Abstract
The open reading frame UL84 of human cytomegalovirus encodes a multifunctional regulatory protein which is required for viral DNA replication and binds with high affinity to the immediate-early transactivator IE2-p86. Although the exact role of pUL84 in DNA replication is unknown, the nuclear localization of this protein is a prerequisite for this function. To investigate whether the activities of pUL84 are modulated by cellular proteins we used the Saccharomyces cerevisiae two-hybrid system to screen a cDNA-library for interacting proteins. Strong interactions were found between pUL84 and four members of the importin α protein family. These interactions could be confirmed in vitro by pull down experiments and in vivo by coimmunoprecipitation analysis from transfected cells. Using in vitro transport assays we showed that the pUL84 nuclear import required importin α, importin β, and Ran, thus following the classical importin-mediated import pathway. Deletion mutagenesis of pUL84 revealed a domain of 282 amino acids which is required for binding to the importin α proteins. Its function as a nuclear localization signal (NLS) was confirmed by fusion to heterologous proteins. Although containing a cluster of basic amino acids similar to classical NLSs, this cluster did not contain the NLS activity. Thus, a complex structure appears to be essential for importin α binding and import activity.
The nuclear envelope divides eukaryotic cells into a nuclear and a cytoplasmic compartment. This segregation requires specific mechanisms for the continuous transport of large numbers of macromolecules between both compartments. A number of viruses, including herpes-, influenza, and retroviruses, replicate in the host cell nucleus, and thus, as for the cellular macromolecules, viral proteins must traverse the nuclear envelope in order to participate in virus replication (reviewed in reference 58).
The nucleocytoplasmic trafficking of proteins occurs through the nuclear pore complex (NPC) and is mediated by an active and selective mechanism that is controlled by saturable transport receptors and the corresponding cis-acting transport signals that are termed nuclear localization signals (NLSs) and nuclear export signals (NESs) (reviewed in reference 15). With respect to NLSs the best-characterized transport sequences comprise one or two short stretches of basic amino acids. These basic, generally lysine-rich signals are typified by the simian virus 40 (SV40) large T-antigen (TAg) NLS (PKKKRKV) or the cellular nucleoplasmin protein NLS (KRPAATKKAGQAKKKK) and are frequently referred to as classical NLSs. These sequences are recognized in the cytoplasm by a heterodimeric import receptor composed of importin α and importin β. Importin α, from which six isoforms have been identified in humans (29), functions as an adapter protein between the NLS and the importin β protein. Thus, importin α binds classical NLS-bearing proteins via its two NLS binding sites in the central area (5, 21) and importin β via its amino-terminally located importin β-binding domain (14, 57). Importin β is the transport receptor that promotes the interaction of the resulting heterotrimer with the NPC and subsequently the translocation into the nucleus (17, 39, 41, 56). Inside the nucleus, the import reaction is terminated by binding of nuclear Ran in its GTP-bound form to importin β, which results in the dissociation of the ternary importin α/β-cargo complex and the subsequent return of importin α and importin β but not the NLS-bearing protein to the cytoplasm (16, 45, 30).
More recently, a variety of nonconforming NLSs that are not particularly rich in lysine residues have been identified in various viral and cellular proteins of diverse function. Some of these NLSs have been characterized as nonconventional importin α-interacting motifs. Examples include the NLSs of the influenza virus NP protein (55), the Borna disease virus p10 protein (62), the cellular transcription factor Stat1 (47, 34, 35), the histone deacetylase 4 (18, 54), and the U1A spliceosome protein (22). The human immunodeficiency virus type 1 Rev and Tat proteins exemplify a subgroup of factors that utilize exclusively importin β for nuclear import with no apparent requirement for the importin α subunit. An arginine-rich NLS has been identified within these viral proteins, mediating the direct interaction with the importin β receptor (52). The human ribonucleoprotein A1 (hnRNP A1) represents another class of cargo proteins, which contains an NLS of 38 amino acids (aa), termed the M9 domain, which is rich in glycine and aromatic residues. This import sequence, which has also export activity, is directly recognized and carried into the nucleus by transportin, one of the importin β-like molecules (37, 44). Finally, additional members of the importin β family have been identified as import receptors for proteins that bear NLSs that do not resemble any of the signals mentioned above (49, 25).
The open reading frame UL84 of the β-herpesvirus human cytomegalovirus (HCMV) encodes an important regulatory protein that is supposed to be active in the cell nucleus (20). Initially, the UL84 protein (pUL84) was identified as a direct interaction partner of the nuclear HCMV regulatory protein IE2-p86, which is a dual-function activator/repressor polypeptide shown to be essential for viral gene expression (33, 51). Studies concerning the functional consequences of the pUL84/pIE2-p86 interaction revealed that pUL84 is able to specifically inhibit the trans-acting potential of IE2-p86 (12). Additional results indicated that pUL84 exerts an essential function in HCMV oriLyt-dependent DNA replication (46), further suggesting that the nuclear localization of this herpesviral regulatory protein is a prerequisite for its proper function. Since no nuclear targeting signals have been identified within pUL84 and previous publications described that pUL84 can be detected both in the cytoplasm and the nucleus of infected human fibroblasts (20, 51), we initiated our studies in order to elucidate the molecular mechanism of pUL84 nuclear trafficking.
Here, we report that pUL84 is a nuclear protein which interacts with at least four members of the importin α protein family in vitro and in vivo. Using in vitro transport assays, this interaction was shown to be essential for nuclear import of pUL84. The corresponding domain, which binds to importin α, was identified as a large region of 282 aa, comprising a cluster of basic amino acids and two leucine-rich sequence elements. Surprisingly, the cluster of basic amino acids was not sufficient for causing nuclear translocation. These data imply that a complex structure is required for interaction with importin α proteins, and consequently, this domain functions as a nonconventional nuclear targeting signal.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides were obtained from Eurogentec (Seraing, Belgium) or Sigma-ARK (Darmstadt, Germany). The sequences of oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study for sequencing, PCR, and mutagenesis
| Oligonucleotide | Sequence |
|---|---|
| NPI5 | GATCGGATCCATGACCACCCCAGGAAAAGAG |
| NPI-3 | GATCGAATTCAAGCTGGAAACCTTCCATAGG |
| 5RCH1Eco | GCATGAATTCATGTCCACCAACGAGAATGCTAAT |
| 3RCH1Xho | AGTCCTCGAGCTAAAAGTTAAAGGTCCCAGG |
| 5UL84-4111 | TGTCAAGCTTATGCCACGCGTCGACCCCA |
| 5UL84Hind404 | ATATAAGCTTAGGATGGGGGAAGCGAACGACGAG |
| 5UL84 | TAAGAATTCATGCCACGCGTCGACCCCAACCTTCGGAAT |
| 5UL84Eco403 | GATCGAATTCTTGGGGGAAGCGAACGACGAG |
| 5UL84Eco676 | AGCTGAATTCGCCGCCACCATGGTGCGCCTATCACTCAATCT |
| 5UL84Eco766 | AGCTGAATTCGCCGCCACCATGGCGTTACACGACTGTCTGGC |
| 5UL84Xba | AGCTTCTAGAATGCCACGCGTCGACCCCAACCTTCGG |
| 5UL84Nhe469 | AGCTGAATTCGCTAGCAAACAAGAGAAGAAAGAAGAGG |
| 5UL84Nhe586 | AGCTGAATTCGCTAGCGTGCGCCCGGCCTTCTCTCTC |
| 5UL84Nhe676 | AGCTGAATTCGCTAGCGTGCGCCTATCACTCAATCTC |
| 5UL84Nhe766 | AGCTGAATTCGCTAGCGCGTTACACGACTGTCTGGCG |
| 5UL84Nhe856 | AGCTGAATTCGCTAGCGTCGCTGATACCGCGGAATCA |
| 5UL84Not-751 | AGCTGCGGCCGCGCATCACAGCGCGTTA |
| 5UL84Xho925 | AGCTGAATTCCTCGAGCTGTGCTGGCATCGAGTG |
| 5UL84Xho1225 | AGCTGAATTCCTCGAGCTGCCTTACTTTGTACCG |
| 5UL84Xho1375 | AGCTGAATTCCTCGAGCGTCAGGGACTCAAAATG |
| 5UL84-201Eco | GATCGAATTCATGTCTCTCTTTCCCGCACGCCCAGGC |
| 3UL84Asp1699 | TCGACTGCAGGGTACCCAGCTTACAGTCTTGCGGT |
| 3UL84Asp1644 | TCGACTGCAGGGTACCGTCCTCCAGCGCGATGGA |
| 3UL84Asp1548 | TCGACTGCAGGGTACCTGTGGACCGCGAGTGTAG |
| 3UL84Asp1539 | TGCTGGTACCTGACAGGCAACCCCGATTCA |
| 3UL84Asp1524 | TCGACTGCAGGGTACCATTCAGCGTATGCTTTGA |
| 3UL84Asp1464 | TCGACTGCAGGGTACCCGTGGCGCCGTTCTCGTC |
| 3UL84Asp1404 | TCGACTGCAGGGTACCGACCGTAACGGGCATTTT |
| 3UL84Asp1342 | TGCTGGTACCGTAAAAGCGGCGGTAGATAC |
| 3FLAG-Xba | GCCCTCTAGAGCTTGTCATCGTCG |
| 3FLAGPSTI | TCGACTGCAGCTTGTCATCGTCGTCCTT |
| 3UL84Asp | TGCTGGTACCTTCGAGATCGCCGCAGACCATGG |
| 3FlagXbaXho | AGCTGTTTAAACCTCGAGTCTAGAGCTTGTCATCGTCGT |
| 3UL84XbaXho1524 | AGCTCTCGAGTCTAGAATTCAGCGTATGCTTTGA |
| 3UL84XbaXho1464 | AGCTCTCGAGTCTAGACGTGGCGCCGTTCTCGTC |
| 3UL84Xba-os | AGCTCTCGAGTCTAGAGAGATCGCCGCAGACCATGGC |
| 3UL84Nhe1524 | AGCTCTGCAGGCTAGCATTCAGCGTATGCTTTGA |
| 3UL84Not-720 | AGCTGCGGCCGCCGGCGTGATGATACGGAGCGC |
| 5UL84-M-C | CGAATCACCGCCGGACGCGACGGCGTCGTCACTCACGC |
| 3UL84-M-nC | GCGTGAGTGACGACGCCGTCGCGTCCGGCGGTGATTC |
Plasmid construction and in vitro mutagenesis.
