Abstract
The role of promoter methylation in the process of cancer cell metastasis has not yet been studied. Recently, methylation of the TPEF (transmembrane protein containing epidermal growth factor and follistatin domain) gene was reported in human colon, gastric, and bladder cancer cells. Using the Methylight assay, TPEF/HPP1 gene methylation was assessed in primary colorectal cancers (n = 47), matched normal colon mucosa, as well as in the liver metastasis of 24 patients with colorectal cancer, and compared to the methylation status of the TIMP-3, APC, DAPK, caveolin-2, and p16 genes. TPEF was frequently methylated in primary colorectal cancers (36 of 47) compared to the normal colon mucosa (1 of 21) (P < .0001). Interestingly, promoter methylation was significantly more frequent in proximal nonrectal cancers (P < .05). Furthermore, a high degree of methylation of the TPEF gene was also observed in liver metastasis (19 of 24). In summary, we observed frequent TPEF methylation in primary colorectal cancers and liver metastases, indicating that epigenetic alterations are not only present in the early phases of carcinogenesis, but are also common in metastatic lesions. The high frequency of TPEF methylation in this series of colorectal cancers underscores the importance of epigenetic changes as targets for the development of molecular tests for cancer diagnosis.
Keywords: methylation, epigenetic, metastasis, promoter, colon cancer
Introduction
In the United States, the annual incidence of colorectal cancer is approximately 150,000, with 56,600 individuals dying from colorectal cancer each year [1,2]. The lifetime risk of colorectal cancer in the general population is about 5% to 6%. Despite intensive efforts in recent years in the screening and early detection of colon cancer, until today, most cases have been diagnosed in an advanced stage with regional or distant metastasis [3,4]. Although therapeutic options include surgery and adjuvant or palliative chemotherapy, most patients die from progression of their cancer within a few months. Identifying the molecular changes that underlie the progression of colon cancer and the formation of metastasis may help to develop new diagnostic and therapeutic options that could improve the overall poor prognosis of these patients [1–4].
The current model of colorectal cancer pathogenesis favors a stepwise progression of adenomas, which includes the development of dysplasia and, finally, signs of invasive cancer. The molecular changes underlying this adenoma-carcinoma sequence include genetic and epigenetic alterations of tumor-suppressor genes (APC, p53, and DCC), activation of oncogenes (K-ras), and inactivation of DNA mismatch repair genes [1]. Recently, further molecular changes and genetic defects have been revealed. Thus, activation of the Wnt signalling pathway not only includes mutations of the APC gene, but may also result from β-catenin mutations [5]. Furthermore, alterations in the TGF-β signalling pathway, together with its signal transducers SMAD4 and SMAD2, have been linked to the development of colon cancer [6].
Apart from mutations, aberrant methylation of CpG islands has been shown to lead to the transcriptional silencing of certain genes that have been previously linked to the pathogenesis of various cancers [7,8]. CpG islands are short sequences, which are rich in CpG dinucleotides and can usually be found in the 5′ region of approximately 50% of all human genes. Methylation of cytosines in these islands leads to the loss of gene expression and has been reported in the inactivation of the X chromosome and genomic imprinting [7,8].
Recently, several groups have also analyzed the methylation of various genes in colorectal cancer and reported transcriptional silencing by promoter methylation for p16INK4, p14ARF, p15INK4b, MGMT, hMLH1, GSTP1, DAPK, CDH1, TIMP-3, and APC, among others [8–10]. In addition, Liang et al. [11] reported the frequent methylation of the TPEF/HPP1 gene in colorectal cancers. Recent studies confirmed that TPEF methylation is a frequent event in other cancers as well, such as gastric, bladder, and gallbladder cancers [11–14]. Thus, apart from mutational inactivation of certain genes, the hypermethylation of several genes contributes significantly to the pathogenesis of primary colorectal cancer. However, the role of promoter methylation in the progression and formation of distant metastasis in colorectal cancer is far less well known.
Materials and Methods
Subjects for Methylight Analysis
Colon tissues were obtained by surgical resection from 47 patients (29 males, 18 females) with colon cancer, with a median age of 66 years (range: 31–93 years), from the tumor and a tumor-free location, which was at least 2 cm distant from the tumor and which was confirmed to be without any tumor cell infiltration by histologic assessment. In all 47 patients, tissue samples from the colon cancer were obtained for molecular analysis; in 21 of these cases, a matched noncancer colon sample was also obtained for molecular analysis after tumor cell infiltration was ruled out by histologic assessment. The metastatic lesions were obtained from 24 patients (13 males, 11 females, median age: 64.5 years, range: 41–79 years) with colorectal cancer, who developed liver metastasis after prior successful colon cancer resection. In one case, the primary colon cancer and a single liver metastasis were resected at the same time in a 74-year-old female patient. Immediately after surgery, tissue samples were put in liquid nitrogen and stored at -80°C until use. Formalin-fixed tissues were processed as previously described and sections were stained with hematoxylin and eosin for histologic evaluation [15]. Tumor stages were assessed using the WHO classification for tumors of the digestive tract, and the study was approved by the Ethics Committee of the University of Magdeburg.
