or the LAT RNA might have direct antisense effects on ICP0 mRNA levels. Finally, LAT might encode a protein that has a regulatory function.
The antisense overlap between the abundant 2.0-kb LAT intron and the 3′ end of the ICP0 transcript has given rise to speculation that the 2.0-kb LAT intron might function as a trans-acting antisense repressor of ICP0 mRNA stability or translation (18, 30, 46). This notion is attractive, because it attributes an RNA-level function to the evolutionarily conserved stable structure of the abundant 2.0-kb LAT. The following data have provided limited evidence in support of this idea. (i) Triple transient-transfection experiments, using simultaneous transfection with three different plasmids, showed that the ability of an ICP0 expression plasmid to trans activate the thymidine kinase promoter was suppressed by the presence of a LAT-expressing plasmid (17). Unfortunately, these experiments are difficult to interpret because multiple variables, which are hard to control, are involved in simultaneous plasmid transfection. (ii) Overexpression of LAT in neuronal cell lines was able to suppress replication of wild-type and LAT-null mutant viruses, in addition to suppressing the expression of ICP0 (30). However, the replication of LAT-expressing viruses was suppressed to a greater degree than LAT-null viruses, implying that LAT may exert additional cis-acting effects. Furthermore, the reduction in IE gene steady-state RNA levels was not restricted to ICP0; the transcripts corresponding to ICP4 and ICP27 were also reduced, implying that a simple antisense mechanism might not be the full explanation.
As an initial effort to elucidate the regulatory events underlying the establishment of latency and the enhanced cytotoxicity of LAT-null viruses, we decided to study putative LAT antisense mechanisms. We reasoned that if the 2.0-kb LAT intron had antisense activity against the ICP0 mRNA, which was simply dependent on interstrand duplex formation between the LAT and the ICP0 RNA, this should be detectable in trans in cultured nonneuronal cells. If, however, the LAT had a complex neuron-specific function involving a cell-type-dependent mechanism, then the trans-acting effect previously demonstrated in neuronal cells should not be detected in nonneuronal cells.
We took the following approaches to dissociating the trans-acting properties of the 2.0-kb LAT intron from the cis-acting properties of the LAT locus. First, we generated multiple cultured 293T cell lines and populations stably expressing the nuclear 2.0-kb LAT intron at various levels, similar to those found during lytic infection. Then, trans-acting effects of cell-encoded LAT on ICP0 expression in these cells were examined by using three different strategies: (i) infection with viruses in which the wild-type LAT loci are intact but which do not express LAT because of derangements in the viral transcriptional program induced by null mutations at the ICP4 and ICP27 loci, (ii) infection with wild-type virus and examination of ICP0 expression at time points before expression of viral LAT, and (iii) transfection with an ICP0 expression plasmid. Using these complementary techniques, we were able to exclude any trans-acting effect of the 2.0-kb LAT intron on the stability or accumulation of ICP0 mRNA or protein in nonneuronal cells. We then examined the cis-acting effects induced by a deletion in the latency-active promoters LAP1 and LAP2 (9). We found that the expression of viral IE genes following infection of wild-type 293T cells with this mutant virus was enhanced with respect to expression in wild-type virus, even at time points before the expression of viral LAT. The delivery of 2.0-kb LAT intron RNA in trans did not alter this phenotype, implying that a cis-acting, non-LAT RNA-dependent effect on viral gene expression was responsible. We conclude that an antisense mechanism involving the 2.0-kb LAT intron is unlikely to account for the changes in viral gene expression, and associated phenotypic alterations, that accompany the deletion of the LAT loci in the context of the viral genome in neuronal cells.
MATERIALS AND METHODS
Plasmids and probes.
pIRES-Bleo-17 was made by inserting the EcoRV-MluI restriction fragment of strain 17 LAT into the EcoRV-BstXI sites of pIRES-Bleo (Clontech). pIRES-Bleo-SD was identical except that the EcoRV-NotI restriction fragment derived from strain 17 LAT contained a mutation within the 2.0-kb consensus splice donor site (AGgtaggt to AGagtaga) (lowercase letters indicate introns, uppercase letters indicate exons, and bold letters indicate bases that were mutated). This mutation was generated as described in a previous study, in which the mutation was shown to abolish 2.0-kb LAT splicing (2). The ICP0 expression plasmid, pEG110, was a kind gift from Patrick Lomonte, Université Claude Bernard Lyon 1, Villeurbanne Cedex, France; it was made by inserting the ICP0 cDNA into the pEGFP-C1 vector (Clontech) (29). Templates for the generation of riboprobes were constructed with pDP18 (Ambion). The ICP0 riboprobe template, pDP18-ICP0, was made by inserting a 255-bp XhoI-KpnI fragment of ICP0 exon 2 into the KpnI-SalI sites of pDP18. The LacZ riboprobe template, PDP18-LacZ, was made by inserting a 365-bp PvuII-EcoRI fragment of the Escherichia coli LacZ gene into the EcoRI (PstI/Klenow blunt-ended) sites of pDP18. The ICP22 riboprobe template was a kind gift from S. Wendell, Pittsburgh, Pa.; it was made by inserting a 210-bp PCR fragment spanning nucleotides 133056 to 133265 of the HSV-1 strain KOS ICP22 gene into pGEM-T (Promega). All constructs were characterized by restriction digestion and prepared at high concentration and purity by using a commercially available maxiprep kit (Qiagen). Sequences of oligonucleotide probes were as follows: LAT-probe, 5′-CCGGACTGACCTGGCCTCTGG-3′; 18S-probe, 5′-GCTATCAATCTGTCAATCCTGTCCGTGTCC-3′.
Viruses.
Wild-type HSV infections were carried out using an HSV-1 laboratory strain, KOS. The LAT mutant, ΔLAP1-2, contains a deletion encompassing LAP1 and LAP2; the construction and characterization of this vector were previously described (9). The construction and characterization of QOZ.HG (genotype ICP4−, ICP27−:HCMV IEp-GFP, β-ICP22, β-ICP47, UL41−:ICP0placZ) was previously described (8); the vector is based on the d106 backbone, which was generated by N. DeLuca (University of Pittsburgh). The genome of QOZ.HG is depicted schematically in Fig. 7A. KOS and ΔLAP1-2 were propagated in Vero cells. QOZ.HG was propagated in complementing 7B cells, which stably express ICP4 and ICP27. All vectors were purified by three rounds of limiting dilution and verified by Southern blot analysis. High-titer and high-purity vector stocks were prepared as described previously (5) and titrated in quadruplicate at each dilution using Vero or 7B cells as described above.
