Abstract
Earlier in vivo studies have shown that the sequential action of the IspG and IspH proteins is essential for the reductive transformation of 2C-methyl-d-erythritol 2,4-cyclodiphosphate into dimethylallyl diphosphate and isopentenyl diphosphate via 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate. A recombinant fusion protein comprising maltose binding protein and IspG protein domains was purified from a recombinant Escherichia coli strain. The purified protein failed to transform 2C-methyl-d-erythritol 2,4-cyclodiphosphate into 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate, but catalytic activity could be restored by the addition of crude cell extract from an ispG-deficient E. coli mutant. This indicates that auxiliary proteins are required, probably as shuttles for redox equivalents. On activation by photoreduced 10-methyl-5-deaza-isoalloxazine, the recombinant protein catalyzed the formation of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate from 2C-methyl-d-erythritol 2,4-cyclodiphosphate at a rate of 1 nmol⋅min−1⋅mg−1. Similarly, activation by photoreduced 10-methyl-5-deaza-isoalloxazine enabled purified IspH protein to catalyze the conversion of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate into a 6:1 mixture of isopentenyl diphosphate and dimethylallyl diphosphate at a rate of 0.4 μmol⋅min−1⋅mg−1. IspH protein could also be activated by a mixture of flavodoxin, flavodoxin reductase, and NADPH at a rate of 3 nmol⋅min−1⋅mg−1. The striking similarities of IspG and IspH protein are discussed, and plausible mechanistic schemes are proposed for the two reactions.
Keywords: gcpE‖lytB‖deazaflavin‖1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase‖1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase
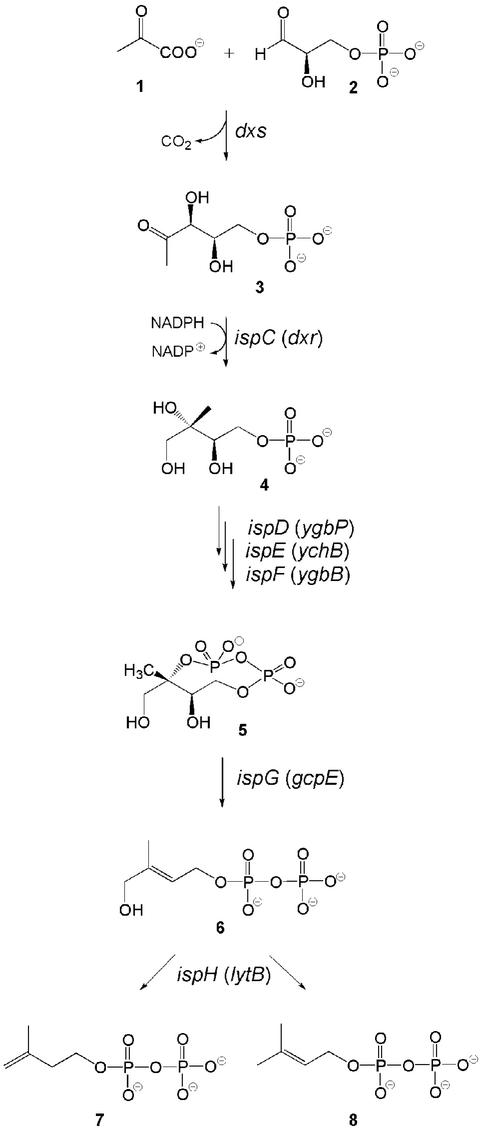
For a period of several decades, the mevalonate pathway has been considered as the unique source for the universal terpene precursors, isopentenyl diphosphate (IPP; 7) and dimethylallyl diphosphate (DMAPP; 8, Fig. 1). The pioneering studies on this pathway by Bloch, Cornforth, Lynen, and their coworkers (for review, see refs. 1–4) served as the basis for the development of cholesterol biosynthesis inhibitors that have a central role in the prevention and treatment of cardiovascular disease (5).
Figure 1.
The deoxyxylulose phosphate pathway of isoprenoid biosynthesis.
Only about a decade ago, independent work by Arigoni, Rohmer, and their respective coworkers proved the existence of a second isoprenoid pathway, which is operative in many eubacteria and in the plastid compartment of higher plants (for review, see refs. 6–8). Subsequent studies demonstrated that Dxs protein catalyzes the condensation of pyruvate (1) with d-glyceraldehyde 3-phosphate (2) affording 1-deoxy-d-xylulose 5-phosphate (3) (9, 10), which is transformed into 2C-methyl-d-erythritol 4-phosphate (4) by a skeletal rearrangement and reduction catalyzed by the IspC protein (Fig. 1) (11). The polyol phosphate is then converted into 2C-methyl-d-erythritol 2,4-cyclodiphosphate (5) by the consecutive action of the IspD, IspE, and IspF proteins (refs. 12–14; for review see ref. 15).
