Abstract
Array comparative genomic hybridization (aCGH) and microarray expression profiling were used to subclassify DNA and RNA alterations associated with differential response to chemotherapy in ovarian cancer. Two to 4 Mb interval arrays were used to map genomic imbalances in 26 sporadic serous ovarian tumors. Cytobands 1p36, 1q42-44, 6p22.1-p21.2, 7q32.1-q34 9q33.3-q34.3, 11p15.2, 13q12.2-q13.1, 13q21.31, 17q11.2, 17q24.2-q25.3, 18q12.2, and 21q21.2-q21.3 were found to be statistically associated with chemotherapy response, and novel regions of loss at 15q11.2-q15.1 and 17q21.32-q21.33 were identified. Gene expression profiles were obtained from a subset of these tumors and identified a group of genes whose differential expression was significantly associated with drug resistance. Within this group, five genes (GAPD, HMGB2, HSC70, GRP58, and HMGB1), previously shown to form a nuclear complex associated with resistance to DNA conformation-altering chemotherapeutic drugs in in vitro systems, may represent a novel class of genes associated with in vivo drug response in ovarian cancer patients. Although RNA expression change indicated only weak DNA copy number dependence, these data illustrate the value of molecular profiling at both the RNA and DNA levels to identify small genomic regions and gene subsets that could be associated with differential chemotherapy response in ovarian cancer.
Keywords: cisplatin, taxol, gene amplification, gene deletion, microarray data mining
Introduction
Ovarian cancer is the second most frequently diagnosed gynecologic malignancy, and causes more deaths than any other cancer of the reproductive system. The lack of reliable methods of early detection and the absence of specific symptoms result in late-stage diagnosis in 70% of patients. Although initial response rates to conventional chemotherapy among advanced stage patients are high, resistance to chemotherapy remains the primary factor accounting for the low 5-year survival in this patient population [1].
Ovarian cancer chemotherapy most commonly involves a first-line combination of platinum-based compounds plus paclitaxel following cytoreductive surgery. Response to chemotherapy varies among patients, and initial treatment response is often the most important consideration in choosing second-line therapies. The role of CA 125 serum levels as a surrogate marker to assess chemotherapy response is well established (reviewed in Ref. [2]). Both the rate of decline as well as the absolute value of CA 125, determined after the first courses of chemotherapy, are generally considered predictors of the final clinical response [3].
Most investigations of drug resistance in ovarian cancer have used anticancer drugs in vitro to select for subclones of cell lines with resistance to the selected agent [4–9]. A disadvantage of these approaches is that the cultured cells used are often genomically unstable and may have acquired in vitro genetic and epigenetic alterations that are not representative of in vivo conditions. In addition, such models primarily address acquired drug resistance, and do not provide direct insights into the expression and genomic alterations associated with intrinsic drug resistance.
In recent years, cytogenetic study of solid tumors has been directed toward the identification of recurrent chromosomal rearrangements and patterns of copy number imbalance that may pinpoint genomic regions involved in cancer initiation, progression, drug resistance, and patients' outcome [10,11]. Molecular cytogenetic methods such as spectral karyotyping and comparative genomic hybridization (CGH) have provided useful insights concerning genomic alterations in ovarian cancer [12,13]. However, because metaphase CGH has a resolving power of 10 to 20 Mb [14], it has not been possible to determine genomic imbalance patterns within cytobands. Recently, genomic and cDNA arrays (reviewed in Ref. [15]) have provided more detailed maps of genomic copy number alterations in tumors and, in due course, will provide comprehensive maps of genomic imbalance in a variety of tumors [16–18]. Moreover, high-resolution maps of copy number imbalance are now being integrated with expression profile data to identify clinically relevant subsets of genes based on concomitant alterations at the DNA and RNA levels [19–23]. Microarray expression profiling has been utilized in a number of recent studies in ovarian cancer (reviewed in Ref. [24]). However, no study to date has performed parallel microarray expression and array comparative genomic hybridization (aCGH) analyses to address genomic imbalance and concurrent expression alterations associated with intrinsic drug resistance in ovarian cancer.
Materials and Methods
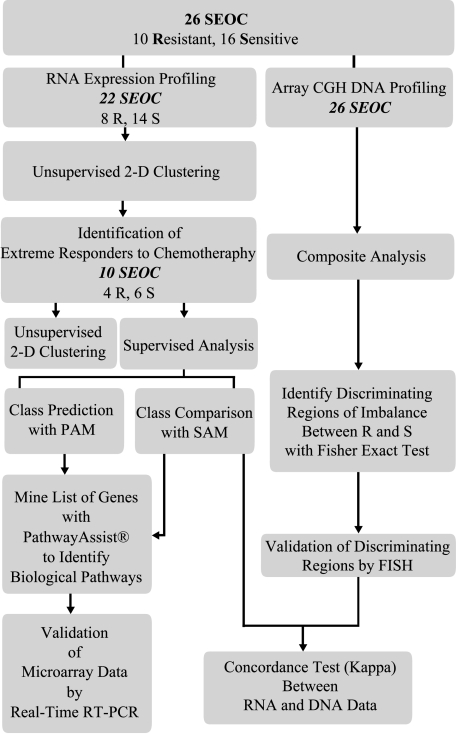
This study was designed in three phases (Figure 1). In the first phase, a 2- to 4-Mb genomic interval aCGH map of genomic imbalance in 26 serous epithelial ovarian cancer (SEOC) tumors was generated. In the second phase, statistical analysis of aCGH data sets was used to identify cytobands in which imbalance was associated with drug resistance. In the third phase, gene expression profiles were obtained from a subset of tumors, patterns of gene expression associated with drug response were identified, and a concordance analysis of the relationship between genomic imbalance and expression levels was performed. Finally, expression microarray prediction analysis was carried out to identify a subset of classifier genes that could predict chemotherapy response in ovarian cancer patients.
Figure 1.
Flowchart of the experimental design.
SEOC Tumor Samples
Snap-frozen tumor tissue samples from 26 sporadic SEOC tumors naïve to chemotherapy were selected from the Toronto Ovarian Tissue Bank and Database. No patient included in this study had a family history of either breast or ovarian cancer. All samples were acquired according to the institutional guidelines of the Research Ethics Board. The tumor specimens selected for this study contained at least 75% tumor content as assessed by the surface area of histology slides corresponding to the snap-frozen tissues (the available clinical data are summarized in Table 1). Patients received standard SEOC chemotherapy (carboplatin + taxol). To be classified as sensitive, CA 125 values from patient tumor samples had to meet two criteria. First, the CA 125 values had to fall to below the normal reference (∼35 U/ml)within three cycles of chemotherapy, regardless of the initial baseline. Second, the values had to remain below the normal reference of a period of at least 6 months from the initiation of chemotherapy. Using these criteria within our group of samples, 16 met the criteria for sensitivity and 10 were thus classified as resistant. Due to the accepted variability of CA 125 values, especially in those classified as resistant, a subset of samples was used for a more detailed class comparison. In this group of six sensitive and four resistant samples, the resistant tumors displayed CA 125 levels that failed to decline below 50% of their original postsurgical values, whereas the selected subset of sensitive samples comparatively displayed the highest rate of decline from initial baseline [3].
