Abstract
A typical vertebrate cell contains several hundred sites of transitional ER (tER). Presumably, tER sites generate elements of the ER–Golgi intermediate compartment (ERGIC), and ERGIC elements then generate Golgi cisternae. Therefore, characterizing the mechanisms that influence tER distribution may shed light on the dynamic behavior of the Golgi. We explored the properties of tER sites using Sec13 as a marker protein. Fluorescence microscopy confirmed that tER sites are long-lived ER subdomains. tER sites proliferate during interphase but lose Sec13 during mitosis. Unlike ERGIC elements, tER sites move very little. Nevertheless, when microtubules are depolymerized with nocodazole, tER sites redistribute rapidly to form clusters next to Golgi structures. Hence, tER sites have the unusual property of being immobile, yet dynamic. These findings can be explained by a model in which new tER sites are created by retrograde membrane traffic from the Golgi. We propose that the tER–Golgi system is organized by mutual feedback between these two compartments.
INTRODUCTION
Proteins are transported through the secretory pathway in membrane-bound carriers (Rothman and Wieland, 1996; Farquhar and Hauri, 1997). The first step in this process is export from the transitional ER (tER) in COPII-coated vesicles (Kuehn and Schekman, 1997; Kaiser and Ferro-Novick, 1998). After pinching off from the tER, COPII vesicles uncoat and then fuse with an acceptor compartment, but the nature of this acceptor remains uncertain. The simplest possibility is that COPII vesicles fuse directly with the cis-Golgi. However, two considerations suggest that the picture is more complex. First, studies of vertebrate cells indicate that secretory material passes through an ER–Golgi intermediate compartment (ERGIC) before reaching the Golgi (Hauri and Schweizer, 1992; Saraste and Kuismanen, 1992; Bannykh and Balch, 1997). Second, accumulating evidence supports a cisternal maturation model for Golgi function (Glick and Malhotra, 1998; Pelham, 1998). This model implies that ERGIC elements and Golgi cisternae are transitory structures that must be continually replaced. Therefore, an attractive idea is that COPII vesicles fuse homotypically to form ERGIC elements, which then coalesce and mature into new cis-Golgi cisternae. To test the maturation model, it is important to characterize the relationships between the early secretory compartments, and to determine whether each of these compartments is a stable structure or a transitory intermediate.
The ERGIC has been studied extensively. In vertebrate cells, ERGIC elements consist of vesicular–tubular membrane clusters (Bannykh and Balch, 1997). Some ERGIC elements are located in the juxtanuclear region, at the cis-face of the Golgi; these structures are probably identical to the “cis-Golgi network“ (Mellman and Simons, 1992; Ladinsky et al., 1999). Other ERGIC elements are found in the cell periphery. Recent findings indicate that ERGIC elements are mobile carriers that deliver secretory material to the juxtanuclear Golgi ribbon (Presley et al., 1997; Scales et al., 1997). The movement of ERGIC elements requires microtubules and is apparently mediated by cytoplasmic dynein (Corthésy-Theulaz et al., 1992; Burkhardt et al., 1997; Presley et al., 1997). Characterization of the ERGIC has been facilitated by a relatively specific marker protein termed ERGIC-53 in human cells or p58 in rodent cells (Hauri et al., 2000).
Less is known about the tER. This compartment was originally defined as a ribosome-free ER subdomain that adjoins the rough ER and contains protrusions resembling budding vesicles (Palade, 1975; Merisko et al., 1986). tER sites were presumed to function in ER export. Subsequent immunolocalization studies revealed that cells have multiple discrete tER sites and that the protrusions at tER sites are nascent COPII vesicles (Orci et al., 1991; Kuge et al., 1994; Shaywitz et al., 1995; Paccaud et al., 1996; Tang et al., 1997; Rossanese et al., 1999; Shugrue et al., 1999). Thus, COPII components serve as convenient markers for the tER. Although COPII vesicle biogenesis is being characterized in detail (Kuehn and Schekman, 1997; Springer et al., 1999), these approaches have not revealed how COPII vesicle budding is restricted to tER sites or how tER sites maintain their identity as subdomains within the general ER.
We favor the interpretation that the tER and the ERGIC are separate, sequential compartments in the secretory pathway. The tER is continuous with the general ER and is marked by the presence of COPII proteins. The ERGIC is the first post-ER compartment; it lacks significant levels of COPII proteins but contains the COPI coat protein complex (Bannykh and Balch, 1997; Scales et al., 1997; Martínez-Menárguez et al., 1999). Despite this clear conceptual distinction, the tER and the ERGIC are sometimes viewed as parts of a single entity. One suggestion is that tER sites are consumed during the formation of ERGIC elements (Presley et al., 1998). A related idea is that tER sites and ERGIC elements remain linked in the form of mobile “export complexes” (Bannykh and Balch, 1997). We find that in fact, tER sites are long-lived structures that are distinct from ERGIC elements. The two compartments can be resolved by fluorescence microscopy. Moreover, unlike ERGIC elements, tER sites do not undergo directed long-range movements.
If the tER is the birthplace of the Golgi, then understanding the distribution and behavior of tER sites may be vital for understanding Golgi organization. Vertebrate tER sites are present throughout the ER network with a concentration in the juxtanuclear Golgi region (Palade, 1975; Bannykh and Balch, 1997; Tang et al., 1997). However, the mechanisms that establish this distribution have not been explored. Here we present evidence that tER sites form preferentially in certain parts of the ER. Specifically, tER sites are concentrated in areas that receive extensive retrograde membrane traffic from the Golgi. If Golgi-to-ER traffic is altered by disrupting microtubules, the pattern of tER sites changes accordingly. During mitosis, the inactivation and reactivation of tER sites parallels the breakdown and reassembly of the Golgi. These observations suggest that tER dynamics and Golgi dynamics are closely linked.
MATERIALS AND METHODS
Expression of Tagged Myt1 and E1 Proteins in HeLa Cells
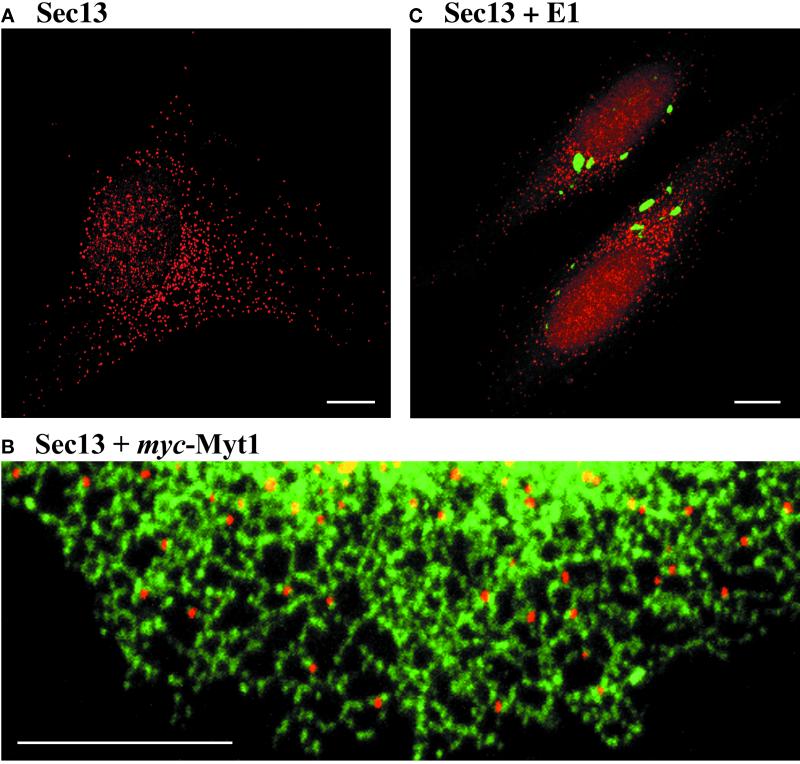
To label the general ER with c-myc-tagged Myt1, the FuGENE 6 kit (Roche Molecular Biochemicals, Indianapolis, IN) was used for the transient transfection of HeLa cells with a plasmid (provided by Paul Mueller, University of Chicago) encoding six copies of the c-myc epitope fused to the N-terminus of Xenopus laevis Myt1 (Mueller et al., 1995). It was reported that human Myt1 is found in both the ER and the Golgi (Liu et al., 1997), but we find that Xenopus Myt1 localizes primarily or exclusively to the ER in HeLa cells. To generate a cell line stably expressing tagged E1 protein, the CellPhect kit (Pharmacia, Piscataway, NJ) was used to transfect HeLa cells simultaneously with two plasmids: pE1-GCT, which encodes a fusion between the Rubella virus E1 protein and the carboxy-terminal 10 residues of VSV-G protein (Hobman et al., 1998), and pBK-CMV (Strategene, La Jolla, CA), which contains a neomycin/G418-resistance cassette. Stable G418-resistant cell lines were examined by immunofluorescence using monoclonal antibody P5D4. As previously described (Hobman et al., 1998), many of the stable transfectants exhibited large E1-containing structures. However, we found that these structures did not label for Sec13. Figure 1C shows a representative transfectant, which was designated clone HeLa-E1.
Figure 1.
Sec13 marks discrete ER subdomains that are distinct from the E1 compartment. (A) HeLa cells were fixed and stained with anti-Sec13 polyclonal antibody followed by Rhodamine Red-X-conjugated anti-rabbit antibody. The image shows a representative cell, which contains several hundred tER sites. (B) HeLa cells transiently expressing myc-tagged Myt1 were examined by double-label immunofluorescence. Sec13 (red) was visualized as in (A), and Myt1-containing ER tubules (green) were visualized using anti-myc monoclonal antibody followed by Cy2-conjugated anti-mouse antibody. This merged image shows a portion of the ER network from the periphery of a cell. All of the tER sites are associated with ER tubules. (C) HeLa-E1 cells, which produce the Rubella virus E1 fusion protein, were examined by double-label immunofluorescence. Sec13 (red) was visualized as in (A), and the E1 fusion protein (green) was visualized using the P5D4 monoclonal antibody followed by Cy2-conjugated anti-mouse antibody. Two cells are present in this merged image, which shows that Sec13 is largely excluded from regions that contain the E1 fusion protein. Scale bars represent 10 μm.
Construction of a CHO Cell Line Expressing Sec13-GFP
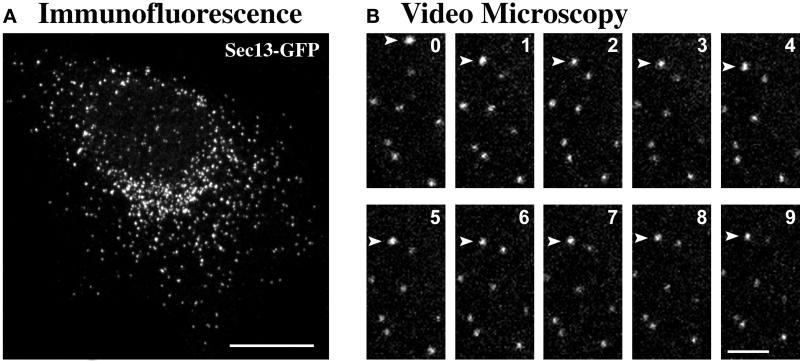
pAA35, which contains the cDNA for human Sec13 (Swaroop et al., 1994), was mutagenized using the QuikChange kit (Stratagene) to replace the stop codon with a BamHI site. The modified Sec13 gene then was excised with EcoRI and BamHI. In parallel, the codon-optimized GFP gene from pEGFP-1 (Clontech, Palo Alto, CA) was excised with BamHI and NotI. pcDNA3.1 (Invitrogen, Carlsbad, CA) was cut with EcoRI and NotI, and a three-way ligation created a Sec13-GFP fusion gene between these sites. This plasmid, termed pcDNA-SEC13-GFP, proved to be suitable for transient expression but not for generating stable transfectants. Therefore, the Sec13-GFP gene was excised with NheI and XhoI and subcloned into the corresponding sites in pBK-CMV to yield pBK-SEC13-GFP. This plasmid was transfected into CHO cells using the CellPhect kit (Pharmacia). Fluorescence microscopy identified a stably expressing transfectant, which was designated CHO-S13G. This clone is routinely cultured in DMEM medium supplemented with 10% fetal calf serum and 200 μg/ml G418.