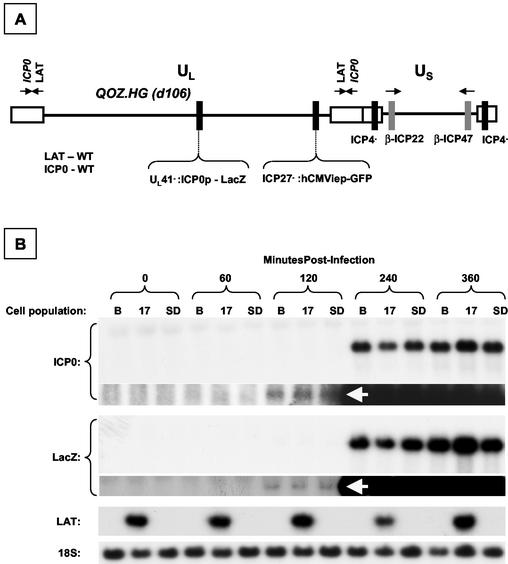
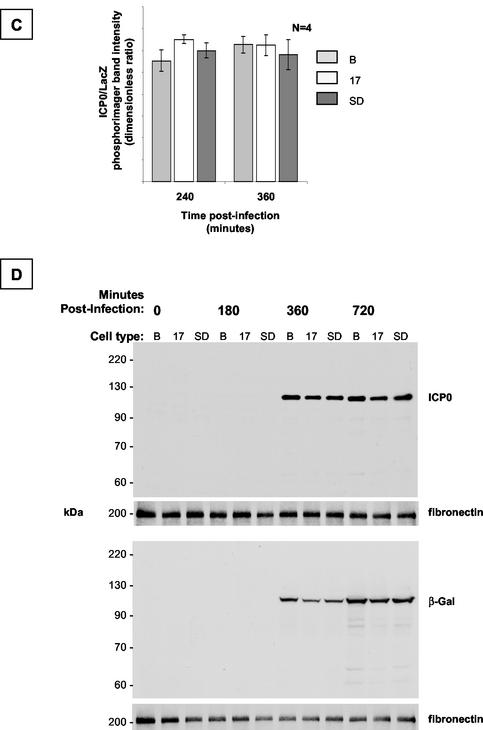
FIG. 7.
Dynamic expression profile of ICP0 and LacZ in LAT-expressing cell populations after infection with QOZ.HG. (A) Schematic depiction of the QOZ.HG genome. The virus contains wild-type LAT and ICP0 loci but does not express LAT, as the viral transcriptional program is disrupted by mutations in the ICP4, -22, and -27 loci. (B) Northern blot hybridization analysis of ICP0, LacZ, and LAT expression in LAT-expressing and control cell populations before and at four time points after infection with QOZ.HG. The second and fourth panels show prolonged exposures of the blots depicted in the first and third panels. ICP0 and LacZ mRNAs appear at 120 min postinfection (arrows) in all cell populations and peak at similar levels. (C) Quantitative analysis of the ICP0/LacZ ratio provides a surrogate marker of ICP0 stability (see the text). This is not altered by the presence or absence of the 2.0-kb LAT at any time point. (D) Western blot hybridization analysis of ICP0 and LacZ protein expression in LAT-expressing cell populations and controls before and after infection with QOZ.HG. The upper two and lower two panels represent duplicate blots that were probed for ICP0 or LacZ and then stripped and reprobed for the fibronectin loading control. B, Bleo.
Cell culture and transfection.
293T cells were maintained in Dulbecco's modified Eagle medium (Gibco-BRL) supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were transfected with 1 μg of plasmid expression construct at 80% confluence in six-well plates with Lipofectamine Plus (Gibco-BRL), following the manufacturer's protocol. Plasmid-expressing cell populations were selected by the addition of 200 μg of bleomycin per ml to the media, which killed 100% of control nontransfected cells. Selected clones were combined and cultured in the ongoing presence of bleomycin. Clonal isolation was carried out using limiting dilution in 96-well plates. Cultured monolayers at 80% confluence were subjected to HSV-1 infection for 1 h. Time points postinfection were counted from the time of addition of HSV-1 to the cell culture medium.
Northern blots.
Total RNA was extracted from cultured cells by using a kit (RNAqueous; Ambion, Austin, Tex.), according to the manufacturer's protocol. RNA was quantified by spectrophotometry at 260 nm. Denaturing electrophoresis was carried out with a 1.2% agarose gel made with proprietary formaldehyde gel buffer (NorthernMax; Ambion) according to the manufacturer's protocol, and electrophoresis was carried out in 1× MOPS (morpholinepropanesulfonic acid) running buffer. Total RNA (5 μg) was dissolved in 5 μl of H2O and 10 μl of loading buffer (NorthernMax) and heated to 65°C for 15 min prior to gel loading. Transfer to a Nytran positively charged nylon membrane (Schleicher & Schuell) was carried out by using proprietary transfer buffer (NorthernMax) and a TurboBlotter apparatus (Schleicher & Schuell). Membranes were stored at −20°C prior to hybridization, which was carried out in Ultrahyb (Ambion). Prehybridization for 2 h was followed by overnight hybridization at 68°C (riboprobes) or 42°C (oligonucleotide probes). Membranes were washed five times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at room temperature over 30 min and then three times in 0.1× SSC-0.1% SDS at 68°C (riboprobes) or 42°C (oligonucleotide probes) over 45 min. Membranes were exposed to film at room temperature for 5 to 120 min (riboprobes and the 18S oligonucleotide probe) or at −80°C overnight with intensifying screens (LAT probe). Band quantification was carried out with a Storm 840 PhosphorImager and ImageQuant software (Molecular Dynamics). Riboprobes were generated by in vitro transcription. Probe templates were linearized by digestion with HindIII (pDP18-LacZ and pDP18-ICP0) or NotI (pGEM-T-ICP22), phenol-chloroform extracted, ethanol salt precipitated, and resuspended at a concentration of 1 μg/μl. The 20-μl transcription reaction mixture contained 1 μg of linearized probe template, 10 U of T3 polymerase, 1× transcription buffer (40 mM Tris-HCl [pH 7.9] at 25°C, 10 mM NaCl, 6 mM MgCl2, 2 mM spermidine, 0.05% Tween-20) and 10 mM dithiothreitol (Riboprobe; Promega), 20 U of recombinant RNase inhibitor (RNasin; Promega), high-purity unlabeled riboguanosine triphosphate (rGTP), rUTP, and rATP (Pharmacia-Amersham) at final concentrations of 500 μM each, and 3,000 Ci of [α-32P]rCTP (NEN Life Science) per mmol at a final concentration of approximately 1.15 μM. After incubation at 37°C for 2 h, the DNA probe template was digested by the addition of 20 μl of DNase I solution (2 U in 1× transcription buffer). Following a further incubation of 1 h, the unincorporated labeled nucleotide was removed by minicolumn chromatography purification (G-25 MicroSpin; Amersham). Oligonucleotide probes were end-labeled with T4 polynucleotide kinase (PNK; New England Biolabs). The 20-μl reaction mixture contained 10 U of PNK, 1× PNK buffer (70 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 5 mM dithiothreitol; New England Biolabs), 20 nM oligonucleotide, 5 μl of [γ-32P]ATP at 6,000 Ci/mmol. After incubation for 1 h, unincorporated label was removed by using a MicroSpin column as described above.