Table 1.
Patient Sample Information.
| Sample Number | Stage | Grade | Surgery | Age | Classification |
| OVCA 3 | IIIB | 3 | SOPT | N/A | R |
| OVCA 8 | IIIC | 2 | SOPT | N/A | S |
| OVCA 33* | IIIB | 2 | SOPT | N/A | S |
| OVCA 38 | III | 2 | SOPT | 66 | R |
| OVCA 46* | IIIB | 3 | N/A | N/A | S |
| OVCA 93 | IIIC | 3 | SOPT | 67 | S |
| OVCA 123* | III | 3 | OPT | 59 | R |
| OVCA 130* | III | 3 | OPT | 46 | R |
| OVCA 161* | IIC | 3 | SOPT | 78 | S |
| OVCA 162* | III | 3 | SOPT | 44 | S |
| OVCA 180 | IIC | 3 | OPT | 88 | S |
| OVCA 209* | III | 1 | OPT | 46 | S |
| OVCA 237 | III | 3 | SOPT | 57 | R |
| OVCA 239 | III | 3 | OPT | 63 | S |
| OVCA 249* | III | 3 | N/A | 52 | R |
| OVCA 261 | IV | 3 | SOPT | 50 | R |
| OVCA 263 | III | 3 | OPT | 44 | S |
| OVCA 304 | III | 1 | SOPT | 57 | R |
| OVCA 329 | III | 3 | SOPT | 59 | S |
| OVCA 354 | III | 2 | SOPT | 65 | S |
| OVCA 363 | III | 3 | SOPT | 55 | S |
| OVCA 365 | III | 3 | SOPT | 68 | R |
| OVCA 371* | III | 3 | N/A | N/A | S |
| OVCA 384 | III | 3 | SOPT | N/A | S |
| OVCA 390 | III | 1 | N/A | 37 | S |
| OVCA 498* | IIIC | 3 | OPT | 52 | S |
N/A: not available; NC: no change; OPT: optimally debulked; SOPT: sub-optimally debulked.
Extreme responders to chemotherapy.
Tissue Arrays
A tissue array comprising 1-mm-diameter bores through tumor-rich areas of formalin-fixed paraffin-embedded (FFPE) tumors was designed following published methods [25] and constructed using a standard arraying device (Beecher Instruments, Sun Prairie, WI). Duplicate tissue cores from each donor block were included in the tissue microarray, and sections (5 µm) were cut from the recipient tissue array block for hematoxylin and eosin staining and interphase fluorescence in situ hybridization (FISH) analysis.
FISH
Interphase FISH was performed on unstained 5-µm sections from the FFPE tissue array using a commercially available Spectrum Green RB1 locus probe mapping to cytoband 13q14 (Vysis, Downers Grove, IL) according to the manufacturer's instructions. Slides were imaged using the Vysis Quips SmartCapture (Vysis) imaging system. The scoring criteria used for the interpretation of FISH results on the FFPE sections have been previously described [19]. Chromosomal gains were assigned when more than 10% of the nuclei exhibited more than two signals.
aCGH
Genomic DNA was obtained from all tumor samples using standard phenol chloroform extraction methods. The normal human reference DNA was comprised of an equimolar mixture of DNA derived from multiple male donors (Promega, Madison, WI). The genomic array slides were obtained from Spectral Genomics (Houston, TX) and comprised 1300 large insert clones (BACs/PACs) spaced ∼2 to 4 Mb apart. Supplier-recommended protocol was used. In brief, 2 µg each of genomic tumor and normal DNA was directly labeled with Cy3-dCTP or Cy5-dCTP (Amersham, Baie D'Urfe, Canada) using random priming. Following hybridization, the microarrays were washed using 50% formamide/2x SSC (20 minutes), 0.1% Igepal/2x SSC (20 minutes), and 0.2x SSC (10 minutes), all prewarmed to 50°C. A final wash with deionized distilled water was carried out. Air-dried microarray slides were scanned with an Axon GenePix 4000A confocal scanner, and fluorescence intensities were quantified with the GenePix Pro 3.0 software (Axon Instruments, Union City, CA). Hybridizations were carried out in duplicate with fluor reversals to ensure that labeling differences did not affect imbalance assignments. Repetitive spots showing >20% variation in their signal ratio were removed prior to value averaging. Details concerning software, normalization, and imbalance assignments have been described previously [16,18] and are available at http://www.utoronto.ca/cancyto/. The analysis software provides data in two formats: 1) the raw normalized data for each feature on the array, and 2) the feature intensity data represented as significant gain (two baseline standard deviations) and loss per individual experiment. This second output was the primary analysis format used in the aCGH portion of the study. To compensate for possible interexperimental variability, data were normalized to show areas of significant gain or loss in relation to each given experiment. Individual array features were assigned a positive value for significant gain and a negative value for significant loss based on 2 SD from a baseline determined for each individual experiment. The baseline threshold for each experiment is determined by the software using the largest chromosomal region of contiguous clones having the smallest deviation in their intensity ratios. Spots with fluorescence intensity ratios of greater than ±2 SD threshold are assigned as copy number imbalance and given a score of +1 for gain and -1 for loss.
For group comparisons, the differences in log2 ratios as well as the Fisher exact test were used to determine whether there was any significant gain or loss of genomic content within particular cytobands between resistant and sensitive tumors. The Fisher exact test utilized three categories (gain, loss, and no change), with the null hypothesis that the relative proportions of each of the three imbalance categories would be expected to be the same in both groups. The statistical package S-Plus was used for these group comparisons. We reported uncorrected P values and used the permutation-based stepdown method to correct the P values for multiple comparisons [26].
