Abstract
During the meiotic cell cycle, a surveillance mechanism called the “pachytene checkpoint” ensures proper chromosome segregation by preventing meiotic progression when recombination and chromosome synapsis are defective. The silencing protein Dot1 (also known as Pch1) is required for checkpoint-mediated pachytene arrest of the zip1 and dmc1 mutants of Saccharomyces cerevisiae. In the absence of DOT1, the zip1 and dmc1 mutants inappropriately progress through meiosis, generating inviable meiotic products. Other components of the pachytene checkpoint include the nucleolar protein Pch2 and the heterochromatin component Sir2. In dot1, disruption of the checkpoint correlates with the loss of concentration of Pch2 and Sir2 in the nucleolus. In addition to its checkpoint function, Dot1 blocks the repair of meiotic double-strand breaks by a Rad54-dependent pathway of recombination between sister chromatids. In vegetative cells, mutation of DOT1 results in delocalization of Sir3 from telomeres, accounting for the impaired telomeric silencing in dot1.
INTRODUCTION
During eukaryotic cell division, faithful transmission of genetic information is ensured by the operation of cell-cycle checkpoints. These surveillance mechanisms arrest or delay cell cycle progression in response to defects in cellular processes, thereby preventing the initiation of late events until earlier events have been successfully completed (Hartwell and Weinert, 1989). In the mitotic cell cycle, several checkpoint controls have been characterized. For example, the “DNA damage checkpoint” responds to genome injuries by arresting the cell cycle at G1/S or G2/M, or slowing progression through S phase, thus allowing time to repair the damage and preventing the replication or segregation of damaged chromosomes (reviewed by Weinert, 1998; Lowndes and Murguia, 2000).
In Saccharomyces cerevisiae, several components of the DNA damage checkpoint have been identified. Rad9 and the Rad24 group of proteins (Rad24, Rad17, Mec3, and Ddc1) are thought to be involved in sensing damage and/or generating a signal in response to damage (reviewed by Weinert, 1998; Lowndes and Murguia, 2000). This signal is transduced through a cascade of protein kinases, including Mec1, Rad53, and Chk1, that act on downstream effectors, ultimately triggering cell cycle arrest (reviewed by Weinert, 1998; Lowndes and Murguia, 2000). A number of checkpoint proteins initially characterized in budding and fission yeasts have counterparts in mammals, demonstrating the conservation of these surveillance mechanisms (Weinert, 1998).
Meiosis is a specialized form of cell division that generates haploid gametes from diploid parental cells in sexually reproducing organisms. Proper distribution of chromosomes to the haploid progeny depends on a coordinated series of interactions between homologous chromosomes (homologs) that occur during meiotic prophase, including reciprocal genetic exchange and the intimate association of homologs in the context of the synaptonemal complex (SC) (reviewed by Roeder, 1997). As in the mitotic cell cycle, checkpoints operate in meiosis to ensure the accurate transmission of genetic information. The same checkpoint controls that arrest progression of the mitotic cell cycle in response to DNA damage, blocks in replication, or defects in spindle integrity also function in meiosis (reviewed by Bailis and Roeder, 2000a). In addition, meiotic cells possess a surveillance mechanism called the “pachytene checkpoint” that monitors events specific to meiosis, such as recombination and chromosome synapsis (i.e., SC formation), that are critical for proper meiotic chromosome segregation (reviewed by Roeder, 1997). In mutants of budding yeast, mouse, and Caenorhabditis elegans that are defective at intermediate steps in recombination and/or synapsis, the pachytene checkpoint triggers arrest at midmeiotic prophase (reviewed by Bailis and Roeder, 2000a). In the case of mouse and worm germ cells, arrest is followed by apoptotic death.
In yeast, meiotic recombination occurs concurrently with chromosome synapsis and is required for SC formation (reviewed by Roeder, 1997). Recombination is initiated by double-strand breaks (DSBs) that occur before synapsis. The breaks are rapidly processed to expose single-stranded tails that then invade homologous sequences usually in a nonsister chromatid. Strand invasion is followed by repair synthesis and branch migration to form double Holliday junctions, around the time of SC formation. Mature recombinants are produced near the end of pachytene as the SC disassembles. During meiotic recombination, a number of intermediates are formed in which DNA molecules are not intact or are interlocked; thus, any attempt to segregate chromosomes before completion of recombination would be deleterious. The pachytene checkpoint prevents meiotic nuclear division in the presence of recombination intermediates (Bailis and Roeder, 2000a).
In S. cerevisiae, dmc1 and zip1 are two well-characterized examples of mutants that undergo checkpoint-mediated arrest at pachytene. Dmc1 is a meiosis-specific homolog of the bacterial RecA strand-exchange enzyme (Bishop et al., 1992). In the dmc1 mutant, synapsis is delayed and DSBs remain unrepaired (Bishop et al., 1992; Rockmill et al., 1995). Zip1 is a major structural component of the central region of the SC (Sym et al., 1993; Sym and Roeder, 1995; Tung and Roeder, 1998; Dong and Roeder, 2000). The zip1 mutant arrests in meiosis with unsynapsed chromosomes and unresolved Holliday junctions (Sym et al., 1993; Storlazzi et al., 1996).
Several components of the pachytene checkpoint have been identified in S. cerevisiae. During meiotic recombination, DSBs and regions of single-stranded DNA occur; checkpoint proteins that respond to these types of lesions in vegetative cells also monitor recombination in meiosis. Thus, a subset of DNA damage checkpoint proteins, including Rad17, Rad24, Mec3, Ddc1, and Mec1, function in the pachytene checkpoint (Lydall et al., 1996; San-Segundo and Roeder, 1999; Hudson, San-Segundo, and Roeder, unpublished data). The pachytene checkpoint also requires the SC-associated proteins Red1 and Mek1 (Xu et al., 1997; Bailis and Roeder, 2000b; Bailis et al., 2000). Checkpoint-dependent cell-cycle arrest at pachytene is achieved by accumulation of hyperphosphorylated Swe1, which presumably inactivates Cdc28 (Leu and Roeder, 1999), and by inhibiting the activity of the Ndt80 transcription factor, which is required for transcription of the CLB1 gene encoding the major cyclin active at meiosis I (MI) (Chu and Herskowitz, 1998; Hepworth et al., 1998).
A genetic screen for pachytene checkpoint mutants uncovered a meiosis-specific protein, Pch2, required for zip1 meiotic arrest (San-Segundo and Roeder, 1999). Pch2 localizes to the ribosomal DNA region (rDNA) and this nucleolar localization depends on the silencing factor Sir2, which is also necessary for pachytene checkpoint function. Silencing is a position-dependent, gene-independent form of repressed chromatin structure that affects large chromosomal domains. In yeast, three regions are subjected to silencing: the telomeres, the HML/HMR silent mating-type loci, and the rDNA array. Whereas telomeric and HML/HMR silencing involves the silent information regulators Sir2, Sir3, and Sir4, only the Sir2 protein is required for rDNA silencing (reviewed by Lustig, 1998).
Presented here is the characterization of another protein, Dot1, identified in our screen for components of the pachytene checkpoint (San-Segundo and Roeder, 1999). The DOT1 gene was independently isolated in a screen for high-copy disruptors of telomeric silencing and shown also to affect HML/HMR and rDNA silencing (Singer et al., 1998), further implicating chromatin silencing in the pachytene checkpoint. In the absence of Dot1, the zip1 and dmc1 mutants fail to arrest; they proceed through meiosis and sporulation to produce inviable spores. In addition to its checkpoint function, Dot1 inhibits the operation of a Rad54-dependent intersister recombination pathway that repairs DSBs in the absence of Dmc1. The nucleolar Pch2 and Sir2 proteins, as well as the telomeric Sir3 protein, are mislocalized in the absence of Dot1.
