Abstract
Initiation of transcription of protein-encoding genes by RNA polymerase II (Pol II) was thought to require transcription factor TFIID, a complex comprised of the TATA box-binding protein (TBP) and TBP-associated factors (TAFIIs). In the presence of TBP-free TAFII complex (TFTC), initiation of Pol II transcription can occur in the absence of TFIID. TFTC containing the GCN5 acetyltransferase acetylates histone H3 in a nucleosomal context. We have identified a 130 kDa subunit of TFTC (SAP130) that shares homology with the large subunit of UV-damaged DNA-binding factor. TFTC preferentially binds UV-irradiated DNA, UV-damaged DNA inhibits TFTC-mediated Pol II transcription and TFTC is recruited in parallel with the nucleotide excision repair protein XP-A to UV-damaged DNA. TFTC preferentially acetylates histone H3 in nucleosomes assembled on UV-damaged DNA. In agreement with this, strong histone H3 acetylation occurs in intact cells after UV irradiation. These results suggest that the access of DNA repair machinery to lesions within chromatin may be facilitated by TFTC via covalent modification of chromatin. Thus, our experiments reveal a molecular link between DNA damage recognition and chromatin modification.
Keywords: histone acetyltransferase/nucleotide excision repair/spliceosome assembly factor/TATA box-binding protein-associated factors (TAFIIs)/xeroderma pigmentosum group E
Introduction
Transcription initiation of protein-encoding genes by RNA polymerase II (Pol II) was thought to require transcription factor TFIID, which is comprised of the TATA box-binding protein (TBP) and a series of TBP-associated factors (TAFIIs) (Tansey and Herr, 1997; Bell and Tora, 1999). However, we have shown recently that initiation of Pol II transcription can occur in the absence of TFIID, in the presence of a novel human (h) multiprotein complex, termed TFTC for TBP-free TAFII complex (Wieczorek et al., 1998). TFTC is able to direct pre-initiation complex assembly on both TATA-containing and TATA-less promoters in vitro. TFTC contains neither TBP nor TBP-like factor, but is composed of several TAFIIs (Wieczorek et al., 1998). The three-dimensional structure of TFTC has been determined, together with that of TFIID, at 3.5 nm resolution by electron microscopy and digital image analysis of single particles (Brand et al., 1999a). Human TFTC resembles a macromolecular clamp that contains five globular domains organized around a solvent-accessible groove of a size suitable to bind DNA. TFIID contains only four domains, which are also organized around a solvent-accessible groove (Andel et al., 1999; Brand et al., 1999a). Comparison of the two three-dimensional models indicates that the structure of TFIID is almost included in that of TFTC, further confirming some of the described functional similarities.
TFTC, similarly to other TBP-free TAFII complexes, including yeast SAGA, hSTAGA and hPCAF/GCN5, contains the histone acetyltransferase (HAT) hGCN5 and is able to acetylate histone H3 in both a free and a nucleosomal context (Grant et al., 1998; Martinez et al., 1998; Ogryzko et al., 1998; Brand et al., 1999b; Brown et al., 2000). The fact that histone acetylation has been linked to the activation of transcription (Kuo and Allis, 1998) suggests that TFTC is recruited to chromatin templates by activators to acetylate histones and potentiate transcriptional initiation (Wieczorek et al., 1998; Brand et al., 1999b). Additional TFTC subunits common to other human TAFII–HAT complexes recently have been identified, including hADA3, hTAFII150, hSPT3, hPAF65β and TRRAP (Brand et al., 1999b).