Cell Lines and 5-Aza-2′-Deoxycytidine Treatment
The metastatic colon cancer cell line, LoVo, and the primary colon cancer cell line, DLD-1, were obtained from the American Type Culture Collection (ATCC; Manassas, VA), and cultured in F-12K and RPMI 1640 media, respectively. Incubation with 5-aza-2′-deoxycytidine was performed as previously described [15]. Briefly, cells were seeded at a density of 1 x 106 cells/60-mm dish. Twenty-four hours later, they were incubated with 5-aza-2′-deoxycytidine (10-6 M) for 24 hours (Sigma Chemical Co., Deisenhofen, Germany); the medium was changed daily for 3 days. After 3 days, total RNA was extracted for assessment of TPEF mRNA levels, as outlined below. The same concentration of dimethyl sulfoxide was also used as a control for nonspecific solvent effect on cells as previously described [15].
Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis
TPEF mRNA levels were determined in the two colon cancer cell lines after incubation with DMSO or 5-aza-2′-deoxycytidine, and in nine colorectal cancer tissues of patients undergoing cancer surgery. Briefly, tissue specimens were homogenized with an ultrasound homogenizer (Ultra-Turrax T25; Janke and Kunkel, Köln, Germany) in the presence of RNAzolB (CINNA/MRC, Cincinnati, OH) containing RNase inhibitors. Total RNA extracted from cell lines and frozen tissues by the acid/guanidinium and phenol/chloroform extraction method was quantified by measuring the optical density at 260 nm and was separated by gel electrophoresis, as previously described [15]. Total RNA (1 µg) was reverse-transcribed at 37°C for 1 hour in a final volume of 20 µl of RT buffer (50 mM Tris-HCl, pH 8.3, 50 mM KCl, 10 mM MgCl2, 0.5 mM spermidine, and 10 mM DTT) containing 4.8 U of AMV reverse transcriptase (Promega, London, UK), 16 U of RNAase inhibitors, 200 pmol of random primers, and 1.0 mM dNTPs (Biomol Feinchemikalien, Hamburg, Germany). The reaction was terminated by incubating the mixture at 95°C for 10 minutes. PCR amplification of the cDNA was performed as previously described by Young et al. [12] using primers HPC and HPZ. A 306-bp fragment encoding a portion of the 5′-UTR and coding sequence of TPEF was identified by gel electrophoresis and ethidium bromide staining. Loading was controlled by coamplification of a fragment of β2 microglobulin, as previously described [15].
DNA Extraction
Genomic DNA was extracted from the cell lines and tissues using the proteinase K digestion method, as previously reported [15].
Methylight Analysis
Genomic DNA was analyzed by the Methylight technique after bisulfite conversion, as previously reported by Eads et al. [16,17]. In this analysis, three oligonucleotides are used in every reaction. Two locus-specific PCR primers flank an oligonucleotide probe with a 5′ fluorescent reporter dye (6FAM) and a 3′ quencher dye (BHQ-1). For this analysis, primers and probes are specifically designed to bind to bisulfite-converted DNA, which generally span 7 to 10 CpG dinucleotides. The gene of interest is then amplified and normalized to a reference set (β-actin (ACTB)) to normalize for input DNA. The specificity of the reactions for methylated DNA is confirmed using human sperm DNA (unmethylated) and CpGenome Universal Methylated DNA (Chemicon [subsidiary of Serologicals] catalog no. S7821; Chemicon, Temecula, CA) (methylated). For standardization, the primers and the probe for analysis of the ACTB gene lack CpG dinucleotides so that amplification is possible regardless of methylation levels. TaqMan PCR reactions were performed in parallel with primers specific for the bisulfite-converted methylated sequence for a particular locus and with the ACTB reference primers. The ratio between the values was calculated in these two TaqMan analyses; using this approach, the degree of methylation at that locus was determined. The extent of methylation at a specific locus was determined by the following formula: [(gene/actb)sample:(gene/actb)SssI-treated genomic DNA] x 100. A cutoff value of 4% gave the best discrimination between normal and cancerous samples, as previously reported [16,17]. Therefore, samples with ≥4% fully methylated molecules were termed methylated, whereas samples with <4% were considered unmethylated. The primer and probe sequences are listed in Table 1 and were used as previously reported by Eads et al. [16,17].
Table 1.
List of Primers and Probes Used for Methylight Analysis [15,16].