A recombinant Escherichia coli strain engineered for expression of the xylB and ispCDEF genes was shown to transform exogenous 13C-labeled 1-deoxy-d-xylulose into endogenous cyclic diphosphate 5 in high yield (16). The additional implementation of a recombinant ispG gene resulted in the in vivo formation of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (6) (16), which was also isolated from an ispH-deficient E. coli mutant (17). Subsequently, the in vitro formation of 6 from 5 by crude cell extracts of E. coli overexpressing ispG was confirmed by radiochemical methods (18). Compound 6 was shown by in vivo as well in vitro experiments to serve as the biosynthetic precursor of IPP (7) and DMAPP (8), which were obtained in a ratio of 6:1 by the catalytic action of the IspH protein (19, 20).
Both the IspG and IspH proteins catalyze reduction steps involving the overall transfer of two electrons and carry three absolutely conserved cysteine residues. We now present additional evidence supporting the striking similarities of the two proteins.
Experimental Procedures
Materials.
[2,2′-13C2]-, [1,3,4-13C3]-, and [2-14C]5 (23 μCi⋅μmol−1; 1 Ci = 37 GBq) were prepared by published procedures (21). [U-13C5]6 was prepared by in vivo biotransformation of 1-deoxy-d-[U-13C5]xylulose using a recombinant E. coli strain (16). [1-3H]6 (86 μCi⋅μmol−1) was synthesized by published procedures (22). Compound 7 was prepared by published procedures (23). [4-14C]7 (57.8 μCi⋅μmol−1) was purchased from NEN. 10-Methyl-5-deaza-isoalloxazine (subsequently designated deazaflavin) was a generous gift of Andrée Marquet, Paris. Recombinant MalE/IspH fusion protein was prepared by published procedures (20).
Microorganisms.
Bacterial strains and plasmids used in this study are summarized in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Radiochemical Assay of IspG Protein Activity.
Method A.
Assay mixtures contained 50 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 20 mM NaF, 1 mM NADH, 200 μM FMN, 1 mM FeCl3, 1 mM Na2S, 0.5 mM pamidronate, 40 μM [2-14C]5 (23 μCi⋅μmol−1), recombinant MalE/IspG fusion protein (180 μg) (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site), and crude cell extract (3 mg) of the E. coli ispG-deficient mutant (see Supporting Materials and Methods) in a total volume of 200 μl. The assay mixtures were incubated at 37°C for 4 h and placed on ice. The reaction was terminated by the addition of 10 μl of 30% (wt/vol) trichloroacetic acid followed by immediate neutralization with 20 μl of 1 M sodium hydroxide. The mixtures were centrifuged, and the supernatants were subjected to ultrafiltration (nanosep 100-kDa membrane, Pall Gelman). Aliquots (50 μl) were analyzed by reversed-phase ion pair HPLC using a Luna C8 column (5 μm, 4 × 250 mm, Phenomenex, Aschaffenburg, Germany). The column was developed with 10 ml of 3% (vol/vol) methanol in 10 mM tetra-n-butylammonium phosphate (pH 6.0), followed by a linear gradient of 2 ml of 3–21% (vol/vol) methanol in 10 mM tetra-n-butylammonium phosphate (pH 6.0), a linear gradient of 13 ml of 21–35% (vol/vol) methanol in 10 mM tetra-n-butylammonium phosphate (pH 6.0), a linear gradient of 10 ml of 35–49% (vol/vol) methanol in 10 mM tetra-n-butylammonium phosphate (pH 6.0), and a linear gradient of 5 ml of 49–56% (vol/vol) methanol in 10 mM tetra-n-butylammonium phosphate (pH 6.0), at a flow rate of 1 ml⋅min−1. The effluent was monitored by online liquid scintillation analysis (Beta-RAM, Biostep, Jahnsdorf, Germany). The retention volumes of 5 and 6 were 20 and 25 ml, respectively. Compounds 7 and 8 were both eluted at a retention volume of 36 ml.
Method B.
All steps were carried out under anaerobic conditions. Assay mixtures contained 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, 40 μM [2-14C]5 (23 μCi⋅μmol−1), 1 mM FeCl3, 1 mM Na2S, and recombinant MalE/IspG fusion protein (180 μg) in a volume of 200 μl. A solution (20 μl) containing 5 mM deazaflavin in dimethyl sulfoxide was added, and the mixture was irradiated with a mercury vapor lamp. During irradiation, the sample was cooled with ice. The samples were processed and analyzed by reversed-phase ion pair HPLC as described in Method A.
Radiochemical Assay of IspH Protein Activity.
Method A.
All steps were carried out under anaerobic conditions. Assay mixtures contained 50 mM Tris⋅HCl (pH 8.0), 7.5 mM DTT, 1.5 mM CoCl2, 5.3 μM [1-3H]6 (86 μCi⋅μmol−1), and 12 μg of MalE/IspH fusion protein in a volume of 200 μl. A solution (20 μl) of 5 mM deazaflavin in dimethyl sulfoxide was added, and the mixture was irradiated as described above. The reaction was terminated by the addition of 10 μl of 30% (wt/vol) trichloroacetic acid followed by immediate neutralization with 20 μl of 1 M sodium hydroxide. The mixtures were centrifuged. The supernatants were subjected to ultrafiltration (nanosep 100 kDa, Pall Gelman) and analyzed by reversed-phase ion pair HPLC using a Luna C8 column (4 × 250 mm, 5 μm), which was developed as described above for the assay of IspG protein activity.
Method B.