MATERIALS AND METHODS
Strains and Plasmids
Yeast strain genotypes are listed in Table 1. DOT1 was cloned as an ∼6.5-kb EcoRI-BglII fragment from S. cerevisiae λ clone 5513 (ATCC 70580) into the EcoRI-BamHI sites of Bluescript SK+ to generate pSS24. DOT1 was disrupted by transformation with pSS30 (dot1::URA3) or pSS44 (dot1::TRP1). To generate the disruption plasmids, an ∼2.8-kb fragment containing DOT1 was amplified by polymerase chain reaction (PCR) by using oligonucleotides ORF26#1 (5′-GGGGGATCCAGGAACACTGAAGAACGGG-3′) and ORF26#2 (5′-GGGGGGCCCAGGTACCTGGTCCACGGCGC-3′) as primers and DNA from λ clone 5513 as template. BamHI and ApaI sites added in the primers are underlined. The PCR fragment was cut with BamHI and ApaI and cloned into the same sites of Bluescript SK+ to generate pSS25. pSS30 was constructed by replacing a HindIII fragment of pSS25 containing most of the DOT1 coding region (from nucleotides − 414 to +1373) with a HindIII fragment (from pR1333) containing URA3. pSS44 was constructed by replacing the BstBI-NheI fragment of pSS25 containing most of the DOT1 coding region (from nucleotides −109 to +1278) with a ClaI-SpeI fragment (from pSS43) containing TRP1. pSS43 was made by cloning an ∼0.85-kb EcoRI-BglII fragment containing TRP1 (but not ARS1) from YRp7 (Struhl et al., 1979) between the EcoRI and BamHI of Bluescript SK+. Both pSS30 and pSS44 were cut with BamHI and KpnI before transformation into yeast. Although dot1 mutants have been shown to be defective in transcriptional silencing of a reporter gene inserted at the HM loci (Singer et al., 1998), we have not detected a mating defect in dot1 haploids; therefore, dot1 homozygous diploids were made directly by mating the haploid parents. A rad54::LEU2 disruption plasmid (pSM31) was provided by D. Schild (Lawrence Berkeley Laboratories, Berkeley). Plasmids used for other gene disruptions have been described previously: zip1::LEU2 (Sym et al., 1993), zip1::LYS2 (Sym and Roeder, 1994), zip1::URA3 (Sym and Roeder, 1995), dmc1::ARG4 (Bishop et al., 1992), and rad24::TRP1 (Lydall and Weinert, 1997). Strains containing SIR3-HA integrated at the ura3-1 locus were provided by E. Hong and B. Rockmill (Yale University). Plasmids pSS63 and pSS64 containing DOT1-HA and DOT1-GFP, respectively, in the high-copy vector pRS424 (Christianson et al., 1992) were constructed by cloning in frame a NotI-NotI fragment carrying three copies of the hemagglutinin (HA) epitope or green fluorescent protein (GFP)-coding sequences (from plasmids p264 and p266, respectively; provided by B. Santos; Yale University) into a NotI site generated by PCR after the second codon of DOT1. Additional details of plasmid and strain constructions are available upon request. Media, sporulation conditions, and determination of the frequency of sporulation and meiotic nuclear division have been described (Sym et al., 1993; Sym and Roeder, 1994; San-Segundo and Roeder, 1999).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| BR2495 | MATa leu2-27 his4-280 trp1-1 arg4-8 thr1-1 ura3-1 ade2-1 cyh10 |
| MATα leu2-3,112 his4-260 trp1-289 ARG4 thr1-4 ura3-1 ade2-1 CYH10 | |
| MY63 | BR2495 but homozygous zip1∷LEU2 |
| DP138 | BR2495 but homozygous dot1∷URA3 |
| DP139 | BR2495 but homozygous zip1∷LEU2 dot1∷URA3 |
| MY152 | BR2495 but homozygous zip1∷URA3 |
| DP381 | BR2495 but homozygous zip1∷URA3 dot1∷TRP1 |
| DP382 | BR2495 but homozygous zip1∷URA3 dot1∷TRP1 rad54∷LEU2 |
| DP201 | BR2495 but homozygous zip1∷LEU2 PCH2-HA |
| DP285 | BR2495 but homozygous zip1∷LEU2 dot1∷TRP1 PCH2-HA |
| BR1919-2N | MATa leu2-3,112 his4-260 trp1-289 ARG4 thr1-4 ura3-1 ade2-1 CYH10 |
| MATα leu2-3,112 his4-260 trp1-289 ARG4 thr1-4 ura3-1 ade2-1 CYH10 | |
| DP365 | BR1919-2N but homozygous sir3∷TRP1 SIR3-HA∷URA3 |
| DP366 | BR1919-2N but homozygous sir3∷TRP1 SIR3-HA∷URA3 dot1∷TRP1 |
| DP185 | MATa CENIII leu2-hisG HIS4 lys2 ho∷LYS2 trp1-H3 ura3 |
| MATα CENIII∷TRP1 leu2-hisG his4-B-LEU2 lys2 ho∷lys2 trp1-H3 ura3 | |
| DP184 | DP185 but homozygous dot1∷URA3 |
| DP246 | DP185 but zip1∷LEU2 |
| zip1∷LYS2 | |
| DP371 | DP185 but zip1∷LEU2 and homozygous dot1∷URA3 |
| zip1∷LYS2 | |
| DP338 | MATa leu2 lys2 ho∷LYS2 trp1 ura3 arg4 |
| MATα leu2 lys2 ho∷LYS2 trp1 ura3 arg4 | |
| DP339 | DP338 but homozygous dmc1∷ARG4 |
| DP341 | DP338 but homozygous dmc1∷ARG4 dot1∷TRP1 |
| DP343 | DP338 but homozygous dmc1∷ARG4 dot1∷TRP1 rad54∷LEU2 |
| DP352 | DP338 but homozygous dmc1∷ARG4 rad24∷TRP1 |
| DP342 | DP338 but homozygous dmc1∷ARG4 rad54∷LEU2 |
Strains DP185, DP184, DP246, DP371, DP338, DP339, DP341, DP343, DP352 and DP342 are isogenic with SK1.
Cytology
Chromosome spreads and immunostaining of Pch2-HA, Sir2, Zip1, and Red1 were performed as described previously (San-Segundo and Roeder, 1999). To detect Sir3-HA and Dot1-HA, mouse monoclonal anti-HA antibody (HA.11; Babco, Richmond, CA) was used at 1:100 and 1:150 dilution, respectively. Rat antitubulin antibody (YOL 1/34; Sera-Lab, Crawley Down, Sussex, United Kingdom) was used at 1:100 dilution. Rabbit anti-Rad51 antibody (a gift of D. Bishop, University of Chicago) was used at 1:400 dilution. Anti-Nsr1 antibodies were described previously (San-Segundo and Roeder, 1999). To observe the Dot1-GFP fluorescent signal combined with 4′-6-diamidino-2-phenylindole (DAPI) staining in whole cells, 0.1 ml of 37% formaldehyde was added to aliquots (0.9 ml) of vegetative or sporulating cultures. After fixation for 15–20 min (mitotic cells) or 45–60 min (meiotic cells) at 30°C with rotation, cells were washed twice with phosphate-buffered saline (PBS) and incubated for 15 min in PBS containing 0.1% Triton X-100 and DAPI (2 μg/ml). Cells were washed twice with PBS, resuspended in a small volume of PBS, and observed at the fluorescence microscope (Nikon Eclipse E800, Plan Apo 100×/1.4 oil objective) equipped with an FITC HYQ filter (Chroma Technology Corporation, Brattleboro, VT) to visualize the GFP signal. Images were captured with a Sensys charge-coupled device camera (Photometrics, Tucson, AZ).
Dityrosine Fluorescence Assay
To examine dityrosine fluorescence as an indicator of sporulation, patches of cells were grown on YPAD plates and replica-plated to sporulation plates overlaid with nitrocellulose filters (Protran BA85; Schleicher & Schuell, Keene, NH). After 1–3 d incubation at 30°C (depending on strain background), dityrosine fluorescence was visualized by illuminating the plates from the top (without the lid) with 302-nm UV light. Photographs were taken using 667 Polaroid film and a Wratten gelatin filter (No. 47B; Kodak, Rochester, NY).
RESULTS
Identification of DOT1
To identify genes involved in the pachytene checkpoint in S. cerevisiae, a screen for mutations that alleviate the meiotic arrest of the zip1 mutant was performed; mutations were introduced by transformation with a transposon-mutagenized yeast genomic library (San-Segundo and Roeder, 1999). In addition to insertions in three other genes (RAD24, DDC1, and PCH2), two transposon insertions were found in the open reading frame YDR440w, which we initially called PCH1 (for pachytene checkpoint). During the course of this study, the same gene was isolated in a screen for high-copy disruptors of telomeric silencing and named DOT1 (Singer et al., 1998). Therefore, the name DOT1 is used throughout this article.