Individual cells of clone CHO-S13G contain different amounts of Sec13-GFP; such variability is common with CHO cells (Paulin et al., 1998). However, the overall expression level in cultures of CHO-S13G is quite consistent, and many of the cells are suitable for fluorescence microscopy. Several observations confirm that Sec13-GFP localizes to tER sites in CHO-S13G cells. First, as shown in Figure 3A, the GFP fluorescence pattern is indistinguishable from the pattern seen in nontransfected cells stained with anti-Sec13 antibody. Second, when fixed cells of clone CHO-S13G are costained with anti-GFP and anti-Sec13 antibodies, the two fluorescence patterns are identical, indicating that Sec13-GFP is present in all of the structures that contain wild-type Sec13 (our unpublished results). Finally, like wild-type Sec13, Sec13-GFP forms Golgi-associated clusters in nocodazole-treated cells (see Figure 4).
Figure 3.
tER sites primarily exhibit undirected short-range movements. Experiments were performed with CHO-S13G cells, which produce Sec13-GFP. (A) Immunofluorescence of a fixed CHO-S13G cell stained with anti-GFP monoclonal antibody followed by Cy2-conjugated anti-mouse antibody. (B) A living cell of clone CHO-S13G was examined by video confocal microscopy at 37°C. Images were captured every 3.3 s for 15 min and then assembled into a movie. Some photobleaching occurred as a result of the imaging. The panels display a region in the cell periphery; for the figure, image frames were selected at ∼1-min intervals from the first 9 min of the video. Eight tER sites are visible. For the most part, these tER sites exhibited only slow movements. An exception is the uppermost tER site (arrowhead), which underwent a single rapid movement of ∼1.2 μm ∼30 s after the beginning of the video. Scale bars represent 10 μm (A) or 2 μm (B).
Figure 4.
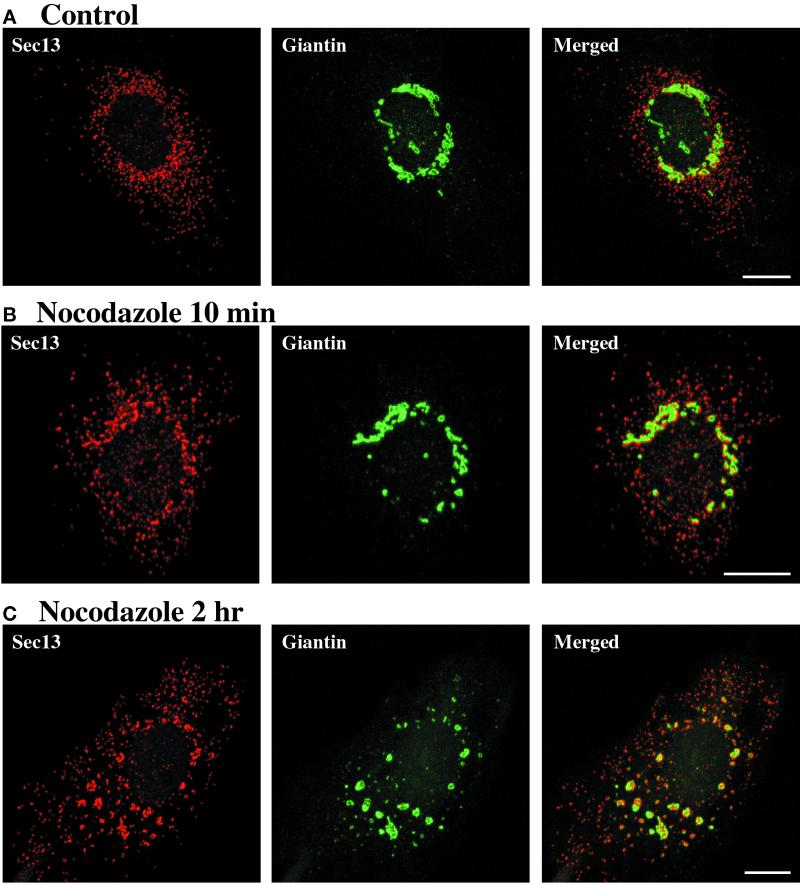
tER sites become clustered near Golgi structures after the addition of nocodazole. (A) A control plate of NRK cells was left at 37°C in the absence of nocodazole. (B and C) NRK cells were incubated on ice for 15 min. Nocodazole was then added to 5 μg/ml, and the cells were quickly warmed to 37°C and incubated for an additional 10 min (B) or 2 h (C). tER sites were visualized by staining for Sec13 as in Figure 1A. The Golgi was visualized using anti-giantin monoclonal antibody followed by Cy2-conjugated anti-mouse antibody. As shown in the merged images, tER sites become clustered near Golgi structures within 10 min after nocodazole addition, and this association persists after 2 h of nocodazole treatment. Scale bars represent 10 μm.
For unknown reasons, in addition to fluorescent tER sites, many of the transfectants obtained with pBK-Sec13-GFP exhibit vacuolar structures that contain a fluorescent non-GFP substance. This problem is rarely observed with the CHO-S13G clone.
Nocodazole Treatment
To depolymerize microtubules, cells were incubated for 15 min on ice and then were warmed to 37°C in the presence of 5 μg/ml nocodazole (Cole et al., 1996). Staining with anti-tubulin antibody confirmed that this method causes a rapid and complete loss of the microtubule array. In contrast to a previous report (Minin, 1997), we find that the ice pretreatment did not alter the final reorganization of the tER–Golgi system. For the experiment shown in Figure 5, nocodazole was added directly to cells that were maintained at 37°C.
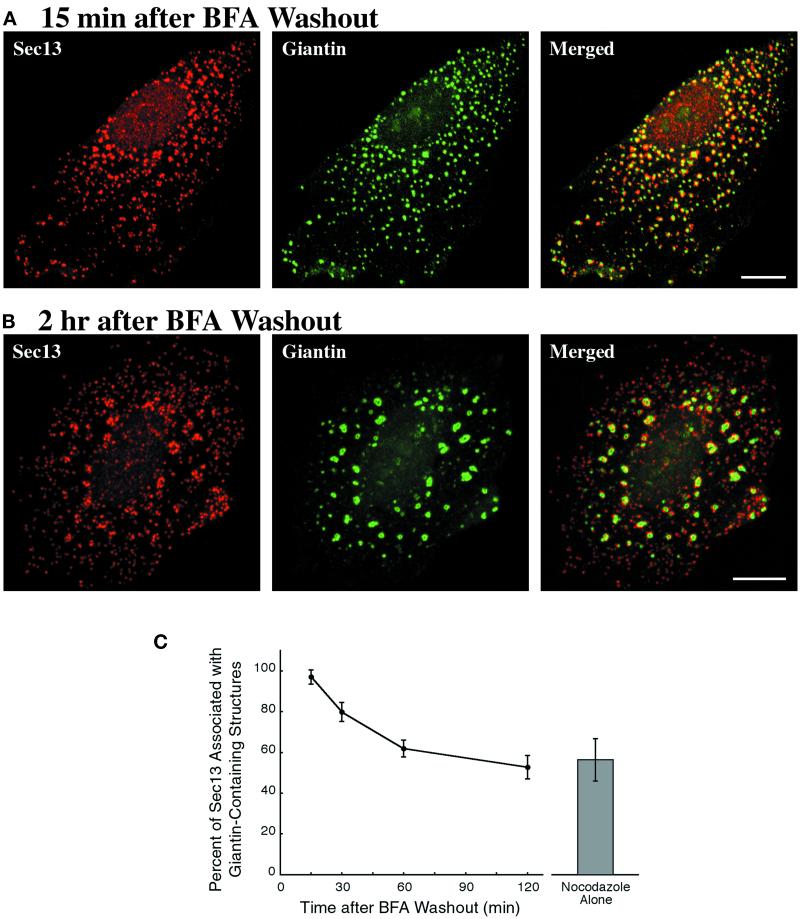
Figure 5.
tER clusters form even when Golgi structures reemerge from the ER. NRK cells were incubated with 5 μg/ml BFA for 1 h; this treatment caused the complete redistribution of giantin into the ER. Nocodazole was then added to 5 μg/ml, and after 30 min the BFA was washed out in the continued presence of nocodazole. Sec13 and giantin were visualized as in Figure 4. (A) Fifteen minutes after BFA removal, small giantin-containing structures are associated with nearly all of the tER sites. (B) Two hours after BFA removal, large giantin-containing structures are associated with clustered tER sites, but many of the individual tER sites lack adjacent giantin staining. Scale bars represent 10 μm. (C) Quantitation of the localization data, as described in Materials and Methods. The graph shows the percentage of the Sec13 staining that is within 0.3 μm of a giantin-containing structure at various times after BFA washout in the presence of nocodazole. The bar on the right gives the corresponding value for NRK cells that were treated with nocodazole alone for 2 h. SDs are indicated.
Antibodies for Immunofluorescence
The following rabbit polyclonal antibodies were used: anti-PDI (StressGen Biotechnologies, Victoria, B.C., Canada) at 1:100; and affinity-purified anti-Sec13 (Tang et al., 1997) at 1:100. A finely punctate nuclear staining was observed with the anti-Sec13 antibody; this nuclear background was more pronounced in HeLa cells than in NRK or CHO cells. The following monoclonal antibodies were used: anti-c-myc (clone 9E10, BAbCO/Covance, Richmond, CA) at 1:200; anti-ERGIC-53 (Schweizer et al., 1988; a kind gift of Hans-Peter Hauri, Biozentrum, Basel, Switzerland) at 1:100; antigiantin (Linstedt and Hauri, 1993; a kind gift of Adam Linstedt, Carnegie-Mellon, Pittsburgh, PA) at 1:100; anti-mannosidase II (clone 53FC3, BAbCO/Covance) at 1:5000; anti-PDI (clone 1D3, Stressgen) at 1:100; anti-VSV-G (clone P5D4, Roche Biochemicals) at 1:100; anti-GFP (a mixture of clones 7.1 and 13.1, Roche Biochemicals) at 1:100; anti-β-tubulin (clone KMX-1, Roche Biochemicals) at 1:100; and anti-galactosyltransferase (Berger et al., 1986; clone GT2/36/118, a kind gift of Eric Berger, University of Zurich, Switzerland) at 1:20. Fluorescent secondary antibodies from donkey (Jackson ImmunoResearch, West Grove, PA) were used at 1:200.