Assay mixtures contained 50 mM Tris⋅HCl (pH 8.0), 2 mM DTT, 5.3 μM [1-3H]6 (86 μCi⋅μmol−1), 2 mM NADPH, 54 μg of flavodoxin (FldA protein), 45 μg of flavodoxin reductase (Fpr protein) (see Supporting Materials and Methods), and 50 μg of MalE/IspH fusion protein in a volume of 150 μl. The mixture was incubated at 37°C for 1 h under aerobic conditions. The reaction was terminated by the addition of 10 μl of 30% (wt/vol) trichloroacetic acid followed by immediate neutralization with 20 μl of 1 M sodium hydroxide. The mixtures were centrifuged and the supernatants were analyzed by reversed-phase HPLC as described above.
Enzymatic Conversion of 13C-Labeled 2C-Methyl-d-Erythritol 2,4-Cyclodiphosphate by IspG Protein in the Presence of Photoreduced Deazaflavin.
All reaction steps were carried out under anaerobic conditions.
Reaction mixtures contained 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, 100
μM [2,2′-13C2]- or
[1,3,4-13C3]-5,
0.5 mM deazaflavin, and 1.8 mg of recombinant MalE/IspG protein in a
total volume of 3 ml. Samples were irradiated for 2 h with a
mercury vapor lamp. During irradiation, the samples were cooled with
ice. The reaction was terminated by the addition of 150 μl of
trichloroacetic acid (30%, wt/vol) and immediate neutralization with
300 μl of 1 M sodium hydroxide. DOWEX 50 WX8 (NH form) was added. The mixtures were centrifuged. The combined
supernatant of four parallel samples was applied to a column of DOWEX
1 × 8 (volume, 400 μl; HCO
form) was added. The mixtures were centrifuged. The combined
supernatant of four parallel samples was applied to a column of DOWEX
1 × 8 (volume, 400 μl; HCO form). The column
was washed with 25 ml of water, and the product was eluted with 1 ml of
2 M NH4HCO3. The mixture
was analyzed by 13C NMR spectroscopy.
form). The column
was washed with 25 ml of water, and the product was eluted with 1 ml of
2 M NH4HCO3. The mixture
was analyzed by 13C NMR spectroscopy.
Enzymatic Conversion of 13C-Labeled 1-Hydroxy-2-Methyl-2-(E)-Butenyl 4-Diphosphate by IspH Protein in the Presence of Photoreduced Deazaflavin.
All reaction steps were carried out under anaerobic conditions by using degassed buffers and solutions. Reaction mixtures containing 50 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 1 mM CoCl2, 1.23 mM [U-13C5]6, 140 μM deazaflavin (dissolved in dimethyl sulfoxide), and 1.84 mg of recombinant MalE/IspH protein in a total volume of 1.5 ml were irradiated for 20 min with a mercury vapor lamp. During irradiation, the sample was cooled with ice. The reaction was terminated by addition of 75 μl of trichloroacetic acid. The samples were immediately neutralized with 1 M sodium hydroxide. DOWEX 50 (WX8, Na+-form) was added. The mixture was centrifuged, and the supernatant was lyophilized. The residue was dissolved in 600 μl of 5% (vol/vol) D2O and analyzed by 13C NMR spectroscopy.
Next, 100 mM Tris⋅HCl, 10 mM MgCl2, and 0.65 mg of recombinant isopentenyl diphosphate isomerase (see Supporting Materials and Methods) were added to the NMR sample. The reaction mixture (total volume, 780 μl) was incubated for 2 h at 37°C and analyzed by 13C NMR spectroscopy.
NMR Spectroscopy.
13C and 1H NMR spectra were recorded at 25°C by using an AVANCE DRX 500 spectrometer from Bruker Instruments (Karlsruhe, Germany).
Results
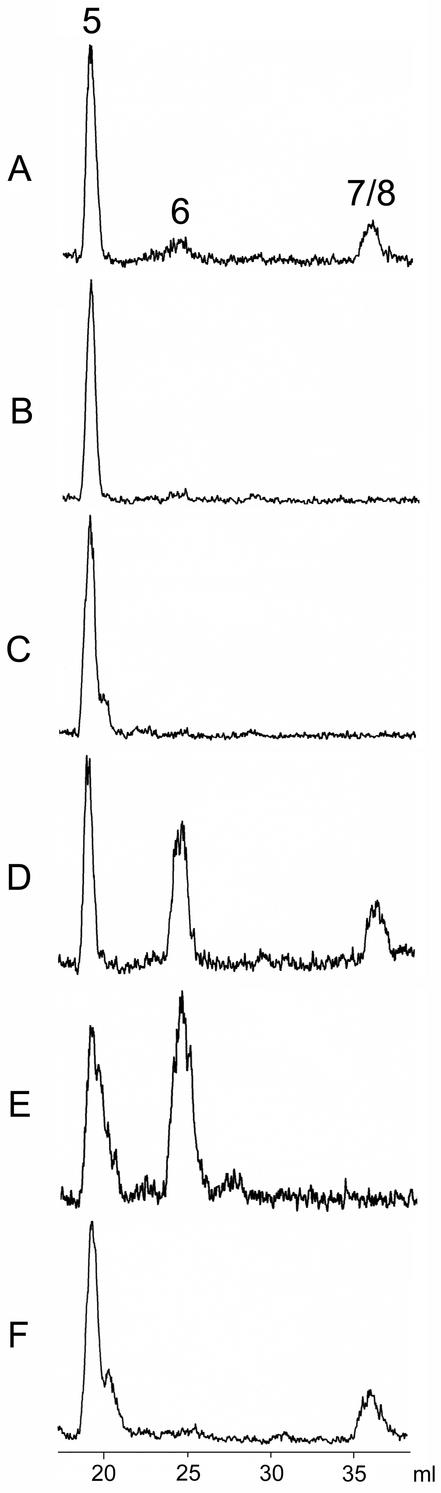
The incubation of reaction mixtures containing E. coli wild-type cell extract with [2-14C]2C-methyl-d-erythritol 2,4-cyclodiphosphate (5) afforded two radioactive product fractions that were characterized by retention volumes of 25 and 36 ml in reversed-phase ion pair HPLC (Fig. 2A). The minor product fraction with a retention volume of 25 ml migrated at the same velocity as an authentic sample of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (6), and the major fraction with a retention volume of 36 ml migrated at the same velocity as IPP (7) and DMAPP (8), which cannot be separated from each other under the experimental conditions (see below). The reaction proceeded best at pH 8.0 with supplements of 200 μM FMN, 1 mM NADH, 1 mM FeCl3, and 1 mM Na2S. The reaction rate based on total protein in the crude cell extract was 2 pmol⋅mg−1⋅min−1.