Western blots and antibodies.
Protein samples were harvested directly from cells in six-well plates. Monolayer samples were washed three times in phosphate-buffered saline (PBS) at room temperature and then lysed in 100 μl of 1× lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.02% sodium azide, and 1% NP-40). Lysates were centrifuged to remove insoluble debris, and the protein concentration of the supernatant was quantified by a Bradford assay (Bio-Rad). Samples of 20 μg of protein were heated to 95°C for 5 min in 1× SDS loading buffer (50 mM Tris [pH 6.8], 5% 2-mercaptoethanol, 10% glycerol, 1% SDS, and 5 μg of bromphenol blue per ml) prior to electrophoresis, which was carried out with a 4% stacking gel and an 8% resolving gel. Transfer to a nitrocellulose membrane (Schleicher & Schuell) was carried out at 150 V, in 1× transfer buffer (25 mM Tris, 192 mM glycine, and 10% methanol) at 4°C for 2 h. Membranes were blocked in 1× TBST (Tris-buffered saline-Tween)-5% milk overnight at 4°C. Primary antibody was incubated for 2 h in 1× TBST-5% milk at room temperature, followed by four washes in 1× TBST over 40 min. Peroxidase-conjugated secondary antibody was incubated in 1× TBST-5% milk at room temperature for 1 h, followed by four washes in 1× TBST over 1 h. Hybridizing bands were visualized by using photographic film after incubation of the membrane with chemiluminescent peroxidase substrate (ECL Western blotting detection reagents; Amersham). The primary antibodies were ICP0 monoclonal antibody (Virusys), LacZ monoclonal antibody (reference G6282; Sigma), and fibronectin polyclonal antibody (reference F3648; Sigma). Secondary antibodies were peroxidase-conjugated rabbit anti-mouse antibody (reference A9044; Sigma) and horseradish peroxidase-conjugated goat anti-rabbit antibody (reference A0545; Sigma).
In situ hybridization.
The riboprobe template for LAT RNA in situ hybridization was generated by inserting the HpaI-SalI fragment of the HSV-1 strain 17 2.0-kb LAT intron into the EcoRV-XhoI sites of the pcDNA3 vector (Invitrogen). The resulting plasmid was linearized by digestion with EcoRI, and LAT-antisense riboprobe was made by in vitro transcription using SP6 RNA polymerase. The probe was labeled by incorporation of digoxigenin-UTP (Roche Boehringer Mannheim) into the transcription reaction mixture, as described previously (8). Cells were plated on one eight-well Lab-Tek chamber slide (Nalge Nunc), such that each slide contained the test cell population and all of the controls in adjacent wells, to ensure that all samples were processed identically. In situ hybridization was performed as described in a previous study (8). Cells were fixed in 4% paraformaldehyde for 10 min, treated with 1% HCl in PBS for 5 min, and washed with PBS. After equilibration in 5× SSC for 5 min, hybridization with digoxigenin-labeled riboprobe was carried out overnight at 68°C in 15 μl of hybridization solution (50% formamide, 5× SSC, 40 μg salmon sperm DNA per ml, 0.5 μg of riboprobe per ml) per well. Following hybridization, the slide was rinsed with 2× SSC and 0.1× SSC at 68°C for 1 h. After blocking with 5% normal goat serum for 1 h, the LAT probe was localized with alkaline phosphatase-conjugated antidigoxigenin antibody (1:5,000; Roche Boehringer Mannheim) and detected with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate solution (Roche Boehringer Mannheim).
RESULTS
Probes.
The positions of probes to the 2.0-kb LAT intron and ICP0 are depicted schematically in Fig. 1. All probes were single stranded to avoid problems of interpretation inherent in RNA strand-specific experiments. Figure 2 shows that each of the probes for ICP0, LacZ, LAT, and 18S rRNA hybridized with a single transcript of the expected size in Northern blot analyses. It was thus possible and admissible to quantify relative transcript abundance by measuring the intensities of specific hybridizing bands with a PhosphorImager and correcting them for gel loading and transfer by using the 18S rRNA signal as a control.
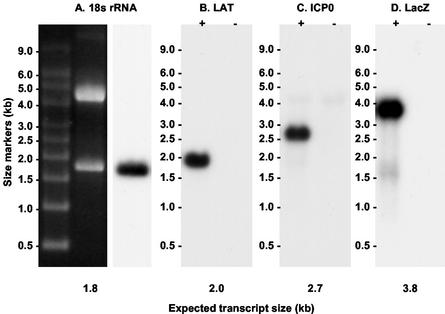
FIG. 2.
Northern blot controls showing that the probes used in this study hybridized specifically with the intended target RNA. (A) 18S RNA oligonucleotide. The RNA sample was from 293T cells. (B) LAT oligonucleotide. RNA was derived from Vero cells infected with HSV-1 strain KOS (+) and uninfected controls (−). (C) ICP0 antisense riboprobe. RNA was from 293T cells infected with QOZ.HG (+) and uninfected controls (−). (D) LacZ antisense riboprobe. RNA samples were as in panel C.
Generation of cell populations and clonal lines stably expressing the 2.0-kb LAT intron.