Preparing Cells for Immunofluorescence Microscopy
In our hands, standard immunofluorescence methods give poor preservation of small structures, particularly tER sites. We obtain better results by fixing the cells with a cold organic solvent (Melan and Sluder, 1992) and then rehydrating them in a solution containing a homobifunctional cross-linker. Using this procedure, the immunofluorescence pattern of Sec13-GFP in fixed CHO-S13G cells (Figure 3A) is indistinguishable from the fluorescence pattern in living CHO-S13G cells. Moreover, the protocol described below has the advantage that multiple “wells” can be analyzed on the same coverslip.
Cells are grown to ∼60% confluence on a sterile No. 1.5 glass coverslip in a tissue culture dish. For fixation, the coverslip is removed with a forceps and dropped into a 50-ml conical tube containing organic solvent at −20°C. We use either methanol or acetone, after determining empirically which solvent is better for a given antigen–antibody combination. The interval between removal from 37°C growth medium and submersion in −20°C solvent is < 5 s. Cells are typically fixed for 10 min but can be left for several days in −20°C solvent.
After fixation, the coverslip is removed from the solvent with a forceps, the excess solvent is drawn off by blotting at one corner, and the coverslip is held vertically in an air stream until dry. “Wells” then are created on the coverslip as follows. A clear laminating sheet is cut to yield a piece that is slightly smaller than the coverslip. Holes that are 5 mm in diameter are introduced with a leather punch, and the perforated laminating plastic is sealed tightly to the surface of the coverslip. Subsequent steps are performed in a humidified chamber. For rehydration, each well is incubated for 30 min with 10 μl of PBS, pH 7.4, plus 0.1% n-octyl-β-d-glucopyranoside (PBSO) containing 100 μM bis(sulfosuccinimidyl) suberate (BS3; Pierce Chemical, Rockford, IL) added fresh from a 10 mM aqueous aliquot stored at −80°C. Each well then is washed three times with a drop of PBSO, and residual BS3 is quenched by incubating for 15 min with 10 μl of 0.1 M ethylenediamine–HCl, pH 7.5. The ethylenediamine is removed by washing three times with PBSO.
For antibody staining, each well is first incubated for 1 h with a drop of blocking buffer (PBSO plus 1% nonfat milk, 1% fish gelatin [Sigma, St. Louis, MO]), and 1% donkey serum.) The well then is incubated for 30–60 min with 10 μl of primary antibody diluted in blocking buffer. After eight washes with blocking buffer, the well is incubated for 30–60 min in the dark with 10 μl of fluorescent secondary antibody diluted in blocking buffer. The secondary antibody solution can be supplemented with the DNA stain Hoechst 33258 (Molecular Probes, Eugene, OR), diluted 1:5000 from a 1% stock solution. Finally, the well is washed 10 times with blocking buffer; all traces of liquid then are aspirated, and 5–10 μl of mounting medium (see below) is added. The coverslip is inverted onto a slide, and the edges are sealed with VALAP (Weiss et al., 1989). Slides can be stored for months at room temperature in the dark.
Mounting medium is prepared by adjusting 10 ml of 10× PBS to pH 9 with Na2CO3, adding 90 ml of glycerol, and then dissolving n-propyl gallate to a final concentration of 5% by bath sonication. Aliquots of 200 μl are stored at −80°C. To stain DNA, the mounting medium can be supplemented with 4 μM TOTO-3 (Molecular Probes).
Confocal Microscopy and Image Analysis
All experiments were performed with a Zeiss (Thornwood, NY) LSM 510 confocal microscope equipped with a 100X 1.4-NA Plan-Apo objective lens and with standard filters for visualizing FITC/Cy2, Rhodamine Red-X, and Cy5/TOTO-3. The laser intensities and amplifier gains were adjusted to prevent saturation of the detectors. When two or more fluorescent markers were imaged in the same cells, each fluorophore was excited and detected separately to avoid the crossover of strong signals. Images in the red and green channels were in register to within less than one pixel. For fixed cells, a confocal Z-stack was collected (20 8-bit images at 1024 × 1024 resolution, 0.35 μm focus intervals, and 1.0 Airy unit). Each Z-stack either was quantified as described below or was projected to form a single image. To maximize the resolution of three-dimensional (3D) objects reconstructed by the software, the Z-stacks used for quantitation were captured at 0.2-μm focus intervals and the images were zoomed to give pixel sizes of ≤80 nm (Centonze and Pawley, 1995). Two-color merged images were generated automatically by the Zeiss software during acquisition. Adobe Photoshop 5.0 (Mountain View, CA) was used to adjust brightness and contrast, and figures were printed on an Epson Stylus Photo inkjet printer (Long Beach, CA). Quantitation of immunofluorescence images was carried out using the Zeiss 3D for LSM software in conjunction with NIH Image (available at: http://rsb.info.nih.gov/nih-image/) and Microsoft Excel (Redmond, WA).
To quantify the colocalization of Sec13-containing spots with ER tubules (Figure 1B), HeLa cells expressing c-myc-tagged Myt1 were costained with anti-myc and anti-Sec13 antibodies, and separate images were collected for the two markers. Peripheral regions that showed a clear reticular Myt1 pattern were highlighted in eight separate cells without regard to the Sec13 signal. The corresponding Sec13 images then were examined, and the Sec13-containing spots in the highlighted regions were identified (307 spots total). The colocalization of these Sec13-containing spots with ER tubules then was scored using the merged images. Colocalization was defined as partial or complete overlap of a Sec13-containing spot with the contour of an ER tubule.
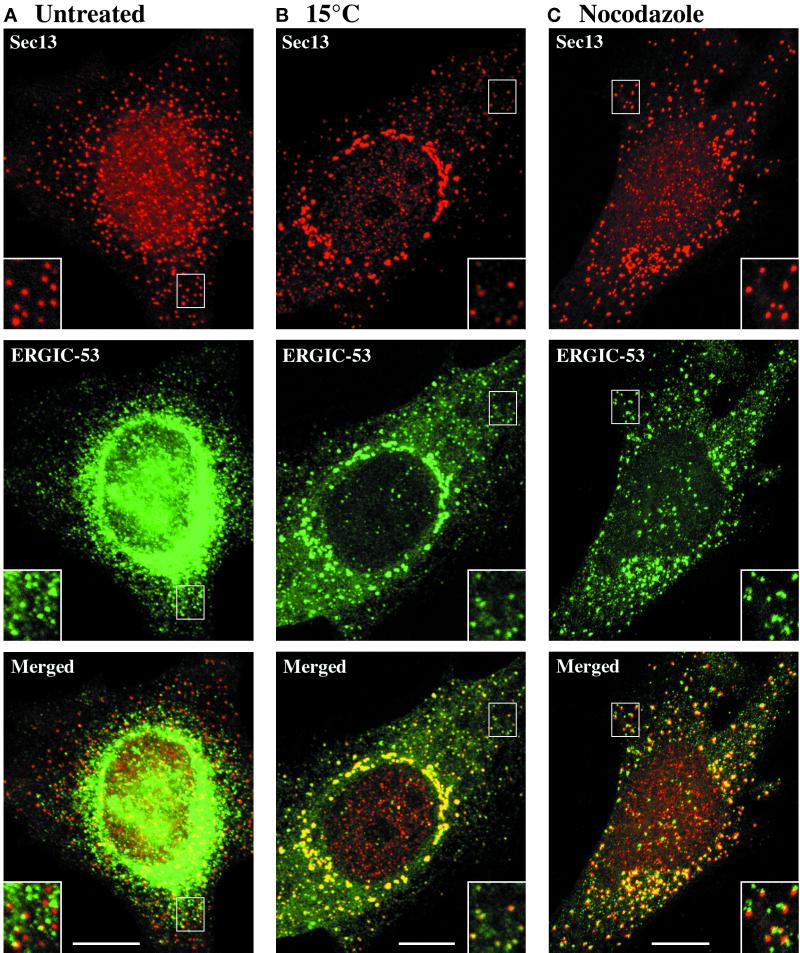
The overlap between Sec13 and ERGIC-53 (Figure 2; Table 1) was quantified as follows. For each marker protein, the 3D for LSM software was used to define a threshold mask that included all of the discernible labeled structures. Localized regions were excluded from the analysis if they contained labeled structures that were ≥ 0.6 μm in diameter. Using the projected images, each Sec13-containing spot was examined to determine whether it was concentric with an ERGIC-53-containing spot. The reciprocal analysis then was performed for the ERGIC-53-containing spots. Two spots were deemed to be concentric if their centers were located within three pixels (0.2 μm) of each other. For a given cell, we calculated the percentage of the total labeled spots that were concentric with spots of the other marker. Ten cells were analyzed for each of the two treatments: incubation at 15°C (Figure 2B) or incubation with nocodazole (Figure 2C).
Figure 2.
The tER and the ERGIC can be resolved in cells grown at 37°C, but are indistinguishable in cells incubated at 15°C. tER sites (red) were visualized by staining HeLa cells for Sec13 as in Figure 1A. ERGIC elements (green) were visualized in the same cells using anti-ERGIC-53 monoclonal antibody followed by Cy2-conjugated anti-mouse antibody. (A) Cells growing normally at 37°C were fixed and processed for immunofluorescence. The dominant ERGIC-53 signal derives from ER-localized molecules, but as shown in the inset, punctate ERGIC elements can be seen next to some of the tER sites. (B) Cells were incubated at 15°C for 3 h before fixation. As shown in the merged image and the inset, the Sec13 and ERGIC-53 staining patterns are almost identical. (C) Cells were incubated on ice for 15 min, then quickly warmed to 37°C in the presence of 5 μg/ml nocodazole for an additional 2 h. As shown in the merged image, tER sites and ERGIC elements are closely associated, but distinct. The inset contains examples of tER and ERGIC structures that are optically well resolved. Scale bars represent 10 μm.
Table 1.
Quantitation of overlap between Sec13 and ERGIC-53
| Treatment | Total Sec13 spots evaluated | Total ERGIC-53 spots evaluated | Average colocalization per cell (%) (n = 10) |
|---|---|---|---|
| 15°C | 2522 | 2435 | 89.3 ± 2.1 |
| Nocodazole | 2595 | 2111 | 10.7 ± 5.3 |
As described in Figure 2, HeLa cells either were incubated at 15°C for 3 h or were treated with nocodazole for 2 h and then were fixed and stained for Sec13 and ERGIC-53. For each of 10 cells, the spots of diameter <0.6 μm were counted separately for the Sec13 and ERGIC-53 images. The merged images then were evaluated for colocalization. Two spots were scored as colocalizing if they were concentric (see Materials and Methods). SDs are indicated.
For the brefeldin A (BFA) washout experiment (Figure 5), the following method was used to estimate the fraction of the tER that was in close proximity to Golgi structures. The analysis involved 10 NRK cells treated with nocodazole alone and 10 cells from each time point after BFA washout. Cells that were clearly separated from their neighbors were chosen by viewing the Hoechst-stained DNA, without regard to the tER or Golgi patterns. The Sec13 and giantin signals then were imaged separately using a predetermined set of microscope parameters. For each cell, 3D masks were defined to include the optically resolvable tER and Golgi structures. The total volume density of the tER signal was measured. Then the edges of the Golgi mask were expanded by 0.3 μm, and the tER signal present within this expanded mask was deleted. The remaining tER signal was subtracted from the total tER signal, and this difference was divided by the total to yield the fraction of the tER that was within 0.3 μm of a Golgi structure.