Figure 2.
Enzymatic conversion of 14C-labeled 5 into 6 and 7/8. Radiochemical assays of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase activity were performed as described in Experimental Procedures. (A–D) Method A. (E and F) Method B. The assay mixtures contained 3 mg of crude cell extract of E. coli cells SK6600 (A); 3 mg of crude cell extract of E. coli cells SK6600ispG∷neoR (B); 180 μg of recombinant MalE/IspG fusion protein (C); 3 mg of crude cell extract of E. coli cells SK6600ispG∷neoR and 180 μg of recombinant MalE/IspG fusion protein (D); 180 μg of recombinant MalE/IspG fusion protein and 0.5 mM photoreduced deazaflavin (E); 180 μg of recombinant MalE/IspG fusion protein, 48 μg of recombinant MalE/IspH fusion protein, and 0.5 mM photoreduced deazaflavin (F).
To construct an ispG mutant, the chromosomal ispG gene of E. coli was replaced by a recombinant ispG gene with an inserted neomycin resistance (neoR) gene specifying neomycin phosphotransferase using the method reported by Kushner and his coworkers (24) (see Supporting Materials and Methods). A minioperon comprising the mk and pmk genes of Saccharomyces cerevisiae (specifying mevalonate and phosphomevalonate kinase) and the dpmd gene of Arabidopsis thaliana (specifying diphosphomevalonate decarboxylase) on a low copy plasmid was implemented into that strain to enable the formation of 7 from exogenous mevalonolactone (see Supporting Materials and Methods). The resulting strain had an absolute mevalonolactone requirement for growth. The insertion of the neoR gene (Fig. 8, which is published as supporting information on the PNAS web site) in the ispG gene was confirmed by PCR analysis and sequence determination of the resulting 2.45-kb fragment (Fig. 9, which is published as supporting information on the PNAS web site).
Western blots of cell extract from wild-type E. coli cells developed with a rabbit antiserum against ispG protein showed an immunoreactive band of ≈40 kDa in agreement with the predicted mass of 40.7 kDa. That band was absent in cell extracts of the ispG mutant (Fig. 10, which is published as supporting information on the PNAS web site). Cell extracts of the ispG mutant strain failed to convert [2-14C]5 into any of the products found with wild-type cell extract (Fig. 2B).
The ispG gene of E. coli was cloned into an expression vector to afford a fusion protein comprising a maltose binding protein domain (MalE) and an IspG protein domain. In a recombinant E. coli strain, that plasmid directed the synthesis of an 83.6-kDa protein at an approximate level of 5% based on total cell protein. The MalE/IspG fusion protein was purified to apparent homogeneity by affinity chromatography using an amylose column and by subsequent size exclusion chromatography (Fig. 11, which is published as supporting information on the PNAS web site). The UV/visible spectrum is displayed in Fig. 12A, which is published as supporting information on the PNAS web site.
Purified, recombinant IspG protein failed to convert [2-14C]5 into 6 under a variety of conditions (Fig. 2C). However, the conversion of [2-14C]5 into 6 took place, albeit at a low rate, in a mixture containing purified, recombinant IspG protein and crude cell extract from the ispG mutant (Fig. 2D). FMN, NADPH, FeCl3, and Na2S were required for catalysis. The rate of product formation based on the recombinant IspG protein was 90 pmol⋅mg−1⋅min−1.
In agreement with a recent report by Rohmer and his coworkers (25), we observed the activation of recombinant IspG protein by photoreduced deazaflavin. In closer detail, a mixture containing 40 μM [2-14C]5, 5 mM DTT, 1 mM FeCl3, 1 mM Na2S, 0.5 mM deazaflavin, and pure recombinant IspG protein was irradiated with white light from a mercury lamp. As shown by radiochromatography, 6 was formed at a rate of 1 nmol⋅mg−1⋅min−1 (Fig. 2E). Product formation was not observed in control experiments without the IspG protein or deazaflavin.