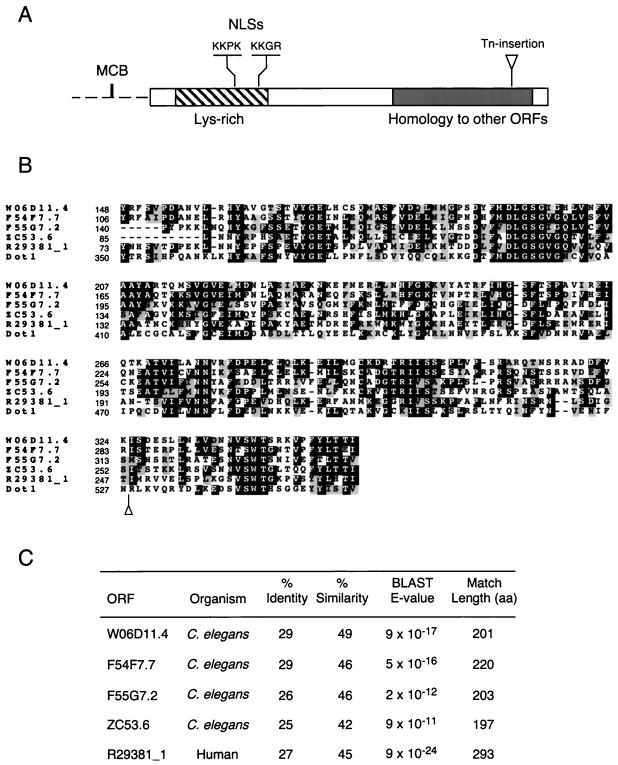
DOT1 encodes a basic (predicted pI 9.7) 582-amino-acid protein with no other homologs in the yeast genome. The N-terminal half of Dot1 is unique and has a high content of lysine residues (Figure 1A). This lysine-rich domain contains at least two consensus monopartite nuclear localization signals (Chelsky et al., 1989; Figure 1A). The C terminus of Dot1 displays significant homology over a region of ∼200 amino acids with putative proteins identified in systematic sequencing of the human and C. elegans genomes (Figure 1). One of the mutant alleles of DOT1 recovered from the screen encodes a truncated protein lacking only the last 55 amino acids, thus removing the last conserved block (Figure 1, A and B). This allele displays a phenotype indistinguishable from that of the dot1 null mutant (see below; our unpublished results).
Figure 1.
DOT1 sequence features. (A) Diagram of the Dot1 protein (rectangle) and DOT1 promoter (discontinuous line). Indicated are a lysine-rich domain (striped box; 21.5% of residues between amino acids 35 and 169 are lysines), two putative NLSs starting at positions 126 and 162, and the region of homology to human and C. elegans ORFs (gray area). The sequence ACGCGTCA at positions −172 to −165 in the DOT1 promoter corresponds to a consensus MCB element (MluI cell cycle box site is underlined). (B) Alignment of the regions of homology from S. cerevisiae Dot1, human ORF R29381_1 and C. elegans ORFs W06D11.4, F54F7.7, F55G7.2, and ZC53.6. Identities are highlighted in black and conservative substitutions in gray. Numbers at the left represent amino acid positions. Alignment and shading were performed with ClustalW 1.7 and Boxshade 3.21, respectively, at the BCM Search Launcher (http://dot.imgen.bcm.tmc.edu:9331/). In A and B, the position of the C-terminal transposon insertion recovered from the mutant screen is marked with a triangle. (C) Percentage identity and similarity of Dot1 to the human and nematode homologs over the indicated length. The significance of the homology is indicated by the BLAST E-values.
Examination of the DOT1 promoter revealed the presence of a “MluI cell cycle box” (Figure 1A). This element is the binding site for the MBF transcription factor, which activates transcription of genes at the G1/S transition in the mitotic cell cycle (Johnston and Lowndes, 1992; Koch and Nasmyth, 1994). Indeed, genome-wide analysis of gene expression during the mitotic cell cycle has shown that transcription of DOT1 is cell-cycle regulated with the peak of expression at G1 (Spellman et al., 1998). In contrast, DOT1 transcription is not regulated during the meiotic cell cycle (Chu et al., 1998).
Dot1 Is Required for Meiotic Prophase Arrest or Delay in the zip1 Mutant
To characterize the role of Dot1 in meiosis, null mutants were constructed and analyzed in diploid strains. The dot1 single mutant displays the wild-type level of sporulation (Figure 2A) and high levels of spore viability and crossing over (Table 2). Thus, Dot1 is not normally required for meiotic progression, recombination, chromosome segregation and spore formation.
Figure 2.
Dot1 is required for checkpoint-induced meiotic arrest (or delay) of zip1 and dmc1 mutants. (A) Mutation of DOT1 alleviates the meiotic arrest of zip1 in BR2495 isogenic strains. Time course of sporulation of strains BR2495 (wild type), MY63 (zip1), DP138 (dot1), and DP139 (zip1 dot1). (B) Mutation of DOT1 bypasses the meiotic delay of zip1 in the SK1 strain background. Dityrosine fluorescence, an indicator of sporulation, was examined after 1 and 2 d on sporulation plates. Strains are DP185 (wild type), DP246 (zip1), and DP371 (zip1 dot1). (C) Distribution of tetrad types. The percentages of tetrads with 4, 3, 2, 1 and 0 viable spores (4-sv, 3-sv, 2-sv, 1-sv, and 0-sv, respectively) are represented. Tetrads (389 and 513) were dissected from DP246 (zip1) and DP371 (zip1 dot1), respectively. (D) Bypass of dmc1 arrest by dot1 and effects of the rad54 mutation. Dityrosine fluorescence was examined after 2 d on sporulation plates. (%) MI + MII represents the fraction of cells that have undergone at least one meiotic nuclear division (i.e., containing two or more DAPI-stained bodies) after 24 h in liquid sporulation medium. (%) Spo. represents the fraction of cells that formed mature or immature asci. The asterisks indicate that most asci are immature. At least 300 cells were scored for each strain. Strains are DP338 (wild type), DP339 (dmc1), DP341 (dmc1 dot1), DP343 (dmc1 dot1 rad54), DP352 (dmc1 rad24), and DP342 (dmc1 rad54).
Table 2.
Spore viability and crossing over in SK1 strains
| Genotype | Spore viability (%) | Crossing over (cM) | Tetrads dissected | Strains |
|---|---|---|---|---|
| Wild type | 98.0 | 50.8 | 215 | DP185 |
| dot1 | 93.5 | 45.0 | 128 | DP184 |
| zip1 | 52.3 | 15.2 | 389 | DP246 |
| zip1 dot1 | 31.5 | 13.6 | 513 | DP371 |
Crossover data are the sum of the map distances in the MAT-CENIII and CENIII-HIS4 intervals on chromosome III. Calculations are based on the following numbers of 4-spore-viable tetrads: 203 for wild type, 112 for dot1, 129 for zip1 and 84 for zip1 dot1.
The sporulation phenotype of the zip1 mutant depends on strain background, though chromosomes fail to synapse in all zip1 strains tested (Sym et al., 1993; Sym and Roeder, 1994). In the BR2495 strain background, zip1 arrests at pachytene, and no mature recombinants are produced (Sym et al., 1993; Roeder, 1997). Deletion of DOT1 completely alleviates the zip1 meiotic arrest; the kinetics and frequency of sporulation in zip1 dot1 are similar to wild type (Table 3 and Figure 2A). However, spore viability is reduced to ∼37% (Table 3), suggesting that some or all of the defects conferred by zip1 persist in the double mutant. In fact, chromosomes do not synapse in zip1 dot1 (see DISCUSSION).
Table 3.
Sporulation and spore viability in BR2495 strains
| Genotype | Sporulation efficiencya (%) | Spore viabilityb (%) | Strains |
|---|---|---|---|
| Wild typec | 51 | 95.3 | BR2495 |
| zip1 | 3 | — | MY152 |
| zip1 dot1 | 57 | 36.9 | DP381d |
| zip1 dot1 rad54 | 54 | 13.7 | DP382e |
Sporulation efficiency was determined by counting the fraction of cells forming asci after 3 days on sporulation plates; 400 cells were scored for each strain.
Spore viability was assessed by tetrad dissection.
Viability data of wild type was presented by San-Segundo and Roeder (1999), and is based on the dissection of 256 tetrads.
Eighty-eight tetrads were dissected for DP381.
Eighty-seven tetrads were dissected for DP382.