The bait plasmid pHM479 for the Saccharomyces cerevisiae two-hybrid screen was generated by isolating the EcoRI/BglII fragment of pHM379 containing the UL84 reading frame (24) followed by insertion into the EcoRI/BamHI-digested vector pGBT9 (Clontech, Palo Alto, Calif.). The GAL4BD-EBNA1 bait plasmid (pGBT9-E1CT) and the yeast importin α1 plasmid pGAD-GH 142 were obtained from N. Fischer (Homburg, Germany) (10). The FLAG-tagged full-length proteins importin α1 (pHM815) and importin α5 (pHM1661) were constructed by PCR amplification using the oligonucleotides 5RCH1Eco/3-RCH1Xho and plasmid pET3a-RCH1 (10) as a template (Impα1) or using the oligonucleotides NPI5/NPI3 and plasmid pSE420 (40) as a template (Impα5). The resulting fragments were inserted into the pCATCH or pSUPER-CATCH vector (13), using the EcoRI/XhoI or BamHI/EcoRI restriction sites, respectively. For eukaryotic expression as FLAG-tagged importin α3 and α4 proteins, full-length importin α3 was isolated as an XhoI fragment from the yeast vector Y84-1 and full-length importin α4 was isolated as a SmaI/XhoI fragment from the yeast vector Y84-26. The fragments were cloned into the XhoI- or EcoRV/XhoI-cut vector pSUPER-CATCH to produce pHM1659 and pHM1657. The prokaryotic expression plasmid for GST-UL84 was a kind gift of B. Plachter (University of Mainz, Mainz, Germany). Plasmids pHM758 and pHM759 expressing wild-type UL84 fused N or C terminally to green fluorescent protein (GFP) were constructed by PCR amplification of UL84 followed by insertion into the GFP-H expression plasmids pHM758 and pHM759, respectively (50). The C-terminal UL84-FLAG deletion series for eukaryotic expression was produced by a PCR-based method using the oligonucleotides 5UL84-4111 and 3UL84Asp1699 to 3UL84Asp1342 and pcDNAUL84 (12) as a template. The fragments were inserted into pcDNAUL84 using the HindIII and Asp718 restriction sites, thereby replacing the full-length UL84 reading frame. The resulting plasmids were termed pHM660 to pHM665 (aa 1 to 468 to aa 1 to 566) and pHM534 (aa 1 to 447) and pHM535 (aa 1 to 513). The UL84 fragments of this deletion series were amplified using the primers 5UL84 and 3FLAG/PSTI and subcloned into the yeast vector pGBT9 via the EcoRI and PstI restriction sites. The resulting yeast plasmids expressing C-terminal UL84 deletion mutants fused in frame to the GAL4-BD were termed pHM697 to pHM702 (aa 1 to 468 to aa 1 to 566) and pHM671 (aa 1 to 447) and pHM672 (aa 1 to 513). Eukaryotic expression plasmids encoding N-terminal UL84 deletion mutants aa 60 to 586 (pHM537), aa 68 to 586 (pHM615), aa 83 to 568 (pHM616), aa 105 to 568 (pHM617), aa 111 to 568 (pHM595), and aa 119 to 568 (pHM596) were described previously (12). Additional UL84 N-terminal deletion mutants aa 135 to 586 (pHM536), aa 201 to 586 (pHM 2030), aa 226 to 586 (pHM831), aa 256 to 586 (pHM832), aa 226 to 508 (pHM833), and aa 256 to 508 (pHM834) were generated by PCR amplification using pcDNAUL84 as a template and oligonucleotides 5UL84Hind404/3UL84Asp, 5UL84-201Eco/3FLAG-Xba, 5UL84Eco676/3FlagXbaIXho, 5UL84Eco766/3FlagXbaIXho, 5UL84Nhe676/3UL84XbaXho1524, and 5UL84Nhe776/3UL84XbaXho1524, respectively. The PCR products were inserted into pcDNAUL84 or pCATCH either via the HindIII/Asp718 or the EcoRI/XbaI restriction sites, thereby replacing the full-length UL84 reading frame. The internal pUL84 deletion mutant UL84Δ241-253 (pHM835) was constructed by PCR amplification of the N-terminal UL84 fragment using the oligonucleotides 5UL84 and 3UL84Not-720. The C-terminal UL84 fragment was amplified using the oligonucleotides 5UL84Not-751 as well as oligonucleotide 3FlagXbaXho. Thereafter, the N-terminal fragment was inserted into the pcDNA3 vector using the EcoRI and NotI restriction sites, followed by ligation with the C-terminal fragment via NotI and XbaI. Site-directed mutagenesis within pcDNAUL84 was performed using a QuikChange site-directed mutagenesis kit as instructed by the manufacturer (Stratagene, Heidelberg, Germany) using the complementary oligonucleotide primers 5UL84-M-C and 3UL84-M-nC, which introduced the alanine replacement mutations indicated in Fig. 7. The resulting plasmid was termed LL359/361AA (pHM1756). The N-terminal UL84 deletion series fused to the GAL4-BD was produced by PCR using pcDNAUL84 as a template and oligonucleotides 3UL84Nhe1524 plus 5UL84Eco403, 5UL84Nhe469 to 5UL84Nhe856, or 5UL84Xho925 to 5UL84Xho1375. The oligonucleotides incorporated EcoRI (5′) and PstI (3′) restriction sites for in-frame insertion of the PCR product into the yeast vector pGBT9, to derive the Gal4BD-UL84 deletion series aa 135 to 508 to aa 459 to 508 (pHM768-771, pHM773-775, and pHM822-823). Plasmids expressing full-length UL84 or UL84 fragments simultaneously fused to GFP and beta-galactosidase (β-Gal) were generated by PCR amplification using pcDNAUL84 as a template and oligonucleotides 5UL84Xba/3UL84Xba-os, 3UL84XbaXho1524 plus 5UL84Nhe469, 5UL84Nhe586, 5UL84Nhe676, 5UL84Nhe766, 5UL84Nhe856, and 5UL84Nhe676/3UL84XbaXho1464. The PCR products were then cloned into the XbaI or NheI/XbaI sites of plasmid pHM761 (50), resulting in plasmids pHM780, pHM782, pHM817 to pHM821, and pHM836. Plasmids expressing the putative pUL84 NLS motifs (aa 160 to 172 and aa 241 to 253) simultaneously fused to GFP and β-Gal (pHM1648 and pHM1671) were generated by inserting the respective oligonucleotides within the NheI and PmlI restriction sites of vector pHM829 (50). Plasmids F-IE2 (pHM705), F-hSPT6 (pHM635), IE1 (pHM124), and IE2 (pHM134), used as controls in coimmunoprecipitation, pull down, or immunolocalization assays, were described previously (12, 24, 59). The DNA sequence of each plasmid construct was confirmed by automated sequence analysis (ABI, Weiterstadt, Germany).
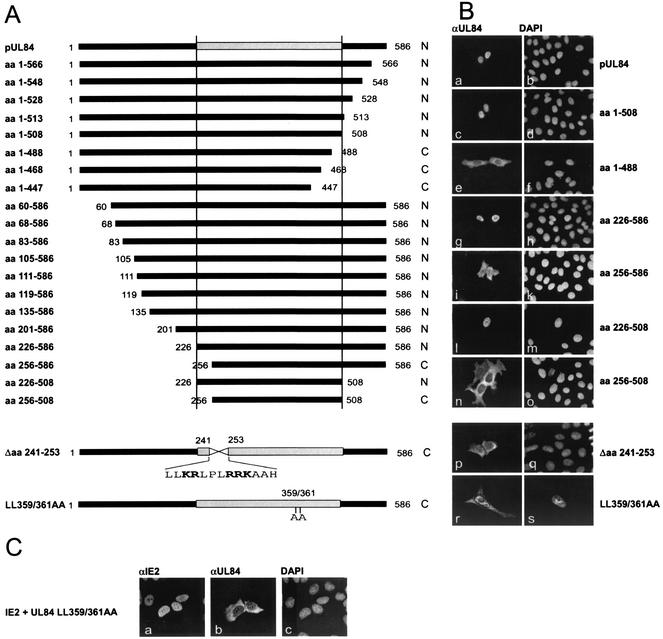
FIG. 7.
Delineation of a domain with NLS activity within the central part of pUL84 by deletion mutagenesis. (A) Schematic diagram illustrating C- and N-terminal pUL84 deletion mutants, an internal pUL84 deletion mutant lacking aa 241 to 253 of pUL84, and a pUL84 double alanine replacement mutant (LL359/361AA). Each mutant was subcloned in frame with the FLAG epitope, which can be detected by an anti-FLAG antibody. The subcellular localization of each pUL84 mutant is indicated by N (nucleus) or C (cytoplasm). The domain required for nuclear localization is shown as a gray box. (B) HeLa cells transfected with plasmids expressing pUL84 mutants were analyzed for subcellular localization of the indicated mutants via indirect immunofluorescence analysis. Abbreviations: αUL84, immunostaining of transfected HeLa cells using the anti-FLAG antibody M2 specific for the FLAG epitope; DAPI, DNA staining of transfected HeLa cells. (C) HeLa cells were cotransfected with plasmids expressing the viral protein IE2 and the pUL84 mutant LL359/361AA. Subsequently a double immunofluorescence analysis was performed with a polyclonal antiserum against the IE2 protein (αIE2) and an MAb directed against the FLAG epitope (αUL84). DAPI, DNA staining of transfected HeLa cells.