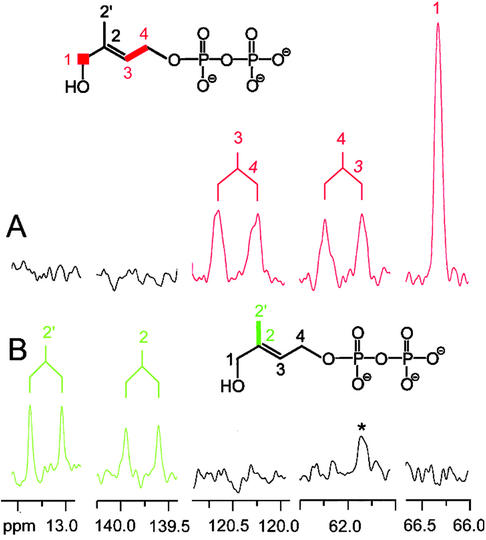
A similar set of experiments was carried out with the help of 13C-labeled substrates. Fig. 3B shows 13C NMR signals observed in a reaction mixture containing IspG protein, deazaflavin, and [2,2′-13C2]5, which had been irradiated with white light for 2 h. The sample showed two doublets at 139.8 and 13.2 ppm reflecting the presence of two 13C-enriched carbon atoms. The doublet signature of both signals is caused by coupling of the respective carbon atoms with a coupling constant of 42 Hz, and the chemical shifts and coupling constants identify the reaction product as [2,2′-13C2]6.
Figure 3.
13C NMR signals of 6 obtained from [1,3,4-13C3]5 (A) and [2,2′-13C2]5 (B). Assay mixtures containing 50 mM Tris⋅HCl (pH 8.0), 0.5 mM deazaflavin, 100 μM 13C-labeled 5, and recombinant MalE/IspG fusion protein were irradiated as described in Experimental Procedures. *, A signal from an impurity.
In a similar experiment with [1,3,4-13C3]5 as substrate, the spectrum of the reaction mixture showed three enriched signals, namely a singlet at 66.3 ppm and two doublets at 120.4 and 62.0 ppm (Fig. 3A), each of which displayed a coupling constant of 50 Hz. Clearly, these signal represent carbon atoms 1, 3, and 4 of [1,3,4-13C3]6 obtained from [1,3,4-13C3]5. Long-range 13C13C coupling and 13C31P coupling are not visible in Fig. 3A as a consequence of line broadening applied during NMR data processing to improve the signal to noise ratio.
Earlier, we had shown that recombinant IspH protein can be activated by unidentified proteins present in the cell extract of wild-type E. coli cells (20). The UV/visible spectrum is displayed in Fig. 12B. We now show that IspH protein can also be activated by a mixture of flavodoxin, flavodoxin reductase, and NADPH (Fig. 13, which is published as supporting information on the PNAS web site). The catalytic rate based on IspH protein (in presence of a large excess of the two auxiliary proteins) was 3 nmol⋅min−1⋅mg−1.
In light of the data with the IspG protein, it was of interest to study whether IspH protein can be also activated by photoreduced deazaflavin. For that purpose, we irradiated a mixture containing 5.3 μM [1-3H]6, 7.5 mM DTT, 1.5 mM CoCl2, recombinant IspH protein, and 0.5 mM deazaflavin. Reversed-phase ion pair HPLC analysis showed a radioactive fraction with the characteristic retention volume of 7/8 (data not shown). The rate of product formation based on the recombinant IspH protein was 0.4 μmol⋅min−1⋅mg−1.
When the HPLC conditions described by Zenk and his coworkers (26) were used, the product peak was observed at a retention volume of 66 ml. The same retention value was also detected for an authentic sample of [4-14C]7 (Fig. 14, which is published as supporting information on the PNAS web site) as well as for a 3:7 mixture of radiolabeled 7 and 8 obtained from [4-14C]7 after incubation with isopentenyl diphosphate isomerase (Fig. 13A). The ratio of 7 and 8 in the equilibrium mixture was confirmed by 1H NMR spectroscopy (Fig. 15, which is published as supporting information on the PNAS web site). This result is at variance with a recent report (26) in which separation of peaks corresponding to radioactive 7 and 8 has been described (see also ref. 27), and we have no explanation for the discrepancy.
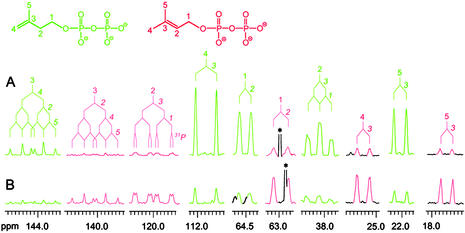
Direct evidence for the simultaneous formation of 7 and 8 was secured in experiments with 13C-labeled substrates. For that purpose, we irradiated mixtures containing [U-13C5]6, deazaflavin, and recombinant IspH protein with white light. The reaction mixture was analyzed by 13C NMR spectroscopy without prior purification. The signals of all carbon atoms of [U-13C5]7 and [U-13C5]8 were observed. They all appear as multiplets because of 13C13C coupling (Fig. 4). The chemical shifts and 13C13C coupling constants agree with the published values (19). The ratio of 7 to 8 in Fig. 4A is about 6:1, in close similarity to the ratio observed in our earlier in vivo and in vitro studies (19, 20). After incubation with recombinant isopentenyl diphosphate isomerase (Idi protein) this sample displays NMR signals whose intensities had been shifted from the original values to the equilibrium value of 3:7 (28) (Fig. 4B).