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Probe Sequence (5′-3′) |
| caveolin-2 | TTTCGGATGGGAACGGTGTA | CTCCCACCGCCGTTACC | 6FAM-CCCGTCCTAACCGTCCGCCCT-BHQ1 |
| DAPK | TCGTCGTCGTTTCGGTTAGTT | CCCTCCGAAACGCTATCGA | 6FAM-CGACCATAAACGCCAACGCCG-BHQ1 |
| TPEF | TTTTTTTTTCGGACGTCGTTG | CCTCTACATACGCCGCGAAT | 6FAM-AATTACCGAAAACATCGACCGA-BHQ1 |
| p16 | TGGAATTTTCGGTTGATTGGTT | AACAACGTCCGCACCTCCT | 6FAM-ACCCGACCCCGAACCGCG-BHQ1 |
| APC | GAACCAAAACGCTCCCCAT | TTATATGTCGGTTACGTGCGTTTATAT | 6FAM-CCCGTCGAAAACCCGCCGATTA-BHQ1 |
| TIMP3 | GCGTCGGAGGTTAAGGTTGTT | CTCTCCAAAATTACCGTACGCG | 6FAM-AACTCGCTCGCCCGCCGAA-BHQ1 |
Statistical Analysis
The percentage of methylated reference (PMR) values of the Methylight assays were dichotomized for statistical purposes, as previously reported by Eads et al. [16,17]. PMR values above 4% were considered as methylation-positive and classified as “1,” whereas PMR levels below 4% were classified as “0” (no methylation). This dichotomization should level off the quantitative impact of different levels of hypermethylation per gene, and allow the crossgene comparison of methylation per gene in colon cancer and metastasis. The different clinicopathologic features, such as location of primary tumor, grade of differentiation, or stage of cancer, were used as nominal variables in the Fisher's exact test or chi-square analysis. Otherwise, Student's t test was used to determine statistical difference. All tests were two-sided, and P < .05 was considered statistically significant [15–17].
Results
Analysis of TPEF Gene Methylation in Primary and Metastatic Colorectal Cancer
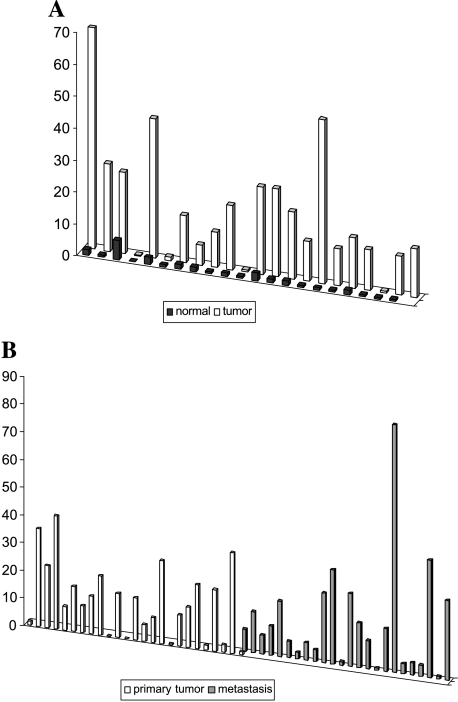
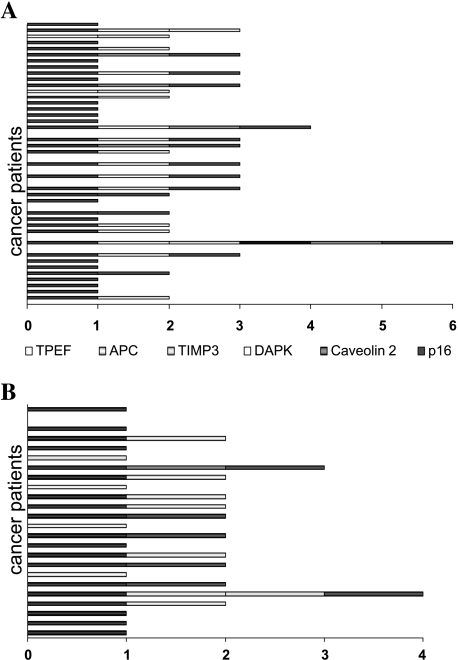
TPEF promoter methylation was analyzed in a set of primary cancers, and matched normal colon mucosa as well as the liver metastasis of 24 patients with colorectal cancer. Using Methylight assays, we also assessed the methylation status of five other genes in our series of primary and metastatic colon cancers. The cutoff of methylation was chosen to be a PMR of >4% (as previously reported by Eads et al. [16,17]) and all samples with a PMR >4% were classified as methylation-positive (“1”), whereas samples with a PMR below 4% were considered methylation-negative (“0”). TPEF promoter methylation was observed in 36 of 47 primary cancers, whereas in the normal colon mucosa, only 1 of 21 cases exhibited TPEF gene methylation (P < .001) (Figure 1A). Furthermore, 19 of 24 liver metastases exhibited TPEF gene methylation (Figure 1B). Next, we analyzed our tissues for gene methylation using a set of genes that had been previously reported by other groups to be associated with either colon cancer pathogenesis or development of cancer metastasis: p16/INK4A, APC, caveolin-2, DAPK, and TIMP3. The results of the methylation analysis of the other genes in primary and colon cancers are given in Table 2. In addition, the numbers were added, giving the total numbers of methylated genes per sample. Using this approach, we observed at least one methylated gene in 39 of 47 primary cancers (Figure 2A). In contrast, only 1 of 21 normal colon mucosa samples exhibited TPEF gene methylation, whereas no other gene was found to be methylated in the normal colon mucosa. In the metastatic lesions all but one of the 24 cases exhibited at least one of these methylated genes: TPEF, TIMP3, or APC (Figure 2B).