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed using GAL4 fusion proteins and S. cerevisiae Y153 as described previously (24). Yeast strain Y153 containing the bait plasmid pHM479 was transformed with a cDNA library derived from human B lymphocytes fused to the GAL4 activation domain in the pACT vector (7). The primary transformants (2.2 × 106) were selected for growth on histidine dropout plates containing 30 mM 3-aminotriazole. His+ colonies were subsequently analyzed for β-Gal activity by filter lift experiments (4). The interaction was then quantified by o-nitrophenyl-β-d-galactopyranoside (ONPG) assays as described earlier (19). Interactor plasmids from colonies positive in both tests were rescued by transformation of competent KC8 bacteria with total yeast DNA (23). For mapping of the pUL84 interaction domain using the yeast two-hybrid system, the respective UL84 deletion mutants within yeast vector pGBT9 were transformed together with the interactor plasmids into yeast strain Y153 and tested as described above.
Cell culture, transfection, and infection.
Primary human foreskin fibroblasts (HFF), HeLa cells, and 293 human kidney cells were cultured as described previously (60, 24). HFF were infected with HCMV (strain AD169) at a multiplicity of infection of 1 to 2 PFU per cell for the indicated time. HeLa and 293 cells were transfected via the calcium phosphate coprecipitation procedure using N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid as described earlier (60). Cells were harvested 48 h after transfection and used for Western blotting, immunoprecipitation, or immunofluorescence analysis.
Western blotting and immunoprecipitation analysis.
For Western blot analysis, transfected cells were lysed, diluted in sodium dodecyl sulfate (SDS)-Laemmli buffer, and boiled at 94°C for 5 min (24). Samples were electrophoresed by SDS-polyacrylamide gel electrophoresis (PAGE) on 8 to 12.5% polyacrylamide gels, and the proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). Western blotting and chemiluminescence detection were performed according to the manufacturer's protocol (ECL Western Detection Kit; Amersham Pharmacia Europe, Freiburg, Germany). Coimmunoprecipitation analysis was performed as described elsewhere (2). Briefly, transfected 293 cells were lysed in 800 μl of NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 5 mM EDTA; 0.5% NP-40; 1 mM phenylmethylsulfonyl fluoride; and 1 mg each of aprotinin, leupeptin, and pepstatin per ml) and incubated for 20 min at 4°C. After centrifugation, the supernatant was incubated with the appropriate antibody for 2 h at 4°C, and thereafter a 50% protein A-Sepharose suspension was added and incubation continued for an additional hour at 4°C. The Sepharose beads were collected and washed five times in NP-40 lysis buffer. Antigen-antibody complexes were recovered by boiling in SDS sample buffer and analyzed by Western blotting.
Purification of GST fusion proteins and pull-down assays.
The purification of GST fusion proteins was described in detail elsewhere (31). For pull-down assays, 5 to 30 μl of glutathione-Sepharose-bound proteins were preincubated for 10 min in 200 μl of ELB+ buffer (250 mM NaCl; 50 mM HEPES, pH 7.0; 0.1% NP-40; 0.5 mM EDTA; 1 mM phenylmethylsulfonyl fluoride; 0.5 mM dithiothreitol) containing bovine serum albumin (BSA) to a final concentration of 1 mg/ml. After addition of 1 to 5 μl of the indicated in vitro-translated, radiolabeled protein which had been generated by using the TNT system (Promega, Heidelberg, Germany), the beads loaded with GST fusion proteins were gently mixed for 2 h at 4°C. The beads were washed five times in 1 ml of ELB+ buffer, pelleted, and boiled in SDS sample buffer. The bound proteins were then resolved using SDS-polyacrylamide gels. The gels were fixed, incubated in Amplify (Amersham Life Science) for 30 min, dried, and subjected to autoradiography.
Antibodies and indirect immunofluorescence analysis.
The polyclonal antiserum against pUL84 of HCMV was described previously (12). The monoclonal antibody 69-66 (directed against pUL69) was obtained from B. Britt (Birmingham, Ala.); the pUL44-specific monoclonal antibody was obtained from B. Plachter (University of Mainz). The anti-FLAG monoclonal antibody (MAb) M2, which is directed against the synthetic FLAG octapeptide N-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-C, was purchased from INTEGRA Bioscience (Fernwald, Germany). The Anti-NPC MAb 414 was obtained from BabCo (Berkeley, Calif.). Anti-mouse and anti-rabbit horseradish peroxidase-, or fluorescein isothiocyanate (FITC)-, Texas Red-, and Cy5-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany). Indirect immunofluorescence analysis was performed according to a standard protocol as described previously (24). After the secondary antibody incubation, cells were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, Calif.) or DABCO and MOWIOL (Sigma Chemical Co., Deisenhofen, Germany, and Hoechst, respectively). Images were analyzed using a Zeiss Axioplan-2 microscope and recorded with a cooled Spot color digital camera (Diagnostic Instruments, Sterling Heights, Mich.). The Meta-Imaging series and Adobe Photoshop package (Universal Imaging Corp., Brandywine, Pa.; Adobe Systems Incorporated) were used for processing.
Nuclear import assays.
Nuclear import assays using permeabilized HeLa cells were exactly performed as described by Kann et al. (27). Briefly, digitonin-permeabilized HeLa cells grown on coverslips were incubated with glutathione S-transferase (GST)-UL84 (15 μg/ml) or GST alone in 20 μl of transport buffer (2 mM magnesium acetate, 20 mM HEPES [pH 7.3], 110 mM potassium acetate, 1 mM EGTA, 5 mM sodium acetate-1 mM dithiothreitol) containing rabbit reticulocyte lysate (RRL) (20 mg/ml; Promega) and aprotinin, leupeptin, and pepstatin (10 μg/ml each; Sigma) for 20 min at 37°C. An ATP-generating system (1 mM ATP, 5 mM creatine phosphate, creatine phosphokinase [20 U/ml]) and, if required, wheat germ agglutinin (WGA) (100 ng/ml; Boehringer Mannheim, Germany) were added. For competition experiments, peptides derived from the NLS of SV-40 TAg (STPPKKKRKRKV) or the hnRNP A1 M9 domain (YNNQSSNFGPMK) were added to final concentrations of up to 1 mM. For transport assays in the absence of cytosolic proteins, BSA was added instead of reticulocyte lysate to the same protein concentration (20 mg/ml). In some experiments the BSA-containing buffer was complemented with the indicated combinations of purified importin α1 (50 ng/μl), importin β (80 ng/μl), or Ran (100 ng/μl). FITC-labeled BSA conjugated with SV40 TAg NLS or hnRNP A1 M9 NS were used as control proteins in transport assays and added to a final concentration of 20 μg/ml. After the transport reaction, cells were washed and fixed onto the coverslips using 3% paraformaldehyde and subsequently subjected to immunofluorescence analyses. Images were analyzed using a Leica DM IRBE confocal microscope using the FITC, tetramethylrhodamine isothiocyanate, and Cy5 settings.
RESULTS
Identification of importin α1, α3, α4, and α5 as cellular interaction partners of the HCMV UL84 protein by yeast two-hybrid experiments.
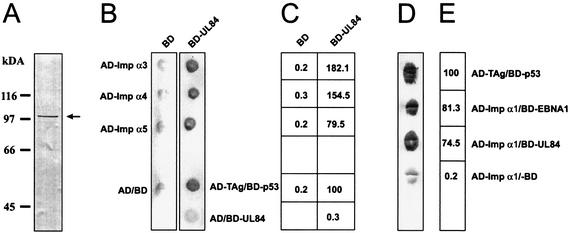
The GAL4-based yeast two-hybrid system was employed in order to identify cellular interaction partners of pUL84 of HCMV. For this, S. cerevisiae Y153 was transformed with the yeast expression plasmid BD-UL84 (pHM479) coding for an in-frame fusion of the UL84 sequence to the GAL4 DNA-binding domain. The presence of the GAL4-UL84 expression plasmid was stably maintained by selection in liquid dropout culture medium lacking tryptophan, and the expression of the respective fusion protein was confirmed by Western blot analysis (Fig. 1A). In order to determine whether the bait protein was able to activate transcription in yeast by itself, β-Gal expression of the yeast strain Y153/BD-UL84 that was transformed with the GAL4 activation domain plasmid pGAD424 was tested by filter lift experiments. No β-Gal expression could be detected with this combination, indicating that GAL4BD-UL84 alone does not activate expression of the reporter genes in yeast (Fig. 1B, AD/BD-UL84). The yeast two-hybrid screen was performed by transformation of yeast strain Y153 containing the BD-UL84 expression plasmid with a cDNA library derived from human B lymphocytes in the vector pACT (7). By using this screening procedure, 2.2 × 106 independent cDNA clones were tested for interaction with pUL84. Twenty-six clones were identified that activated histidine and β-Gal reporter gene expression in the presence of the BD-UL84 fusion protein. The specificity of the interaction was confirmed by retransforming the putative UL84-interacting cellular clones into Y153 strains containing pGBT9 vector only or BD-UL84. Only those library plasmids demonstrating a requirement of BD-UL84 for activation of both reporter genes were characterized further by automated sequencing and a search for homologies in the National Center for Biotechnology Information (Washington, D.C.) databases. Here, we report the identification of importin α3/Quip1, importin α4/hSRP1γ, and importin α5/hSRP1 as specific interaction partners of the UL84 protein (Fig. 1B, lane BD-UL84). For these interaction partners, more than one copy was found in the yeast two-hybrid screen, indicating a sufficient complexity of the cDNA library and the specificity of the interaction with pUL84. Cotransformation experiments of the respective interactor clones and the empty pGBT9 vector excluded the activation of the reporter genes by the importin α proteins in the absence of a bait protein (Fig. 1B, lane BD). Additionally, liquid β-Gal assays (ONPG assays) were performed in order to quantify the strength of interaction (Fig. 1C). Interestingly, the interaction of importin α3 and importin α4, which were selected as full-length cDNAs in the yeast two-hybrid screen, turned out to be even stronger than the interaction between p53 and the SV40 T-antigen, which served as a positive control.