Figure 4.
13C NMR signals of 7 (green) and 8 (red) generated by irradiation from [U-13C5]6 in the presence of MalE/IspH fusion protein and deazaflavin (A) and after incubation of the latter sample with Idi protein (B). The 13C coupling patterns are indicated. *, A signal of an impurity.
In addition, white light irradiation of reaction mixtures containing both IspG and IspH protein together with [2-14C]5 and deazaflavin afforded predominantly a radioactive fraction characterized by a retention volume of 36 ml (i.e., the retention volume of both 7 and 8 as documented above) (Fig. 2F).
Discussion
The IspG protein of E. coli is involved in the mechanistically intriguing conversion of 2C-methyl-d-erythritol 2,4-cyclodiphosphate (5) into 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (6). We have now expressed in a recombinant E. coli strain a fusion protein encompassing IspG and a maltose binding domain and demonstrated that, whereas the purified protein is inactive, activity can be restored by the addition of a crude cell extract from an ispG-deficient mutant of E. coli. This requirement of auxiliary proteins parallels the one previously detected for the IspH protein, which catalyzes the subsequent transformation of 6 into a mixture of IPP (7) and DMAPP (8) (20). The plausible assumption that such auxiliary proteins catalyze the shuttling of redox equivalents has been corroborated in this work by showing that IspH protein can be activated by the joint action of flavodoxin and flavodoxin reductase in the presence of NADPH. Similar results have been reported in the meantime for the activation of the IspG protein (25).
The IspG and IspH proteins share further remarkable similarities; they both carry three strictly conserved cysteine residues and display in their UV/visible spectra similar absorption bands centered at ≈415 nm (see Fig. 12), which are compatible with the presence of [4Fe-4S]2+ clusters (see refs. 29–32). Additional evidence for the presence of such a prosthetic group in an artificially restored form of IspG has been provided recently (25), together with a radiochemical demonstration that the reconstructed holoprotein can be activated efficiently, as can a number of other proteins with similar prosthetic groups (33–35), by the semiquinone radical generated via photoreduction of deazaflavins in the presence of Tris⋅HCl, thiols, and other electron donors (36).
As described in detail in Experimental Procedures, we have confirmed and extended these results by using both 14C- and 13C-labeled substrates in experiments with a purified recombinant IspG protein, which did not require prior reconstruction of its Fe-S cluster. The observed low rate of 1 nmol⋅mg−1⋅min−1 may well reflect a partial aerobic disruption of the holoenzyme during the isolation procedure. Most rewardingly, a similar but more efficient activation by the same photo-generated radical was observed for the otherwise inert form of the recombinant IspH protein. Both [1-3H]6 and uniformly 13C-labeled forms of 6 were converted into products at a rate of 0.4 μmol⋅min−1⋅mg−1. In the experiments with the tritiated substrate, the inseparability of the radioactivity peaks corresponding to the tritiated forms of 7 and 8 did not allow an assessment of their relative rate of formation; however, a value of 6:1 for this ratio was cleanly evidenced by the relative intensities of the corresponding 13C-NMR signals in the spectrum of the mixture generated from [U-13C5]-labeled 6. This value, which is in close agreement with earlier findings of in vivo and in vitro experiments (19, 20), must represent the outcome of a kinetic control, as demonstrated by the fact that it shifted to the expected equilibrium value of 3:7 (29) on treatment of the solution with the recombinant isopentenyl diphosphate isomerase from E. coli. Not surprisingly, a mixture containing IspG protein, IspH protein, 14C-labeled 5, and photoreduced deazaflavin afforded predominantly a radioactive peak with a retention volume of 7/8. In keeping with the results of previously reported in vivo experiments (19), this finding confirms that the IspH protein operates at a higher rate than the IspG protein.
The two reactions catalyzed by the IspG respectively IspH proteins are reductive processes involving the overall transfer of two electron equivalents and resulting in the cleavage of nonactivated C—OH bonds. The bulk of the now available evidence is consistent with the idea that the two proteins are endowed with prosthetic groups that are able to accept single electrons from an appropriate external source and transfer them stepwise to the respective substrates; there is solid evidence that in the case of IspG the oxygen-sensitive prosthetic group is identical with a [4Fe-4S]2+ cluster (25) and convincing, albeit less direct evidence that a similar group is involved in catalysis by the IspH protein. This, in turn, implies the involvement of radical intermediates in each of the reactions catalyzed by the two proteins. As far as the IspG protein is concerned, such a requirement rules out the previously considered alternative of a purely ionic mechanism patterned after the mode of action of vitamin K epoxyquinone reductase (16). The postulated analogy with anaerobic ribonucleotide reductase (16, 18) is invalidated by the demonstrated absence of S-adenosylmethionine as a radical initiator, and the suggestion of a resemblance with the mechanism of ascarylose biosynthesis (18) must be considered irrelevant, inasmuch as in the latter process the reductive elimination of the critical hydroxy group rests entirely on ionic steps catalyzed by the essential pyridoxamine cofactor and the radical part of the reaction deals exclusively with the subsequent stepwise reduction of the oxidized cofactor (for review, see ref. 37).