Figure 1.
(A) TPEF gene methylation in the normal colon mucosa and matched colon cancers. (B) TPEF gene methylation in primary colon cancer and colon cancer metastasis.
Table 2.
Summary of Results from the Analysis of Gene Methylation in Primary Cancer and Metastasis.
| Gene | Normal (n = 21) | Tumor (n = 47) | Metastasis (n = 24) | Class |
| TPEF | 1/21 | 36/47* | 19/24 | I |
| p16 | 0/21 | 15/47† | 6/24 | I |
| APC | 0/21 | 10/47† | 10/24‡ | II |
| TIMP3 | 1/21 | 11/47 | 2/24 | III |
| DAPK | 0/21 | 1/47 | 0/24 | III |
| caveolin-2 | 0/21 | 5/47 | 1/24 | III |
Class refers to the classification of gene methylation in liver metastasis, as outlined in the text.
Normal versus tumor: P < .0001.
Normal versus tumor: P < .002.
Tumor versus metastasis: P = .045.
Figure 2.
Number of methylated genes per patient with primary colorectal cancer (A) and in metastasis (B).
Association of Gene Methylation in Colon Cancer with Clinicopathologic Features of Colon Cancer
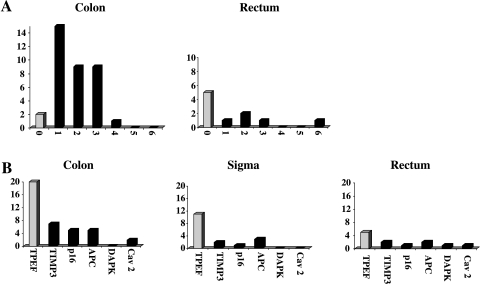
To assess a potential association of the presence of methylation with the location of the primary tumor, we classified our colorectal cancers into two groups: rectal cancers (n = 10) and nonrectal cancers (n = 36). Using Fisher's exact test, we found that there was a statistically significant difference in the presence of methylation with regard to the location of the primary tumor. Although rectal cancers exhibited no methylation at all in five cases and methylation of at least one gene in five cases, the vast majority of colon cancers (34 of 36) exhibited methylation in at least one gene (P < .001). Thus, from this analysis, we can assume that methylation is significantly more frequent in proximal parts (i.e., nonrectal cancers of the large intestines; Figure 3A).
Figure 3.
Analysis of methylation with regard to clinicopathologic features of colorectal cancers. (A) In cancers of the colon including the sigmoid, methylation was significantly more frequent than in cancers of the rectum only. y-axis: number of patients; x-axis: number of methylated genes. (B) TPEF methylation was significantly more frequent in cancers of the colon including the sigmoid. Number of patients analyzed: colon cancer (without sigmoid colon): n = 23, sigmoid cancer: n = 13, rectal cancer: n = 10. One patient with recurrent cancer was not included in this analysis. y-axis: number of patients; x-axis: number of methylated genes.
We analyzed not only the association between the location of the primary and the overall presence of gene methylation per patient, but also analyzed each single gene with regard to the association of location and gene methylation. Interestingly, TPEF promoter methylation was—as opposed to all other genes analyzed—linked to the location of the primary tumor in the colon and was, thus, more frequently methylated in colon cancers (31 of 36) compared to rectal cancers (5 of 10) (P = .023) (Figure 3B).
Next, we analyzed the frequency of TPEF methylation in our series of colorectal cancers with regard to clinical parameters of tumor progression and differentiation. However, the presence of TPEF methylation in our series of colorectal cancers was independent of the T-stage and the presence of lymph node metastasis and/or distant metastasis. Furthermore, no association between TPEF methylation and overall tumor stage (UICC) and/or grade of differentiation was found. In addition, TPEF methylation was independent of age and gender of the cancer patients.