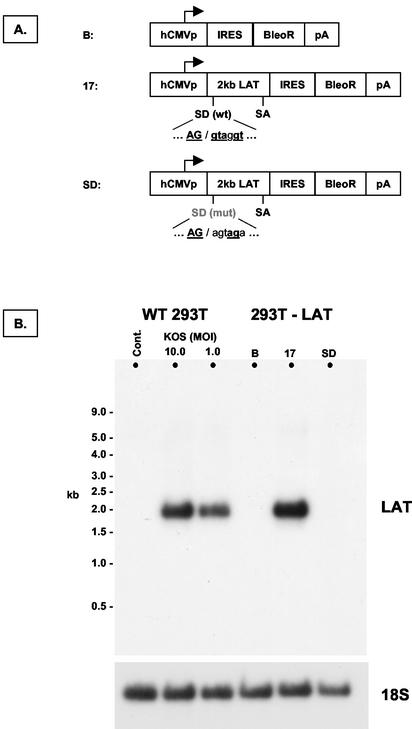
We decided to isolate any direct trans-acting effect of the 2.0-kb LAT intron on ICP0 mRNA by generating nonneuronal cell populations that express the 2.0-kb LAT intron. The EcoRV-MluI fragment of strain 17 LAT, encompassing the 2.0-kb intron, was cloned into an expression vector, pIRES-Bleo (Clontech). Three constructs were made (Fig. 3A). The 17 construct contained the wild-type sequence. The Bleo construct comprised unmodified vector and served as a negative control. The SD construct contained a LAT sequence similar to that of the 17 construct, except that the conserved 2.0-kb LAT splice donor signal was mutated. The mutation, which consisted of two 3-bp deletions, was based on that described in a previous study, in which it was shown to abolish 2.0-kb LAT splicing in the context of the entire LAT locus during viral infection (2). The SD plasmid thus contains almost the same sequences as the 17 plasmid but does not give rise to 2.0-kb intron splicing. The pIRES-Bleo vector contains an internal ribosome entry site 3′ of the cloning site containing the LAT sequence, followed by an expression cassette for the bleomycin resistance gene. The 2.0-kb LAT would thus be spliced as an intron from the mRNA encoding the selectable marker. Following transfection of 293T cells with the three LAT constructs, cultures were selected for bleomycin resistance. This revealed many hundreds of resistant colonies in each sample. Some experiments used mixed selected cell populations, avoiding the problems associated with the idiosyncratic properties of single clonal cell lines. For other experiments, we selected, characterized, and expanded multiple clonal cell lines in order to assess the effects of LAT expression level.
FIG. 3.
Construction of cell lines and populations that stably express the 2.0-kb LAT intron. (A) Schematic depiction of transgene expression cassettes introduced into cells by transfection prior to selection. Abbreviations: hCMVp, major IE promoter of human cytomegalovirus; IRES, internal ribosome entry site from encephalomyocarditis virus; BleoR, bleomycin resistance gene. (B) Northern blot hybridization analysis of control 293T cells (cont.), 293T cells 18 h after infection with HSV-1 strain KOS, and cell populations produced with the expression cassettes shown in panel A (293T-LAT). The blot was sequentially hybridized to probes for LAT and 18S rRNA. The 17 cell population expresses the 2.0-kb LAT intron at levels approximating those seen after wild-type infection at an MOI of 10. B, Bleo.
Characterization of cell populations and clonal lines stably expressing the 2.0-kb LAT intron.
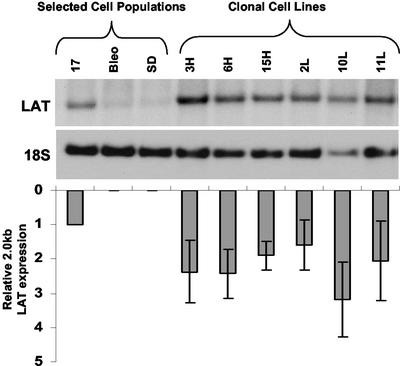
Total RNA was harvested from the cell populations and subjected to Northern blot analysis (Fig. 3B). A 2.0-kb transcript hybridized with the single-stranded LAT oligonucleotide probe shown in Fig. 2. The electrophoretic mobility of the band was identical to that seen when RNA from cells infected with wild-type HSV-1 was hybridized with the same probe. This transcript was not present in cells expressing a LAT sequence carrying a mutation in the 2.0-kb splice donor. These features identified the hybridizing transcript as the 2.0-kb LAT intron. The expression level of the LAT 2.0-kb intron in this selected cell population was similar to that seen after infection of 293T cells with wild-type HSV strain KOS. A series of eight different clonal cell lines was isolated from the 17 population by limiting dilution culture and expanded under continued selective pressure. The Northern blot hybridization profile of these cell lines is shown in Fig. 4. Each of the clonal lines used in subsequent experiments expressed LAT at higher levels than the selected 17 population from which it was derived.
FIG. 4.
Northern hybridization analysis of clonal cell lines derived from the 17 cell population. The blot was sequentially hybridized with LAT and 18S rRNA loading control probes. The graph shows the level of LAT expression in each of the clonal cell lines relative to controls and the parent cell population. Error bars indicate standard error of the mean for three measurements.
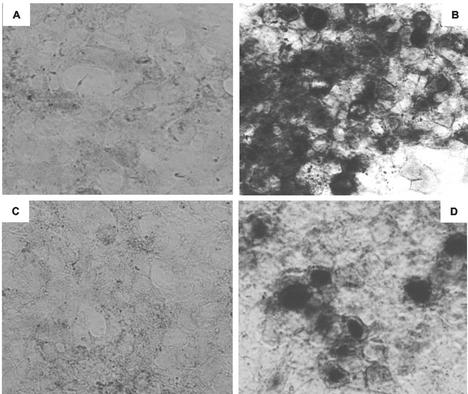
The subcellular localization of the 2.0-kb LAT intron was examined by using RNA in situ hybridization (Fig. 5). Cells from the selected Bleo and SD populations were cultured on four-well slides, alongside cells from the selected 17 population and wild-type 293T cells that had been infected with wild-type HSV strain KOS. The slides were processed for in situ hybridization, such that all of the cell samples were exposed to identical reagents under identical conditions. As predicted from Northern hybridization experiments, no detectable signal was observed in the Bleo or SD cell populations (Fig. 5A and C). The 17 cell population showed expression of LAT in the nuclei of >90% of the selected cell population (Fig. 5B). The appearances were identical to those arising from infection with wild-type HSV strain KOS (Fig. 5D). Elimination of the nuclear permeabilization step from the protocol also revealed a smaller amount of LAT within the cytoplasm of the selected cell population (data not shown).
FIG. 5.