Many proteins essential for spliceosome formation and splicing have been identified (Kramer, 1996; Will and Luhrmann, 1997). Among the best characterized of these factors are the components of the U2 snRNP. In mammalian cells, 17S U2 snRNP can be assembled from 12S snRNP and two essential splicing complexes SF3a and SF3b (Brosi et al., 1993a,b). All of the components of SF3a and three components of the SF3b (SAP49, SAP145 and SAP155) have been cloned previously (Brosi et al., 1993a,b; Kramer, 1996). Recently, the fourth component of SF3b has been identified, and termed SAP130 (Das et al., 1999). In parallel, the purification of the yeast U2 snRNP revealed a novel pre-mRNA splicing factor required for pre-spliceosome assembly, called yRse1p, which is the yeast homologue of SAP130 (Caspary et al., 1999). Furthermore, SAP130 (or KIAA0017) was also purified from nuclear interchromatin granule clusters (IGCs) in which components of the pre-mRNA splicing machinery are localized (Mintz et al., 1999). More recently, SAP130 has also been identified in a novel N-CoR corepressor complex (Underhill et al., 2000). Interestingly, SAP130 has striking sequence similarity over its entire length to the large subunit of UV-damaged DNA-binding protein (UV-DDB p127), a protein associated with the hereditary disease xeroderma pigmentosum group E, a human disease characterized by defective nucleotide excision repair (NER) and an increased incidence of skin cancer (Keeney et al., 1994; and references therein). SAP130 also shows significant homology to the 160 kDa subunit of cleavage polyadenylation specificity factor (CPSF160) (Caspary et al., 1999; Das et al., 1999; Mintz et al., 1999) and the fission yeast Rik1p gene silencing protein (Neuwald and Poleksic, 2000). Moreover, it appears that these proteins belong to the UV-DDB-127 family (p127, SAP130, Rse1p, CPSF160, Cft1p and Rik1p) that share a common structure composed of two or three β-propeller domains, each containing 16–21 repeats (Neuwald and Poleksic, 2000). These repeats are similar in structure to the WD40 repeats present in the WD40 family of β-propeller-containing proteins (Neer et al., 1994; Smith et al., 1999).
Microsequencing of a novel subunit of TFTC has allowed us to identify a 130 kDa polypeptide that is identical to SAP130 (Das et al., 1999) and KIAA0017 (Nomura et al., 1994; Mintz et al., 1999). Since SAP130 resembles the large subunit of UV-DDB p127, which has been implicated in the repair of DNA lesions in chromatin (Rapic Otrin et al., 1998), we examined the ability of TFTC to recognize DNA damage. Our results demonstrate that TFTC is able to bind UV-damaged DNA in vitro in both a free and a nucleosomal context. Moreover, we show that as a consequence of this increased binding, TFTC preferentially acetylates nucleosomes assembled on UV-irradiated DNA. Thus, our experiments provide a direct molecular link between DNA damage recognition and histone acetylation.
Results
The splicing factor SAP130 is a subunit of TFTC
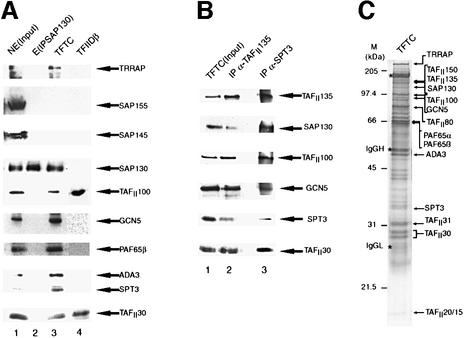
To characterize unidentified polypeptides of the TFTC complex further, we microsequenced an ∼130 kDa TFTC component and obtained three peptide sequences (Materials and methods). Database searches using these peptides identified a human protein (KIAA0017; Nomura et al., 1994) encoded by a 4261 bp open reading frame which is identical to the recently identified human splicing factor known by several names: SAP130 (SAP for spliceosome-associated factor), human Rse1 or KIAA0017 (Nomura et al., 1994; Caspary et al., 1999; Das et al., 1999; Mintz et al., 1999). Hereafter we call the identified ∼130 kDa polypeptide SAP130. An anti-SAP130 mouse monoclonal antibody (mAb) directed against a 20 amino acid peptide of SAP130 (Materials and methods) was generated. This antibody specifically recognized a polypeptide with an apparent mol. wt of ∼130 kDa in the purified TFTC complex (Figure 1A, lane 3), but not in the purified TFIID complex (lane 4), further confirming the specific association of the identified SAP130 protein with the TFTC complex.
Fig. 1. SAP130 is a component of the TFTC complex. (A) A 50 µg aliquot of HeLa cell nuclear extract (NE; lane 1), 15 µl of immuno purified and eluted SAP130 from HeLa NE [E(IPSAP130); lane 2], 15 µl of immunopurified TFTC (lane 3) and TFIIDβ (lane 4) were separated by SDS–PAGE, blotted on a nitrocellulose filter and examined for the presence of the indicated proteins using the corresponding antibodies. (B) TFTC (lane 1, and see C) was re-immunoprecipitated (IP) with either an anti-TAFII135 mAb (lane 2) or an anti-hSPT3 mAb (lane 3), the mAb–protein G-bound proteins were washed extensively, separated by SDS–PAGE, blotted on a nitrocellulose filter and examined for the presence of the indicated proteins by using the corresponding antibodies. (C) A 15 µl aliquot of TFTC was separated by SDS–PAGE and analysed by silver staining.