FIG. 1.
Specific interaction between HCMV pUL84 and importins α1, α3, α4, and α5 in yeast. (A) Western blot analysis of yeast Y153 cell extracts after transformation with the yeast expression plasmid BD-UL84 coding for an in-frame fusion of the UL84 sequence to the GAL4 DNA-binding domain using a pUL84 specific antiserum. (B) Qualitative analysis of interactions between pUL84 and importins α3, α4, and α5 as determined in filter lift experiments after staining for β-Gal activity. Yeast cells were transformed with two separate expression plasmids, one of which encoded either BD-UL84 or the GAL4 DNA-binding domain alone (BD). The second plasmid encoded either importins α3, α4, and α5 as fusions with the GAL4 activation domain (AD-Imp α3, AD-Impα4, AD-Impα5) or the GAL4 activation domain alone (AD). The association of murine p53 (encoded by plasmid pVA3) and SV40 TAg (plasmid pTD1) served as a positive control. (AD-TAg/BD-p53). (C) Quantitation of the association between the proteins indicated in panel B as determined by liquid β-Gal assays (ONPG assays). Cotransformation experiments were performed as described for panel B. β-Gal activity was assayed from liquid cultures in at least three independent experiments, each with duplicate samples. The β-Gal activity of yeast cells transformed with a vector combination encoding murine p53 and SV40 TAg was set as 100. (D) Qualitative analysis of the interaction between pUL84 and importin α1 as determined by transformation and filter lift experiments as described in panel B. The association between importin α1 and EBNA1 served as an additional positive control (AD-Impα1/BD-EBNA-1). (E) Quantitation of the protein associations shown in panel D via ONPG assays as described in panel C.
Importin α proteins function as adapter molecules which bind NLS-containing proteins and the nuclear import receptor importin β (reviewed in reference 15). In humans, six isoforms for importin α have been described so far. Based on the similarity of their primary structures, the members of the importin α protein family can be grouped into three main subfamilies: (i) an importin α3-like subfamily consisting of importin α3 and α4; (ii) an importin α5-like subfamily consisting of importin α5, α6, and α7; (iii) and an importin α1 subfamily with importin α1 as the only member (28, 29). In our yeast two-hybrid screen we identified only members of the importin α3- and the α5-like subfamilies as interaction partners of pUL84. Therefore, we asked whether pUL84 is also able to interact with a member of the third importin α subfamily represented by importin α1/RCH-1. For this, the yeast strain Y153 was transformed with AD-Impα1, containing aa 38 to 529 of importin α1 in fusion with the GAL4 activation domain (obtained from N. Fischer) and BD-UL84. A yeast strain transformed with AD-Impα1 and the Epstein-Barr virus protein EBNA1, fused to the GAL4 DNA-binding domain, served as a positive control (10) (Fig. 1D). Since we observed comparable activation of the reporter genes His3 and lacZ in the AD-Impα1/BDUL84 transformed strain and the AD-Impα1/BD-EBNA1 positive control (Fig. 1E), we concluded that pUL84 actually binds to importin α1. Thus, we showed that importin α isoforms of all three subgroups have distinct pUL84 binding specificities.
GST pull down analysis reveals a direct interaction between pUL84 and at least one member of every importin α subfamily.
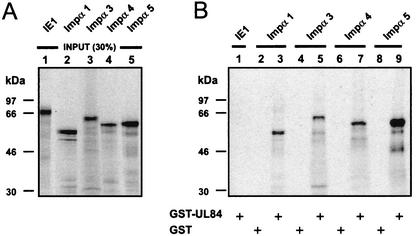
To confirm whether the observed interactions of pUL84 with the indicated importin α proteins could also be observed in an independent experimental approach, GST pull down analyses were performed. For this purpose, we fused the coding sequence of UL84 to the C terminus of GST and purified the fusion protein from Escherichia coli (GST-UL84). The cellular proteins importin α1, α3, α4, and α5 and the viral protein IE1-p72 (IE1) were in vitro translated in reticulocyte lysates in the presence of [35S]methionine (Fig. 2A). The radiolabeled proteins were then incubated with the bacterially expressed GST-UL84 fusion or with GST alone, which served as a negative control. As shown in Fig. 2B, lanes 3, 5, 7 and 9, all tested importin α proteins were able to interact strongly with the GST-UL84 fusion protein. No interaction was observed with GST alone (Fig. 2B, lanes 2, 4, 6, and 8). In addition, the GST-UL84 protein was not able to bind to IE1 (Fig. 2B, lane 1), arguing against a nonspecific interaction of pUL84 with importin α proteins. These data showed that pUL84 interacts with the indicated importin α proteins in an in vitro binding assay, confirming the results obtained by the yeast two-hybrid screen.
FIG. 2.
pUL84 interacts physically with importins α1, α3, α4, and α5 in GST pull down experiments. (A) In vitro-translated 35S-labeled IE1-p72 (lane 1) and wild-type importins α1, α3, α4, and α5 (lanes 2 to 5) were used for pull down assays. After SDS-PAGE the proteins were depicted by autoradiography. Lanes 1 to 5 contain input proteins (30% of the amount used in the pull down analyses). (B) Proteins are shown that were recovered after GST pull down analysis. Lanes 1, 3, 5, 7, and 9, wild-type UL84 in fusion with GST (GST-UL84) or GST alone (GST; lanes 2, 4, 6, and 8) was incubated with in vitro-translated IE1-p72 and importins α1, α3, α4, and α5, respectively. After extensive washing, the bound proteins were resolved by SDS-PAGE, and an autoradiograph of the gel is shown.
Coimmunoprecipitation of pUL84 with four members of the importin α protein family confirms the in vivo interaction of both proteins after transient transfection of cells.
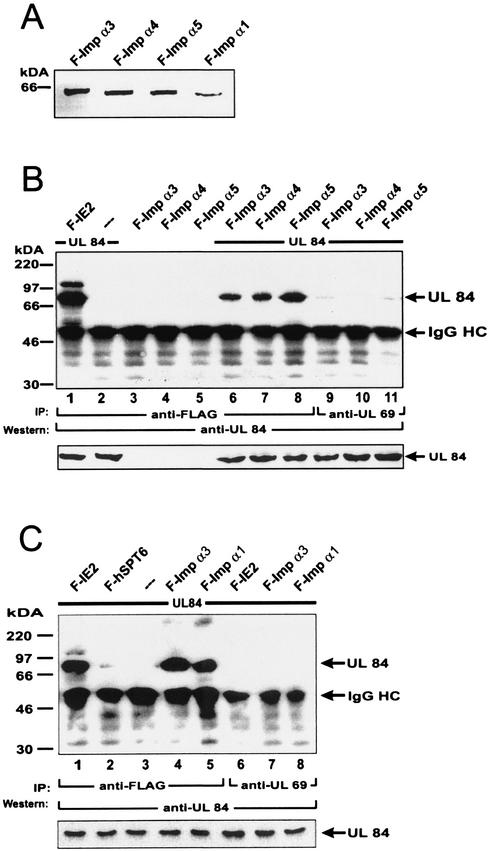
Although the yeast two-hybrid screen and the in vitro interaction experiments indicated a strong binding of pUL84 to importin α proteins, we sought to confirm this within the context of a mammalian cell. In order to be able to immunoprecipitate the various importin α proteins from mammalian cell extracts we constructed eukaryotic vectors, expressing the indicated importin α proteins in fusion with the FLAG epitope (Fig. 3A). Cotransfection experiments in 293 cells were performed using expression vectors for the indicated FLAG-tagged importin α proteins and pUL84, followed by immunoprecipitation with an anti-FLAG antibody. For the detection of coprecipitated pUL84, a specific antiserum was used in Western blot analyses (12). Initially, we tried to confirm the interaction of pUL84 and the importin α family members that were isolated in our yeast two-hybrid screen (Fig. 3B). For this purpose we coexpressed pUL84 together with FLAG-tagged importin α3, α4, and α5 in 293 cells. After immunoprecipitation and Western blot analysis, a strong signal corresponding to pUL84 could be observed in reactions with anti-FLAG antibody (Fig. 3B, lanes 6, 7, and 8). This reaction was specific, since no significant signals were present when each of the proteins was expressed alone (Fig. 3B, lanes 2, 3, 4, and 5) or when an MAb against the viral UL69 protein (anti UL69) was used for precipitation (Fig. 3B, lanes 9, 10, and 11). The interaction of pUL84 with the FLAG-tagged IE2-p86 transactivator of HCMV served as a positive control in these experiments (Fig. 3B and C, lanes 1) (12). Identical results were obtained when we analyzed the interaction of pUL84 with importin α1 in coimmunoprecipitation experiments (Fig. 3C). In addition, after cotransfection of pUL84 together with the FLAG-tagged cellular protein hSPT6 (59) and coimmunoprecipitation with the anti-FLAG antibody, no pUL84 was detectable (Fig. 3C, lane 2), thus further confirming the specificity of the importin α-interactions (Fig. 3C, lanes 4 and 5). On the basis of these results we concluded that pUL84 is able to interact with at least 4 importin α proteins in the background of a mammalian cell.
FIG. 3.