In the SK1 strain background, the zip1 mutant sporulates, but MI is delayed and Holliday junctions persist longer than in wild type (Sym and Roeder, 1994; Storlazzi et al., 1996); however, recombination intermediates are eventually resolved, and ∼50% of the spores produced are viable (Table 2; Sym and Roeder, 1994). DOT1 is required for the meiotic delay of zip1 in the SK1 strain background; zip1 dot1 sporulates with wild-type kinetics (Figure 2B; our unpublished results). SK1 strains were used to compare recombination and chromosome segregation in the zip1 and zip1 dot1 mutants. The reduction in spore viability in zip1 (Table 2; Sym and Roeder, 1994) is mostly due to pairs of homologous chromosomes segregating to the same pole at MI because of a failure to cross over (Sym and Roeder, 1994). Relief of the meiotic delay of zip1 by a dot1 mutation leads to a further decrease in spore viability from ∼52 to ∼31% (Table 2). However, crossing over in zip1 dot1 is not significantly different from zip1, at least for the chromosome III intervals examined (Table 2). Homolog nondisjunction at meiosis I in zip1 is manifested by a nonrandom pattern of spore death (Sym and Roeder, 1994), with 0-, 2-, and 4-spore-viable tetrads predominating (Figure 2C). The same pattern of spore death is exhibited by zip1 dot1 (Figure 2C), but the proportion of 0-spore-viable tetrads is increased at the expense of tetrads with four viable spores (Figure 2C). Thus, in the absence of DOT1, zip1 fails to arrest or delay in meiosis, resulting in chromosome missegregation and the formation of aneuploid meiotic products.
Dot1 Prevents Cell Cycle Progression and Rad54-dependent Intersister Recombination in the dmc1 Mutant
In SK1 strains, the dmc1 mutant arrests in meiosis with unrepaired DSBs. To determine whether Dot1 is required for this checkpoint-dependent arrest, dmc1 dot1 double mutants were examined. Deletion of DOT1 relieves the meiotic arrest of dmc1. The dmc1 dot1 double mutant undergoes high levels of meiotic nuclear division and sporulation (Figure 2D); however, spore viability is very low (∼5% for dmc1 dot1 vs. >95% for wild type; 110 and 227 tetrads dissected, respectively).
Strikingly, a high proportion of the dmc1 dot1 meiotic products are enclosed in asci-containing mature spores similar to those of wild type, as evidenced by microscopic examination (our unpublished results) and formation of dityrosine (Figure 2D), a fluorescent component of spore walls (Briza et al., 1990). In contrast, the dmc1 rad24 double mutant proceeds through the meiotic divisions with unrepaired DSBs, resulting in the formation of nuclei with fragmented DNA and few asci with mature spores (Lydall et al., 1996), manifested by the almost complete absence of dityrosine fluorescence (Figure 2D). The fact that dmc1 dot1 forms morphologically normal spores suggests that a significant fraction of meiotic DSBs in dmc1 are repaired in the absence of Dot1.
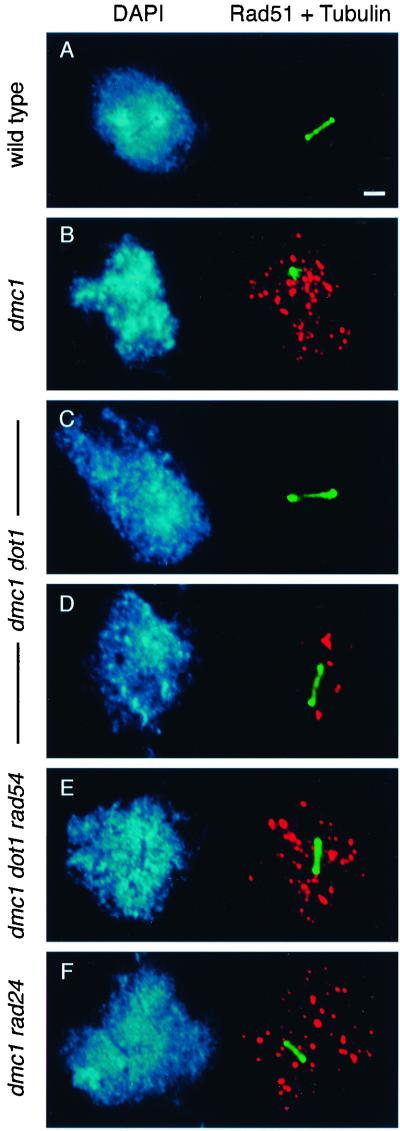
To examine the presence of recombination intermediates relative to meiotic progression, the cytological assay described by Lydall et al. (1996) was used. This method is based on the use of anti-Rad51 antibodies to detect recombination intermediates (presumably unrepaired DSBs) in combination with tubulin staining to identify cells that have initiated MI. In wild type, all Rad51 foci have disappeared when the MI spindle is formed (Lydall et al., 1996; Figure 3A). In the arrested dmc1 mutant, a single tubulin-stained focus characteristic of duplicated but unseparated spindle pole bodies is present, and numerous Rad51 foci persist (Table 4 and Figure 3B). Nuclei in the dmc1 dot1 mutant fall into three classes (Table 4). Some nuclei (38%) are similar to those of wild type (i.e., MI spindle and no Rad51 foci), indicating that all DSBs have been repaired (Figure 3C). In other nuclei (56%), a few Rad51 foci persist concomitantly with the elongated spindle (Figure 3D). Occasionally (6%), MI nuclei containing 20–35 Rad51 foci are observed (Table 4; our unpublished results). Thus, whereas ∼63 Rad51 foci accumulate in the dmc1 single mutant, only approximately four foci (on average) persist in dmc1 dot1 nuclei undergoing MI (Table 4). These results indicate that most DSBs are repaired when dmc1 arrest is bypassed by mutation of DOT1; in ∼40% of cells, all DSBs are repaired. Because most dmc1 dot1 spores are dead, this repair presumably occurs using sister chromatids as donors instead of homologs (interhomolog recombination would ensure proper disjunction at meiosis I).
Figure 3.
Cytological analysis of unrepaired DSBs and spindle elongation. Spread meiotic nuclei from strains DP338 (wild type; A), DP339 (dmc1; B), DP341 (dmc1 dot1; C and D), DP343 (dmc1 dot1 rad54; E), and DP352 (dmc1 rad24; F), stained with DAPI (left) and anti-Rad51 (red) and anti-tubulin (green) antibodies (right). Spreads were prepared after 6 h in sporulation medium. Bar, 2 μm.
Table 4.
Quantitation of Rad51 foci associated with MI spindle
| Genotype | Nuclei (%) | Rad51 foci (No.) | MI spindle | Nuclei scored | Strain |
|---|---|---|---|---|---|
| dmc1 | 100 | 62.7 ± 7.0 | − | 20 | DP339 |
| dmc1 dot1a | 38 | 0 | + | DP341 | |
| 56 | 4.0 ± 3.0 | + | 50 | ||
| 6 | 25.6 ± 6.0 | + | |||
| Averagea | 3.8 ± 6.4 | ||||
| dmc1 dot1 rad54 | 100 | 56.7 ± 10.5 | + | 22 | DP343 |
| dmc1 rad24 | 100 | 58.0 ± 12.9 | + | 20 | DP352 |
| zip1 | 100 | 25.1 ± 4.3 | − | 15 | MY152 |
| zip1 dot1 | 100 | 0 | + | 20 | DP381 |
| zip1 dot1 rad54b | 43 | 0 | + | 30 | DP382 |
| 56 | 2.3 ± 1.6 | + | |||
| Averageb | 1.3 ± 1.6 |
MI Nuclei of dmc1 dot1 are grouped into three classes. The overall number of Rad51 foci per nucleus is also represented (average).
MI Nuclei of zip1 dot1 rad54 are grouped into two classes. The overall number of Rad51 foci per nucleus is also represented (average).
Several studies indicate that meiotic DSBs in the dmc1 mutant can be repaired by a Rad54-dependent pathway of recombination between sister chromatids, if RED1 is mutated or cells are returned to growth medium (Schwacha and Kleckner, 1997; Xu et al., 1997; Arbel et al., 1999; Bishop et al., 1999). To test whether this alternative pathway is active in dmc1 dot1, the effects of a rad54 mutation were examined. The dmc1 dot1 rad54 triple mutant proceeds through the meiotic nuclear divisions (Figure 2D); however, the triple mutant is unable to make mature spores (Figure 2D; our unpublished results). Furthermore, numerous Rad51 foci are detected in dmc1 dot1 rad54 cells that have exited prophase and elongated the MI spindle (Table 4 and Figure 3E). In this respect, dmc1 dot1 rad54 is similar to dmc1 rad24, which enters MI with unrepaired DSBs (Table 4 and Figure 3F; Lydall et al., 1996). Thus, the Rad54-dependent pathway is active in dmc1 dot1, but blocking repair of DSBs by a rad54 mutation does not prevent meiotic progression.