TPEF mRNA Levels and Gene Methylation in Colorectal Cancer
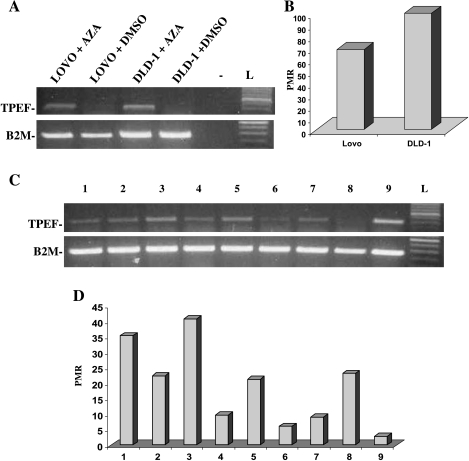
Next, we determined the expression of TPEF in colon cancer cell lines and colon cancer tissues. Both colon cancer cell lines, LoVo and DLD-1, did not exhibit TPEF expression (Figure 4A). Methylight analysis of the TPEF gene revealed a high degree of methylation in both cancer cell lines (Figure 4B). Accordingly, incubation of the two cancer cell lines with the methylation inhibitor, 5-aza-2′-deoxycytidine, led to the restoration of TPEF mRNA levels in both cell lines (Figure 4A). In addition, we also determined the levels of TPEF mRNA in a subset of primary colorectal cancers. Using total RNA and RT-PCR analysis, we identified various levels of TPEF mRNA in these cancer tissues (Figure 4C). Interestingly, Methylight analysis revealed a high frequency of gene methylation in this subset of cancers (Figure 4D). However, we observed no clear correlation of TPEF gene methylation with loss of expression in the majority of these cancer cases. Apart from cancer cells, the tissue specimens that were analyzed by RT-PCR analysis were also composed of other noncancer cells, including endothelial cells, fibroblasts, epithelial cells, and others. Therefore, other cells that may have expressed TPEF may have contributed to the overall level of TPEF mRNA in these cases. Nonetheless, one case (number 9) with a low level of TPEF methylation exhibited a strong degree of TPEF mRNA expression (Figure 4, C and D).
Figure 4.
(A) TPEF mRNA levels in two colon cancer cell lines (LoVo and DLD-1) after incubation with DMSO or the methylation inhibitor, 5-aza-2′-deoxycytidine (AZA). Treatment with 5-aza-2′-deoxycytidine restored TPEF mRNA levels in both cell lines. (-) Negative control; L, DNA ladder. β2 Microglobulin levels were determined to allow comparison of TPEF mRNA levels (B2M). (B.) Methylight analysis revealed high levels of TPEF gene methylation in both cancer cell lines. (C) RT-PCR analysis of TPEF expression in nine human colorectal cancers revealed varying levels of TPEF mRNA. L: DNA ladder. β2 Microglobulin levels were determined to allow comparison of TPEF mRNA levels (B2M). (D) Methylight analysis revealed a high degree of TPEF gene methylation in the majority of colorectal cancers. PMR, percentage of methylated reference; numbers correspond to the cases in panel C.
Discussion
Using methylation-sensitive arbitrarily primed PCR (MS-APPCR), a hypermethylated DNA fragment was isolated, which was later found to be part of a gene encoding a transmembrane protein containing EGF and follistatin domains, thus, termed TPEF [11,12]. The gene maps to chromosome 2q33, which is frequently identified as a region of loss of heterozygosity (LOH) in various cancers, such as esophageal cancer, colon, prostate, and breast cancers. Further analysis revealed that TPEF, which is also known as HPP1, is expressed in the normal colon mucosa; however, in colon cancers, the expression is frequently lost. Interestingly, the loss of TPEF/HPP1 expression in colon cancer is associated with hypermethylation of the 5′ region of the gene [12,18]. Further analysis revealed that TPEF is frequently hypermethylated in various other cancer cell lines and in primary bladder, gallbladder, and gastric cancers [11–14,18]. The exact function of TPEF is currently unknown; however, from structural analysis, it has been suggested that TPEF may function as an inhibitor of growth factors [11]. The more detailed analysis of TPEF gene methylation in colon cancer and its precursor lesions revealed that promoter methylation, and thus its transcriptional silencing, are present in hyperplastic and adenomatous polyps, as well as in dysplasia of the colon mucosa associated with ulcerative colitis, which indicates that TPEF gene methylation is an early epigenetic alteration in colorectal carcinogenesis [12,18].