Expression Microarrays
RNA was extracted using Trizol (Invitrogen Canada, Burlington, Ontario, Canada). RNA quality and concentration were verified using an Agilent Bioanalyzer (Agilent BioTechnologies, Palo Alto, CA). High-quality RNA was obtained from 22 of 26 tumors. Standard optimized protocols and a full description of the cDNA arrays used in this study can be found at the University Health Network (UHN) Microarray Centre (http://www.microarrays.ca). Ten micrograms of ovarian tumor total RNA or Human Universal Reference (HUR) RNA (Stratagene, La Jolla, CA) was reverse-transcribed with Superscript II reverse transcriptase (Invitrogen Canada) while incorporating Cy3-dCTP or Cy5-2dCTP (NEN, Boston, MA). The fluorescently labeled cDNA were cohybridized overnight at 37°C to human 19K UHN microarrays comprising 19,200 sequence-verified cDNA fragments spotted in duplicate. Each of the 22 samples was assayed with dye reversal microarray hybridizations (to control for potential labeling differences) for a total of 44 hybridizations. Microarrays were scanned by a confocal laser reader (ScanArray 4000; Packard BioScience, Meriden, CT) after stringent washes. Quantification was carried out using GenePix Pro 3.0 (Axon Instruments). Low-quality spots were filtered using GenePix Pro 3.0 and by visual examination of the images. Paired dye reversal correlations were examined by unsupervised cluster analysis. Samples displaying correlation less than .65 were repeated (data not shown).
Expression Microarray Data Analysis
Data warehousing, filtering, and normalization were performed using the GeneTraffic software (version 2.7, Iobion; Stratagene). Hybridizations were annotated according to the Minimum Information About a Microarray Experiment (MIAME) guidelines (http://www.mged.org/Workgroups/MIAME/miame.html and Ref. [27]). The initial data set was filtered to exclude spots flagged in the quantification process, spots whose raw intensity was less than the local background in either one of the two channels, spots that had an intensity-to-background ratio of less than 2, and spots whose raw intensity was less than 500. Locally Weighted Scatter Plot Smoother (LOWESS) normalization by subarray (for background, see http://www.stat.berkeley.edu/users/terry/zarray/Html/normspie.html and GeneTraffic 2.7 Manual, Iobion; Stratagene) was used for normalization between the arrays. Expression array data are available at http://www.utoronto.ca/cancyto/OVCA2004NEO/.
Unsupervised two-dimensional hierarchical clustering was carried out as described in Ref. [28], using Cluster 2.01 available at http://rana.lbl.gov/EisenSoftware.htm, on gene expression values that were present in at least 80% of the tumors (10,806 genes). Gene expression ratios were median-centered across all samples and arrays before agglomerative average linkage clustering using uncentered Pearson correlation. All observations for a given item were weighted equally. Clustering results were visualized using the Treeview software available at http://rana.lbl.gov/EisenSoftware.htm. Significance analysis of microarrays (SAM) (Ref. [29] and http://www-stat.stanford.edu/∼tibs/PAM/) and prediction analysis of microarrays (PAM) (Ref. [30] and http://www-stat.stanford.edu/∼tibs/PAM/) were performed using the software available and published methods.
Validation by Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Two micrograms of total RNA from six sensitive and four resistant ovarian tumors classified as extreme responders according to the patients' CA 125 profiles was reverse-transcribed in a 100-µl reaction mixture comprising 5.5 mM MgC12, 500 µM of each dNTP, 2.5 µM random hexamers, 0.4 U/µl RNase inhibitor, and 3.125 U/µl MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA) under the following conditions: 25°C for 10 minutes, 48°C for 30 minutes, and 95°C for 5 minutes. Real-time relative quantitative PCR was performed in triplicate using the ABI PRISM 7900HT Sequence Detection system (Applied Biosystems) according to the manufacturer's instructions. A subset of genes was chosen for validation using commercially available Assays-on-Demand probe primer sets with provided master mix (Applied Biosystems). The following PCR conditions were used: 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Human cyclophilin A was used as an endogenous control because it resulted in minimum variation throughout the samples and has been previously used to validate cancer microarray expression data by real-time RT-PCR [31]. The initial copy numbers of RNA targets can be quantified using real-time PCR analysis based on threshold cycle (Ct) determinations. Ct is defined as the cycle at which a statistically significant increase in fluorescence (above background signal contributed by the fluorescence-labeled oligonucleotides within the PCR reaction) is detected. The threshold cycle is inversely proportional to the log of the initial copy number. The Ct value of human cyclophilin A was subtracted from each Ct value of OVCA or HUR sample for normalization and the ratio of OVCA tumor:HUR RNA expression was calculated so that real-time RT-PCR and microarray data could be compared.
Results
In this study, aCGH analysis improved the resolution of such regions including 9q21.11-q33.1 and 11p15.1-pter, as well as identified novel regions of loss at 15q11.2-q15.1 and 17q21.32-q21.33 that have not been reported in ovarian cancer. When imbalance profiles of resistant tumors were compared to sensitive tumors, 13 regions of the genome were strongly associated with differentiating responses. Parallel expression analysis by cDNA microarrays revealed a nuclear complex comprised of GAPD, HMGB2, HSC70, GRP58, and HMGB1 whose RNA levels were lower in the resistant tumors in comparison to the sensitive group.
Overall aCGH Analysis of 26 SEOC Tumors
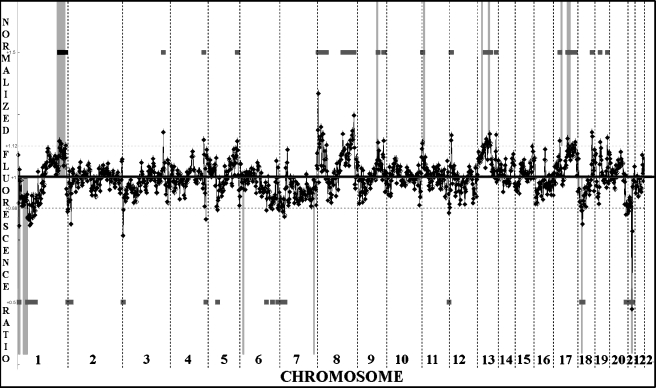
The patterns of genomic imbalances of DNA overrepresentation and underrepresentation at 2- to 4-Mb intervals in 26 SEOC tumors were identified by aCGH (Figure 2). All imbalance data from individual aCGH profiles of each tumor are published as supporting information at http://www.utoronto.ca/cancyto/OVCA2004NEO/. Losses at 1p, 4q, 6q, 8p, 9q, 13q, 16q, 17p, and 18q were present. Gains at 1q, 3q, 8q, 12p, and 20q were also detected, but no focal high copy number gene amplification was evident within this study group. To validate the imbalances detected at 13q14 by aCGH, interphase FISH analysis was performed using a 13q14-specific probe (RB1 gene) (data not shown). In 17 of 23 samples studied by interphase FISH, imbalances were in agreement with aCGH. For three samples, alterations in ploidy levels or cellular heterogeneity within tissue sections were identified. The remaining three samples could not be scored as a result of poor signal intensity.
Figure 2.
Summary of all aCGH findings using 26 SEOC samples. Overall gains and losses as determined by mean values for individual features are shown to the right of each chromosome ideogram as green and red bars, respectively. In this analysis, closely linked BACs that consistently exhibited fluorescence intensities that deviated by 2 SD or more were used to generate the average imbalance profile (traced in red).