RNA in situ hybridization analysis of LAT expression in 293T cell populations. (A) Negative control cells expressing the bleomycin resistance gene only (Bleo); (B) cells from the 17 population expressing the 2-kb LAT intron; (C) cells from the SD population, in which 2.0-kb LAT splicing is prevented by a mutation in the splice donor consensus; (D) positive control 293T cells, 18 h after infection with HSV-1 strain KOS at an MOI of 1. Dark staining in panels B and D shows that the LAT probe specifically hybridizes to the nuclei of infected cells and those expressing the 2.0-kb LAT intron.
These analyses show that the selected cell populations express the 2.0-kb LAT intron at levels similar to those arising during wild-type infection and that the resulting stable intron is located in a similar subcellular compartment. These cell populations and the derived clonal lines should therefore be useful for investigation of the possible trans-acting effects of the 2.0-kb LAT intron.
Measurement of wild-type HSV-1 replication in the presence of LAT.
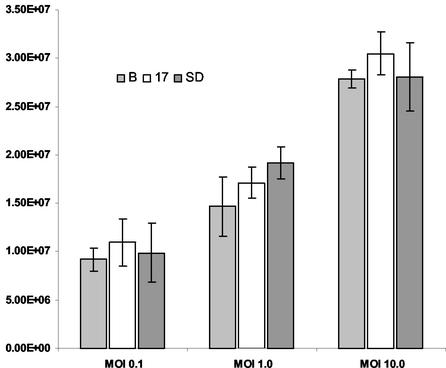
We tested the hypothesis that the 2.0-kb LAT intron acts in trans to repress viral replication. Parallel samples of the 293T cell populations described above were infected at a multiplicity of infection (MOI) of 0.1, 1.0, and 10.0 with wild-type HSV-1 strain KOS. Virus was harvested from the cultures at 18 h postinfection and titrated on permissive Vero cells. The number of infectious particles was determined in each sample at different dilutions. There was no difference in the titer of virus passaged in any of the three cell populations, indicating that the 2.0-kb LAT intron does not repress viral replication in trans in these nonneuronal cells (Fig. 6).
FIG. 6.
Replication of HSV-1 strain KOS in LAT-expressing cell populations. Multiple samples of each cell population were infected at three different multiplicities. The resulting virus-containing medium was titrated on permissive Vero cells, and the number of PFU per milliliter of medium was determined. HSV KOS replicated equally well in all three cell populations at all three MOI. B, Bleo.
Measurement of ICP0 accumulation during infection of LAT-expressing cells.
The generation of cell lines allowed us to supply the LAT 2.0-kb intron in trans, but to examine trans-acting effects of the LAT on ICP0 expression, it was necessary to eliminate cis-derived expression of LAT, without altering the genetic context of the ICP0 gene. We achieved this in three ways: (i) using a virus in which the LAT locus and ICP0 gene are present in their entirety but in which the viral transcriptional program is disrupted by mutations in the genes encoding other IE genes, such that the LAT genes are not expressed; (ii) using wild-type virus but examining IE gene expression at time points before LAT is expressed, and (iii) using an ICP0 expression plasmid which does not contain any other HSV-1 sequences.
(i) QOZ.HG (Fig. 7A) is a derivative of the previously described mutant d106 (38). d106 has deletions of both copies of the gene encoding ICP4, the major transcriptional regulator of HSV-1. In addition, d106 has a deletion of the ICP27 gene, which is replaced by a green fluorescent protein expression cassette and has mutations in the promoters of the ICP22 and ICP47 genes that render them unresponsive to VP16. The mutations of ICP4 and ICP27 prevent progression of viral gene expression to the E and L phases and thus prevent LAT expression from the virus in nonneuronal cells. QOZ.HG does express LAT in neuronal cells, in keeping with its neuron-specific promoter activity (unpublished observation). The QOZ.HG derivative of d106 has a deletion of the UL41 gene, which is replaced with the bacterial β-galactosidase gene under transcriptional regulation of the ICP0 promoter (8). As transcription of both ICP0 and LacZ is regulated similarly in this virus, any difference in the ratio of the two transcripts from sample to sample must arise from differences in the stability or splicing and transport of one transcript or the other. This serves as a control for small differences in the percentage of cells infected in each sample and in the viral copy number in each cell. The ICP0/LacZ ratio thus provides a surrogate marker of ICP0 mRNA stability.
The LAT-expressing cell populations and controls were infected with QOZ.HG at an MOI of 3. Samples were harvested before infection and at 60, 120, 240, and 360 min postinfection. RNA was subjected to Northern blot analysis with probes to ICP0, LacZ, and LAT and with 18S rRNA as a loading control (Fig. 7B). LAT was not expressed from QOZ.HG during this experiment, as evidenced by the absence of a hybridizing 2.0-kb LAT band in either of the non-LAT-expressing (Bleo or SD) cell populations at any time point. The first appearance of both LacZ and ICP0 mRNA was noted at 120 min postinfection in all three of the cell populations, by prolonged exposure of the blots. mRNA encoding both ICP0 and LacZ was present in abundance in all three cell populations by 240 min postinfection. Minor differences apparent in the levels of ICP0 and LacZ mRNA between samples in Fig. 7B were caused by loading artifacts, as shown by examination of the loading control. The experiment was repeated four times, and quantitative analysis was undertaken by measuring the relative levels of LacZ and ICP0 mRNA in each of the samples at 240 and 360 min (Fig. 7C). This showed no significant difference in the ratio of ICP0 to LacZ between the different cell populations and time points, indicating that ICP0 mRNA stability was not affected by the presence or absence of the 2.0-kb LAT intron. Furthermore, we did not see an accumulation of ICP0 primary transcripts, excluding the possibility that the 2.0-kb LAT intron interferes with splicing of the ICP0 pre-mRNA or the formation of mature mRNA. We considered the possibility that LAT prevented translation of ICP0 mRNA without affecting the steady-state levels of the mRNA. Western blot hybridization analysis excluded this possibility; ICP0 accumulation at 6 and 12 h postinfection was identical in all three selected cell populations (Fig. 7D).