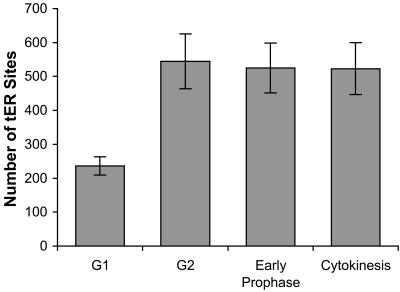
To measure tER site proliferation (Figure 6), an unsynchronized population of NRK cells was stained for Sec13 as in Figure 1A. The same cells also were incubated with anti-tubulin monoclonal antibody followed by Cy2-conjugated anti-mouse antibody, and with TOTO-3 to stain DNA. Ten cells each from G1, G2, early prophase, and cytokinesis were used for the analysis. G1 cells were identified by examining the microtubule array (Andersen et al., 1978). G2 cells were identified by measuring the volume density of the TOTO-3 fluorescence signal from the nucleus; a cell was considered to be in G2 if the TOTO-3 signal was between 1.7 and 2.3 times the average signal from the G1 cells. Cells in early prophase or cytokinesis were identified by examining their chromosomes (McIntosh and Koonce, 1989). After a cell was assigned to a stage of the cell cycle, the number of tER sites was measured as follows. A 3D mask was defined to include all of the optically resolvable structures in the Z-stack representing the Sec13 signal. The number of individual tER sites then was determined by automated counting of the structures in the mask, followed by manual correction for structures that contained multiple tER sites.
Figure 6.
tER sites proliferate during interphase. As described in Materials and Methods, fixed and stained cells from an unsynchronized NRK culture were identified as being in G1, G2, early prophase, or cytokinesis. After a cell was assigned to a stage of the cell cycle, its tER sites were counted. Bars indicate the average number of tER sites per cell. SDs are indicated.
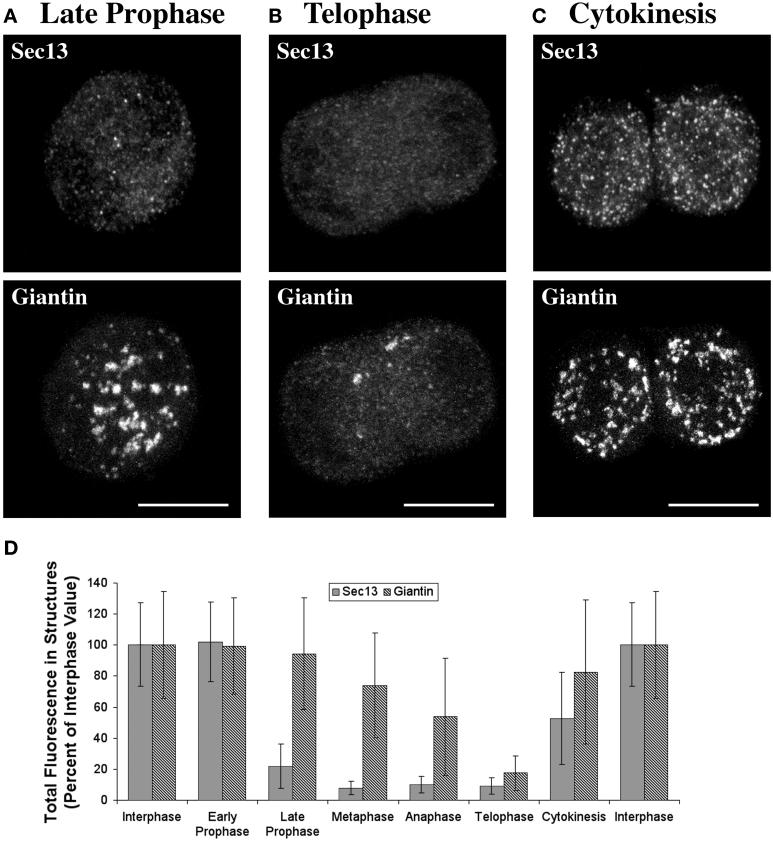
The mitotic breakdown of tER and Golgi structures (Figure 7) was quantified as follows. All of the cells in a given experiment were photographed from a single coverslip using identical microscope settings. Twenty interphase CHO cells and 20 cells from each stage of mitosis were chosen by viewing the Hoechst-stained DNA (McIntosh and Koonce, 1989) without regard to the tER or Golgi patterns. The tER and Golgi images then were collected in separate Z-stacks. For each cell, 3D masks were defined to include the optically resolvable tER and Golgi structures; in practice, such structures exhibited an apparent diameter of ≥0.2 μm and an average fluorescence intensity at least threefold over background. We then measured the volume densities of the tER and Golgi signals defined by the masks. This quantitation method seems to be reliable, and the results are consistent with our qualitative impressions. One potential caveat is that the immunofluorescence procedure might give inaccurate results with rounded mitotic cells. To control for this possibility, we attempted to quantify the total fluorescence emanating from the cells; however, the antibody labeling generates a low but finite cytoplasmic background, and the total volume density of this background signal was too large to be reliably subtracted. As an alternative, we relied on internal controls to verify the accuracy of the quantitation. For example, cellular morphology is similar during telophase and cytokinesis, yet the fluorescence signals from optically resolvable tER and Golgi structures are much stronger during cytokinesis (Figure 7D).
Figure 7.
The tER and the Golgi undergo parallel transformations during mitosis. An unsynchronized population of CHO cells was fixed and stained for Sec13 and giantin as in Figure 4. In addition, chromosomes were stained with Hoechst dye to determine the stage of the cell cycle. (A–C) Representative cells from late prophase (A), telophase (B), and cytokinesis (C). Sec13 staining of tER sites is greatly reduced in the late prophase and telophase cells but is strong in the cell undergoing cytokinesis. Giantin staining reveals that multiple large Golgi fragments are present in the late prophase cell and in the cell undergoing cytokinesis, but only a few optically resolvable structures are present in the telophase cell. (D) Quantitation of the mitosis data, as described in Materials and Methods. The total cellular intensity of the Sec13 or giantin signal from optically resolvable structures is plotted, with the interphase values being defined as 100%. SDs are indicated. The large deviations reflect a substantial cell-to-cell variability in the tER and Golgi patterns at each stage of the cell cycle.
Living cells were viewed with the aid of a Bioptechs (Butler, PA) ΔTC3 system, which maintained the growth chamber and objective at a fixed temperature. Cells were grown in DMEM plus 10% fetal calf serum in a ΔTC3 dish. To promote the spreading of the cells on the glass, the medium was supplemented with a 1:50 dilution each of 0.01% poly-l-lysine solution and 0.1% type 1 collagen solution (both from Sigma) before seeding the cells into the dish. For microscopy, the medium was buffered with 25 mM Na+-HEPES, pH 7.5. The GFP chromophore was excited with the 488-nm laser line and was visualized with a 505–550-nm bandpass filter. Fluorescent structures were viewed in a single image plane at 512 × 512 resolution with a zoom factor of two and with the pinhole adjusted to yield 1.2 Airy units. Images were captured at 3.3-s intervals and then were assembled into movies using Apple (Cupertino, CA) QuickTime Pro software. To visualize tER dynamics (Figure 3B), CHO-S13G cells were imaged at 37°C. To visualize ERGIC dynamics, CHO cells plated in a Bioptechs ΔTC3 dish were transfected with 1 μg of pVSVGtsO45-GFP (Presley et al., 1997) using the FuGENE 6 kit (Roche), and were incubated at 40°C for 34 h before being transferred to the microscope stage. The cells had grown to ∼75% of confluence, and roughly 5% of the cells had a bright ER pattern when viewed at 40°C. Strongly expressing cells were imaged as the temperature was reduced to 32°C. Consistent with previous reports (Presley et al., 1997; Scales et al., 1997), multiple fluorescent ERGIC elements could be seen moving from the cell periphery to the juxtanuclear Golgi region.
RESULTS
Sec13 Is a Marker for tER Sites
To study the dynamics of the vertebrate tER, we needed a marker for this compartment. The only known proteins that localize specifically to tER sites are components of the COPII coat. A well-characterized subunit of the COPII coat is Sec13 (Pryer et al., 1993; Kuehn and Schekman, 1997). When visualized by immunoelectron microscopy, Sec13 is detected at tER sites with little or no labeling of other secretory compartments (Shaywitz et al., 1995; Tang et al., 1997; Martínez-Menárguez et al., 1999; Rossanese et al., 1999). Therefore, we obtained an aliquot of a strongly reactive antibody against human Sec13 (Tang et al., 1997). In addition, we developed an improved immunofluorescence protocol that yields excellent preservation of intracellular structures (see Materials and Methods). When this protocol is used to label HeLa cells with anti-Sec13 antibody, each cell is seen to contain several hundred cytoplasmic spots, which presumably represent individual tER sites (Figure 1A).
Is Sec13 a specific marker for the tER? Genetic studies indicate that yeast sec13 mutants are defective not only in transporting proteins out of the ER, but also in sorting proteins for export from the Golgi (Roberg et al., 1997). However, this sorting effect is probably indirect because no Sec13 has been detected on the Golgi in any cell type. Other experiments suggest that yeast Sec13 plays a role in nuclear pore assembly (Siniossoglou et al., 1996). Although this issue has not been addressed in vertebrate cells, we consistently find that in addition to labeling cytoplasmic structures, the anti-Sec13 antibody gives a finely punctate nuclear staining that may reflect a nuclear pool of Sec13 (Figure 1A) (see also Shugrue et al., 1999). This nuclear background sometimes obscures putative tER signals that emanate from above or below the nucleus but otherwise has not interfered with our analysis. Apart from the nuclear background, the localization pattern of vertebrate Sec13 is indistinguishable from that of other COPII coat proteins (Paccaud et al., 1996; Tang et al., 1997, 1999). Recent studies have confirmed that antibodies against the Sec31 subunit of COPII recognize the same cytoplasmic structures that contain Sec13 (Shugrue et al., 1999; Tang et al., 2000).
To demonstrate that the fluorescent spots labeled by anti-Sec13 antibody correspond to ER subdomains, we visualized Sec13 in HeLa cells that were costained to reveal the ER. These cells had been transiently transfected with a myc-epitope-tagged version of Xenopus Myt1, a mitotic kinase that is integrated into the ER membrane (Mueller et al., 1995; Liu et al., 1997). Cells overexpressing tagged Myt1 have apparently normal morphology and can be stained with an anti-myc monoclonal antibody to highlight the ER. A network of ER tubules is visible in flattened peripheral regions of these cells (Figure 1B). As expected, the Sec13-containing structures are closely associated with Myt1-containing ER tubules (Figure 1B). To quantify this pattern, we examined each of the peripheral Sec13 spots in eight different cells (307 spots total; see Materials and Methods) and found that all of these spots colocalized with ER tubules. Thus, the combined data indicate that Sec13 is a specific marker for tER sites.