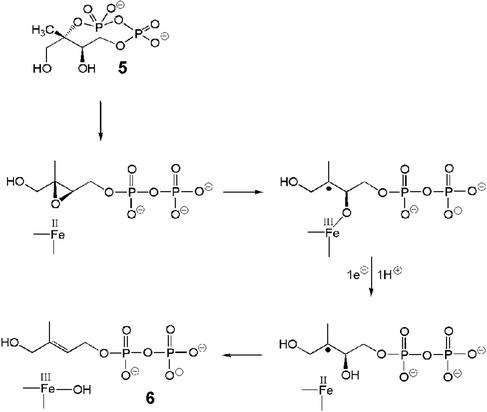
The well documented ability of the reduced form of a synthetic [4Fe-4S]2+ cluster to deoxygenate epoxides to the corresponding olefins (38) suggests that the IspG-catalyzed reaction may take place as indicated in the simplified hypothetical scheme of Fig. 5, in which the epoxide derived from 5 by hydroxyl-assisted solvolysis of the cyclopyrophosphate ring acts as a substrate for the reductive process. An additional bonus of this scheme with respect to an alternative mechanistic proposal (25) is that the geometry of the epoxide intermediate, dictated by the configuration of the two chiral centers in the precursor, is closely related to that of the final product, thus providing a welcome chemical rationale for the exclusive formation of the (E) isomer.
Figure 5.
Proposed mechanism for the conversion of 5 into 6 catalyzed by IspG protein.
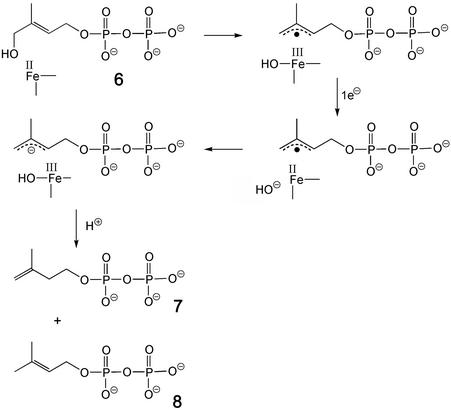
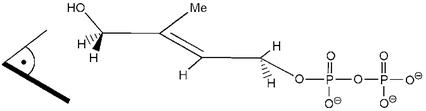
As for the reaction catalyzed by the IspH protein, all of the available experimental evidence is compatible with the operation of the scheme outlined in Fig. 6, which can be considered as a biological counterpart of the Birch reduction of allylic alcohols with lithium in liquid ammonia. It is relevant to note that the enzyme can control the outcome of the reaction by enforcing on the substrate the specific conformation illustrated in Fig. 7, which for obvious stereoelectronic reasons favors the elimination of the hydroxyl group while not allowing the alternative elimination of pyrophospate, the otherwise better leaving group.
Figure 6.
Proposed mechanism for the conversion of 6 into 7/8 catalyzed by IspH protein.
Figure 7.
Postulated conformation of 6 at the active site of the IspH protein. The indicated reference plane corresponds to the nodal plane of the olefinic bond.
Supplementary Material
Acknowledgments
We thank Prof. Andrée Marquet for a generous gift of the deazaflavin, Fritz Wendling and Katrin Gärtner for skillful assistance, and Angelika Werner for expert help with the preparation of the manuscript. We thank the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Hans-Fischer Gesellschaft for support. Financial support by Novartis International AG, Basel (to D.A.) is gratefully acknowledged.
Abbreviations
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
Note Added in Proof
Results matching the ones described in this work have been published in the meantime by the Jomaa group for the IspG protein from Thermus thermophilus (39) and for the IspH protein from Aquifex aerolicus (40).