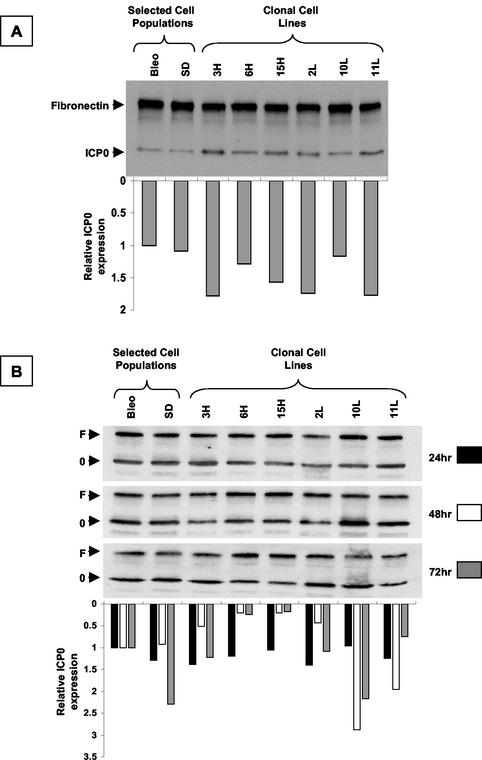
(ii) We next examined the accumulation of ICP0 protein in a series of LAT-expressing clonal cell populations derived from the selected 293T cell population (Fig. 8A). The cells were infected alongside control cells with wild-type HSV-1 strain KOS at an MOI of 3. Protein was extracted at 5 h postinfection, before the viral LAT gene was expressed. The samples were subjected to electrophoresis and transfer and probed simultaneously with antibodies to ICP0 and a protein loading control, fibronectin. No systematic or appreciable differences in the expression of ICP0 between the different cell lines and controls following infection with wild-type virus were identified.
FIG. 8.
Western blot hybridization analysis of ICP0 expression in LAT-expressing clonal cell lines. The blots were simultaneously probed with antibodies for ICP0 (0) and fibronectin (F) as a loading control. Quantification of the densitometric ICP0/fibronectin ratio is shown below each lane of the gel. (A) Cells were infected with HSV-1 strain KOS and harvested 5 h postinfection, before viral LAT expression. (B) Cells were transfected with an ICP0 expression plasmid and harvested at the time points shown. There is no systematic difference in ICP0 expression between the different cell lines and controls at any time point following either infection or transfection. As might be expected from a transient-transfection assay, there is more variability at later time points in panel B.
(iii) To confirm the results obtained by methods 1 and 2, we performed plasmid transfections, which completely dissociate the presence of LAT in trans from any possible cis-acting effect in the context of the viral genome (Fig. 8B). An ICP0 expression plasmid in which the ICP0 cDNA is under the transcriptional control of the cytomegalovirus IE promoter, was used to transfect the 2.0-kb LAT-expressing cell lines and controls. ICP0 protein levels were determined by Western blot hybridization analysis at various time points after the transfections. ICP0 expression in the LAT-expressing clonal cell lines and controls was similar at early time points following the transfection. At later time points, there was more variability, as might be expected in a transient-transfection assay. However, there was no relationship between the ICP0 expression level and the LAT expression level at any time point.
IE gene expression following infection of 293T cells with a LAP1-2 deletion virus.
In order to study the cis-acting effects of deletions of the LAT locus on the expression of IE genes following viral infection in culture, we utilized a previously described virus bearing deletions for both LAP1 and LAP2 (9). This virus contains a deletion encompassing the PstI sites at positions −800 and −3 with respect to the 5′ end of the 2.0-kb LAT intron. Although the deletion greatly reduces expression of LAT, the entire sequences encoding the 2.0-kb LAT intron and ICP0 gene are spared.
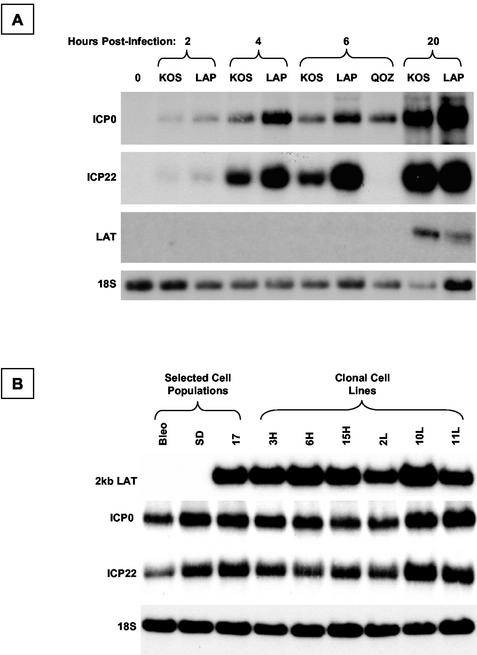
Figure 9A shows a representative Northern blot analysis of gene expression following infection of 293T cells with wild-type HSV strain KOS and the LAP deletion derivative. The viral preparations were done in parallel, and the resulting stocks were titrated in parallel in Vero cells in culture. Identical numbers of PFU of virus were added to 293T cells, and the samples were lysed at 2, 4, 6, and 20 h postinfection. Total RNA was extracted and subjected to Northern blot hybridization analysis. No LAT expression occurs from either virus at early time points following infection. At 20 h, the ΔLAP1-2 virus expresses substantially less 2.0-kb LAT than the wild-type virus. At all time points postinfection, the ICP0 and ICP22 mRNAs are clearly more abundant in cell samples infected with LAP deletion viruses. Thus, the LAP deletion results in overexpression of viral IE genes before any expression of LAT occurs.
FIG. 9.
Analysis of IE gene expression from the HSV-1 LAT-null mutant ΔLAP1-2. (A) Northern blot analysis of viral gene expression from wild-type HSV-1 (strain KOS), ΔLAP1-2, and QOZ.HG at different time points following infection of 293T cells. The expression of ICP0, ICP22, and LAT is shown by sequential hybridization of a Northern blot with different probes; 18S rRNA acts as a loading control. (B) Northern blot hybridization analysis of ICP0 and ICP22 mRNA expression in LAT-expressing cell populations, clonal LAT-expressing cell lines, and controls after infection with ΔLAP1-2 at an MOI of 1. There is no evidence that ICP0 mRNA accumulation is suppressed in the presence of LAT in trans, even though the LAT-null deletion was shown to increase ICP0 mRNA expression (A), presumably by acting in cis.
LAT in trans does not complement the absence of LAT in cis.
Finally, we examined the effect of the 2.0-kb LAT intron, delivered in trans, on the expression of IE genes following infection with ΔLAP1-2, to test the hypothesis that complementation would occur and that IE gene expression would be reduced by the presence of the 2.0-kb LAT intron in the cells (Fig. 9B). Cells were infected, and total RNA was extracted at 6 h postinfection, before the endogenous LAT genes are expressed. The samples were subjected to Northern blot hybridization analysis using probes to ICP0, ICP22, LAT, and a loading control, 18S rRNA. After adjustment for loading artifacts, no differences in ICP0 and ICP22 expression between LAT-expressing and control cells were detected.