Presumably, COPII coat proteins such as Sec13 are concentrated not only on nascent vesicles, but also on fully formed COPII vesicles and in cytosolic areas near tER sites. Therefore, it seems likely that a fluorescent spot of Sec13 staining represents a tER site plus an immediately adjacent volume of cytoplasm. This interpretation fits with immunoelectron microscopy studies (Shaywitz et al., 1995; Tang et al., 1997; Martínez-Menárguez et al., 1999). Therefore, an ideal tER marker would be a membrane protein that was concentrated in tER sites but excluded from COPII vesicles. In the budding yeast Pichia pastoris, such a marker is provided by Sec12, a transmembrane protein that initiates COPII vesicle assembly (Kuehn and Schekman, 1997; Rossanese et al., 1999). Unfortunately, vertebrate Sec12 has not yet been characterized. SNARE proteins such as rsec22b and rbet1 are membrane bound, but they localize to both the tER and the ERGIC (Hay et al., 1998; Chao et al., 1999; Martínez-Menárguez et al., 1999; Zhang et al., 1999). The same is true for the tsO45 mutant of VSV-G protein (Balch et al., 1994). A potentially specific membrane marker for the tER is the Rubella virus E1 glycoprotein. When E1 is overexpressed in the absence of its partner E2 subunit, the cells accumulate large tubuloreticular membrane structures, and it has been suggested that these structures represent hypertrophied tER sites (Hobman et al., 1992, 1998). However, we find that E1-containing membranes lack detectable Sec13 (Figure 1C). Thus, although the E1 compartment is interesting and possibly novel, it does not seem to represent an exaggerated form of the tER. In conclusion, because no integral membrane protein of the vertebrate tER has been identified, COPII subunits remain the best available markers for tER sites.
The tER Is Distinct from the ERGIC at 37°C
The relationship between the tER and the ERGIC often has been ambiguous. For example, when the antibody against human Sec13 originally was characterized, the results were interpreted to mean that Sec13 was a component of the ERGIC (Tang et al., 1997). By contrast, functional studies have implicated Sec13 as part of the COPII coat, which assembles on tER-derived vesicles but apparently not on ERGIC membranes (Bannykh and Balch, 1997; Kuehn and Schekman, 1997; Scales et al., 1997; Martínez-Menárguez et al., 1999). One reason for this uncertainty is that the tER and the ERGIC show similar distributions—both compartments are found throughout the cytoplasm but are concentrated in a juxtanuclear region (Saraste and Svensson, 1991; Bannykh et al., 1996). However, if these two compartments are truly distinct, they should be distinguishable by confocal microscopy. We therefore set out to resolve the tER and the ERGIC by double-label immunofluorescence.
Our optical system can completely resolve two structures that are separated by 0.25 μm in the XY-plane and can partially resolve structures that are even closer together (Inoué, 1995). The inherent Z-resolution of the confocal system is severalfold lower, an effect that is exaggerated by the tendency of organic solvent fixation to shrink cells in the Z-direction. As a result, we obtain the best resolution with adjacent structures that shows minimal overlap in the Z-direction. The structures that satisfy this criterion are usually small, because adjacent structures that are large and topologically complex tend to overlap extensively in the Z-direction. Hence, we have focused our attention on labeled structures of diameter < 0.6 μm.
To visualize tER sites in conjunction with the ERGIC, we double-labeled HeLa cells with anti-Sec13 antibody and with a monoclonal antibody against ERGIC-53 (Schweizer et al., 1988; Schindler et al., 1993). In normally growing cells, most of the ERGIC-53 molecules actually reside in the ER (Klumperman et al., 1998), but punctate structures that contain ERGIC-53 can be visualized in peripheral regions of the cell (Figure 2A). A subset of these ERGIC-53-containing structures is presumably newly formed ERGIC elements because they are associated with tER sites. Despite this close association, tER sites and ERGIC elements usually can be distinguished from one another, especially when viewed at high magnification (Figure 2A; see inset). In some cases, the structures marked by Sec13 and ERGIC-53 appear to be completely separate. In other cases, the two signals show partial overlap, which may reflect either close apposition of separate structures or distinct subdomains of a continuous membrane. Regardless of the precise topology, Sec13 and ERGIC-53 are concentrated in different locations.
A limitation of this experiment is that many of the ERGIC-53-containing structures are not visibly associated with tER sites, and vice versa (Figure 2A). The likely reason for this low steady-state association is that ERGIC elements move away from tER sites along microtubules (Presley et al., 1997; Scales et al., 1997). Thus, to facilitate a quantitative analysis, we sought conditions that would accumulate ERGIC elements at a stage before their movement along microtubules.
The most common way to accumulate ERGIC elements is to incubate cells at 15–16°C for several hours (Hauri and Schweizer, 1992; Saraste and Kuismanen, 1992). This treatment induces the proliferation of a “15°C compartment,” which is generally assumed to represent the ERGIC. Therefore, we treated HeLa cells for 3 h at 15°C. Under these conditions, ERGIC-53 becomes concentrated in structures that are associated with Sec13-containing structures (Figure 2B; see inset). However, this association is more intimate than in cells grown at 37°C; indeed, the staining patterns of ERGIC-53 and Sec13 overlap almost completely in 15°C-treated cells (Table 1). Based on the additional results described below, we suggest that 15°C treatment causes the formation of a hybrid tER/ERGIC compartment. This effect evidently has fostered confusion about the relationship between the tER and the ERGIC. To verify that the tER and the ERGIC are distinct at 37°C, we needed an alternative method to accumulate ERGIC elements.
ERGIC elements normally move along microtubules to the Golgi, so depolymerizing microtubules with nocodazole causes ERGIC elements to accumulate near their sites of origin (Lippincott-Schwartz et al., 1990; Saraste and Svensson, 1991; Cole et al., 1996; Presley et al., 1997; Scales et al., 1997). In HeLa cells that have been treated with nocodazole for 2 h, most of the ERGIC-53 is found in ERGIC elements (Figure 2C). These ERGIC elements are located near virtually all of the Sec13 spots, confirming that the Sec13 spots represent functional tER sites. As in untreated cells, ERGIC elements in nocodazole-treated cells usually can be distinguished from the adjacent tER sites when viewed at high magnification (Figure 2C; see inset). This conclusion was verified by quantifying the colocalization of small structures that were stained for either ERGIC-53 or Sec13 (Table 1). The combined data suggest that the tER and the ERGIC are fused at 15°C but exist as distinct compartments at 37°C.
If our interpretation is accurate, then tER sites are relatively immobile structures that give rise to mobile ERGIC elements. We tested this idea using video microscopy. In previous experiments with a fluorescent secretory protein, ERGIC elements were seen to move from the cell periphery to the juxtanuclear Golgi (Presley et al., 1997; Scales et al., 1997). This result can be reproduced with our experimental setup (see Materials and Methods). A centripetal motion of ERGIC elements also has been observed using a fluorescently tagged version of p58, the rat homolog of ERGIC-53 (T. Roberts and J. Lippincott-Schwartz, personal communication). By contrast, we predicted that peripheral tER sites would not undergo such directed long-range movements. To visualize tER dynamics, GFP was fused to the C-terminus of human Sec13; analogous Sec13p–GFP fusions have been shown to be functional in budding yeasts (Rossanese et al., 1999). A CHO cell line stably expressing Sec13–GFP gives a fluorescence pattern similar to that seen using anti-Sec13 antibody (Figure 3A). Individual tER sites in these cells can be monitored by video microscopy, often for many minutes at a time (Figure 3B). On rare occasions, a tER site shows a single rapid movement of a micron or more, probably due to a rearrangement of the underlying ER network (Terasaki, 1990; Waterman-Storer and Salmon, 1998). However, the vast majority of the movements are short-range, and they exhibit no obvious directionality (Figure 3B). Disruption of microtubules does not substantially alter the short-range movements of tER sites (our unpublished results), suggesting that tER sites may simply be diffusing within the plane of the ER. These findings confirm that the tER and the ERGIC have different properties.
tER Sites Cluster Near the Golgi in Nocodazole-Treated Cells
Although individual tER sites are nearly immobile, the overall tER distribution in the cell can change rapidly. In particular, after the addition of nocodazole, much of the Sec13 reorganizes from its normal pattern of distinct tER sites (Figure 4A) to a pattern that includes larger structures (Figure 4, B and C) (see also Shugrue et al., 1999). This effect is detectable within 10 min after nocodazole addition and persists after 2 h of nocodazole treatment. Individual confocal sections suggest that the large Sec13-containing structures consist of multiple tER sites. We cannot be certain whether these tER sites remain as discrete units or whether they become interconnected; but for convenience, we will refer to the large Sec13-containing structures as “tER clusters.” This clustering of tER sites does not reflect a broader reorganization of the ER, because nocodazole treatment has no significant effect on general ER structure under our experimental conditions.
Is tER clustering linked to Golgi reorganization? Nocodazole is known to induce fragmentation of the Golgi (Kreis et al., 1997). To visualize tER sites in conjunction with the Golgi, we double-labeled NRK cells with anti-Sec13 antibody and with a monoclonal antibody against giantin (Linstedt and Hauri, 1993). In untreated cells, tER sites are most abundant in the general vicinity of the Golgi (Figure 4A). Nocodazole treatment dramatically enhances this tER–Golgi association (Figure 4, B and C). Even after a 10-min nocodazole treatment, when the giantin distribution is largely unchanged, virtually all of the Golgi structures are juxtaposed to tER clusters (Figure 4B). After 2 h in nocodazole, giantin is found in multiple Golgi structures throughout the cytoplasm, and each of these Golgi structures is associated with a tER cluster of corresponding size (Figure 4C). Although the tER clusters are broadly similar in shape to the Golgi structures, the staining patterns of Sec13 and giantin are clearly distinct. Similar results were obtained when the Golgi of HeLa or CHO cells was visualized with anti-giantin antibody, or when the Golgi of NRK cells was visualized with a monoclonal antibody against mannosidase II (our unpublished results).
Previous workers reported that Golgi stacks are located next to ERGIC elements in nocodazole-treated cells (Saraste and Svensson, 1991; Cole et al., 1996). Because the tER and the ERGIC are closely apposed in these cells (see Figure 2C), it is not surprising to observe Golgi structures near tER sites. An unexpected finding is that most of the nonclustered tER sites lack associated Golgi structures (Figure 4C), even after prolonged incubations in nocodazole (up to 12 h). Similar results were seen with other Golgi markers, including mannosidase II in NRK cells and galactosyltransferase in HeLa cells (our unpublished results). These data bear on current views of Golgi dynamics. After the addition of nocodazole, the secretory compartments eventually reach a new steady-state distribution, which has been thought to consist of a single Golgi stack next to each tER site (Rogalski and Singer, 1984; Thyberg and Moskalewski, 1985; Cole et al., 1996). By contrast, our results suggest that the steady-state distribution in nocodazole-treated cells includes intermediate-sized Golgi structures next to clustered tER sites.
The persistence of intermediate-sized Golgi structures could be explained by assuming that our nocodazole treatment gives an incomplete effect. To exclude this possibility, we first redistributed giantin and other Golgi proteins into the ER using BFA (Klausner et al., 1992). We then induced Golgi reassembly by washing out the BFA in the presence of nocodazole. At 15 min after BFA washout, giantin is found in many small spots, each of which is associated with one or a few tER sites (Figure 5A). At later time points, most of the giantin is present in larger Golgi structures next to clustered tER sites (Figure 5B). The nonclustered tER sites progressively lose their associated giantin staining, yielding a pattern similar to that seen after treatment with nocodazole alone (Figure 5, B and C). Hence, regardless of whether the Golgi starts out intact or completely disrupted, the final effect of nocodazole is to generate intermediate-sized Golgi structures and clustered tER sites.