DNA Copy Number Changes Associated with Differential Treatment Response
The samples were divided into sensitive and resistant groups, as described earlier, for a more detailed analysis of the patterns of genomic imbalances associated with differential responses to chemotherapy (Figure 3). Based on the number of BAC clones subject to imbalance, in comparison to the total number of clones analyzed, a group analysis of the overall percentage of the genome altered indicated that the resistant group had an increased level of genomic imbalance (8.3% gain, 2.4% loss) compared to the sensitive group (4.4% gain, 1.1% loss) (data not shown). Moreover, consistent with this elevated percentage of genomic change, twice as many imbalances were identified in the resistant group (55 losses/gains) compared to the sensitive group (28 losses/gains). The Fisher exact test was used to compare the resistant and sensitive groups in three categories (gain, loss, and no change) to determine which contiguous genomic regions were statistically concordant with differential treatment response (Table 2, Figure 3). Three particular regions of imbalance are identified as 13q12.2-13q13, 1p36.33, and 17q11.2.
Figure 3.
Whole genome plot of the relative difference in normalized log2 average ratios between the 10 resistant and 16 sensitive samples. Positive (above baseline) and negative (below baseline) deflections of the profile indicate the mean overrepresentation and underrepresentation of the region in resistant versus sensitive samples, respectively. The vertical pale gray bars correspond to the contiguous array features differentially identified using the Fisher exact test (see Table 2), with bars above the profile indicating overrepresentation and bars below indicating underrepresentation. Horizontal dark gray bars above (gain) and below (loss) the profile highlight the features with fluorescence intensity ratios greater than threshold.
Table 2.
Fisher Exact Test: Differential Cytoband Regions Between Resistant and Sensitive Groups.
| Cytoband | Mb Size (Mb Range) | P Range* | Gene of Interest within Cytoband (Cytoband)† | Relative Copy Nb Change (R/S)‡ | Copy Nb-Resistant Versus Normal§ | Copy Nb-Sensitive Versus Normal§ |
| 1p36.33 | 0.515 (1.164–1.679) | 0.049–0.056 | TP73 (1p36.3) | U | - | NC |
| 1p36.13 | 1.448 (19.705-18.257) | 0.0009–0.04 | U | - | NC | |
| 1q42-44 | 20.518 (222.771-243.289) | 0.001–0.06 | LGALS8 (1q43) | O | + | NC |
| 6p22.1-p21.2 | 11.491(28.342–39.833) | 0.019-0.09 | BAK1 (6p21.31) | U | NC | + |
| 7q32.1-q34 | 11.873 (126.656–138.529) | 0.012–0.029 | BRAF (7q34) | U | NC | + |
| 9q33.3-q34.3 | 13.285 (122.613–135.898) | 0.006–0.09 | O | NC | - | |
| 11p15.2 | 1.559 (14.206–15.765) | 0.043-0.043 | RRAS2 (11p15.2) | O | NC | - |
| 13q12.2-q13.1 | 5.22 (25.719–30.939) | 0.004–0.0062 | BRCA2 (13q12.3) | O | NC | - |
| 13q21.31 | 0.385 (61.538–61.923) | 0.025–0.046 | O | NC | - | |
| 17q11.2 | 1.421 (30.848–32.269) | 0.017–0.059 | NF1 (17q11.2) | O | NC | - |
| 17q24.2-q25.3 | 13.974 (64.589–78.563) | 0.021-0.15 | BIRC5 (17q25.3) | O | NC | - |
| 18q12.2 | 1.143 (31.407–32.550) | 0.011–0.043 | U | - | NC | |
| 21q21.2-q21.3 | 3.982 (23.885–27.867) | 0.0085–0.080 | U | - | NC | |
(+) Gain in copy number when compared to normal; (-) loss in copy number when compared to normal.
Uncorrected P values from the Fisher exact test for the features representing the region.
BAK1: BCL2-like 7 protein cell death inhibitor 1 apoptosis regulator BAK Bcl-2 homologous antagonist/killer; BIRC5: baculoviral IAP repeat-containing protein 5, survivin; BRAF: v-raf murine sarcoma viral oncogene homolog B1; BRCA2: breast cancer 2, early onset; LGALS8: lectin, galactoside-binding, soluble, 8 (galectin 8); NF1: neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, and Watson disease); RRAS2: related RAS viral (r-ras) oncogene homolog 2; TP73: tumor protein p73.
U: underrepresented copies in resistant compared to the sensitive group; O: overrepresented copies in the resistant compared to the sensitive group.
NC: no change in copy number when compared to normal.
Microarray Expression Profiling
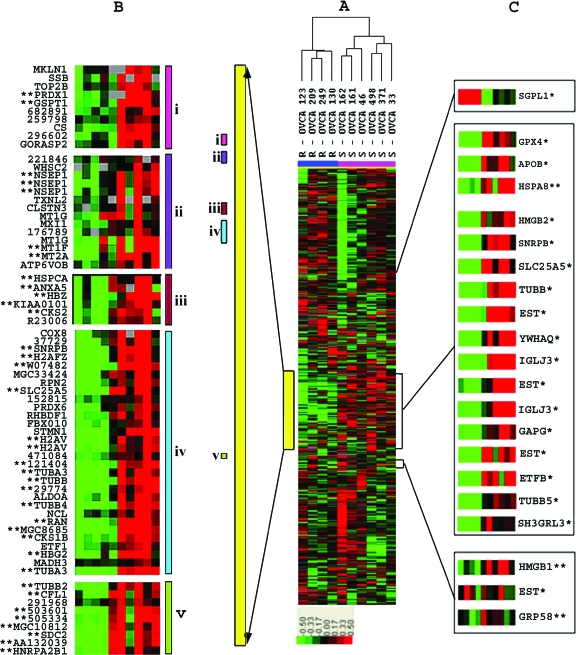
Unsupervised two-dimensional hierarchical clustering (Figure W1) was performed using expression data derived from 22 of 26 SEOC tumor cohorts classified as sensitive or resistant based on their CA 125 patterns. The clustering pattern observed did not clearly separate tumors based on response to chemotherapy and, consistent with these findings, supervised analysis using SAM only identified a limited number of genes differentially expressed in this group comparison (data not shown). A subset of 10 tumor samples was then selected from patients exhibiting the most extreme differences in CA 125 response. Unsupervised two-dimensional hierarchical clustering (Figure 4) clearly stratified this subset into a resistant and a sensitive group. A cluster of 1301 genes (highlighted in yellow) largely overlapped with those of a similar-sized gene cluster apparent in the hierarchical clustering performed on the complete sample cohort (highlighted in yellow in Figure W1).