Despite recent progress in the understanding of the pathogenesis of adenomas and carcinomas of the colon and their genetic and molecular changes, the genetic and epigenetic changes underlying the development of metastasis are less well understood. It is, however, generally well accepted that the process of invasion and proteolysis of the extracellular matrix, as well as infiltration of the vascular basement membrane, involve adhesive proteins, such as members of the family of integrin receptors, the cadherins, the immunoglobulin superfamily, the laminin-binding protein, and the CD44 receptor [19]. More recently, other groups have compared the genetic and molecular changes in metastatic lesions to the changes found in primary colorectal cancers. Thus, Kleeff et al. [20] reported the loss of DOC-2, a candidate tumor-suppressor gene, both in primary and metastatic colorectal cancers. Furthermore, Zauber et al. reported that, in their series of 42 colorectal cancers, Ki-ras mutations in the primary cancers were identical in all of the 42 paired primary and synchronous metastatic lesions. Similarly, LOH at the APC locus was identical for 39 paired carcinomas and synchronous metastases [21]. However, other groups have found genetic and molecular changes in metastatic colon cancers, which are not present in the primary cancers. Thus, the development of LOH of chromosome 3p in colorectal metastasis has been reported [22]. In addition, using comparative genomic hybridization, several alterations that were unique to metastastic lesions (-9q, -11q, and -17q) were found in liver metastases [23]. Furthermore, Saha et al. [24] reported the overexpression of the PRL-3 tyrosine phosphatase in the metastasis of colorectal cancers using global expression profiling by SAGE technology.
To our knowledge, our study is the first to also address the presence of gene methylation in metastatic colorectal cancers using a panel of six genes—including TPEF—that have previously been linked either to colon cancer pathogenesis or metastatic development: TPEF/HPP1, p16/INK4A, APC, caveolin-2, DAPK, and TIMP3 [9,11,25–28]. Gene methylation was analyzed by the highly sensitive Methylight assay and, using this assay, we found several genes to be methylated in both metastatic lesions and primary cancers, as well as genes that were neither methylated in metastasis nor in primary colorectal cancers. Although TPEF exhibited a high frequency of methylation in primary colon cancer [11,12], p16 and APC were less frequently methylated and no relevant methylation was observed for caveolin-2, DAPK, and TIMP3. Of the 47 analyzed primary colon cancers, 39 exhibited methylation of at least one of these three genes: TPEF, APC, or p16. Although TPEF gene methylation was also very frequent in metastatic lesions of colorectal cancer patients, APC gene methylation increased in metastatic lesions compared to primary cancers. Based on these findings, we can classify the patterns of methylation in liver metastasis in three groups: class I genes—high degree of methylation in primary tumor and liver metastasis (TPEF and p16); class II genes—higher degree of methylation in metastasis compared to primary tumor (APC); and class genes III—low degree of, or even no, methylation in either primary tumor or metastasis (caveolin-2, DAPK, and TIMP3). Interestingly, all but two of the 24 metastases exhibited methylation of either the TPEF or APC gene, indicating that these two genes may be valuable for the methylation-specific detection of liver metastasis in colon cancer.
When cancers were grouped according to the location of the primary cancer, overall gene methylation was more frequently present in nonrectal cancers compared to cancers of the rectum—an observation that has been reported by other groups as well [29]. Furthermore, TPEF gene methylation was significantly more frequent in cancers of the colon compared to cancers of the rectum, supporting previous studies reporting a high degree of methylation in proximal colon cancers.
Recent studies by Liang et al. [11], Young et al. [12], and Sato et al. [18] indicate that TPEF gene methylation is associated with reduction or loss of TPEF expression in colon cancers. In our study, both colon cancer cell lines—one from a primary colon cancer (DLD-1) and a further metastastic cancer cell line (LoVo)—did not exhibit TPEF expression by RT-PCR analysis; however, after treatment with the methylation inhibitor 5-aza-2′-deoxycytidine, expression was restored. In the colon cancer tissues, we observed varying degrees of TPEF expression, despite the fact that TPEF gene methylation was present in the vast majority of cases. Although some cases exhibited a close correlation of TPEF gene methylation and loss of expression, the lack of association in the tissues as opposed to the cancer cell lines may result from the source of tissues used for RT-PCR analysis. Thus, apart from cancer cells, these tissue specimens were composed of noncancerous epithelial cells, endothelial cells, smooth muscle cells, and fibroblasts, among others. Because previous studies by Young et al. [12] have shown that these cells may express TPEF at various levels, they may have contributed to the overall TPEF mRNA levels that were measured by RT-PCR analysis, despite the fact that TPEF methylation was present in the cancer cells in these tissue specimens.