Analysis of the interaction between pUL84 and importins α3, α4, α5, and α1 by coimmunoprecipitation from transfected cells. (A) Western blot analysis of 293 cell extracts after transfection with eukaryotic expression plasmids coding for in-frame fusions of wild-type importin α1, α3, α4, and α5 to the FLAG epitope using the anti-FLAG MAb. (B) 293 cells were transfected with expression vectors encoding pUL84; FLAG-importin α3, α4, and α5; or FLAG-IE2-p86 (F-IE2) as indicated. After cell lysis, immunoprecipitations (IP) were performed using the anti-FLAG MAb (lanes 1 to 8) or the unspecific UL69 MAb 69-66 as control antibody (lanes 9 to 11). Precipitates were separated by SDS-PAGE, and coprecipitated pUL84 was detected by Western blot analysis using the pUL84 specific antiserum. Lanes: 1, transfection with a combination of vectors encoding FLAG IE2-p86 and pUL84; 2, transfection with the pUL84 expression vector alone; 3 to 5, transfection with the indicated FLAG-Importin α expression vectors alone; 6 to 11, transfection with a combination of vectors encoding the indicated FLAG-importin α proteins and pUL84. The sizes of the molecular mass markers are indicated on the left; the position of pUL84 and the immunoglobulin heavy chain (IgG Hc) are shown on the right. The expanded section at the bottom of panel B is a Western blot using pUL84 antiserum that shows relative levels of pUL84. (C) 293 cells were transfected with expression vectors encoding pUL84, FLAG IE2-p86, FLAG-hSPT6, and FLAG-importin α3 and α1 as indicated and analyzed via immunoprecipitation analyses as described in panel B. Immunoprecipitations were performed using the anti FLAG MAb (lanes 1 to 5) or the unspecific UL69 MAb 69-66 as control antibody (lanes 6 to 8). Lanes: 1 and 6, transfection with a combination ofvectors encoding FLAG IE2-p86 and pUL84; 2, transfection with a combination of vectors encoding FLAG hSPT6 and pUL84; 3, transfection with the pUL84 expression vector alone; 4, 5, 7, and 8, transfection with a combination of vectors encoding the indicated FLAG-importin α proteins and pUL84. The expanded section at the bottom of panel C is a Western blot using pUL84 antiserum that shows relative levels of pUL84.
pUL84 is located within the nucleus of both infected and transfected cells.
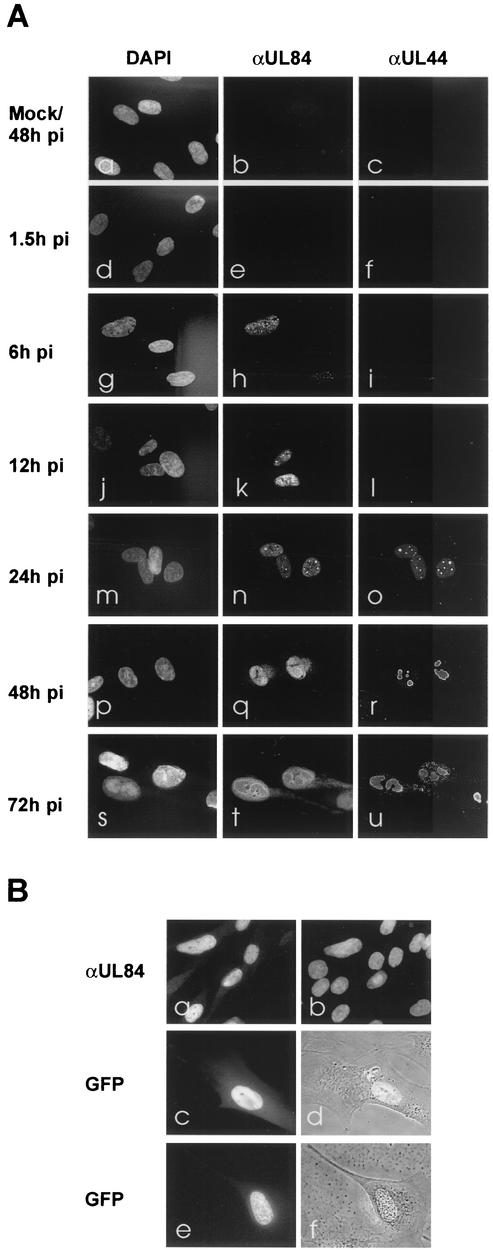
Previous publications described that pUL84 can be detected both within the nucleus and the cytoplasm of HCMV-infected fibroblast cells (20). To confirm these data with our HCMV strain, we infected primary human fibroblasts with HCMV AD169. pUL84 localization was determined at various times after infection via indirect immunofluorescence microscopy by using a specific rabbit antiserum. Since pUL84 has been described to be required for viral DNA replication, we performed a costaining with an MAb against the replication processivity factor pUL44, which has previously been identified as a component of viral replication centers (46). As shown in Fig. 4A, panels h, k, n, q, and t, pUL84 could predominantly be found within the nuclei of infected fibroblasts during all phases of the viral replication cycle. No signal could be detected with mock-infected fibroblasts or when the preimmune serum was used for staining (Fig. 4A, panels a and b; data not shown). Interestingly, although the prereplicative globular structures that could be detected at 24 h postinfection with anti-pUL44 showed some colocalization with pUL84, we were not able to observe an exclusive colocalization of pUL84 with viral replication compartments during later times of the replicative cycle (Fig. 4A, panels m to u).
FIG. 4.
Nuclear localization of pUL84 in infected and transfected cells. (A) Immunofluorescence analysis of HCMV-infected fibroblast cells. Mock (a to c) or HCMV AD169 (d to u) infected HFF were fixed at the indicated time post infection (pi) and costained using a specific anti-UL84 antiserum (b, e, h, k, n, q, and t; αUL84) and a pUL44-specific MAb (c, f, i, l, o, r, and u; αUL44). DAPI, DNA staining ofinfected cells (left column [a, d, g, j, m, p, and s]). (B) Immunofluo- rescence analysis of transiently transfected HFF. HFF were transfected with expression vectors encoding pUL84 (a and b), pUL84 in fusion with the GFP at the C terminus (c and d), or pUL84 in fusion with GFP at the N terminus (e and f). Transfected cells were fixed and immunostained using the specific rabbit antiserum against pUL84 (αUL84) (a), or living cells were analyzed for GFP expression by direct fluorescence microscopy (c to f). (b) DAPI DNA staining of the transfected HFF. (d and f) phase-contrast microscopy of the GFP-expressing cells.
Since pUL84 binds very tightly to the IE2-p86 transactivator of HCMV, which contains two NLS, this interaction might play a role for nuclear localization of pUL84 during viral infection. To exclude this possibility, primary human fibroblasts were transfected with the pUL84 expression vector pcDNAUL84, and this was followed by fixation of transfected cells and immunostaining using the specific rabbit antiserum against pUL84. A strong nuclear staining was observed in this experiment, which argues against a role of additional viral proteins for nuclear localization of pUL84 (Fig. 4B, panels a and b). To finally exclude any artifacts from fixation and staining, we constructed plasmids pHM758 and pHM759, which expressed pUL84 in fusion with the GFP either at the C or the N terminus, respectively. Fibroblasts were then transfected either with pHM758 or pHM759, and living cells were analyzed for GFP expression by fluorescence microscopy. This revealed a strong nuclear fluorescence for both fusion proteins (Fig. 4B, panels c to f). In addition, weak cytoplasmic fluorescence could be detected in cells that expressed high amounts of the respective UL84 fusion protein (Fig. 4, panels c and e). In summary, these data clearly demonstrate that pUL84 shows a predominantly nuclear localization after both transfection and infection which is independent from additional viral proteins.
The UL84 protein contains no classical NLS.
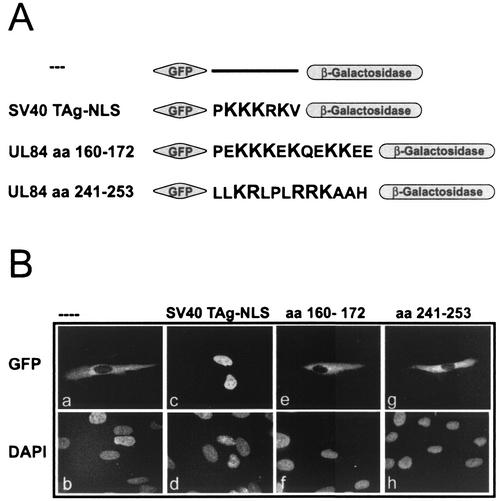
It is well known that the binding site for importin α proteins on a nucleophilic polypeptide is usually represented by a short stretch of basic amino acids. To define the NLS responsible for pUL84 nuclear import, we scanned the pUL84 primary sequence and noticed two clusters of basic amino acids which may serve as NLSs (aa 160 to 172 and aa 241 to 253). To evaluate the potential nuclear import function of these motifs, the respective pUL84 domains were inserted into the NLS mapping vector pHM829 (Fig. 5A) (50). This vector allows the expression of proteins that are simultaneously fused to GFP and to β-Gal. The GFP part encoded by this vector functions as a fluorescent tag, whereas the β-Gal part increases the molecular weight of the fusion protein, thus preventing passive diffusion of smaller fusion proteins into the nucleus. As demonstrated previously (50) this allows the unambiguous identification of the NLS activity of a protein domain due to the strictly cytoplasmic staining of a GFP-β-Gal fusion without NLS (Fig. 5B, panels a and b). Interestingly, none of the basic amino acid clusters derived from pUL84 was able to translocate the corresponding fusion protein into the nucleus after transfection of human fibroblasts (Fig. 5B, panels e to h) and HeLa cells (data not shown). In contrast, the positive control containing the NLS of the SV40 TAg showed an exclusively nuclear staining (Fig. 5B, panel c and d). These data suggested that both putative NLSs of pUL84 are not functional, leading to the assumption that pUL84 should possess an NLS which is distinct from the classical type.