References
- 1.Bach T J. Lipids. 1995;30:191–202. doi: 10.1007/BF02537822. [DOI] [PubMed] [Google Scholar]
- 2.Bloch K. Steroids. 1992;57:378–382. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- 3.Bochar D A, Friesen J A, Stauffacher C V, Rodwell V W. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 15–44. [Google Scholar]
- 4.Qureshi N, Porter J W. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 47–94. [Google Scholar]
- 5.Slater E, MacDonalds J S. Drugs. 1988;36:72–82. doi: 10.2165/00003495-198800363-00016. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz M, Arigoni D. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 367–399. [Google Scholar]
- 7.Rohmer M. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 45–68. [Google Scholar]
- 8.Eisenreich W, Schwarz M, Cartayade A, Arigoni D, Zenk M H, Bacher A. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 9.Sprenger G A, Schörken U, Wiegert T, Grolle S, deGraaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk M H. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lüttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr C A, Fellermeier M, Sagner S, Zenk M H, Bacher A, Eisenreich W. Proc Natl Acad Sci USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herz S, Wungsintaweekul J, Schuhr C A, Hecht S, Lüttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk M H, Bacher A, Rohdich F. Proc Natl Acad Sci USA. 2000;97:2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohdich F, Kis K, Bacher A, Eisenreich W. Curr Opin Chem Biol. 2001;5:535–540. doi: 10.1016/s1367-5931(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 16.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Proc Natl Acad Sci USA. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck F, Jomaa H. FEBS Lett. 2001;499:37–40. doi: 10.1016/s0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 18.Wolff M, Seemann M, Grosdemange-Billiard C, Tritsch D, Campos N, Rodriguez-Concepcion M, Boronat A, Rohmer M. Tetrahedron Lett. 2002;43:2555–2559. [Google Scholar]
- 19.Rohdich F, Hecht S, Gärtner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W. Proc Natl Acad Sci USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adam P, Hecht S, Eisenreich W, Kaiser J, Gräwert T, Arigoni D, Bacher A, Rohdich F. Proc Natl Acad Sci USA. 2002;99:12108–12113. doi: 10.1073/pnas.182412599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuhr C A, Hecht S, Kis K, Eisenreich W, Wungsintaweekul J, Bacher A, Rohdich F. Eur J Org Chem. 2001;2001:3221–3226. doi: 10.1021/jo015890v. [DOI] [PubMed] [Google Scholar]
- 22.Hecht S, Amslinger S, Jauch J, Kis K, Trentinaglia V, Adam P, Eisenreich W, Bacher A, Rohdich F. Tetrahedron Lett. 2002;43:8929–8933. [Google Scholar]
- 23.Davisson V J, Woodside A B, Neal T R, Stremler K E, Muehlbacher M, Poulter C D. J Org Chem. 1986;51:4768–4779. [Google Scholar]
- 24.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann M, Bui B T S, Wolff M, Tritsch D, Campos N, Boronat A, Marquet A, Rohmer M. Angew Chem Int Ed. 2002;41:4337–4339. doi: 10.1002/1521-3773(20021115)41:22<4337::AID-ANIE4337>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Gao W, Loeser R, Rachke M, Dessoy M A, Fulhorst M, Alpermann H, Wessjohann L A, Zenk M H. Angew Chem Int Ed. 2002;41:2604–2607. doi: 10.1002/1521-3773(20020715)41:14<2604::AID-ANIE2604>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 27.McCaskill D, Croteau R. Anal Biochem. 1993;215:142–149. doi: 10.1006/abio.1993.1566. [DOI] [PubMed] [Google Scholar]
- 28.Street I P, Christensen D J, Poulter C D. J Am Chem Soc. 1990;112:8577–8578. [Google Scholar]
- 29.Külzer R, Pils T, Kappl R, Hüttermann J, Knappe J. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 30.Duin E C, Lafferty M E, Crouse B R, Allen R M, Sanyal I, Flint D H, Johnson M K. Biochemistry. 1997;36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 31.Nakamaru-Ogiso E, Yano T, Ohnishi T, Yagi T. J Biol Chem. 2002;277:1680–1688. doi: 10.1074/jbc.M108796200. [DOI] [PubMed] [Google Scholar]
- 32.Johnson M K, Robinson A E, Thomson A J. In: Iron-Sulfur Proteins. Spiro T D, editor. New York: Wiley Interscience; 1982. pp. 367–406. [Google Scholar]
- 33.Conradt H, Hohmann-Berger M, Hohmann H P, Blaschkowski H P, Knappe J. Arch Biochem Biophys. 1984;228:133–142. doi: 10.1016/0003-9861(84)90054-7. [DOI] [PubMed] [Google Scholar]
- 34.Bianchi V, Eliasson R, Fontecave M, Mulliez E, Hoover D M, Matthews R G, Reichard P. Biochem Biophys Res Commun. 1993;197:792–797. doi: 10.1006/bbrc.1993.2548. [DOI] [PubMed] [Google Scholar]
- 35.Méjan A, Bui B T S, Florentin D, Ploux O, Izumi Y, Marquet A. Biochem Biophys Res Commun. 1995;217:1231–1237. doi: 10.1006/bbrc.1995.2900. [DOI] [PubMed] [Google Scholar]
- 36.Massey V, Hemmerich P. Biochemistry. 1978;17:9–17. doi: 10.1021/bi00594a002. [DOI] [PubMed] [Google Scholar]
- 37.He X, Liu H-W. Curr Opin Chem Biol. 2002;6:590–597. doi: 10.1016/s1367-5931(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 38. Itoh, T., Nagano, T., Sato, M. & Hirobe, M. (1989) Tetrahedron Lett.30,
- 39.Kollas A-K, Duin E C, Eberl M, Altincicek B, Hintz M, Reichenberg A, Henschker D, Henne A, Steinbrecher J, Ostrovsky D N, et al. FEBS Lett. 2002;532:432–436. doi: 10.1016/s0014-5793(02)03725-0. [DOI] [PubMed] [Google Scholar]
- 40.Altincicek B, Duin E C, Reichenberg A, Hedderich R, Kollas A-K, Hintz M, Wagner S, Wiesner J, Beck E, Jomaa H. FEBS Lett. 2002;532:437–440. doi: 10.1016/s0014-5793(02)03726-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.