Repair of DSBs in zip1 dot1 Is Largely Independent of Rad54
In contrast to dmc1, which accumulates DSBs, the zip1 mutant accumulates primarily unresolved Holliday junctions; a small fraction (∼10%) of DSBs also remain unrepaired (Storlazzi et al., 1996). Although the spore viability of the zip1 dot1 double mutant is significantly lower than that of wild type, the moderate fraction of viable spores produced (Tables 2 and 3) indicates that DSBs have been repaired. To test whether the repair of DSBs in zip1 dot1 depends on Rad54, a zip1 dot1 rad54 strain was analyzed. The triple mutant sporulates with the same efficiency as wild type or zip1 dot1 (Table 3) and generates mature spores (our unpublished results), suggesting that most (if not all) DSBs are repaired even in the absence of Rad54.
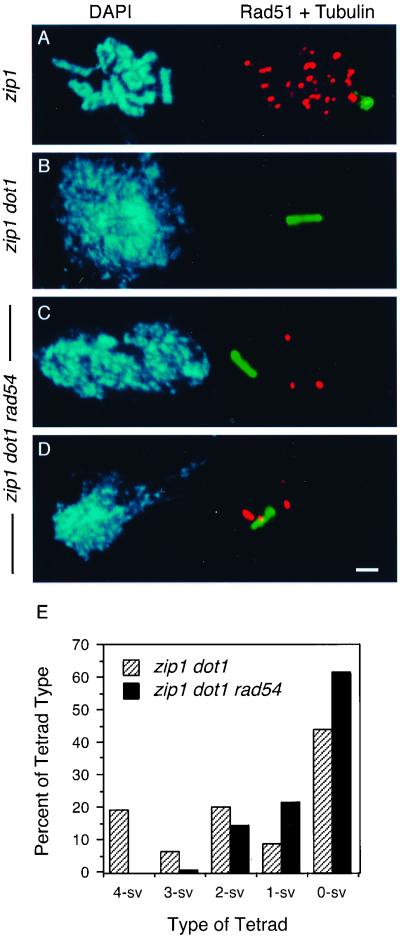
To confirm this conclusion, Rad51 and tubulin staining was carried out. In the arrested zip1 mutant, no spindle is formed and ∼25 Rad51 foci are present (Table 4 and Figure 4A). In zip1 dot1 nuclei that have entered MI, all Rad51 foci have disappeared (Table 4 and Figure 4B). In the zip1 dot1 rad54 triple mutant, two kinds of MI nuclei are found: ∼43% are like zip1 dot1 (i.e., no Rad51 foci coexist with the spindle; Table 4; our unpublished results), whereas ∼56% nuclei still contain a few Rad51 in conjunction with a spindle (Table 4 and Figure 4, C and D). Consistent with the small number of unrepaired DSBs present in zip1 dot1 rad54 (Table 4), the triple mutant exhibits a further decrease in spore viability compared with zip1 dot1 (Table 3) and loss of the distinctive 4-, 2-, and 0-spore-viable pattern of spore death (Figure 4E). Thus, unlike the situation in dmc1 dot1, only a minor fraction of the DSBs accumulated by zip1 requires Rad54 for repair when meiotic arrest is relieved by a dot1 mutation.
Figure 4.
Repair of DSBs in zip1 dot1 is largely independent of Rad54. (A–D) Cytological analysis of ongoing recombination and entry into MI, assessed by the presence of Rad51 foci and spindle elongation, respectively. Spread meiotic nuclei from strains MY152 (zip1; A), DP381 (zip1 dot1; B), and DP382 (zip1 dot1 rad54; C and D), stained with DAPI (left) and anti-Rad51 (red) and anti-tubulin (green) antibodies (right). Spread nuclei were prepared after 16 h in sporulation medium. Bar, 2 μm. (E) Distribution of tetrad types. The percentages of tetrads with 4, 3, 2, 1, and 0 viable spores (4-sv, 3-sv, 2-sv, 1-sv, and 0-sv, respectively) are represented. Tetrads (88 and 87) were dissected from strains DP381 (zip1 dot1) and DP382 (zip1 dot1 rad54), respectively.
Dot1 Is a Nuclear Protein That Associates with Chromosomes
To study the cellular localization of Dot1, the protein was tagged at the N terminus with GFP or with three copies of the HA epitope. Both fusion proteins are fully functional (our unpublished results).
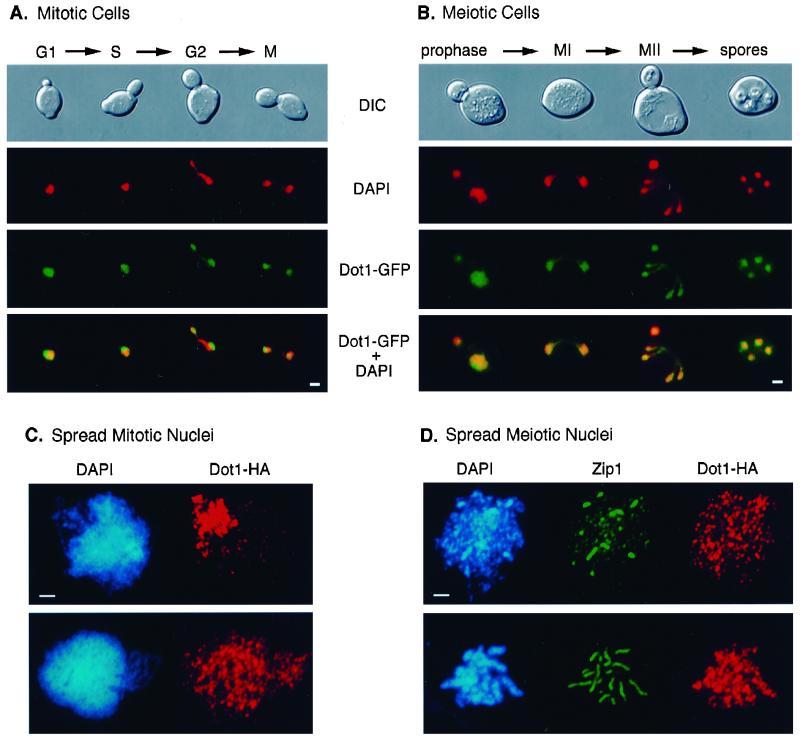
In mitotic diploid cells containing a high-copy plasmid carrying DOT1-GFP, the Dot1-GFP signal is detected in the nucleus, but is often enriched toward one side of the nucleus in a region that stains faintly with the DNA-binding dye DAPI (Figure 5A) and corresponds to the nucleolus (Pintard et al., 2000). Dot1-GFP is found in cells at different stages of the mitotic cell cycle. The same pattern of nuclear localization with nucleolar accumulation is present in vegetative cells expressing Dot1-GFP from a low-copy plasmid, suggesting that this localization is not an artifact of overexpression (our unpublished results). In cells at different stages of meiotic progression, Dot1-GFP is also found in the nucleus, but accumulation in the nucleolus is not evident until meiosis has been completed and spores are being formed (Figure 5B). In meiotic cells, visualization of Dot1-GFP depends on the use of a high-copy plasmid.
Figure 5.
Nuclear localization of Dot1. (A and B) Cells from strain DP138 (dot1) transformed with pSS64 (2 μ DOT1-GFP) growing vegetatively (A) or at different times in sporulation medium (B) were fixed, stained with DAPI, and observed at the fluorescence microscope. The merged images of DAPI (red) and Dot1-GFP (green) are shown in the bottom panels. Overlap appears yellow. The corresponding differential interference contrast images are also presented (top). The approximate cell-cycle stage was estimated based on bud size and nuclear morphology and position. In B, only the large mother cells have entered meiosis; the small buds should not be considered. (C) Spread nuclei from strain DP138 (dot1) transformed with pSS63 (2 μ DOT1-HA) growing vegetatively, stained with DAPI (blue) and anti-HA antibodies (red). (D) Spread leptotene/zygotene (top row) and pachytene (bottom row) nuclei from strain DP138 (dot1) containing pSS63 (2 μ DOT1-HA) stained with DAPI (blue) and anti-Zip1 (green) and anti-HA (red) antibodies. Bar, 2 μm.