What causes tER sites to cluster near the Golgi in nocodazole-treated cells? tER sites move very little, so presumably they do not migrate to the Golgi structures. A more likely explanation is that Golgi structures stimulate the localized formation of new tER sites. This tER proliferation may be induced by retrograde Golgi-to-ER membrane traffic. In nocodazole-treated cells, all of the retrograde traffic is short range, so tER sites will proliferate in the immediate vicinity of Golgi structures (see Discussion).
tER Sites Proliferate during Interphase but Lose Sec13 During Mitosis
Our proposed mechanism for tER clustering is plausible only if new tER sites can form during interphase. An alternative is that tER sites might duplicate during mitosis, as has been suggested for fungal hyphae (Bracker et al., 1996). To address this issue, we measured the average number of tER sites in NRK cells at four points in the cell cycle: G1; G2; early prophase (the first phase of mitosis); and cytokinesis (the last phase of mitosis) (Figure 6). Cells undergoing cytokinesis have the same number of tER sites as early prophase cells. By contrast, G2 cells have about twice as many tER sites as G1 cells. These data indicate that in vertebrate cells, tER sites proliferate during interphase.
During mitosis, membrane traffic is inhibited and the Golgi breaks down (Warren, 1993; Rabouille and Warren, 1997; Thyberg and Moskalewski, 1998). We therefore suspected that the tER–Golgi relationship might be altered in mitotic cells. Early electron micrographs indicated that mitotic Golgi fragments are often found next to tER sites (Lucocq et al., 1989). Surprisingly, our immunofluorescence studies have not revealed a juxtaposition of tER and Golgi structures during any phase of mitosis (Figure 7) (see also Farmaki et al., 1999). On the other hand, rounded mitotic cells contain a nearly uniform distribution of tER sites, making it difficult to ascertain whether a given Golgi element is specifically associated with one or more tER sites (Figure 7C). Additional data will be needed to answer this question.
Even though the tER distribution in mitotic cells is uninformative, the tER sites themselves show an interesting change. An electron microscopy study reported that less Sec13 is associated with tER sites during mitosis than during interphase (Farmaki et al., 1999). In support of this conclusion, we find that tER sites exhibit less intense Sec13 staining during mitosis. This reduction is moderate in NRK and HeLa cells (our unpublished results) but quite strong in CHO cells (Figure 7). We quantified the Sec13 signals obtained from optically resolvable structures in mitotic CHO cells (Figure 7D). The punctate Sec13 staining drops abruptly when cells enter late prophase and recovers during cytokinesis.
Do mitosis-induced changes in the tER correlate with changes in the Golgi? We used giantin as a marker for Golgi reorganization in mitotic CHO cells. Golgi breakdown occurs in two stages (Thyberg and Moskalewski, 1992; Rabouille and Warren, 1997; Zaal et al., 1999). The first stage is fragmentation; beginning in prophase, the Golgi ribbon separates into multiple discrete fragments (Figure 7A). Golgi fragmentation coincides with the reduction in tER-associated Sec13. The second stage of Golgi breakdown is dispersal; by the time cells reach telophase, most of the giantin is no longer present in optically resolvable structures, but is spread diffusely throughout the cytoplasm (Figure 7B). This Golgi dispersal is quantified in Figure 7D. Multiple Golgi fragments reappear during cytokinesis (Lucocq et al., 1989; Thyberg and Moskalewski, 1992; Shima et al., 1997; Zaal et al., 1999) simultaneously with the recovery of tER-associated Sec13 (Figure 7, C and D). Similar results were seen using the Golgi markers mannosidase II in NRK cells and galactosyltransferase in HeLa cells, although the processes of Golgi fragmentation, dispersal, and reassembly occur at slightly different phases of mitosis for different cell types (our unpublished results). In each cell type examined, the breakdown and reassembly of the Golgi parallel the decline and recovery of tER-associated Sec13.
DISCUSSION
The tER seems to play a central role in organizing the early secretory compartments (Bannykh and Balch, 1997; Glick and Malhotra, 1998; Rossanese et al., 1999). Although the tER was identified several decades ago (Palade, 1975), it subsequently received little attention due to a lack of specific markers. This deficiency has been remedied by the identification of COPII proteins (Kuehn and Schekman, 1997). Here we have used Sec13, a component of the COPII coat, to investigate the properties of the tER (Figure 1). The analysis was facilitated by an improved immunofluorescence method that reliably preserves tER sites and other fragile structures.
Our first goal was to establish that the tER is distinct from the ERGIC. Peripherally generated ERGIC elements have been shown to move inward along microtubules to the Golgi (Saraste and Svensson, 1991; Presley et al., 1997; Scales et al., 1997). Disrupting microtubules with nocodazole blocks this inward movement and causes ERGIC elements to accumulate near tER sites. However, the Sec13-containing structures in nocodazole-treated cells can be resolved optically from the ERGIC elements (Figure 2C and Table 1), implying that the tER and the ERGIC are distinct compartments under these conditions. During normal growth in the presence of microtubules, mobile ERGIC elements presumably form next to immobile tER sites. To test this interpretation, we rendered tER sites fluorescent by expressing a Sec13–GFP fusion protein. Video microscopy revealed that tER sites are long lived and that they primarily exhibit slow, short-range movements (Figure 3). We did not observe the type of directed long-range movements that are characteristic of the ERGIC (Presley et al., 1997; Scales et al., 1997). A similar conclusion was reached in a video microscopy study of GFP-labeled rbet1 and rsec22b; some of the fluorescent structures were mobile and probably represented ERGIC elements, but a subset of the structures were relatively immobile and probably represented tER sites (Chao et al., 1999). These results argue against the notion that each tER site generates a single ERGIC element and then translocates together with the ERGIC element along microtubules. It seems more likely that each tER site gives rise to a series of ERGIC elements.
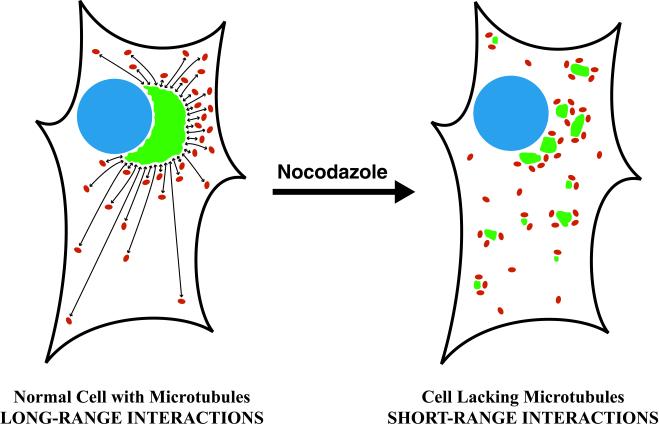
Our second goal was to investigate the factors that specify the distribution of tER sites. It has been consistently observed that tER sites are concentrated in the vicinity of the juxtanuclear Golgi (Figures 1A and 4A) (Palade, 1975; Bannykh and Balch, 1997; Tang et al., 1997). One potential reason for this concentration is that a large fraction of the total ER is found near the nucleus (Terasaki et al., 1986). In addition, our data indicate that the Golgi influences the distribution of tER sites. This effect becomes obvious in nocodazole-treated cells, which contain clustered and/or fused tER sites that are closely apposed to Golgi structures (Figure 4). Why do tER sites cluster near the Golgi when microtubules are disrupted? We infer that preexisting Golgi structures cause a local expansion of the tER. The evidence for this assertion is that tER clusters appear within minutes after nocodazole addition, well before the change in Golgi morphology (Figure 4B). We propose that tER sites proliferate near the Golgi due to an elevated concentration of recycling proteins in the ER. Candidate recycling proteins include ERGIC-53, the KDEL receptor, members of the p23/p24 family, various SNAREs, and possibly resident proteins of the Golgi stack (Tang et al., 1993; Nickel et al., 1997; Cole et al., 1998; Hay et al., 1998; Klumperman et al., 1998; Girod et al., 1999). Nocodazole treatment blocks long-range retrograde traffic of these proteins to the ER (Lippincott-Schwartz et al., 1995; Sciaky et al., 1997); presumably, the result is enhanced short-range retrograde traffic and a consequent increase in the local concentration of recycling proteins in the ER (Figure 8).
Figure 8.
Model of a proposed mutual feedback relationship between the tER and the Golgi. (Left) In normal cells, tER sites (red) and Golgi structures (green) exchange material by microtubule-directed transport (bidirectional arrows). ERGIC elements arise at tER sites and move inward to the juxtanuclear Golgi, while retrograde traffic from the Golgi moves outward toward the cell periphery. Retrograde transport events terminate either at tER sites (as drawn) or at random locations on the ER. This recycling of proteins to the ER causes tER sites to proliferate. Because most of the retrograde transport events terminate near their point of origin, ER membranes in the Golgi region contain a high concentration of recycling proteins and a correspondingly high density of tER sites. (Right) When microtubules are disrupted with nocodazole, each Golgi structure receives input only from adjacent tER sites, and the number of these sites determines the size and stability of the Golgi structure. Conversely, membrane recycling from a given Golgi structure induces a localized proliferation of the tER. This positive feedback loop generates intermediate-sized Golgi structures that are next to tER clusters.
In cells that contain microtubules, Golgi-to-ER retrograde traffic is driven by microtubule-dependent motors of the kinesin superfamily (Lippincott-Schwartz, 1998; Allan and Schroer, 1999). Most of the retrograde transport events probably terminate near their point of origin, so ER membranes in the vicinity of the Golgi should have high levels of recycling proteins and a correspondingly high density of tER sites (Figure 8). Peripheral tER sites then would be generated by the subset of retrograde transport events that terminate in the cell periphery. This model provides a functional explanation for the observed tER patterns in normal and nocodazole-treated cells. The central conclusion is that tER sites do not actively move within the cell; rather, they proliferate in ER regions that contain high levels of recycling proteins.
Because the tER gives rise to downstream compartments, characterizing changes in the tER should help to clarify the processes that cause reorganization of the ERGIC and Golgi. This study is the first comprehensive analysis of tER dynamics, so our interpretations are tentative. However, a number of insights have emerged.
Implications for 15°C Treatment
When cells are incubated at 15°C, most of the Sec13-containing structures also label for ERGIC-53 (Figure 2B and Table 1). We conclude that at 15°C the tER and the ERGIC fuse to form a nonphysiologic hybrid compartment. This interpretation fits with a previous electron microscopy study, which found that when vertebrate cells are incubated at reduced temperatures, the ERGIC is linked to the ER by tubular connections (Stinchcombe et al., 1995). The hybrid tER– ERGIC compartment previously has been designated the “15°C compartment” (Hauri and Schweizer, 1992; Saraste and Kuismanen, 1992). Because the 15°C compartment accumulates proteins that recycle rapidly through the ER (Schweizer et al., 1990; Tang et al., 1993; Nickel et al., 1997; Hay et al., 1998), low-temperature treatment is a convenient way to characterize such proteins. However, the cause of the 15°C transport block has not been identified. We suggest that this transport block results from the failure of the ERGIC to separate from the tER.