Figure 4.
Analysis of the 10 extreme responders using unsupervised hierarchical clustering. (A) The relative expression patterns of genes that are color-coded in red (up), green (down), black (no change), or grey (data missing) clearly stratifies the sensitive (S; pink) and resistant (R; blue) samples into two major nodes. A major gene cluster that includes most genes identified by SAM analysis is highlighted with a vertical yellow bar. (B) A magnified view of subclusters that include large numbers of genes belonging to functional categories of 1) nucleus/DNA binding; 2) nucleus/metal ion binding; 3) cell cycle/cyclin-dependent protein kinase; 4) microtubule/cytoskeleton; or 5) nucleus/actin cytoskeleton engineering. (C) Relative position and expression patterns of genes identified by PAM analysis. Genes identified by PAM analysis are indicated by an asterisk (*). Genes identified only by SAM analysis are indicated by two asterisks (**).
Identification of Differentially Expressed Genes Associated with Treatment Response
The large discriminating gene cluster identified in Figure 4 contains a majority of the statistically significant expression changers identified by SAM analysis, and this large cluster contains child nodes with a preponderance of genes involved in: 1) nucleus/DNA binding; 2) nucleus/metal ion binding; 3) cell cycle/cyclin-dependent protein kinase; 4) microtubule/cytoskeleton; and 5) nucleus/actin cytoskeleton engineering.
Class comparison by statistical supervised analysis using SAM identified 173 clones (corresponding to 123 unique identified genes), which were statistically differentially expressed between the two classes with a fold change (FC) difference of at least 2 and a false discovery rate (FDR) <1%. The complete list of differentially expressed genes identified is available in supplementary materials (Table W1).
Microarray Prediction Analysis
We employed the “nearest shrunken centroid” methodology [30] that employs leave-one-out cross validation (LOOCV) to identify the most relevant classifier genes capable of predicting chemotherapy resistance in SEOC patients. When applied to the present expression data set from the 10 extreme cases, the PAM algorithm identified a set of 22 genes and ESTs that could predict with 100% accuracy the class of the test sample during LOOCV on this patient sample set. The complete list of these clones is presented in Table 3.
Table 3.
List of Genes Identified by PAM Analysis That Discriminate between Resistant and Sensitive Groups.
| Clone ID or Accession Number* | Fold Change† | Name | Symbol | Cytoband |
| Up‡ | ||||
| BM976649 | 2.01892 | Sphingosine-1-phosphate lyase 1 | SGPL1 | 10q21 |
| Down§ | ||||
| 162892 | 10.39 | Highly similar to Ig lambda chain C regions | N/A | N/A |
| 182721 | 6.24 | Immunoglobulin lambda joining 3 | IGLJ3 | 22q11.1-q11.2 |
| 135961 | 4.99 | Solute carrier family 25 | SLC25A5 | Xq24-q26 |
| 5922013 | 4.84 | Tubulin, beta polypeptide | TUBB | 6p25 |
| 300017 | 4.18 | Apolipoprotein B | APOB | 2p24-p23 |
| 4876644 | 4.08 | Tubulin, beta 5 | TUBB5 | 19p13.3 |
| 261822 | 3.79 | Homo sapiens, clone IMAGE: 5728597 | N/A | 22q11.23 |
| AL536766 | 3.72 | Tubulin, beta polypeptide paralog | MGC8685 | 6p25 |
| 5806720 | 3.31 | Glyceraldehyde-3-phosphate dehydrogenase | GAPD | 12p13 |
| 5740157 | 3.26 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein |
YWHAQ | 2p25.2-p25.1 |
| 5556148 | 3.23 | Small nuclear ribonucleoprotein polypeptides B and B1 | SNRPB | 20p13 |
| 504180 | 2.75 | H. sapiens, clone IMAGE: 5742072, mRNA | N/A | N/A |
| 267145 | 2.74 | High-mobility group box 2 | HMGB2 | 4q31 |
| 471144 | 2.64 | Electron transfer flavoprotein, beta polypeptide | ETFB | 19q13.3 |
| 270849 | 2.48 | H. sapiens-transcribed sequences | N/A | N/A |
| AL558551 | 2.48 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) | GPX4 | 19p13.3 |
| 5932330 | 2.2 | SH3 domain binding glutamic acid-rich protein like 3 | SH3BGRL3 | 1p35-p34.3 |
N/A: not available.
Four of 22 PAM clones are not listed above because they were redundant or not sequence-verified.
Relative fold change between the two classes (resistant/sensitive).
List of genes whose transcript levels are greater in the resistant group compared to the sensitive group.
List of genes whose transcript levels are lower in the resistant group compared to the sensitive group.
Data Mining
The possible functional roles of 15 of 22 clones were examined. Seven clones could not be further analyzed because ambiguity in their DNA sequence prevented their proper annotation. By applying a data mining software (PathwayAssist, Iobion Informatics; Stratagene) to the list of 22 clones identified by PAM, a nuclear complex comprised of two predictive genes identified by PAM (GAPD and HMGB2) and three additional genes identified by SAM (HSC70, GRP58, and HMGB1) was revealed. This nuclear complex was previously reported as being involved in resistance to DNA conformation-altering chemotherapeutic drugs [32].
To validate the expression findings derived from microarray analysis, the levels of expression of GAPD, HMGB2, HSC70, GRP58, and HMGB1 were measured by real-time RT-PCR. The results obtained are shown in Figure W2 and both real-time RT-PCR and microarray data agree in the direction of expression (i.e., up or down). All five genes displayed lower expression levels in resistant samples than in sensitive samples.
Correlation between Overall Pattern Gene Expression and DNA Copy Number
The level of agreement between expression and copy number changes was tested with the simple κ coefficient (Table W2). Overall comparison between expression level and aCGH copy numbers, including the no changers, showed 91% agreement. If expressed genes identified as differential in the two groups were solely considered, 8.2% of the genes agreed with copy number differences. When corrected for chance agreement for the two methods by Cohen's κ, a slight agreement between the two data sets (κ^ = 0.0230; 0 < κ^ < 1) was indicated but was not statistically significant (κ^* = 0.1316; κ^* < 1.96).