DISCUSSION
Taken together, these experiments suggest that the 2.0-kb LAT intron does not exert a trans-acting effect on the expression of ICP0 in nonneuronal cells in culture. Our observations do not concur with earlier reported evidence that LAT may function in trans to inhibit IE gene expression in neuronal cells (30). The difference between this report and our work may relate to our use of nonneuronal cells. If antisense regulation of ICP0 expression occurred through hybridization of the nuclear LAT intron with ICP0 mRNA, it might be expected that this would occur in any cell type. The absence of a trans-acting effect of the 2.0-kb LAT intron RNA on ICP0 expression in 293T cells might result from one of several putative mechanisms.
First, a cell-type specific antisense mechanism could be invoked. It is well established that the effect of an RNA targeting reagent in suppressing gene expression may be critically dependent on the cellular location of the reagent with respect to the targeted transcript. For example, targeting a ribozyme to the nucleolus greatly enhances cleavage of a nucleolar RNA (39), whereas cytoplasmic RNAs may be targeted by a suitably localized ribozyme (27). Antisense regulation is more complicated; the cellular location of the resulting RNA duplex seems to determine both the catabolic pathway taken by the duplex and the response of the cell to the presence of dsRNA (26). It has recently been shown that the posttranscriptional regulation of ICP0 is different in neuronal and nonneuronal cell types (8). This raises the possibility that some aspect of posttranscriptional processing or transport of the ICP0 transcript, which is unique to neurons, makes it susceptible to antisense repression by LAT in a cell-specific manner. Alternatively, the cell-type-dependent splicing of the 1.5-kb LAT intron might account for differences between previous studies and ours. If the 1.5-kb intron were an effective antisense repressor of ICP0, then the detection of trans-acting LAT-dependent suppression of ICP0 mRNA would depend on the neuron-specific splicing event that generates the smaller lariat.
The second possible type of mechanism to explain the discrepancy between our results and those obtained in neuronal cell lines invokes a cell-type-specific, nonantisense mechanism. One possibility would be that the 2.0-kb LAT sequence is translated in neurons, resulting in a protein that functioned, directly or indirectly, as a viral transcriptional regulator. Recent work has provided limited evidence that the 2.0-kb LAT intron may encode a protein. The LAT is associated with the polyribosome fraction of lytically and latently infected cells (20), and an open reading frame (ORF) within the 2.0-kb LAT intron appears to give rise to a protein that enhances viral growth when overexpressed outside the context of the 2.0-kb lariat (47). Stable overexpression of this LAT ORF in cultured cell lines appears to allow sustained expression of a reporter gene driven by the cytomegalovirus IE promoter in the context of a HSV genome, which is null for ICP0, ICP4, and ICP27 (48). Examination of the properties of the epitope-tagged protein provides preliminary evidence that this LAT ORF-encoded protein may be nuclear, phosphorylated, and localized to viral replication compartments. The protein does not appear to directly trans activate the promoters of IE genes (10), and its action on viral replication is directly opposite to that reported from expression of the 2.0-kb intron in cultured neurons (30). In fact, it is not at all clear that this ORF can be translated in the context of the lariat 2.0-kb LAT intron. Although earlier work described polyclonal antisera directed against a LAT-encoded peptide which detected a protein in latently infected cells (14), this finding has never been reproduced (28). Furthermore, insertion of marker genes into the intron does not allow their efficient translation unless aberrant splicing events are induced by the foreign sequence (10, 28). Finally, selective deletions of the LAT ORFs do not appear to induce an abnormal phenotype in the virus (16). The role of 2.0-kb-LAT-encoded proteins thus remains uncertain, as does whatever putative mechanism dictates its selective translation in neurons. Other possible non-antisense-dependent trans-acting mechanisms whereby LAT might affect IE gene expression include functions at an RNA level, which do not depend on hybridization between the LAT intron and the ICP0 mRNA. Specific LAT-binding RNA or protein species within neurons, whose activity is modulated in some way by the LAT intron, could be possible.
Recent work has focused on the role of LAT in protecting neurons from apoptosis following viral infection (33). The regions of the LAT implicated in these effects map to the first 1.5 kb of the LAT and do not include any sequences that are complementary to the ICP0 transcript or any conserved ORFs (1, 23). This strongly suggests that the antiapoptotic mechanism of LAT does not depend on an anti-ICP0 antisense mechanism. Furthermore, it has recently been demonstrated that the reactivation phenotype of a LAT-null virus can be restored to wild-type levels by the expression of a heterologous antiapoptotic gene (34). This suggests that the IE gene dysregulation occurring after LAT deletion may be dissociable from the antiapoptotic function of LAT and that the latter is associated with the impaired latency establishment and reactivation phenotype seen in LAT-null viruses. These data provide limited insight into the function of the stable 2.0-kb LAT intron, only part of which appears to contribute to the noncoding sequences necessary and sufficient for the putative antiapoptotic function of the LAT gene.
Finally, we studied the effects of deletion of the latency promoters to examine whether the effects of LAT expression in cis were different from those in trans. Although ICP0 and ICP22 expression from the LAP deletion mutant ΔLAP1-2 was enhanced with respect to control virus, this effect was seen at time points well before the detectable expression of LAT. It is possible that the LAP1-2 deletion affects elements within the virus that result in IE gene transcriptional dysregulation via a mechanism that does not depend on expression of the LAT RNA, for example by deleting cis-acting negative transcriptional regulatory sequences. This hypothesis is consistent with previously reported data, in which LAP deletion had a significant cis-acting effect on IE gene expression (7, 19, 30). The results dictate caution in the interpretation of data obtained with LAP deletion mutants.
The present study has not elucidated the function of the conserved 2.0-kb LAT intron or the mechanism whereby LAT affects the expression of viral IE genes in trans in neuronal cells (30). Our studies on trans-acting mechanisms do not demonstrate involvement of the 2.0-kb LAT intron in the regulation of ICP0 mRNA accumulation in nonneuronal cells. We conclude that the mechanisms for the previously described trans-acting effects of LAT are cell type specific and are unlikely to be mediated by an antisense mechanism involving the 2.0-kb intron.
Acknowledgments
E.A.B. and C.-S.H. contributed equally to this work.
We thank Steve Wendell and Darren Wolfe for generously sharing plasmid resources and Xiao-Ping Chen for technical help for in situ hybridization.
This work was supported by the following grants: 5R01 GM34534-17 from the NIH/NIGMS, DAMD17-98-1-8626 from the U.S. Army, 5P01 DK44935 from the NIH, 1PO1 NS40923 from the NIH, and 1R01 NS44323 from the NIH/NINDS.