To determine whether Dot1 is free in the nucleoplasm or associated with chromosomes, nuclei from vegetative and meiotic cells carrying DOT1-HA on a high-copy plasmid were surface spread and analyzed by indirect immunofluorescence by using anti-HA antibodies. In spread mitotic nuclei, two patterns of Dot1-HA localization are found. In ∼29% of nuclei, Dot1 is homogeneously distributed throughout chromatin (Figure 5C, bottom). In the remaining nuclei, Dot1 is clearly enriched in the region of the nucleus corresponding to the nucleolus (Figure 5C, top). Nucleolar localization of Dot1 was confirmed by double staining for Dot1-HA and the nucleolar protein Nsr1 (our unpublished results). In spread meiotic nuclei, Dot1 is uniformly dispersed throughout the whole DAPI-stained area during all stages of meiotic prophase (Figure 5D; our unpublished results). These results indicate that Dot1 binds directly or indirectly to chromosomes. However, only a small fraction (∼5%) of spread nuclei (especially from meiotic cells) displays Dot1 staining, suggesting that the interaction of Dot1 with chromosomes is weak and/or sensitive to the spreading procedure.
Dot1 Is Required for Localization of Sir2 and Pch2
The checkpoint defect of zip1 dot1 (Tables 2 and 3) is similar to that of zip1 pch2 and zip1 sir2 (San-Segundo and Roeder, 1999; our unpublished results). Furthermore, like Sir2, Dot1 is involved in chromatin silencing (Singer et al., 1998). These observations suggest that Pch2, Sir2, and Dot1 act at the same step in the pachytene checkpoint pathway and raise the possibility that proper localization of Pch2 and/or Sir2 requires Dot1.
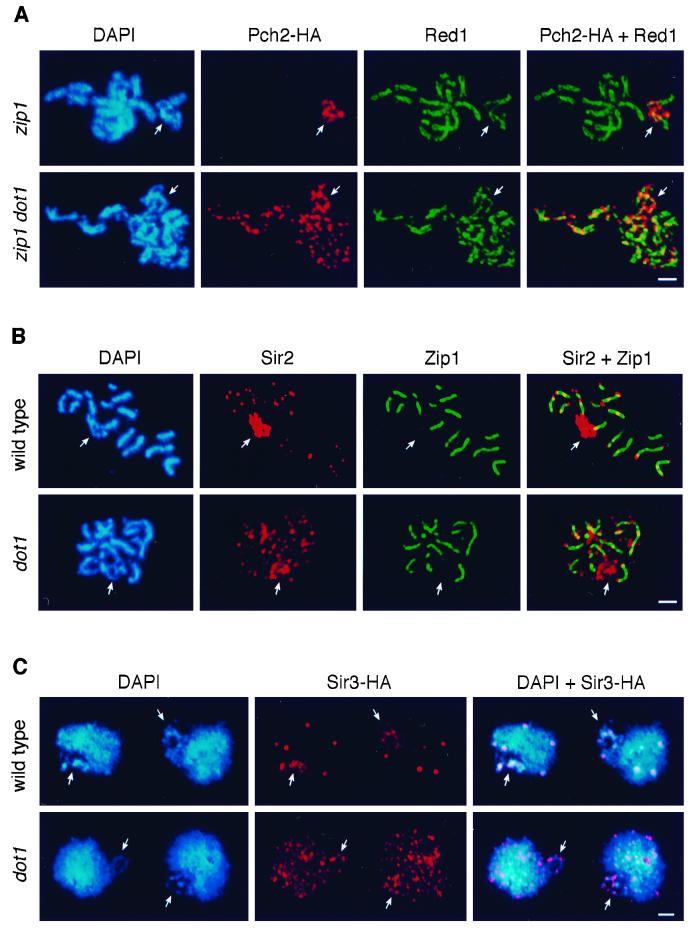
To address this question, localization of Pch2 was examined in spread meiotic chromosomes of the zip1 dot1 mutant. In zip1, the Pch2 protein is detected only in the rDNA region (Figure 6A, top; San-Segundo and Roeder, 1999). In contrast, in the zip1 dot1 double mutant, Pch2 is fairly evenly distributed throughout chromatin (Figure 6A, bottom). Because Pch2 localization depends on Sir2, the possibility that Sir2 is also mislocalized in the absence of Dot1 was explored. In wild-type meiotic chromosomes, Sir2 accumulates in the nucleolus and is also present in foci mostly located at telomeric positions (Figure 6B, top; San-Segundo and Roeder, 1999). In contrast, in the dot1 mutant, a significant fraction of the Sir2 protein is dispersed to other chromosomal locations (Figure 6B, bottom). The localization pattern of Dot1 in mitotic cells is unaffected by a sir2 mutation (our unpublished results). The localization dependence implies the following order of action for these proteins: Dot1 → Sir2 → Pch2.
Figure 6.
Pch2, Sir2, and Sir3 are mislocalized in the absence of DOT1. (A) Spread meiotic nuclei from DP201 (zip1 PCH2-HA, top) and DP285 (zip1 dot1 PCH2-HA, bottom) stained with DAPI (blue) and anti-HA (red) and anti-Red1 (green) antibodies. (B) Spread pachytene nuclei from BR2495 (wild type, top) and DP138 (dot1, bottom) stained with DAPI (blue) and anti-Sir2 (red) and anti-Zip1 (green) antibodies. (C) Spread mitotic nuclei from DP365 (SIR3-HA, top) and DP366 (dot1 SIR3-HA, bottom) stained with DAPI (blue) and anti-HA (red) antibodies. In all panels, arrows point to the nucleolar region. Bar, 2 μm.
Sir3 Is Mislocalized in the dot1 Mutant
In addition to the meiotic checkpoint function described here, Dot1 has been shown to be important for telomeric silencing in vegetative cells (Singer et al., 1998). The observation that Sir2 localization is altered in the dot1 mutant raised the possibility that the defect in telomeric silencing is caused by the failure to recruit and/or stabilize the Sir protein complex at telomeres. To explore this hypothesis, the localization of Sir3 was studied in spread mitotic nuclei of wild type and dot1, using a functional HA-tagged version of Sir3. In wild type, a few very strong Sir3 foci are present that likely represent clusters of telomeres (Gotta et al., 1996; Figure 6C, top). Also, some Sir3-HA is detected in the rDNA region (the possibility of low-level binding of Sir3 to the rDNA has been previously reported; Gotta et al., 1997). However, in the dot1 mutant, Sir3 is dispersed all over chromatin (Figure 6C, bottom). A few stronger foci can be observed that perhaps correspond to telomeric clusters, but they clearly contain less Sir3 protein than those in wild type. Sir3 is also mislocalized in meiotic chromosomes from the dot1 mutant (our unpublished results). Thus, localization of Sir3 (and Sir2) is altered in the absence of the Dot1 protein, presumably accounting for the defect in telomeric silencing in the dot1 mutant.
DISCUSSION
Dot1 Is Required for the Pachytene Checkpoint and for Preventing Repair of DSBs by Intersister Recombination
The results presented here indicate that the product of the DOT1 gene of S. cerevisiae is necessary for function of the pachytene checkpoint. The dot1 single mutant shows no apparent defect in spore viability, recombination, or sporulation, arguing that DOT1 is not required during an unperturbed meiosis. In contrast, DOT1 is essential to trigger meiotic prophase arrest (or delay) when the SC fails to assemble due to lack of Zip1. The defects in chromosome synapsis (see Red1 staining in Figure 6A) and interhomolog exchange characteristic of zip1 are still manifest, or even enhanced, in the zip1 dot1 double mutant. Thus, relief of the meiotic block by the dot1 mutation is not due to suppression of the defects that trigger checkpoint-induced arrest, but rather is due to disruption of the checkpoint per se.
The inappropriate meiotic progression of zip1 dot1 results in chromosome missegregation and the formation of aneuploid spores. The pattern of spore death in zip1 dot1 resembles that of the zip1 single mutant, suggesting that the cause of spore death in both cases is homolog nondisjunction at MI (Sym and Roeder, 1994). The fraction of viable spores produced by zip1 dot1 (∼35%) is higher than expected if chromosomes segregate before recombination intermediates are resolved (segregation before completion of recombination would result in chromosome breakage and extensive spore death). Thus, most or all Holliday junctions and DSBs that accumulate in zip1 must become resolved/repaired as the arrest is bypassed by a dot1 mutation. A similar situation (∼40% spore viability) is found when zip1 meiotic arrest is relieved by mutation of SWE1, the major downstream target of the pachytene checkpoint (Leu and Roeder, 1999). Swe1 inhibits cyclin-dependent kinase activity by phosphorylating tyrosine 19 of Cdc28 (Booher et al., 1993). It is unlikely that Swe1 function directly affects the recombination machinery; however, recombination is completed in zip1 swe1 (Leu and Roeder, 1999) as in zip1 dot1. These observations raise the possibility that the accumulation of recombination intermediates in zip1 is the consequence, rather than the cause, of the meiotic arrest. The defect in SC assembly may be the primary cause of checkpoint-induced arrest in zip1 strains.