Discussion
In keeping with classic cytogenetic studies and metaphase-based CGH findings, the imbalance profiles identified in this study are complex and characterized by low-level gains and losses that affect all chromosomes except chromosome 10. The consensus pattern of imbalance was, in general, consistent with findings previously reported in ovarian cancer by metaphase CGH (reviewed in Ref. [33] and available from CGH databases: http://www.ncbi.nlm.nih.gov/; http://www.progenetix.net; http://amba.charite.de/fksch/cghdatabase/index.htm; and http://www.helsinki.fi/cmg/cgh_data.htm). aCGH permitted identification of cytoband imbalances (Table W3) within the larger genomic intervals identified in published metaphase CGH studies [34,35] and allelic imbalance findings [36,37]. For example, losses at distal 1p have been consistently reported, but aCGH localizes the minimal region of consistent loss to 1p36.11-pter in 18/26 tumors. Similarly, the recurrent gain at 6p was localized to two intervals at 6p21.1-p21.31 and 6p22.1-pter. Moreover, aCGH identified small focal genomic imbalances not previously detected by metaphase CGH, such as losses at 15q11.2-q15.1 and 17q21.32-q21.33. Imbalances such as gains at 1q11-q25.3, 3q22.3-qter, 8q11-qter, and 20q12-qter have been previously detected in a wide variety of epithelial tumors [33] including ovarian cancer, but improved resolution has been obtained in other areas including losses at 9q21.11-q33.1 and 11p15.1-pter.
Resistant tumors were found to have twice as much genomic imbalance as sensitive tumors. These data suggest that the resistant group of SEOC tumors would have a greater capacity to adapt to the selective pressures of chemotherapy by virtue of their elevated frequency of genomic rearrangement in comparison to the sensitive group.
The interval 13q12.2-13q13.1, which comprises ∼400 kb of DNA, contains the BRCA2 gene (13q12.3), a tumor-suppressor gene known to be mutated in a high percentage of hereditary ovarian cancers [38]. This region was subject to loss in 72% of the 16 sensitive tumors. It is possible that acquired loss of BRCA2 and cognate cellular repair functions could enhance susceptibility to chemotherapy. In this context, Kudoh et al. [34] investigated ovarian tumors resistant or sensitive to chemotherapy using metaphase CGH, and found that the region 13q12-14 was more often gained in the resistant ovarian cancer in comparison to sensitive tumors.
The ∼500-kb region, 1p36.33, was found to be underrepresented in resistant tumors relative to sensitive tumors (Table 2). This small region of chromosome 1 contains the TP73 gene, and it has recently been reported that deregulation and overexpression of specific p73 isoforms are associated with reduced overall survival in ovarian cancer [39]. By analogy with p53, it is plausible that genomic imbalance (loss or gain) of 1p36.33 may alter the spectrum of TP73 isoforms and influence treatment response. Similarly, the NF1 gene situated in 17q11.2 has also been studied previously in ovarian cancer cell lines [40]. The authors of this study reported that overexpression and imbalance of type II and type I isoforms of NF1 were associated with differentiation arrest and, potentially, treatment response. The observation of overrepresentation of 17q11.2 in resistant tumors may implicate a role for NF1 in chemotherapy resistance.
Unsupervised two-dimensional hierarchical clustering (Figure 4) analyses of expression microarray data using a subset of 10 tumor samples exhibiting the most extreme differences in CA 125 response revealed a large cluster of 1301 genes (highlighted in yellow) that was particularly important in the segregation of these two groups. Interestingly, the genes in this cluster largely overlap with those of a similar-sized gene cluster apparent in the hierarchical clustering performed on the complete sample cohort (highlighted in yellow in Figure W1). However, it is likely that, due to the relative heterogeneity of the 22 samples, the influence of this cluster was insufficient in completely stratifying the two groups.
Class comparison by SAM identified 173 clones, which included 123 unique identified genes (Table W1). Importantly, there was a significant difference (P < .05) in the proportion of genes involved in DNA binding, regulation of transcription, cell cycle and growth, and metal ion binding between the 173 differentially expressed genes and the gene population on the H19K microarray (data not shown). An in-depth analysis of the 123 genes differentially expressed between sensitive samples and resistant samples is beyond the scope of this study. We have focused instead on the subset of these genes that can predict chemotherapy response as described below.
We employed the “nearest shrunken centroid” methodology [30] to identify the most relevant classifier genes capable of predicting chemotherapy resistance in SEOC patients. This approach was used by Tibshirani et al. [30] on microarray data obtained by Khan et al. [41] on blue cell tumors of childhood and by Golub et al. [42] on leukemia. Tibshirani et al. demonstrated that this method was superior to both a neural network method and to the approach taken by Golub et al. [42]. The complete methodology of PAM is described in details in Ref. [30] and uses a LOOCV, a strategy particularly useful when identifying classifiers genes in smaller sample sizes [43]. When applied to the present expression data set from the 10 extreme cases, the PAM algorithm identified a set of 22 genes and ESTs that could predict with 100% accuracy the class of the test sample during LOOCV. The complete list of these clones is presented in Table 3.
Of the 22 clones identified by PAM analysis, possible functional roles for 15 of the clones were examined (seven had ambiguous DNA sequence, which prevented their proper annotation). β-Tubulin subtypes accounted for 3 of 15 discriminating identified genes differentially expressed between the sensitive and resistance groups. This is of interest because taxanes are thought to function by stabilizing microtubules—a process that eventually leads to apoptosis. Previous studies have indicated that resistance to taxanes may be a consequence of altering the relative amount of the various subtypes of β-tubulin, thereby decreasing the efficiency of microtubule stabilization by taxol (reviewed in Ref. [44]). In these studies, altered levels of the different β-tubulin isotypes were observed as a consequence of acquiring resistance to chemotherapy. Because the SEOC tumors in the present study were naïve to chemotherapy, the results presented here suggest that, before chemotherapy, some patients may already express varying levels of β-tubulin isotypes, which could result in differential response to taxol.
With PathwayAssist, a software that identifies links between the user's genes of interest based on mining Pub-Med's abstracts and public biologic databases, a nuclear complex comprised of two predictive genes identified by PAM (GAPD and HMGB2) and three additional genes (HSC70, GRP58, and HMGB1) was revealed. This nuclear complex was previously reported as being involved in resistance to DNA conformation-altering chemotherapeutic drugs [32]. In contrast, Sugimura et al. [45] reported increased in vitro expression of HSC70 in a human ovarian adenocarcinoma cell line rendered resistant to paclitaxel. Concordant with our results, Vargas-Roig et al. [46] reported decreased HSC70 levels in breast cancer patient tumors resistant to DNA-targeting drugs. In addition, it was shown that the potency of platinum-related drugs could be increased by inducing HMGB1 transcription [47].