FIG. 5.
pUL84 contains no classical NLS. (A) Schematic representation of the classical SV40 TAg-NLS and two putative UL84-NLS which were inserted into NLS mapping vector pHM761 (50). The basic amino acids within the putative NLS sequences are shown in a larger typeface. The resulting expression vectors encode GFP in fusion with the indicated motifs and β-Gal. (B) HFF transfected with plasmids expressing the indicated GFP-β-Gal fusion proteins were analyzed for GFP expression by fluorescence microscopy (a, c, e, and g). DAPI, DNA staining of transfected HFF (b, d, f, and h).
Delineation of the importin α interaction domain within pUL84.
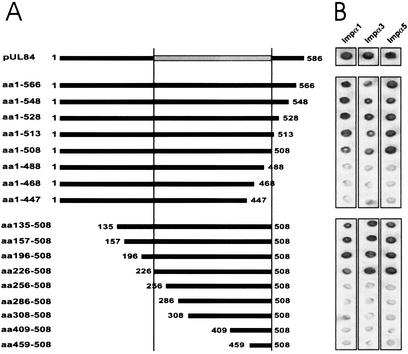
In order to characterize the sequences required for nuclear translocation, we first wanted to delineate the domain within pUL84 which mediates binding to importin α proteins. For this purpose we first constructed a series of C-terminal UL84 deletion mutants termed aa 1 to 566 to aa 1 to 447 as fusions to the GAL4 DNA binding domain. These mutants were analyzed in the yeast two-hybrid system for their interaction with one member of each importin α subgroup. As shown in Fig. 6, the deletion mutant lacking 98 C-terminal amino acids (aa 1 to 488) lost its binding activity, while the mutant lacking 78 C-terminal residues (aa 1 to 508) was still able to interact with all importin α proteins tested (Fig. 6). Due to these results, we then constructed N- and C-terminal deletion mutants (aa 135 to 508 to aa 459 to 508) to define the amino-terminal sequences involved in binding. This revealed that the first 225 aa of pUL84 is dispensable for binding, since pUL84 deletion mutant aa 226 to 508 gave a strong reaction in the filter lift assay. However, mutant aa 256 to 508 was no longer able to interact (Fig. 6). Thus, we mapped the importin α-binding domain to the central region of pUL84 between aa 226 and 508. Interestingly, we could not observe differences in the binding of various importin α subfamily members in these experiments, indicating that all importin α subtypes tested require the same domain for binding to pUL84.
FIG. 6.
Mapping of the importin α1, α3, and α5 interaction domain within pUL84 using the yeast two-hybrid system. (A) Schematic representation of C- and N-terminal deletion mutants of pUL84 generated as in-frame fusions to the GAL 4 DNA-binding domain. (B) Yeast cells were transfected with a combination of vectors encoding the pUL84 deletion mutant fused to the GAL 4 DNA-binding domain (see panel A) and one of the indicated wild-type importin α proteins fused to the GAL 4 activation domain. Yeast colonies were selected for the presence of both plasmids with dropout media lacking tryptophan and leucine. Subsequently, the colonies were analyzed for the expression of β-Gal by filter lift assays.
The importin-α interaction domain of pUL84 constitutes a nonconventional NLS.
In order to test whether the identified importin α-interaction domain of pUL84 is sufficient for mediating the nuclear localization of this protein, a series of N- and C-terminal deletion mutants of pUL84 was generated within the eukaryotic expression vector pcDNA3 (Fig. 7). In addition, an internal deletion mutant of pUL84 was constructed which lacked aa 241 to 253 of the importin α binding domain (Fig. 7A, Δaa 241-253). Each mutant was subcloned in frame with the FLAG epitope, which can be detected by an anti-FLAG antibody. Nonpermissive HeLa cells were then transfected with the respective pUL84 expression plasmids followed by indirect immunofluorescence analysis using a FLAG-specific MAb. Figure 7 summarizes the results of immunolocalization experiments. It shows that the domain which has been identified as the importin α interaction domain is sufficient for nuclear localization of pUL84. In accordance with the importin α interaction in the yeast two-hybrid screen, further truncations at either the C or N terminus prevented nuclear localization (Fig. 7B, panels e, f, n, and o). After transfection of the internal pUL84 deletion mutant, in which the basic amino acid cluster of the importin α binding domain was deleted, we observed an exclusively cytoplasmic localization (Fig. 7B, panels p and q). Thus, although the basic cluster itself apparently did not serve as an NLS, we concluded that it is involved in the nuclear localization of pUL84. In order to investigate whether additional sequences of the importin α interaction domain are important for nuclear localization, UL84 mutant LL359/361AA was constructed by site-directed mutagenesis which contains amino acid exchanges at positions 359 and 361 of pUL84. As shown in Fig. 7B, panels r and s, this mutant showed a predominantly cytoplasmic localization. This further supports the assumption that the cluster of basic amino acids is not sufficient for nuclear localization but additional sequences are required to generate a functional UL84-NLS. Identical results were obtained when primary human fibroblasts or COS7 cells were used for transfection, which argues against a cell-type-specific effect observed in these experiments (data not shown). Furthermore, we asked whether the IE2 protein, which binds with high affinity to pUL84, might be able to induce a nuclear localization of an NLS-deficient UL84 mutant. In order to answer this question, HeLa cells were cotransfected with the pUL84 mutant LL359/361AA and the IE2 expression plasmid pHM134. However, after costaining of doubly transfected cells, pUL84 was still detected in the cytoplasm. This suggests that IE2 is not able to induce a relocalization of pUL84 in the absence of a functional pUL84 NLS further supporting the biological relevance of this NLS domain.
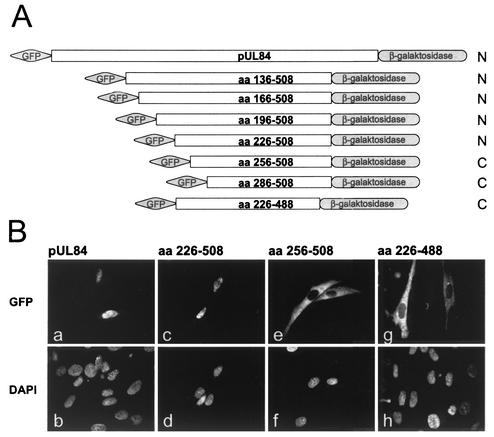
To prove whether the identified domain of pUL84 is able to mediate nuclear localization when fused to a heterologous protein, we inserted fragments of UL84 into our NLS mapping vector (Fig. 8A). When aa 226 to 508 of pUL84 was expressed in fusion with GFP and β-Gal, the resulting fusion protein was detected within the nucleus of transfected human fibroblasts (Fig. 8B, panels c and d). This could also be observed for full-length pUL84 (Fig. 8B, panels a and b). Consistent with the lack of importin α interaction and the cytosolic localization of pUL84 deletion mutants, aa 256 to 508 and aa 226 to 488 were insufficient to induce the nuclear accumulation of the respective fusion proteins (Fig. 8B, panels e and f and panels g and h, respectively). Thus, these experiments identified a large domain within pUL84 comprising aa 226 to 508 which is able to act as an NLS.
FIG. 8.
Nuclear import of heterologous proteins by a distinct pUL84 subdomain. (A) Schematic representation of wild-type pUL84 and N- and C-terminal pUL84 deletion mutants inserted into the NLS mapping vector pHM761 (50). The subcellular localization of each pUL84-derived fusion protein is indicated by N (nucleus) or C (cytoplasm). (B) The subcellular localization of the resulting GFP β-Gal fusion proteins was analyzed as described in the legend of Fig. 5. DAPI, DNA staining of transfected HFF (b, d, f, and h).
pUL84 uses the importin α/β pathway for nuclear import.
The presented experiments demonstrated that pUL84 contains a nuclear localization domain and that this domain interacts with different isoforms of the importin α protein family. However, these assays did not necessarily mean that the importin α/β pathway caused the nuclear import of pUL84. Thus, we wanted to obtain proof that pUL84 is transported into the nucleus in an importin α/β-dependent manner. To answer this, in vitro nuclear import assays using digitonin-permeabilized HeLa cells were performed.
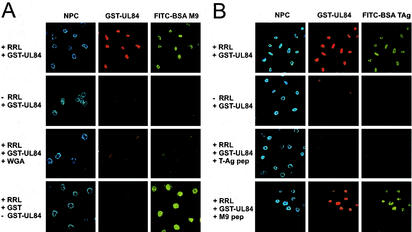
Initially, we wanted to determine whether a GST-UL84 fusion protein was imported into the nuclei of HeLa cells in this experimental system. For this purpose the plasma membrane of HeLa cells grown on coverslips was permeabilized using digitonin and the cytosol was washed out. E. coli-purified GST-UL84 was added in the presence or absence of RRL as a source of cytosolic factors (1). The lysates were complemented with an ATP-generating system. FITC-BSA linked to the M9 domain transport signal of hnRNP A1 (48) was added as an internal positive control for nuclear import. This substrate was chosen in the initial experiments because its import does not require importin α, thus not interfering with the putative importin α-dependent import of GST-UL84. After fixation, GST-UL84 and the nuclear membrane were visualized via confocal immunofluorescence microscopy using pUL84-specific antibodies and antibodies against the NPC followed by Texas Red- and Cy5-coupled secondary antibodies; BSA M9 was detected via its FITC conjugate. A strong uptake of GST-UL84 and the internal positive control into the nuclei of HeLa cells could be observed in the presence of reticulocyte lysate (Fig. 9A, +RRL). In contrast, when the reticulocyte lysate was replaced by a buffer containing BSA, no nuclear import was detected (Fig. 9A, −RRL), indicating the necessity of cellular transport factors. This nuclear import was specific since preincubation of cells with WGA, a lectin that binds to N-acetylglucosamine-containing proteins of the NPC (9), prevented detectable nuclear import of both import substrates (Fig. 9A, +RRL+WGA). As a further negative control, an import reaction in which GST-UL84 was replaced by GST alone was performed (Fig. 9A, +RRL, -GST-UL84, +GST). Taken together, these findings showed that GST-UL84 is imported into the nuclei of permeabilized cells in the presence of cytosolic factors.