Interestingly, only one of the matched normal colon mucosa tissue samples exhibited TPEF gene methylation, indicating that the methylation of the TPEF promoter is an epigenetic alteration, which is specific for the transformation of the epithelial cells of the colon and which may be, therefore, considered a valuable marker for the early detection of neoplastic or preneoplastic lesions of the colon. This observation confirms previous reports by Sabbioni et al. [30] who demonstrated TPEF gene methylation in the tissue and serum of patients with colorectal cancer. The biologic significance of TPEF methylation in the normal mucosa, however, remains unclear. Although we found no significant methylation of TPEF in the normal mucosa in all but one case, Young et al. [12] reported low levels of TPEF methylation in the normal colonic mucosa in a large set of patients. However, a higher degree of methylation was observed only in a few individuals [12]. These divergent results may result from methodological differences because in the study by Young et al., the COBRA assay was used to screen larger tissue series, whereas we and other groups used either methylation-specific PCR or the Methylight assay. Furthermore, the biologic significance of low levels of methylation with regard to the effect on gene expression is still under discussion. Nonetheless, a subgroup of patients presented with TPEF methylation in the colon mucosa independent of age, sex, or site of colon. The biologic role of TPEF methylation in the normal mucosa is, however, still under investigation, especially because the function of TPEF is not fully understood either. TPEF/HPP1 may present both as a transmembranous or soluble molecule, with an EGF module and two follistatin modules in the extracellular domain. Furthermore, it contains a potential G protein-activating motif in the cytoplasmic domain. Thus, TPEF may function both as growth factor and/or receptor, and recent studies indicate that tyrosine phosphorylation of erbB4 in gastric cancer cells may be induced by TPEF [31]. Furthermore, TPEF may also prolong the survival of neural cells [32]. Together, these data indicate that TPEF may have an important role in proliferation, differentiation, and apoptosis. The biologic role of TPEF methylation in the normal mucosa may indicate either the presence of a field effect of the colonic mucosa in colon cancers because these samples were obtained from the tumor-free region of colon cancers, or an age-dependent effect that may imply an age-dependent loss of TPEF-mediated differentiation, which could contribute to colon cancer pathogenesis. Finally, because TPEF is expressed in pericryptal myofibroblasts in the colon mucosa, loss of TPEF expression in cancers or the tumor-free area next to the cancers may also result from the disappearance of these fibroblasts in the course of cancer pathogenesis [12].
In summary, our analysis revealed that TPEF gene methylation is a frequent event in colorectal cancer pathogenesis; it is present in the majority of colorectal cancer metastasis; and TPEF gene methylation is associated with gene silencing in colon cancer cell lines. Therefore, this epigenetic alteration may contribute both to the pathogenesis of colorectal cancer and the progression and formation of metastasis in colorectal cancer. Together with a further set of genes, TPEF methylation may allow the identification of primary and metastatic colorectal cancers indicating that methylation-based diagnostic tests may be helpful in the identification of this and other malignancies and, thus, may improve the detection and overall prognosis of patients with these cancers.
Footnotes
This work was supported by a grant from the DFG (Eb 187/4-1) and by the Heisenberg Programme of the DFG (Eb 187/5-1) awarded to M.E. Further support to M.E. was granted by the Land Sachsen-Anhalt (3488A/0103M), the Forschungszentrum Immunologie (FZI) of the Medical Faculty of the Otto-von-Guericke University Magdeburg, and the Werner-Creutzfeldt-Stipend of the Deutsche Gesellschaft für Verdauungs-und Stoffwechsel erkrankungen.
References
- 1.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–865. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 3.Nelson H, Petrelli N, Carlin A, Couture J, Fleshman J, Guillem J, Miedema B, Ota D, Sargent D. Guidelines 2000 for colon and rectal cancer surgery. J Natl Cancer Inst. 2001;93:583–596. doi: 10.1093/jnci/93.8.583. [DOI] [PubMed] [Google Scholar]
- 4.Zink S, Kayser G, Gabius HJ, Kayser K. Survival, disease-free interval, and associated tumor features in patients with colon/rectal carcinomas and their resected intra-pulmonary metastases. Eur J Cardiothorac Surg. 2001;19:908–913. doi: 10.1016/s1010-7940(01)00724-2. [DOI] [PubMed] [Google Scholar]