Although HSC70, GRP58, and HMGB1 were not identified by PAM as predictive of drug resistance, HSC70 was identified by SAM as significantly differentially expressed between resistant and sensitive samples (Table W1). GRP58 and HMGB1 were also found to be differentially expressed between the two groups, but with FCs of 1.7 and 1.3, respectively, and with a slightly higher FDR (5–10%). The present finding, that all five genes are significantly downregulated in patients resistant to chemotherapy, strongly suggests their involvement in ovarian cancer resistance to cisplatin-related drugs. Although overall comparison between expression level and aCGH copy numbers, including the no changers, showed 91% agreement, when expressed genes identified as differential in the two groups were solely considered, 8.2% of the genes agreed with copy number differences. This level of agreement was not statistically significant (κ^* = 0.1316; κ^* < 1.96) by Cohen's κ method. Because the general distribution of the clones on the H19K arrays was not significantly different from the distribution of genes in the genome (based on Build 34b, version 2), lack of concordance cannot be due to poor representation of the genome in the H19K clone set. Previous studies that examined concordance between expression changes and genomic imbalance have been conflicting [23,48,49], eluding to varying degrees of epigenetic regulation in different cancers. The class comparison presented in this report was applied to a homogeneous group of advanced stage SEOC tissues that were prospectively identified as resistant or sensitive to chemotherapy. Although a subset within this study group identified a set of genes predictive for extreme nonresponsiveness, this same subset did not provide any further information at the DNA level. It is possible that partial chemotherapy response may be determined based on copy number differences at the DNA level. However, extreme nonresponsiveness may be mediated by different processes more dependent on RNA expression.
In conclusion, these data illustrate the value of molecular profiling at both the RNA and DNA levels to identify small genomic regions, and gene subsets that could be associated with differential chemotherapy response in ovarian cancer.
Supplementary Material
Acknowledgements
We are grateful to Amit Oza for helpful discussions and consultation regarding the interpretation of CA 125 responses. We thank all patients for their participation in the study.
Abbreviations
- aCGH
array comparative genomic hybridization
- FFPE
formalin-fixed paraffin-embedded
- GAPD
glyceraldehyde phosphate dehydrogenase
- GRP58
glucose-regulatory protein 58
- HMGB1
high-mobility group beta 1
- HMGB2
high-mobility group beta 2
- HSC70
heat shock cognate protein 70
- PAM
prediction analysis of microarrays
- SAM
significance analysis of microarrays
- SEOC
serous epithelial ovarian cancer
Footnotes
This study was supported by a grant award from the Ontario Cancer Research Network (funded through the Government of Ontario), Genome Canada, and the Ovarian Fashion Show Committee (Princess Margaret Hospital). M.B. was supported by the Kristi Pia Memorial Fellowship. The Toronto Ovarian Tissue Bank and Database was funded, in part, by the National Ovarian Cancer Association and the St. George's Society.
Present address: Canadian Breast Cancer Foundation - Ontario Chapter, 790 Bay Street, Suite 1000, Toronto, Ontario, M5G 1N8, Canada.
This article refers to supplementary material, which is designated by “W” (ie, Table W1, Figure W1) and is available online at www.bcdecker.com.
References
- 1.McGuire WP, III, Markman M. Primary ovarian cancer chemotherapy: current standards of care. Br J Cancer. 2003;89(Suppl 3):S3–S8. doi: 10.1038/sj.bjc.6601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verheijen RH, von Mensdorff-Pouilly S, van Kamp GJ, Kenemans P. CA 125: fundamental and clinical aspects. Semin Cancer Biol. 1999;9:117–124. doi: 10.1006/scbi.1998.0114. [DOI] [PubMed] [Google Scholar]
- 3.Rustin GJ. Use of CA-125 to assess response to new agents in ovarian cancer trials. J Clin Oncol. 2003;21:187–193. doi: 10.1200/JCO.2003.01.223. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto K, Kikuchi Y, Kudoh K, Hirata J, Kita T, Nagata I. Treatment with paclitaxel alone rather than combination with paclitaxel and cisplatin may be selective for cisplatin-resistant ovarian carcinoma. Jpn J Clin Oncol. 2000;30:446–449. [PubMed] [Google Scholar]
- 5.Yamamoto K, Kikuchi Y, Kudoh K, Nagata I. Modulation of cisplatin sensitivity by taxol in cisplatin-sensitive and -resistant human ovarian carcinoma cell lines. J Cancer Res Clin Oncol. 2000;126:168–172. doi: 10.1007/s004320050027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maubant S, Cruet-Hennequart S, Poulain L, Carreiras F, Sichel F, Luis J, Staedel C, Gauduchon P. Altered adhesion properties and alphav integrin expression in a cisplatin-resistant human ovarian carcinoma cell line. Int J Cancer. 2002;97:186–194. doi: 10.1002/ijc.1600. [DOI] [PubMed] [Google Scholar]
- 7.Rogers PM, Beale PJ, Al-Moundhri M, Boxall F, Patterson L, Valenti M, Raynaud F, Hobbs S, Johnston S, Kelland LR. Overexpression of BclXL in a human ovarian carcinoma cell line: paradoxic effects on chemosensitivity in vitro versus in vivo. Int J Cancer. 2002;97:858–863. doi: 10.1002/ijc.10132. [DOI] [PubMed] [Google Scholar]
- 8.Kumar A, Soprano DR, Parekh HK. Cross-resistance to the synthetic retinoid CD437 in a paclitaxel-resistant human ovarian carcinoma cell line is independent of the overexpression of retinoic acid receptor-gamma. Cancer Res. 2001;61:7552–7555. [PubMed] [Google Scholar]
- 9.Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, Tomida A, Funato T, Yokomizo A, Naito S, Imoto I, et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.can-3263-2. [DOI] [PubMed] [Google Scholar]
- 10.Taetle R, Aickin M, Yang JM, Panda L, Emerson J, Roe D, Adair L, Thompson F, Liu Y, Wisner L, et al. Chromosome abnormalities in ovarian adenocarcinoma: I. Nonrandom chromosome abnormalities from 244 cases. Genes Chromosomes Cancer. 1999;25:290–300. [PubMed] [Google Scholar]
- 11.Mertens F, Johansson B, Hoglund M, Mitelman F. Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res. 1997;57:2765–2780. [PubMed] [Google Scholar]
- 12.Bayani J, Brenton JD, Macgregor PF, Beheshti B, Albert M, Nallainathan D, Karaskova J, Rosen B, Murphy J, Laframboise S, et al. Parallel analysis of sporadic primary ovarian carcinomas by spectral karyotyping, comparative genomic hybridization, and expression microarrays. Cancer Res. 2002;62:3466–3476. [PubMed] [Google Scholar]