Implications for Nocodazole-Induced Golgi Fragmentation
In nocodazole-treated cells, tER clusters are closely apposed to Golgi structures (Figure 4). This finding sheds light on the process of nocodazole-induced Golgi reorganization. It has been proposed that the end product of nocodazole treatment is a single Golgi stack next to each individual tER site (Rogalski and Singer, 1984; Thyberg and Moskalewski, 1985; Cole et al., 1996). However, our images are more consistent with reports that intermediate-sized Golgi structures persist in nocodazole-treated cells (Yang and Storrie, 1998; Polishchuk et al., 1999). We observe such intermediate-sized structures even after long incubations in nocodazole, and even after the Golgi has been temporarily fused with the ER using BFA (Figure 5) (see also Polishchuk et al., 1999). The majority of the Golgi structures in nocodazole-treated cells are associated with tER clusters; single tER sites usually lack associated Golgi staining. Although tER sites that are distant from Golgi structures should still export proteins to the ERGIC (Figure 2C), these exported proteins presumably do not transit further through the secretory pathway. Thus, the functional tER–Golgi system in nocodazole-treated cells may consist mainly of clustered tER sites next to intermediate-sized Golgi structures.
Our data highlight a caveat for interpreting the effects of nocodazole. Mounting evidence suggests that nocodazole treatment causes Golgi structures to form de novo from the ER (Cole et al., 1996; Storrie et al., 1998; Yang and Storrie, 1998; Drecktrah and Brown, 1999; Polishchuk et al., 1999), although this interpretation is controversial (Shima et al., 1998). The proximity of Golgi and tER markers in nocodazole-treated cells fits with the idea that Golgi proteins pass through the ER and then emerge at tER sites to generate new Golgi stacks (Saraste and Svensson, 1991; Cole et al., 1996). However, our data indicate that the opposite process also occurs; preexisting Golgi structures promote the localized formation of new tER sites. Therefore, nocodazole-induced Golgi reorganization probably involves the de novo formation of both Golgi structures and tER sites.
Implications for Mitotic Reorganization of the Secretory Pathway
In vertebrate cells, the Golgi disassembles during mitosis (Warren, 1993). Reasoning that this change in the Golgi might be linked to tER function, we set out to compare the mitotic rearrangements of the Golgi and the tER. Our first aim was to characterize mitotic Golgi breakdown. This process occurs in two stages: during prophase the Golgi ribbon splits into multiple fragments (Figure 7A), and during later phases of mitosis these fragments break down further (Rabouille and Warren, 1997; Acharya et al., 1998; Thyberg and Moskalewski, 1998; Zaal et al., 1999). However, it is still unclear what type of structure represents the “end point” of mitotic Golgi breakdown. One group has reported that the Golgi is transformed into a combination of tubulovesicular clusters and small vesicles (Lucocq et al., 1989; Shima et al., 1997). Another study examined cells that had been arrested in mitosis by prolonged nocodazole treatment; under these conditions, Golgi markers were completely dispersed, apparently in the form of small vesicles (Jesch and Linstedt, 1998). Finally, video microscopy experiments have revived an earlier suggestion that the Golgi fuses with the ER during mitosis (Thyberg and Moskalewski, 1992; Zaal et al., 1999). To help clarify this issue, we performed a quantitative analysis using the improved immunofluorescence method. Our strategy was to examine endogenous Golgi proteins in unperturbed cultures, thereby avoiding potential pitfalls of using GFP fusions or cell synchronization. This experiment revealed that during the second stage of mitotic Golgi breakdown, most of the Golgi protein molecules disperse into structures that are not resolved by immunofluorescence microscopy (Figure 7, B and D). The Golgi dispersal that we see is consistent either with the formation of small vesicles or with the redistribution of Golgi proteins into the ER. However, this dispersal is incomplete, so that some Golgi fragments persist throughout mitosis in a majority of the cells (Figure 7, B and D). During cytokinesis the Golgi begins to reassemble (Rabouille and Warren, 1997; Thyberg and Moskalewski, 1998), and most of the Golgi protein molecules reside once again in optically resolvable structures (Figure 7, C and D). The key point is that Golgi morphology is variable at every stage of mitosis; for example, most telophase cells contain very few optically resolvable Golgi structures, but some telophase cells contain multiple fragments. This variability has undoubtedly fueled the debates about mitotic Golgi reorganization.
Once we had developed a reliable approach for quantifying the Golgi signals from mitotic cells, we examined tER sites in the same cells. tER sites show reduced Sec13 immunoreactivity during mitosis, presumably because ER-to-Golgi transport is inhibited (Featherstone et al., 1985; Farmaki et al., 1999). We measured the levels of tER-associated Sec13 at various stages of mitosis (Figure 7). Sec13 staining declines during prophase and then recovers during cytokinesis. The decline in Sec13 staining coincides with the initial fragmentation stage of Golgi breakdown (Figure 7, A and D), and the recovery of Sec13 staining coincides with the reassembly of the Golgi during cytokinesis (Figure 7, C and D). How do we explain this parallel reorganization of the tER and the Golgi? Perhaps tER function and Golgi structure are regulated independently by mitotic kinases (Acharya et al., 1998; Lowe et al., 1998). Another possibility is that changes in the tER affect Golgi structure. In budding yeasts, the activity and distribution of the tER can influence Golgi organization (Wooding and Pelham, 1998; Rossanese et al., 1999; Morin-Ganet et al., 2000). By analogy, the inhibition of tER activity during vertebrate mitosis may promote disintegration of the Golgi.
CONCLUSION
We propose that the tER–Golgi system is maintained by mutual feedback (Figure 8). In the forward direction, tER sites apparently generate ERGIC elements, which then generate new Golgi cisternae (Bannykh and Balch, 1997; Glick and Malhotra, 1998). Hence, tER function should be needed to maintain the Golgi. Consistent with this idea, the Golgi breaks down when tER function is blocked during mitosis (Figure 7). Moreover, in nocodazole-treated cells, the small Golgi structures that are associated with single tER sites are less stable than the larger Golgi structures that are associated with multiple tER sites (Figure 4; Polishchuk et al., 1999). These data suggest that vertebrate cells require input from multiple tER sites to preserve a stable Golgi unit.
In the retrograde direction, feedback from the Golgi induces proliferation of the tER (Figure 4). We suggest that the elevation of short-range Golgi-to-ER traffic causes the localized creation of new tER sites. As predicted by this model, tER sites can proliferate during interphase (Figure 6). tER proliferation is likely due to proteins that recycle from the Golgi to the ER. Many recycling proteins interact directly with COPII components (Springer et al., 1999), so tER expansion might result simply from the nucleation of COPII vesicle assembly. Alternatively, one or more recycling proteins may have a specialized role in generating tER sites (Lavoie et al., 1999).
This integrated view of the tER–Golgi system is particularly useful for explaining the effects of nocodazole treatment. In the absence of microtubules, the Golgi tends to undergo progressive fragmentation. Counteracting this tendency is a positive feedback loop: a given Golgi structure induces the localized proliferation of tER sites, which in turn enlarge and stabilize the Golgi structure. The net result is a dynamic balance that favors intermediate-sized Golgi structures and clustered tER sites (Figure 8).
For the future, a variety of approaches will extend our understanding of the tER. The budding yeast Pichia pastoris contains discrete tER sites (Rossanese et al., 1999) and is suitable both for genetic studies and for video microscopy. Phenomena that are specific to vertebrate cells can be explored using dual-color fluorescence imaging of live cells (Ellenberg et al., 1999). This method will be useful for addressing the following questions. (1) Do multiple ERGIC elements form in succession at each tER site? (2) In nocodazole-treated cells, do tER sites form next to preexisting Golgi structures, or vice versa? (3) As cells exit mitosis, do Golgi structures coalesce next to tER sites? Such experiments will further illuminate the dynamics of the vertebrate tER–Golgi system.
ACKNOWLEDGMENTS
Special thanks to Bor Luen Tang and Wanjin Hong for supplying affinity-purified anti-Sec13 antibody. Thanks for additional reagents to Adam Linstedt, Hans-Peter Hauri, Eric Berger, Tom Hobman, Marilyn Farquhar, Paul Mueller, and Anand Swaroop. Helpful discussion was provided by Theresa Roberts, Jennifer Lippincott-Schwartz, Jan Burkhardt and the members of the Glick lab. We are grateful to Craig Lassy and Tim Karr for assistance with confocal microscopy. A.T.H. was supported by National Institutes of Health training grant 5–20942 and National Science Foundation grant MCB-9875939. B.S.G. was supported by National Science Foundation grant MCB-9875939, a Pilot and Feasibility Study Award from the Diabetes Research Foundation, a Basil O'Connor Starter Scholar Research Award (5-FY96–1138) from the March of Dimes Birth Defects Foundation, and a grant from the Pew Charitable Trusts.
Abbreviations used:
- BFA
brefeldin A
- COP
coat protein
- ERGIC
ER-Golgi intermediate compartment
- GFP
green fluorescent protein
- PDI
protein disulfide isomerase
- tER
transitional ER
- VSV-G
vesicular stomatitis virus G protein
Note added in proof.
While this manuscript was under revision, a report appeared describing video microscopy of fluorescently tagged tER and ERGIC markers (Stephens et al., 2000). Those data accord with our conclusion that tER sites are long-lived, relatively immobile ER subdomains that give rise to mobile ERGIC elements.