Checkpoint-dependent arrest in the dmc1 mutant also requires DOT1. Physical assays of recombination have demonstrated that the dmc1 mutant accumulates processed DSBs (Bishop et al., 1992). Cytologically, the presence of recombination intermediates is manifested by the persistence of multiple Rad51-containing complexes on chromosomes. Although most spores produced by dmc1 dot1 are dead, immunostaining with anti-Rad51 antibodies revealed that a significant fraction of dmc1 dot1 cells that progress into MI have repaired the DSBs (i.e., Rad51 foci have disappeared). The high levels of spore inviability of dmc1 dot1 can be explained if the resolution of recombination does not involve homologous chromosomes and, therefore, does not ensure proper segregation.
It has been recently shown that dmc1 cells can use a Rad54-dependent pathway for repairing DSBs by using sister chromatids instead of homologs as templates for repair (Arbel et al., 1999; Bishop et al., 1999). Indeed, Rad51 foci persist in the dmc1 dot1 rad54 triple mutant, indicating that repair of DSBs in dmc1 dot1 requires RAD54. However, despite the presence of unresolved recombination intermediates, the triple mutant proceeds through meiosis. The spores produced are immature and probably contain fragmented chromosomes, similar to the situation in dmc1 rad24. The dmc1 rad54 double mutant does arrest (Figure 2D), demonstrating that the failure of dmc1 dot1 rad54 to arrest is due to the lack of Dot1 and not the lack of Rad54 function. These data argue that Dot1 performs two distinct functions. Dot1 executes a genuine checkpoint function, preventing meiotic cell cycle progression when recombination is incomplete. In addition, Dot1 blocks a Rad54-dependent pathway that repairs DSBs by intersister recombination when Dmc1 is absent.
Unlike dmc1, the formation and disappearance of meiotic DSBs in the zip1 mutant is essentially normal, except that a small fraction of DSBs (∼10%) remains unrepaired (Storlazzi et al., 1996; Xu et al., 1997). In agreement with these findings, ∼25 foci of Rad51 persist in nuclei of the arrested zip1 mutant. When zip1 arrest is relieved by the dot1 mutation, Rad51 foci disappear, consistent with the notion that recombination intermediates are resolved (see above). However, for the most part, this repair does not require Rad54, because only a very small number of Rad51 foci persist when zip1 dot1 rad54 cells enter MI. Assuming that the Rad54-dependent pathway is mostly involved in intersister recombination (Arbel et al., 1999), this result suggests that most DSBs accumulated in zip1 are already “committed” to undergo repair by using a nonsister chromatid. In contrast, in the dmc1 mutant, the preference for interhomolog recombination is lost (Schwacha and Kleckner, 1997), and DSBs can be repaired by sister-sister recombination when the block to the pathway is relieved by mutation of DOT1 (this work) or RED1 (Xu et al., 1997; Bishop et al., 1999). The small number of Rad54-dependent (likely intersister) recombination events that occur in zip1 dot1 may account for the further reduction in spore viability compared with zip1.
Comparison of Dot1 Function with That of Other Pachytene Checkpoint Proteins
The meiosis-specific proteins Red1 and Mek1 are also involved in the pachytene checkpoint (Xu et al., 1997; Bailis and Roeder, 2000b). Red1 is a component of the lateral elements of the SC (Smith and Roeder, 1997), and Mek1 is a kinase that phosphorylates Red1 (Bailis and Roeder, 1998; de los Santos and Hollingsworth, 1999). Mutation of DOT1 or RED1 has similar effects when combined with dmc1: both bypass meiotic arrest, and both channel the repair of DSBs into a Rad54-dependent pathway (this work; Xu et al., 1997; Bishop et al., 1999). It has been proposed that Red1 (and Mek1) promote a meiosis-specific chromosomal context in which interhomolog recombination is monitored (Xu et al., 1997; Bailis et al., 2000). Recent findings indicate that Mek1-dependent phosphorylation of Red1, triggered by the presence of recombination intermediates, activates the pachytene checkpoint (Bailis and Roeder, 2000b). Dot1 appears to be a chromatin-associated protein that might also contribute to chromosomal context. However, Dot1 is not meiosis specific; in addition, the phenotypes of dot1 and red1 single mutants are clearly different, suggesting that the proteins perform different functions. Whereas the red1 mutant is severely defective in chromosome synapsis, recombination, chromosome segregation, and the production of viable spores (Rockmill and Roeder, 1988, 1990), dot1 is not defective in these processes.
Another set of proteins required for checkpoint-dependent arrest of zip1 and dmc1 are the DNA damage checkpoint proteins Rad24, Rad17, Mec1, and Ddc1 (Lydall et al., 1996; San-Segundo and Roeder, 1999). It is generally believed that these proteins are involved in the detection of broken DNA both in mitotic and meiotic cells (Lydall and Weinert, 1995; Lydall et al., 1996; Longhese et al., 1998; Weinert, 1998). The distribution of Dot1 throughout chromatin is consistent with such a role. However, the meiotic phenotypes of dot1 and DNA damage checkpoint mutants differ in several aspects. Whereas DOT1 is not required during an unperturbed meiosis, the rad24, rad17, and mec1-1 single mutants show decreased crossing over, increased ectopic recombination, increased unequal sister-chromatid exchange, defective chromosome synapsis, and reduced spore viability (Lydall and Weinert, 1995; Lydall et al., 1996; Grushcow et al., 1999; Thompson and Stahl, 1999). Both DOT1 and RAD24 are required for dmc1 arrest, but the outcome of bypassing arrest is different. The dmc1 dot1 mutant completes meiosis with most DSBs repaired via a Rad54-dependent pathway generating mature, though largely inviable, spores. In contrast, the Rad54 pathway appears to remain inactive in dmc1 rad24, resulting in progression through meiosis with unrepaired DSBs and the formation of immature spores.
Recent evidence indicates that the ultimate consequence of activation of the pachytene checkpoint is inhibition of cyclin-dependent kinase activity by Swe1-mediated phosphorylation of Cdc28 and limiting Ndt80-dependent transcription of CLB1 (Chu and Herskowitz, 1998; Hepworth et al., 1998; Leu and Roeder, 1999). A zip1 swe1 mutant sporulates to wild-type levels but after a significant delay; this delay is overcome by simultaneous overexpression of CLB1 (Leu and Roeder, 1999). Because zip1 dot1 sporulates with wild-type efficiency and kinetics, this implies that Dot1 function lies upstream of the bifurcation of the checkpoint pathway into the two regulatory branches of cyclin-dependent kinase activity.
Localization of Pch2, Sir2, and Sir3 Depends on Dot1
The predominantly nucleolar proteins Pch2 and Sir2 are also required for the pachytene checkpoint (San-Segundo and Roeder, 1999), revealing an unexpected link between chromatin silencing and control of meiotic progression. Like Dot1, Pch2 and Sir2 are dispensable during a normal meiosis, but they are essential to prevent progression of meiosis in the absence of Zip1. Several lines of evidence suggest that these three proteins act at the same step in the pachytene checkpoint pathway. First, the meiotic phenotypes of zip1 dot1 are similar to those of zip1 pch2 or zip1 sir2; all three double mutants sporulate with wild-type kinetics, resulting in similar levels and patterns of spore viability (San-Segundo and Roeder, 1999; this work; our unpublished results). Second, like Sir2, Dot1 is involved in chromatin silencing (Singer et al., 1998). Third, proper localization of Pch2 and Sir2 depends on Dot1. Singer et al. (1998) have shown that both Dot1 overproduction and DOT1 deletion disrupt telomeric silencing, suggesting that Dot1 is part of a protein complex in which the proper balance of components is critical for silencing. Indeed, the absence of Dot1 results in mislocalization of the Sir3 protein, which is an essential component of a multiprotein complex required for telomeric silencing (Grunstein, 1998; Lustig, 1998). Furthermore, overexpression of DOT1 in zip1 cells results in partial bypass of the meiotic arrest (our unpublished results). These considerations lead to the speculation that Dot1 is a critical component of protein complexes assembled in heterochromatic regions (nucleolus and/or telomeres) with functions in checkpoint and/or silencing processes. In vegetative cells, Dot1 accumulates in the nucleolus. In meiotic nuclei, Dot1 is also present, but not enriched, in the rDNA region. However, the localization data for Dot1 in spread nuclei should be considered with caution because they are based on overproduction of the protein. The fact that Dot1 does not seem to be localized exclusively in heterochromatic regions is not incompatible with a role in silencing; other essential components of silenced chromatin, such as histones H3 and H4, and Rap1, are also present in both silenced and nonsilenced regions (Shore, 1994; Grunstein, 1998).