FIG. 9.
Active import of recombinant GST-UL84 protein in digitonin-treated cells is inhibited by a peptide derived from the oligobasic SV40 TAg-NLS. GST (+GST) and wild-type GST-UL84 (+GST-UL84) fusion proteins were purified from bacterial lysates and subjected to in vitro transport assays. For this, HeLa cells were permeabilized with digitonin and incubated with the indicated GST proteins in the presence of RRL (+RRL) as a source of native nuclear import factors and ATP at 37°C. Additionally, the FITC-BSA SV40 TAg-NLS (FITC-BSA TAg) or FITC-BSA M9 (FITC-BSA M9) conjugates were added as internal controls as indicated. The cells were processed for microscopic analysis, and the NPCs were stained with an NPC-specific antibody and a Cy5-conjugated secondary immunoglobulin G (NPC). (A) Import of GST-UL84 was analyzed by confocal microscopy using the specific pUL84 antiserum (+RRL, +GST-UL84) and FITC-conjugated secondary immunoglobulin G. In control reactions nuclear import of GST-UL84 was assessed when incubated at 37°C in the absence of either cytosolic factors (−RRL, +GST-UL84) or after preincubation of the cytosolic fraction with WGA (+RRL, +GST-UL84, +WGA). Additionally, a control reaction was performed in which GST-UL84 was replaced by GST (+RRL, +GST, −GST-UL84). (B) Nuclear import of GST-UL84 was assessed as described in panel A. For competition of import, reactions were complemented with unconjugated SV40 TAg-NLS peptide (+RRL, +GST-UL84, +TAg pep) or with M9 peptide (+RRL, +GST-UL84, +M9 pep) as indicated.
To elucidate whether importin α/β is involved in the nuclear uptake of GST-UL84 in digitonin-permeabilized nuclei, we examined whether known peptide binding targets for selected nuclear import factors would be able to competitively inhibit this nuclear uptake when present in excess. Similar approaches have been used successfully to differentiate between transport receptors involved in nuclear import of a given protein (53). The inhibitors chosen were the SV40 TAg-NLS, which binds to importin α directly (47), and the M9 NLS/NES, which binds to the import receptor transportin (3, 44). As substrate proteins we used GST-UL84 and FITC-BSA linked to the SV40 TAg-NLS, the latter serving as an internal positive control for importin α-dependent nuclear import. As shown in Fig. 9B, both substrates were imported into the nuclei of digitonin-permeabilized HeLa cells when RRL was added as a source of native import factors (Fig. 9B, +RRL). No import of GST-UL84 and of the control conjugate could be observed in the absence of cytosolic factors (Fig. 9B, −RRL). The addition of an excess of the SV40 TAg-NLS peptide blocked the uptake of both GST-UL84 and, as predicted, FITC-BSA TAg (Fig. 9B, +RRL, +TAg pep). The inhibition was specific since the M9 peptide, which directly binds to transportin, affected neither SV40 TAg-NLS-dependent nuclear import nor the nuclear translocation of GST-UL84 (Fig. 9B, +RRL, +M9pep). These findings may indicate that both proteins, GST-UL84 and the SV40 TAg-NLS BSA conjugate, compete with the TAg peptide for importin α binding. However, on the basis of these data, we cannot exclude the possibility that the subsequent formation of a complex of TAg-NLS peptide, importin α, and importin β reduced the amount of available importin β. Taken together, the inhibition showed that the importin pathway is used and, since the inhibition was complete, that no other significant import pathways are used in parallel.
Nuclear import of pUL84 is mediated by importin α.
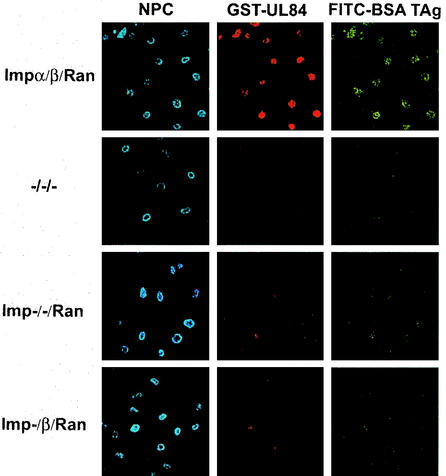
The previous transport assay revealed that pUL84 uses the importin α/β pathway for nuclear translocation. However, the data did not exclude the possibility that only importin β is required and that the import does not involve other cytosolic proteins bridging between pUL84 and the import receptors. To answer these questions we performed nuclear import assays in which the cytosolic extract was replaced by a buffer complemented with recombinant importins α and β. Since the GTPase Ran is required to terminate the nuclear import, Ran was added to the reaction mixture. As shown in Fig. 10, the recombinant importins together with Ran were able to mediate the in vitro nuclear uptake of the FITC-BSA TAg positive control and of the GST-UL84 fusion protein, confirming the use of the importin α/β pathway (Fig. 10, Impα/β/Ran). To dissect the pathway, the assay was performed using these three components in the indicated combinations. Neither Ran alone nor Ran in combination with importin β could promote nuclear import of GST-UL84 or the FITC-BSA TAg control (Fig. 10, Imp−/−/Ran and Imp−/β/Ran). Thus, the requirement of both importins suggested that an interaction of importin α to the newly defined importin α binding domain on pUL84 was necessary for bridging pUL84 to the import receptor importin β.
FIG. 10.
pUL84 nuclear uptake is mediated via the importin α/β/Ran pathway. Digitonin-permeabilized HeLa cells were incubated in a buffer containing recombinant GST-UL84 and FITC-BSA TAg. Recombinant purified import factors (importin α [Imp α], Imp β, and Ran) were added as indicated. Import reactions were carried out as described in the legend of Fig. 9.
DISCUSSION
The study presented here is based on the assumption that the activities and/or the subcellular localization of the viral regulatory protein pUL84 might be modulated by proteins of the host cell. Therefore, by performing a yeast two-hybrid screen we searched for cellular proteins which interact with pUL84 and identified four members of the importin α protein family as strong interaction partners of this viral protein. We utilized GST pull down experiments and coimmunoprecipitation analyses to verify that pUL84 interacts with the full-length importin α isoforms 1, 3, 4, and 5. Importin α proteins function as adapter molecules bridging NLS-containing import cargo proteins to the import receptor importin β, which actually carries the importin α-cargo complex from the cytoplasm into the nucleus. Thus far, six genes coding for importin α proteins have been identified in humans. On the basis of sequence similarities these proteins can be classified into three distinct subfamilies, namely, α1, α3, and α5 (15, 29). The presence of multiple isoforms of importin α in humans suggests either functional differences between these factors and/or a tissue-specific expression pattern. Although a strict tissue-specific expression was only reported for importin α6, which has so far been exclusively found in testis, expression levels of the other importin α proteins show a considerable cell-type-dependent variation (26, 29). With respect to functional differences, a number of studies have reported that certain substrates can be bound and transported by particular members of the importin α family but not by others (38, 42, 47). In contrast, we report strong binding of pUL84 to four importin α isoforms covering all importin α subgroups. At present, the possible existence of different efficiencies of the various importin α isoforms in mediating pUL84 import cannot be excluded entirely. Nevertheless, all our data either from the yeast two-hybrid screen or from GST pull down experiments or from coimmunoprecipitation analysis suggest that all importin α subgroups may function as equivalent NLS receptors for the UL84 protein. This might be a strategy to ensure efficient nuclear import of pUL84 in the broad spectrum of cell types that are infected by HCMV in vivo.
In general, importin α proteins are involved in nuclear import of proteins bearing a classical NLS of the basic type (32). Basic type NLSs show a relatively poor consensus sequence [K(K/R)X(K/R)], with a general feature of a short stretch of amino acids that contain a high proportion of positively charged amino acids (43). Two motifs with homology to classical NLSs were identified within the pUL84 primary sequence by computer analysis. However, we observed that GFP-β-Gal fusion proteins containing each of the putative NLS motifs individually were clearly cytoplasmic (Fig. 5), indicating that the respective motifs on their own are not capable of mediating the nuclear import of large cytosolic proteins. Since this observation was obtained with different cell types, it reflects a general phenomenon rather than a cell-type-specific effect. Furthermore, the proposed classical NLS sequence comprising aa 160 to 171 of pUL84 was entirely dispensable for the nuclear localization of a pUL84 deletion mutant (Fig. 7B, panels g and h). The lack of import capacity of these basic clusters was surprising since a recent publication by Xu et al. showed that one of these putative NLS sequences (aa 160 to 171, PEKKKEKQEKK) is responsible for nuclear localization of pUL84 (63). It must be considered, however, that the MPEKKKEKQEKK-GFP fusion protein that Xu et al. used has a molecular mass of only 30 kDa, thus being below the diffusion threshold of the NPC of 50 to 60 kDa (32). Therefore, it appears possible that the GFP fusion protein entered the nucleus by diffusion and that the basic domain caused an intranuclear retention, possibly via an interaction with chromatin.
Thus, we tried to identify the importin α binding site on pUL84 and correlated the findings with the nuclear import capacity of truncated pUL84 proteins. First, we assayed the interaction of pUL84 with one member of each importin α subfamily in the yeast two-hybrid system by using pUL84 deletion mutants. This produced the intriguing result that all importin α isoforms require an identical 282-aa central domain of the UL84 protein for binding. Apparently, this long domain differs from classical importin α binding motifs, which comprise only a couple of amino acids. Then, the capacity of this importin α binding domain to mediate nuclear import was confirmed by transfection of pUL84 deletion mutants into eukaryotic cells. An important piece of evidence that a sequence acts as an NLS comes from fusing the potential motif with a nonkaryophilic protein. Thus, we expressed the 282-aa central domain of pUL84 and two truncated domains as fusion proteins with GFP/β-Gal. The observation that, again, the central domain was sufficient to cause nuclear localization of the fusion protein whereas the truncated domains failed gave additional proof that importin α binding correlated with the import capacity.