- 5.Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 6.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1276–1277. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2004;4:351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Corn PG, Baylin SB, Herman JGA. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 10.Toyota M, Issa JP. The role of DNA hypermethylation in human neoplasia. Electrophoresis. 2000;21:329–333. doi: 10.1002/(SICI)1522-2683(20000101)21:2<329::AID-ELPS329>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Liang G, Robertson KD, Talmadge C, Sumegi J, Jones PA. The gene for a novel transmembrane protein containing epidermal growth factor and follistatin domains is frequently hypermethylated in human tumor cells. Cancer Res. 2000;60:4907–4912. [PubMed] [Google Scholar]
- 12.Young J, Biden KG, Simms LA, Huggard P, Karamatic R, Eyre HJ, Sutherland GR, Herath N, Barker M, Anderson GJ, et al. HPP1: a transmembrane protein-encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci USA. 2001;98:265–270. doi: 10.1073/pnas.98.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geddert H, Kiel S, Iskender E, Flori AR, Krieg T, Vossen S, Gabbert HE, Sarbia M. Correlation of hMLH1 and HPP1 hypermethylation in gastric, but not in esophageal and cardiac adenocarcinoma. Int J Cancer. 2004;110:208–211. doi: 10.1002/ijc.20058. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi T, Shivapurkar N, Riquelme E, Shigematsu H, Reddy J, Suzuki M, Miyajima K, Zhou X, Bekele BN, Gazdar AF, et al. Aberrant hypermethylation of multiple genes in gallbladder carcinoma and chronic cholecystitis. Clin Cancer Res. 2004;10:6126–6133. doi: 10.1158/1078-0432.CCR-04-0579. [DOI] [PubMed] [Google Scholar]
- 15.Ebert MP, Yu J, Hoffmann J, Rocco A, Rocken C, Kahmann S, Muller O, Korc M, Sung JJ, Malfertheiner P. Loss of beta-catenin expression in metastatic gastric cancer. J Clin Oncol. 2003;21:1708–1714. doi: 10.1200/JCO.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 17.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long JH, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 18.Sato F, Shibata D, Harpaz N, Xu Y, Yin J, Mori Y, Wang S, Olaru A, Deacu E, Selaru FM, et al. Aberrant methylation of the HPP1 gene in ulcerative colitis-associated colorectal carcinoma. Cancer Res. 2002;62:6820–6822. [PubMed] [Google Scholar]
- 19.Berman RS, Portera CA, Ellis LM. Biology of liver metastases. Cancer Treat Res. 2001;109:183–206. doi: 10.1007/978-1-4757-3371-6_10. [DOI] [PubMed] [Google Scholar]
- 20.Kleeff J, Huang Y, Mok SC, Zimmermann A, Friess H, Buchler MW. Down-regulation of DOC-2 in colorectal cancer points to its role as a tumor suppressor in this malignancy. Dis Colon Rectum. 2002;45:1242–1248. doi: 10.1007/s10350-004-6399-2. [DOI] [PubMed] [Google Scholar]
- 21.Zauber P, Sabbath-Solitare M, Marotta SP, Bishop DT. Molecular changes in the Ki-ras and APC genes in primary colorectal carcinoma and synchronous metastases compared with the findings in accompanying adenomas. J Clin Pathol. 2003;56:137–140. doi: 10.1136/mp.56.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaker H, Graf M, Rieker RJ, Otto HF. Comparison of losses of heterozygosity and replication errors in primary colorectal carcinomas and corresponding liver metastases. J Pathol. 1999;188:258–262. doi: 10.1002/(SICI)1096-9896(199907)188:3<258::AID-PATH350>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Al-Mulla F, Keith WN, Pickford IR, Going JJ, Birnie GD. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(sici)1098-2264(199904)24:4<306::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 25.Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, Schorderet D, Bosman FT, Chaubert P. Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab Invest. 2001;81:217–229. doi: 10.1038/labinvest.3780230. [DOI] [PubMed] [Google Scholar]
- 26.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 27.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggests a suppressor role in kidney, brain and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 28.Velentza AV, Schumacher AM, Weiss C, Egli M, Watterson DM. A protein kinase associated with apoptosis and tumor suppression: structure, activity, and discovery of peptide substrates. J Biol Chem. 2001;276:38956–38965. doi: 10.1074/jbc.M104273200. [DOI] [PubMed] [Google Scholar]
- 29.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabbioni S, Miotto E, Veronese A, Sattin E, Gramantieri L, Bolondi L, Calin GA, Gafa R, Lanza G, Carli G, et al. Multigene methylation analysis of gastrointestinal tumors: TPEF emerges as a frequent tumor-specific aberrantly methylated marker that can be detected in peripheral blood. Mol Diagn. 2003;7:201–207. doi: 10.1007/BF03260039. [DOI] [PubMed] [Google Scholar]
- 31.Uchida T, Wada K, Akamatsu T, Yonezawa M, Noguchi H, Mizoguchi A, Kasuga M, Sakamoto C. A novel epidermal growth factor-like molecule containing two follistatin modules stimulates tyrosine phosphorylation of erbB4 in MKN28 gastric cancer cells. Biochem Biophys Res Commun. 1999;266:593–602. doi: 10.1006/bbrc.1999.1873. [DOI] [PubMed] [Google Scholar]
- 32.Horie M, Mitsumoto Y, Kyushiki H, Kanemoto N, Watanabe A, Taniguchi Y, Nishino N, Okamoto T, Kondo M, Mori T, et al. Identification and characterization of TMEFF2, a novel survival factor for hippocampal and mesenphalic neurons. Genomics. 2000;67:146–152. doi: 10.1006/geno.2000.6228. [DOI] [PubMed] [Google Scholar]