REFERENCES
- 1.Ahmed, M., M. Lock, C. G. Miller, and N. W. Fraser. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvira, M. R., W. F. Goins, J. B. Cohen, and J. C. Glorioso. 1999. Genetic studies exposing the splicing events involved in herpes simplex virus type 1 latency-associated transcript production during lytic and latent infection. J. Virol. 73:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618-630. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, D. C., J. M. Hill, G. Devi Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, E. A., S. Huang, W. F. Goins, and J. C. Glorioso. 2003. Use of the herpes simplex viral genome to construct gene therapy vectors. Methods Mol. Med. 76:1-31. [DOI] [PubMed] [Google Scholar]
- 6.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., J. Li, M. Mata, J. Goss, D. Wolfe, J. C. Glorioso, and D. J. Fink. 2000. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J. Virol. 74:10132-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, X., M. C. Schmidt, W. F. Goins, and J. C. Glorioso. 1995. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J. Virol. 69:7899-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, R. S., S. K. Thomas, D. P. Thomas, and D. S. Latchman. 1998. The herpes simplex virus 2 kb latency associated transcript (LAT) leader sequence allows efficient expression of downstream proteins which is enhanced in neuronal cells: possible function of LAT ORFs. J. Gen. Virol. 79:3019-3026. [DOI] [PubMed] [Google Scholar]
- 11.Deatly, A. M., J. G. Spivack, E. Lavi, and N. W. Fraser. 1987. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc. Natl. Acad. Sci. USA 84:3204-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devi Rao, G. B., S. A. Goodart, L. M. Hecht, R. Rochford, M. K. Rice, and E. K. Wagner. 1991. Relationship between polyadenylated and nonpolyadenylated herpes simplex virus type 1 latency-associated transcripts. J. Virol. 65:2179-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson, A. T., F. Sederati, G. Devi Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63:3844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doerig, C., L. I. Pizer, and C. L. Wilcox. 1991. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J. Virol. 65:2724-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drolet, B. S., G. C. Perng, R. J. Villosis, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1999. Expression of the first 811 nucleotides of the herpes simplex virus type 1 latency-associated transcript (LAT) partially restores wild-type spontaneous reactivation to a LAT-null mutant. Virology 253:96-106. [DOI] [PubMed] [Google Scholar]
- 16.Fareed, M. U., and J. G. Spivack. 1994. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J. Virol. 68:8071-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, M. J., T. P. Margolis, W. A. Gomes, and L. T. Feldman. 1994. Effect of the transcription start region of the herpes simplex virus type 1 latency-associated transcript promoter on expression of productively infected neurons in vivo. J. Virol. 68:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldenberg, D., N. Mador, M. J. Ball, A. Panet, and I. Steiner. 1997. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. J. Virol. 71:2897-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 22.Ho, D. Y., and E. S. Mocarski. 1989. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc. Natl. Acad. Sci. USA 86:7596-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inman, M., G. C. Perng, G. Henderson, H. Ghiasi, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javier, R. T., J. G. Stevens, V. B. Dissette, and E. K. Wagner. 1988. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology 166:254-257. [DOI] [PubMed] [Google Scholar]
- 25.Krummenacher, C., J. M. Zabolotny, and N. W. Fraser. 1997. Selection of a nonconsensus branch point is influenced by an RNA stem-loop structure and is important to confer stability to the herpes simplex virus 2-kilobase latency-associated transcript. J. Virol. 71:5849-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwabara, T., M. Warashina, S. Koseki, M. Sano, J. Ohkawa, K. Nakayama, and K. Taira. 2001. Significantly higher activity of a cytoplasmic hammerhead ribozyme than a corresponding nuclear counterpart: engineered tRNAs with an extended 3′ end can be exported efficiently and specifically to the cytoplasm in mammalian cells. Nucleic Acids Res. 29:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lock, M., C. Miller, and N. W. Fraser. 2001. Analysis of protein expression from within the region encoding the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1. J. Virol. 75:3413-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mador, N., D. Goldenberg, O. Cohen, A. Panet, and I. Steiner. 1998. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J. Virol. 72:5067-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perng, G. C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J. Virol. 70:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perng, G. C., C. Jones, J. Ciacci Zanella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 34.Perng, G. C., B. Maguen, L. Jin, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rock, D. L., A. B. Nesburn, H. Ghiasi, J. Ong, T. L. Lewis, J. R. Lokensgard, and S. L. Wechsler. 1987. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J. Virol. 61:3820-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodahl, E., and L. Haarr. 1997. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J. Virol. 71:1703-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 38.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samarsky, D. A., G. Ferbeyre, E. Bertrand, R. H. Singer, R. Cedergren, and M. J. Fournier. 1999. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. USA 96:6609-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66:2157-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedarati, F., K. M. Izumi, E. K. Wagner, and J. G. Stevens. 1989. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J. Virol. 63:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spivack, J. G., and N. W. Fraser. 1988. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J. Virol. 62:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spivack, J. G., G. M. Woods, and N. W. Fraser. 1991. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT): translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J. Virol. 65:6800-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steiner, I., J. G. Spivack, R. P. Lirette, S. M. Brown, A. R. MacLean, J. H. Subak Sharpe, and N. W. Fraser. 1989. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 8:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens, J. G., E. K. Wagner, G. B. Devi Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056-1059. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, S. K., G. Gough, D. S. Latchman, and R. S. Coffin. 1999. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. J. Virol. 73:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, S. K., C. E. Lilley, D. S. Latchman, and R. S. Coffin. 2002. A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome. J. Virol. 76:4056-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, R. L., and N. M. Sawtell. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J. Virol. 71:5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, E. K., W. M. Flanagan, G. Devi Rao, Y. F. Zhang, J. M. Hill, K. P. Anderson, and J. G. Stevens. 1988. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J. Virol. 62:4577-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wechsler, S. L., A. B. Nesburn, R. Watson, S. M. Slanina, and H. Ghiasi. 1988. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J. Virol. 62:4051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, T. T., Y. H. Su, T. M. Block, and J. M. Taylor. 1998. Atypical splicing of the latency-associated transcripts of herpes simplex type 1. Virology 243:140-149. [DOI] [PubMed] [Google Scholar]
- 54.Wu, T. T., Y. H. Su, T. M. Block, and J. M. Taylor. 1996. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J. Virol. 70:5962-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zabolotny, J. M., C. Krummenacher, and N. W. Fraser. 1997. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J. Virol. 71:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]