As outlined in the introduction, nonconventional importin - dependent NLS motifs have been described for various viral or cellular proteins such as the influenza virus NP protein, the Borna disease virus p10 protein, or the histone deacetylase 4 (18, 55, 62). None of them appears to be similar or identical to the NLS domain identified in HCMV pUL84. However, it has recently been reported that transcription factor STAT1 contains a nontransferable, lysine-rich NLS that is dependent on protein dimerization (8, 35, 36). It was proposed that STAT1 dimerization triggers a conformational change in the molecule that reveals a functional importin α-dependent NLS (34). A similar model might be true for the nonconventional pUL84 NLS since (i) a motif resembling a leucine zipper is located within the pUL84 NLS-domain (20), (ii) pUL84 is capable of forming dimers or multimers (Lischka et al., unpublished data), and (iii) the pUL84 NLS domain contains a lysine-rich element between aa 241 to 253 which is required, however not sufficient, for nuclear localization (Fig. 7). Interestingly, a leucine-rich, nuclear export sequence, which is required for nucleocytoplasmic shuttling, is located in the immediate vicinity of the dimerization-dependent NLS of STAT1. Sequence inspection of the nonconventional NLS of pUL84 revealed the presence of two leucine-rich motifs that match the consensus for leucine-rich export sequences (Fig. 11). It will therefore be of interest to investigate whether these motifs constitute functional nuclear export sequences and, consequently, whether pUL84 is a nucleocytoplasmic shuttling protein.
FIG. 11.
Schematic representation of the pUL84 NLS/importin α-binding domain. A putative, but nontransferable classical NLS is shown on a black background, and two putative NES motifs are shown on a gray background.
Although the interaction of pUL84 with importin α proteins suggested that nuclear import occurs via the classical importin α/β pathway, there are examples of proteins such as the human immunodeficiency virus type 1 integrase for which an interaction with importin α proteins was initially reported (11). However, a subsequent detailed analysis of the import pathway revealed that neither importin α nor importin β was required for efficient nuclear translocation (6). Thus, we performed in vitro transport experiments in order to exactly define the nuclear import pathway used by the HCMV regulatory protein pUL84. By using the digitonin-permeabilized cell assay it was confirmed that pUL84 rapidly accumulates in the nucleus of a eukaryotic cell. Our results revealed that the nuclear translocation of pUL84 occurs via an active receptor-mediated transport mechanism through the NPC since (i) the molecular mass of the GST-UL84 import substrate of about 100 kDa is far above the 50- to 60-kDa limit for free diffusion through the NPCs, (ii) the translocation depended on the addition of cytosolic factors, and (iii) the pUL84 transport was inhibited by the lectin WGA, which binds specific nucleoporins, thus blocking transport processes without affecting diffusion (9, 61, 64).
As demonstrated by the competition of pUL84 nuclear import by the SV40 TAg-NLS, the importin α/β import pathway was exclusively or at least predominantly used for nuclear translocation of pUL84. Replacing the RRL which served as the source of cytosolic proteins in the assay with isolated transport factors showed that both importin α and importin β were required for nuclear localization of pUL84. According to the current model of importin α/β-mediated transport, the import was dependent upon the GTPase Ran, which terminates the import reaction (16).
In summary, all these data suggest that the UL84 protein contains an NLS which mediates nuclear import by the well-characterized importin α/β pathway. However, although following this established pathway, the NLS itself is nonconventional. As demonstrated by deletion analysis it requires a stretch of basic amino acids but apparently within a complex overall structure which may depend on dimerization of the protein. Although there is no final characterization of the pUL84 NLS, these data support the hypothesis derived from the import of STAT1 that not only primary sequences but complex foldings may generate an NLS (34).
The exact functions of pUL84 during the viral replication cycle remain still unclear. Previous studies postulated an essential role of pUL84 for lytic viral DNA replication as deduced from results obtained by transient replication assays (46). Although nuclear translocation appears to be a prerequisite for this, it is interesting that we could not detect an exclusive colocalization of pUL84 with viral replication centers during the late phase of viral replication, as we could previously observe for the UL69 protein of HCMV (Fig. 4) (60). This, again, differs from results of a recent publication by Xu et al., who reported a colocalization of pUL84 with pUL44 and IE2 proteins after transfection of fibroblasts with a vector expressing a UL84-GFP fusion protein followed by superinfection with HCMV (63). In contrast, we used a UL84-specific rabbit serum for detection of unmodified UL84 in fibroblasts that were infected with HCMV, strain AD169. Clearly, additional experiments are required to fully clarify the function of this HCMV protein. However, the detection of a strong interaction with importin α proteins together with the presence of potential nuclear export sequences within a nonconventional importin α interaction domain may indicate that nucleocytoplasmic transport plays an important role for the functions of pUL84.
Acknowledgments
We thank Bodo Plachter (University of Mainz) for providing the GST-UL84 expression vector and the pUL44-specific MAb and Martina Kronschnabl and Manfred Marschall for fruitful comments.
This work was supported by grants of the DFG to T.S. (SFB473) and to M.K. (SFB 535) and by the IZKF Erlangen.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 3.Bonifaci, N., J. Moroianu, A. Radu, and G. Blobel. 1997. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94:5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 5.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 6.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 7.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 8.Fagerlund, R., K. Melen, L. Kinnunen, and I. Julkunen. 2002. Argine/lysine-rich NLSs mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 16:30072-30078. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, D. R., D. D. Newmeyer, T. M. Price, and D. J. Forbes. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, N., E. Kremmer, G. Lautscham, N. Mueller-Lantzsch, and F. A. Grasser. 1997. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin alpha2. J. Biol. Chem. 272:3999-4005. [DOI] [PubMed] [Google Scholar]
- 11.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgiev, O., J. P. Bourquin, M. Gstaiger, L. Knoepfel, W. Schaffner, and C. Hovens. 1996. Two versatile eukaryotic vectors permitting epitope tagging, radiolabelling and nuclear localisation of expressed proteins. Gene 168:165-167. [DOI] [PubMed] [Google Scholar]
- 14.Gorlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich, D., N. Pante, U. Kutay, U. Aebi, and F. R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584-5594. [PMC free article] [PubMed] [Google Scholar]
- 17.Gorlich, D., S. Prehn, R. A. Laskey, and E. Hartmann. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 79:767-778. [DOI] [PubMed] [Google Scholar]
- 18.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 20.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 66:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold, A., R. Truant, H. Wiegand, and B. R. Cullen. 1998. Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hieda, M., T. Tachibana, M. Fukumoto, and Y. Yoneda. 2001. Nuclear import of the U1A splicesome protein is mediated by importin alpha/beta and Ran in living mammalian cells. J. Biol. Chem. 276:16824-16832. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamei, Y., S. Yuba, T. Nakayama, and Y. Yoneda. 1999. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 47:363-372. [DOI] [PubMed] [Google Scholar]
- 27.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 29.Kohler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Gorlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90:1061-1071. [DOI] [PubMed] [Google Scholar]
- 31.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride, K. M., G. Banninger, C. McDonald, and N. C. Reich. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melen, K., L. Kinnunen, and I. Julkunen. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276:16447-16455. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, T., A. Begitt, I. Lodige, M. van Rossum, and U. Vinkemeier. 2002. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto, Y., N. Imamoto, T. Sekimoto, T. Tachibana, T. Seki, S. Tada, T. Enomoto, and Y. Yoneda. 1997. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem. 272:26375-26381. [DOI] [PubMed] [Google Scholar]
- 39.Moroianu, J., G. Blobel, and A. Radu. 1995. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92:2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moroianu, J., G. Blobel, and A. Radu. 1996. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. USA 93:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moroianu, J., M. Hijikata, G. Blobel, and A. Radu. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 92:6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 43.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 44.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 45.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683-692. [DOI] [PubMed] [Google Scholar]
- 46.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, M. R., and W. C. Greene. 1992. Characterization of a novel nuclear localization signal in the HTLV-I tax transactivator protein. Virology 187:316-320. [DOI] [PubMed] [Google Scholar]
- 50.Sorg, G., and T. Stamminger. 1999. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to beta-galactosidase. BioTechniques 26:858-862. [DOI] [PubMed] [Google Scholar]
- 51.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 19:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truant, R., Y. Kang, and B. R. Cullen. 1999. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J. Biol. Chem. 274:32167-32171. [DOI] [PubMed] [Google Scholar]
- 54.Wang, A. H., and X. J. Yang. 2001. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21:5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 57.Weis, K., U. Ryder, and A. I. Lamond. 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15:1818-1825. [PMC free article] [PubMed] [Google Scholar]
- 58.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell. Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 59.Winkler, M., S. T. aus Dem, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolff, B., M. C. Willingham, and J. A. Hanover. 1988. Nuclear protein import: specificity for transport across the nuclear pore. Exp. Cell Res. 178:318-334. [DOI] [PubMed] [Google Scholar]
- 62.Wolff, T., G. Unterstab, G. Heins, J. A. Richt, and M. Kann. 2002. Characterization of an unusual importin alpha binding motif in the Borna disease virus p10 protein that directs nuclear import. J. Biol. Chem. 277:12151-12157. [DOI] [PubMed] [Google Scholar]
- 63.Xu, Y., K. S. Colletti, and G. S. Pari. 2002. Human Cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoneda, Y., N. Imamoto-Sonobe, M. Yamaizumi, and T. Uchida. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586-595. [DOI] [PubMed] [Google Scholar]