- 13.Bernardini M, Weberpals J, Squire JA. The use of cytogenetics in understanding ovarian cancer. Biomed Pharmacother. 2004;58:17–23. doi: 10.1016/j.biopha.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Parente F, Gaudray P, Carle GF, Turc-Carel C. Experimental assessment of the detection limit of genomic amplification by comparative genomic hybridization CGH. Cytogenet Cell Genet. 1997;78:65–68. doi: 10.1159/000134632. [DOI] [PubMed] [Google Scholar]
- 15.Mantripragada KK, Buckley PG, de Stahl TD, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Beheshti B, Braude I, Marrano P, Thorner P, Zielenska M, Squire JA. Chromosomal localization of DNA amplifications in neuroblastoma tumors using cDNA microarray comparative genomic hybridization. Neoplasia. 2003;5:53–62. doi: 10.1016/s1476-5586(03)80017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 18.Squire JA, Pei J, Marrano P, Beheshti B, Bayani J, Lim G, Moldovan L, Zielenska M. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- 19.Squire JA, Bayani J, Luk C, Unwin L, Tokunaga J, MacMillan C, Irish J, Brown D, Gullane P, Kamel-Reid S. Molecular cytogenetic analysis of head and neck squamous cell carcinoma: by comparative genomic hybridization, spectral karyotyping, and expression array analysis. Head Neck. 2002;24:874–887. doi: 10.1002/hed.10122. [DOI] [PubMed] [Google Scholar]
- 20.Heiskanen M, Kononen J, Barlund M, Torhorst J, Sauter G, Kallioniemi A, Kallioniemi O. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal Cell Pathol. 2001;22:229–234. doi: 10.1155/2001/981218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritz B, Schubert F, Wrobel G, Schwaenen C, Wessendorf S, Nessling M, Korz C, Rieker RJ, Montgomery K, Kucherlapati R, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- 22.Clark J, Edwards S, John M, Flohr P, Gordon T, Maillard K, Giddings I, Brown C, Bagherzadeh A, Campbell C, et al. Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer. 2002;34:104–114. doi: 10.1002/gcc.10039. [DOI] [PubMed] [Google Scholar]
- 23.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Page C, Provencher D, Maugard CM, Ouellet V, Mes-Masson AM. Signature of a silent killer: expression profiling in epithelial ovarian cancer. Expert Rev Mol Diagn. 2004;4:157–167. doi: 10.1586/14737159.4.2.157. [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Mirlacher M, Sauter G. Tissue microarrays. Biotechniques. 2004;36:100–105. doi: 10.2144/04361RV01. [DOI] [PubMed] [Google Scholar]
- 26.Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. New York: Wiley-Interscience; 1993. [Google Scholar]
- 27.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum Information About a Microarray Experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trogan E, Choudhury RP, Dansky HM, Rong JX, Breslow JL, Fisher EA. Laser capture microdissection analysis of gene expression in macrophages from atherosclerotic lesions of apolipoprotein E-deficient mice. Proc Natl Acad Sci USA. 2002;99:2234–2239. doi: 10.1073/pnas.042683999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–106. [PubMed] [Google Scholar]
- 33.Struski S, Doco-Fenzy M, Cornillet-Lefebvre P. Compilation of published comparative genomic hybridization studies. Cancer Genet Cytogenet. 2002;135:63–90. doi: 10.1016/s0165-4608(01)00624-0. [DOI] [PubMed] [Google Scholar]
- 34.Kudoh K, Takano M, Koshikawa T, Hirai , Yoshida S, Mano Y, Yamamoto K, Ishii K, Kita T, Kikuchi Y, et al. Gains of 1q21-q22 and 13q12-q14 are potential indicators for resistance to cisplatin-based chemotherapy in ovarian cancer patients. Clin Cancer Res. 1999;5:2526–2531. [PubMed] [Google Scholar]
- 35.Staebler A, Heselmeyer-Haddad K, Bell K, Riopel M, Perlman E, Ried T, Kurman RJ. Micropapillary serous carcinoma of the ovary has distinct patterns of chromosomal imbalances by comparative genomic hybridization compared with atypical proliferative serous tumors and serous carcinomas. Hum Pathol. 2002;33:47–59. doi: 10.1053/hupa.2002.30212. [DOI] [PubMed] [Google Scholar]
- 36.Launonen V, Mannermaa A, Stenback F, Kosma VM, Puistola U, Huusko P, Anttila M, Bloigu R, Saarikoski S, Kauppila A, et al. Loss of heterozygosity at chromosomes 3, 6, 8, 11, 16, and 17 in ovarian cancer: correlation to clinicopathological variables. Cancer Genet Cytogenet. 2000;122:49–54. doi: 10.1016/s0165-4608(00)00279-x. [DOI] [PubMed] [Google Scholar]
- 37.Dent J, Hall GD, Wilkinson N, Perren TJ, Richmond I, Markham AF, Bell SM, Bell SM. Cytogenetic alterations in ovarian clear cell carcinoma detected by comparative genomic hybridisation. Br J Cancer. 2003;88:1578–1583. doi: 10.1038/sj.bjc.6600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhei E, Bogomolniy F, Federici MG, Maresco DL, Offit K, Robson ME, Saigo PE, Boyd J. Molecular genetic characterization of BRCA1- and BRCA2-linked hereditary ovarian cancers. Cancer Res. 1998;58:3193–3196. [PubMed] [Google Scholar]
- 39.Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- 40.Iyengar TD, Ng S, Lau CC, Welch WR, Bell DA, Berkowitz RS, Mok SC. Differential expression of NF1 type I and type II isoforms in sporadic borderline and invasive epithelial ovarian tumors. Oncogene. 1999;18:257–262. doi: 10.1038/sj.onc.1202294. [DOI] [PubMed] [Google Scholar]
- 41.Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 42.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 43.Simon R. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br J Cancer. 2003;89:1599–1604. doi: 10.1038/sj.bjc.6601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimura M, Sagae S, Ishioka S, Nishioka Y, Tsukada K, Kudo R. Mechanisms of paclitaxel-induced apoptosis in an ovarian cancer cell line and its paclitaxel-resistant clone. Oncology. 2004;66:53–61. doi: 10.1159/000076335. [DOI] [PubMed] [Google Scholar]
- 46.Vargas-Roig LM, Gago FE, Tello O, Aznar JC, Ciocca DR. Heat shock protein expression and drug resistance in breast cancer patients treated with induction chemotherapy. Int J Cancer. 1998;79:468–475. doi: 10.1002/(sici)1097-0215(19981023)79:5<468::aid-ijc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 47.He Q, Liang CH, Lippard SJ. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc Natl Acad Sci USA. 2000;97:5768–5772. doi: 10.1073/pnas.100108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips JL, Hayward SW, Wang Y, Vasselli J, Pavlovich C, Padilla-Nash H, Pezullo JR, Ghadimi BM, Grossfeld GD, Rivera A, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 2001;61:8143–8149. [PubMed] [Google Scholar]
- 49.Platzer P, Upender MB, Wilson K, Willis J, Lutterbaugh J, Nosrati A, Willson JK, Mack D, Ried T, Markowitz S. Silence of chromosomal amplifications in colon cancer. Cancer Res. 2002;62:1134–1138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.