REFERENCES
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- Allan VJ, Schroer TA. Membrane motors. Curr Opin Cell Biol. 1999;11:476–482. doi: 10.1016/s0955-0674(99)80068-4. [DOI] [PubMed] [Google Scholar]
- Andersen B, Osborn M, Weber K. Specific visualization of the distribution of the calcium dependent regulatory protein of cyclic nucleotide phosphodiesterase (modulator protein) in tissue culture cells by immuofluorescence microscopy: mitosis and intercellular bridge. Cytobiologie. 1978;17:354–364. [PubMed] [Google Scholar]
- Balch WE, McCaffery JM, Plutner H, Farquhar MG. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Balch WE. Membrane dynamics at the endoplasmic reticulum-Golgi interface. J Cell Biol. 1997;138:1–4. doi: 10.1083/jcb.138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EG, Aegerter E, Mandel T, Hauri H-P. Monoclonal antibodies to soluble, human milk galactosyltransferase (lactose synthase A protein) Carbohydr Res. 1986;149:23–33. doi: 10.1016/s0008-6215(00)90366-5. [DOI] [PubMed] [Google Scholar]
- Bracker CE, Morré DJ, Grove SN. Structure, differentiation and multiplication of Golgi apparatus in fungal hyphae. Protoplasma. 1996;194:250–274. [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centonze V, Pawley J. Tutorial on practical confocal microscopy and use of the confocal test specimen. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. 2nd ed. New York: Plenum Press; 1995. pp. 549–569. [Google Scholar]
- Chao DS, Hay JC, Winnick S, Prekeris R, Klumperman J, Scheller RH. SNARE membrane trafficking dynamics in vivo. J Cell Biol. 1999;144:869–881. doi: 10.1083/jcb.144.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthésy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Brown WJ. Phospholipase A2 antagonists inhibit nocodazole-induced Golgi ministack formation: evidence of an ER intermediate and constitutive cycling. Mol Biol Cell. 1999;10:4021–4032. doi: 10.1091/mbc.10.12.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Lippincott-Schwartz J, Presley JF. Dual-color imaging with GFP variants. Trends Cell Biol. 1999;9:52–56. doi: 10.1016/s0962-8924(98)01420-2. [DOI] [PubMed] [Google Scholar]
- Farmaki T, Ponnambalam S, Prescott AR, Clausen H, Tang B-L, Hong W, Lucocq JM. Forward and retrograde trafficking in mitotic animal cells. ER-Golgi transport arrest restricts protein export from the ER into COPII-coated structures. J Cell Sci. 1999;112:589–600. doi: 10.1242/jcs.112.5.589. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Hauri H-P. Protein sorting and vesicular traffic in the Golgi apparatus. In: Berger EG, Roth J, editors. The Golgi Apparatus. Basel, Switzerland: Birkhäuser Verlag; 1997. pp. 63–129. [Google Scholar]
- Featherstone C, Griffiths G, Warren G. Newly synthesized G protein of vesicular stomatitis virus is not transported to the Golgi complex in mitotic cells. J Cell Biol. 1985;101:2036–2046. doi: 10.1083/jcb.101.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, Lord JM, Nilsson T, Pepperkok R. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- Glick BS, Malhotra V. The curious status of the Golgi apparatus. Cell. 1998;95:883–889. doi: 10.1016/s0092-8674(00)81713-4. [DOI] [PubMed] [Google Scholar]
- Hauri H-P, Kappeler F, Andersson H, Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- Hauri H-P, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman TC, Woodward L, Farquhar MG. The rubella virus E1 glycoprotein is arrested in a novel, post-ER pre-Golgi compartment. J Cell Biol. 1992;118:795–812. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman TC, Zhao B, Chan H, Farquhar MG. Immunoisolation and characterization of a subdomain of the endoplasmic reticulum that concentrates proteins involved in COPII vesicle biogenesis. Mol Biol Cell. 1998;9:1265–1278. doi: 10.1091/mbc.9.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué S. Foundations of confocal scanned imaging in light microscopy. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. 2nd ed. New York: Plenum Press; 1995. pp. 1–17. [Google Scholar]
- Jesch SA, Linstedt AD. The Golgi and endoplasmic reticulum remain independent during mitosis in HeLa cells. Mol Biol Cell. 1998;9:623–635. doi: 10.1091/mbc.9.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Ferro-Novick S. Transport from the endoplasmic reticulum to the Golgi. Curr Opin Cell Biol. 1998;10:477–482. doi: 10.1016/s0955-0674(98)80062-8. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Schweizer A, Clausen H, Tang BL, Hong W, Oorschot V, Hauri H-P. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- Kreis TE, Goodson HV, Perez F, Rönnholm R. Golgi apparatus-cytoskeleton interactions. In: Berger EG, Roth J, editors. The Golgi Apparatus. Basel, Switzerland: Birkhäuser Verlag; 1997. pp. 179–193. [Google Scholar]
- Kuehn MJ, Schekman R. COPII and secretory cargo capture into transport vesicles. Curr Opin Cell Biol. 1997;9:477–483. doi: 10.1016/s0955-0674(97)80022-1. [DOI] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C, Paiement J, Dominguez M, Roy L, Dahan S, Gushue JN, Bergeron JJM. Roles for α2p24 and COPI in endoplasmic reticulum cargo exit site formation. J Cell Biol. 1999;146:285–299. doi: 10.1083/jcb.146.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri H-P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J. Cytoskeletal proteins and Golgi dynamics. Curr Opin Cell Biol. 1998;10:52–59. doi: 10.1016/s0955-0674(98)80086-0. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri H-P, Yuan LC, Klausner RD. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Lucocq JM, Berger EG, Warren G. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J Cell Biol. 1989;109:463–474. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Menárguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Koonce MP. Mitosis. Science. 1989;246:622–628. doi: 10.1126/science.2683078. [DOI] [PubMed] [Google Scholar]
- Melan MA, Sluder G. Redistribution and differential extraction of soluble proteins in permeabilized cultured cells: implications for immunofluorescence microscopy. J Cell Sci. 1992;101:731–743. doi: 10.1242/jcs.101.4.731. [DOI] [PubMed] [Google Scholar]
- Mellman I, Simons K. The Golgi complex: in vitro veritas? Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merisko EM, Fletcher M, Palade GE. The reorganization of the Golgi complex in anoxic pancreatic acinar cells. Pancreas. 1986;1:95–109. doi: 10.1097/00006676-198603000-00001. [DOI] [PubMed] [Google Scholar]
- Minin AA. Dispersal of Golgi apparatus in nocodazole-treated fibroblasts is a kinesin-driven process. J Cell Sci. 1997;110:2495–2505. doi: 10.1242/jcs.110.19.2495. [DOI] [PubMed] [Google Scholar]
- Morin-Ganet M-N, Rambourg A, Deitz SB, Franzusoff A, Képès F. Morphogenesis and dynamics of the yeast Golgi apparatus. Traffic. 2000;1:56–68. doi: 10.1034/j.1600-0854.2000.010109.x. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Nickel W, Sohn K, Bunning C, Wieland FT. p23, a major COPI-vesicle membrane protein, constitutively cycles through the early secretory pathway. Proc Natl Acad Sci USA. 1997;94:11393–11398. doi: 10.1073/pnas.94.21.11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Meda P, Holcomb C, Moore H-P, Hicke L, Schekman R. Mammalian Sec23p homologue is restricted to the endoplasmic reticulum transitional cytoplasm. Proc Natl Acad Sci USA. 1991;88:8611–8615. doi: 10.1073/pnas.88.19.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccaud J-P, Reith W, Carpentier J-L, Ravazzola M, Amherdt M, Schekman R, Orci L. Cloning and functional characterization of mammalian homologues of the COPII component Sec23. Mol Biol Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Paulin RP, Ho T, Blzer HJ, Holliday R. Gene silencing by DNA methylation and dual inheritance in Chinese hamster ovary cells. Genetics. 1998;149:1081–1088. doi: 10.1093/genetics/149.2.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. Getting through the Golgi complex. Trends Cell Biol. 1998;8:45–49. doi: 10.1016/s0962-8924(97)01185-9. [DOI] [PubMed] [Google Scholar]
- Polishchuk RS, Polishchuk EV, Mironov AA. Coalescence of Golgi fragments in microtubule-deprived living cells. Eur J Cell Biol. 1999;78:170–185. doi: 10.1016/S0171-9335(99)80096-X. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Presley JF, Smith C, Hirschberg K, Miller C, Cole NB, Zaal KJM, Lippincott-Schwartz J. Golgi membrane dynamics. Mol Biol Cell. 1998;9:1617–1626. doi: 10.1091/mbc.9.7.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Warren G. Changes in the architecture of the Golgi apparatus during mitosis. In: Berger EG, Roth J, editors. The Golgi Apparatus. Basel, Switzerland: Birkhäuser Verlag; 1997. pp. 195–217. [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski AA, Singer SJ. Associations of elements of the Golgi apparatus with microtubules. J Cell Biol. 1984;99:1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Saraste J, Kuismanen E. Pathways of protein sorting and membrane traffic between the rough endoplasmic reticulum and the Golgi complex. Semin Cell Biol. 1992;3:343–355. doi: 10.1016/1043-4682(92)90020-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Schindler R, Itin C, Zerial M, Lottspeich F, Hauri H-P. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur J Cell Biol. 1993;61:1–9. [PubMed] [Google Scholar]
- Schweizer A, Fransen JAM, Baechi T, Ginsel L, Hauri H-P. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Fransen JAM, Matter K, Kreis TE, Ginsel L, Hauri H-P. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- Sciaky N, Presley J, Smith C, Zaal KJM, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol. 1995;128:769–777. doi: 10.1083/jcb.128.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugrue CA, Kolen ER, Peters H, Czernik A, Kaiser C, Matovcik L, Hubbard AL, Gorelick F. Identification of the putative mammalian orthologue of Sec31P, a component of the COPII coat. J Cell Sci. 1999;112:4547–4556. doi: 10.1242/jcs.112.24.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Springer S, Spang A, Schekman R. A primer on vesicle budding. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Lin-Marq N, Pagano A, Pepperkok R, Paccaud J-P. COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites. J Cell Sci. 2000;113:2177–2185. doi: 10.1242/jcs.113.12.2177. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Nomoto H, Cutler DF, Hopkins CR. Anterograde and retrograde traffic between the rough endoplasmic reticulum and the Golgi complex. J Cell Biol. 1995;131:1387–1401. doi: 10.1083/jcb.131.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B, White J, Röttger S, Stelzer EHK, Suganuma T, Nilsson T. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Yang-Feng TL, Liu W, Gieser L, Barrow LL, Chen K-C, Agarwal N, Meisler MH, Smith DI. Molecular characterization of a novel human gene, SEC13R, related to the yeast secretory pathway gene SEC13, and mapping to a conserved linkage group on human chromosome 3p24–p25 and mouse chromosome 6. Hum Mol Genet. 1994;3:1281–1286. doi: 10.1093/hmg/3.8.1281. [DOI] [PubMed] [Google Scholar]
- Tang BL, Kausalya J, Low DY, Lock ML, Hong W. A family of mammalian proteins homologous to yeast Sec24p. Biochem Biophys Res Commun. 1999;258:679–684. doi: 10.1006/bbrc.1999.0574. [DOI] [PubMed] [Google Scholar]
- Tang BL, Peter F, Krijnse-Locker J, Low S, Griffiths G, Hong W. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol Cell Biol. 1997;17:256–266. doi: 10.1128/mcb.17.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Wong SH, Qi XL, Low SH, Hong W. Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J Cell Biol. 1993;120:325–328. doi: 10.1083/jcb.120.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Zhang T, Low DYH, Wong ET, Horstmann H, Hong W. Mammalian homologues of yeast Sec31p: an ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J Biol Chem. 2000;275:13597–13604. doi: 10.1074/jbc.275.18.13597. [DOI] [PubMed] [Google Scholar]
- Terasaki M. Recent progress on structural interactions of the endoplasmic reticulum. Cell Motil Cytoskeleton. 1990;15:71–75. doi: 10.1002/cm.970150203. [DOI] [PubMed] [Google Scholar]
- Terasaki M, Chen LB, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Microtubules and the organization of the Golgi complex. Exp Cell Res. 1985;159:1–16. doi: 10.1016/s0014-4827(85)80032-x. [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Reorganization of the Golgi complex in association with mitosis: redistribution of mannosidase II to the endoplasmic reticulum and effects of brefeldin A. J Submicrosc Cytol Pathol. 1992;24:495–508. [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Partitioning of cytoplasmic organelles during mitosis with special reference to the Golgi complex. Microsc Res Tech. 1998;40:354–368. doi: 10.1002/(SICI)1097-0029(19980301)40:5<354::AID-JEMT3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- Weiss DG, Maile W, Wick RA. Video microscopy. In: Lacey AJ, editor. Light Microscopy in Biology: A Practical Approach. Oxford, UK: IRL Press; 1989. pp. 221–278. [Google Scholar]
- Wooding S, Pelham HRB. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Storrie B. Scattered Golgi elements during microtubule disruption are initially enriched in trans-Golgi proteins. Mol Biol Cell. 1998;9:191–207. doi: 10.1091/mbc.9.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal KJ, Smith CL, Polishchuk R S, Altan N, Cole NB, Ellenberg J, Hirschberg K, Presley J F, Roberts TH, Siggia E, Phair RD, Lippincott-Schwartz J. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wong SH, Tang BL, Xu Y, Hong W. Morphological and functional association of Sec22b/ERS-24 with the pre-Golgi intermediate compartment. Mol Biol Cell. 1999;10:435–453. doi: 10.1091/mbc.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]