Considerations about Heterochromatin, Checkpoints, and DNA Damage Responses
The silencing factor Sir2 is required for meiotic arrest of the zip1 mutant, demonstrating a connection between chromatin silencing and the pachytene checkpoint. The checkpoint defect of sir2 is presumably due to the failure to localize the meiosis-specific Pch2 protein to the nucleolus (San-Segundo and Roeder, 1999). These findings are extended by the results presented here demonstrating a role for another silencing protein, Dot1, in meiotic checkpoint control. In the absence of Dot1, the nucleolar confinement of Sir2 and Pch2 is lost, and the pachytene checkpoint is inactive. The involvement of nucleolar proteins in checkpoint mechanisms is not an exclusive feature of meiotic cells. The human Rad17 protein (homolog of the S. cerevisiae Rad24 checkpoint protein) localizes to the nucleolus in mitotic cells and is redistributed throughout the nucleus upon UV irradiation (Chang et al., 1999). In budding yeast, the cellular localization of most DNA damage checkpoint proteins, including Rad24, remains to be established.
The precise role of the nucleolus in the pachytene checkpoint is not yet understood. However, recent studies have demonstrated a role for the nucleolus in another cell-cycle regulatory process, namely the exit from mitosis (reviewed by Garcia and Pillus, 1999). The Cdc14 phosphatase is sequestered in the nucleolus during most of the mitotic cell cycle by association with the Net1/Cfi1 protein, but Cdc14 is released from the nucleolus in late anaphase, dispersing throughout the cell and triggering mitotic exit (Shou et al., 1999; Visintin et al., 1999). Importantly, localization of Sir2 to the rDNA also requires Net1, and the net1 mutant is defective in rDNA silencing (Straight et al., 1999). It will be of interest to investigate whether Net1, and perhaps Cdc14, are implicated in the pachytene checkpoint.
Several observations also point to a link between telomeric heterochromatin and cellular responses to DSBs/DNA damage in both mitotic and meiotic cells. The Pch2 protein is normally found in the nucleolus, but it localizes to telomeres under some circumstances (SIR4 overexpression or rDNA deletion). Under these conditions, Pch2 still provides at least partial checkpoint function (San-Segundo and Roeder, 1999), arguing that some feature of heterochromatin is important for the pachytene checkpoint. A physical interaction between Mec3 (a DNA damage checkpoint protein) and Set1 (a protein required for telomeric silencing) has been reported (Corda et al., 1999). In addition, analysis of telomeric silencing, telomere length and DNA repair revealed genetic interactions between mec3 and set1 mutants (Corda et al., 1999). In fission yeast, mutations in the rad26+, rad1+, rad17+, and rad3+ checkpoint genes (the last three genes are homologs of S. cerevisiae RAD17, RAD24, and MEC1, respectively), result in telomere shortening (Dahlen et al., 1998) and, in the case of the rad3 mutant, reduced telomeric silencing (Matsuura et al., 1999). A mutation in MEC1 also leads to shorter telomeres (Ritchie et al., 1999) and impaired telomeric silencing (Craven and Petes, 2000). The Sir proteins have not been implicated in the DNA damage checkpoint, but they do facilitate (directly or indirectly) the repair of DSBs (reviewed by Haber, 1999). The Sir proteins, as well as the Ku complex, are redistributed from telomeres to the sites of DSBs. This relocalization requires the function of DNA damage checkpoint proteins and results in decreased telomeric silencing (Martin et al., 1999; McAnish et al., 1999; Mills et al., 1999). The Sir2-related Hst3 and Hst4 proteins of S. cerevisiae and Hst4 of Schizosaccharomyces pombe also contribute to radiation resistance (Brachmann et al., 1995; Freeman-Cook et al., 1999). In consonance with all these observations, the dot1 mutant, which shows reduced telomeric silencing and altered telomere length (Singer et al., 1998), also displays defects in the localization of Sir proteins and in the meiotic checkpoint that responds to unrepaired DSBs. Moreover, mutation of DOT1 increases the DNA damage sensitivity of the rad24 checkpoint mutant (San-Segundo and Roeder, unpublished observations), suggesting that Dot1 also participates in the DNA damage response in vegetative cells.
The connection between telomere function and cell-cycle checkpoints is not restricted to yeast. Human cells deficient in the ATM checkpoint protein undergo telomere shortening (Metcalfe et al., 1996). Mutation of the mrt-2 gene of C. elegans (homolog of RAD17 in S. cerevisiae) results in defects in telomere maintenance and defective responses to the accumulation of meiotic recombination intermediates and DNA damage (Ahmed and Hodgkin, 2000; Gartner et al., 2000). It is tempting to speculate that the metazoan proteins that share homology with Dot1 (Figure 1) have roles in heterochromatin assembly, telomere metabolism, meiotic checkpoint control, and/or cellular responses to damaged DNA.
ACKNOWLEDGMENTS
We thank Julie Bailis, Erica Hong, Janet Novak, and Beth Rockmill for comments on the manuscript. We also thank Elisa Stone and Lorraine Pillus for anti-Sir2 serum, Doug Bishop for anti-Rad51 antibody, David Schild for plasmid pSM31, and Ted Weinert for a rad24 disruption plasmid. P.A.S. was supported in part by a postdoctoral fellowship from the Ministerio de Educación y Ciencia of Spain. This work was supported by National Institutes of Health grant GM-28904 (to G.S.R.) and by the Howard Hughes Medical Institute.
Abbreviations used:
- DAPI
4′-6-diamidino-2-phenylindole
- DSB
double-strand break
- GFP
green fluorescent protein
- HA
hemagglutinin
- MI
meiosis I
- NLS
nuclear localization signal
- PBS
phosphate-buffered saline
- rDNA
ribosomal DNA
- SC
synaptonemal complex
REFERENCES
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or. TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. The pachytene checkpoint. Trends Genet. 2000a;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell. 2000b;101:211–221. doi: 10.1016/S0092-8674(00)80831-4. [DOI] [PubMed] [Google Scholar]
- Bailis JM, Smith AV, Roeder GS. Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Mol Cell Biol. 2000;20:4838–4848. doi: 10.1128/mcb.20.13.4838-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cell. 1999;4:425–443. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Briza P, Breitenbach M, Ellinger A, Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Chang M-S, Sasaki H, Campbell MS, Kraeft S-K, Sutherland R, Yang C-Y, Liu Y, Auclair D, Hao L, Sonoda H, Ferland LH, Chen LB. HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation. J Biol Chem. 1999;274:36544–36549. doi: 10.1074/jbc.274.51.36544. [DOI] [PubMed] [Google Scholar]
- Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–703. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, Gilson E, Geli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:611–621. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hollingsworth NM. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J Biol Chem. 1999;274:1783–1790. doi: 10.1074/jbc.274.3.1783. [DOI] [PubMed] [Google Scholar]
- Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, Pillus L. The Schizosaccharomyces pombe hst4+ gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Sir-Ku-itous routes to make ends meet. Cell. 1999;97:829–832. doi: 10.1016/s0092-8674(00)80795-3. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Lowndes NF. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992;20:2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Leu J-Y, Roeder GS. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol Cell. 1999;4:805–814. doi: 10.1016/s1097-2765(00)80390-1. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Lustig AJ. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Naito T, Ishikawa F. Genetic control of telomere integrity in Schizosaccharomyces pombe: rad3+ and tel1+ are parts of two regulatory networks independent of the downstream protein kinases chk1+ and cds1+ Genetics. 1999;152:1501–1512. doi: 10.1093/genetics/152.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnish AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- Metcalfe JA, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd PJ, Taylor AM. Accelerated telomere shortening in ataxia telangiectasia. Nat Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Pintard L, Kressler D, Lapeyre B. Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenoysyl-l-methionine in vitro. Mol Cell Biol. 2000;20:1370–1381. doi: 10.1128/mcb.20.4.1370-1381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci USA. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- San-Segundo P, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1136. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: a protein regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZWS, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Struhl K, Stinchcomb DT, Scherer S, Davis RW. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J Cell Biol. 1995;128:455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis: isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K-S, Roeder GS. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]