Since SAP130 has been identified as a component of the SF3b–SAP spliceosome complex together with other SAPs, i.e. SAP49, SAP145 and SAP155 (Das et al., 1999), we verified whether the other SAP subunits are present in the TFTC complex (Figure 1A, lane 3). Anti-SAP145 and anti-SAP155 antibodies recognized these proteins in a HeLa cell nuclear extract (NE), but not in the TFTC complex (Figure 1A, lanes 1 and 3), indicating that these SAPs are absent from TFTC.
To investigate the different nuclear pools of SAP130 that may be free or associated with complexes including TFTC and SF3b, SAP130 was immunoprecipitated from HeLa cell NE using an anti-SAP130 mAb. The anti-SAP130 mAb-bound proteins were eluted with the corresponding epitope peptide and tested for the presence of components of the SF3b, TFTC and/or TFIID complexes. Whereas SAP130 was easily detected in the eluate [E (IPSAP130); Figure 1A, lane 2], none of the other proteins tested (subunits from SF3b, TFTC and/or TFIID) was associated with SAP130 in this fraction (Figure 1A, lane 2). These results are consistent with the previously published observation that antibodies raised against a C-terminal epitope of SAP130 are not able to immunoprecipitate the SF3b complex (Das et al., 1999). Thus, the immunoprecipitated pool of SAP130 is not associated with subunits of SF3b, TFTC or TFIID complexes.
To examine the stable association of SAP130 with TFTC further, the TFTC complex (shown in Figure 1C; and hereafter used in all experiments where labelled as TFTC) was re-immunoprecipitated with either an anti-hTAFII135 mAb or an anti-hSPT3 mAb. The resin-bound proteins were washed extensively and the anti-hTAFII135 mAb- or the anti-hSPT3 mAb-bound proteins were tested by western blotting for the presence of components of TFTC (Figure 1B). In both cases, SAP130 co-immunoprecipitated with both hTAFII135 and hSPT3, together with other known TFTC components, namely hGCN5, hTAFII100 and hTAFII30 (Figure 1B, lanes 2 and 3). Thus, SAP130 is a stable component of the TFTC complex. The fact that we were able to re-immunoprecipitate subunits of the TFTC complex with an anti-TAFII135 antibody (Figure 1B, lane 2) and that the anti-hSPT3 antibodies also co-immunoprecipitated TAFII135 (lane 3) together confirm unequivocally that TAFII135 is a stable compon ent of TFTC (Wieczorek et al., 1998; Brand et al., 1999b). However, the clear differences in the stoichiometry of the different proteins in the TFTC complex composition after either anti-TAFII135 or anti-hSPT3 immunoprecipitation (Figure 1B) show, in agreement with our previous results (Brand et al., 1999a,b), that multiple TBP-free TAFII HAT complexes exist.
The SAP130-containing TFTC complex does not affect splicing in vitro
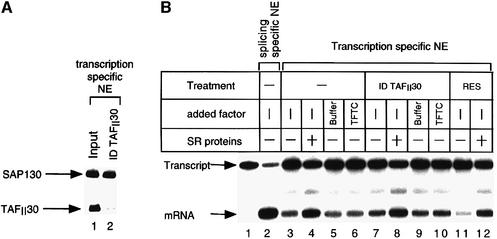
To test whether the SAP130-containing TFTC complex could play a role in correct splicing of pre-mRNAs, a HeLa NE (Dignam et al., 1983) was immunodepleted with an anti-TAFII30 mAb, which reacts with all TAFII30-containing complexes, including TFTC (Wieczorek et al., 1998). The elimination of the vast majority of TFTC from the extract (Figure 2A) had no effect on its splicing capacity in either the presence or absence of SR proteins (serine–arginine-rich splicing factors) (Figure 2B). Moreover, addition of TFTC to either the untreated (Figure 2B, lane 6) or TFTC-depleted extract (ID TAFII30; lane 10) did not increase splicing efficiency. These results exclude the possibility that TFTC may interfere, via either SAP130 or another subunit, during the normal course of spliceosome formation. Notably, in the NE, when taking into consideration the efficiency (95%) of the TAFII30 depletion, the SAP130 depletion represents only ∼5% of the total nuclear SAP130 population (Figure 2A, compare lanes 1 and 2). As from the two TAFII30-containing complexes SAP130 is only present in TFTC, but not in TFIID (Figure 1A), this result suggests that only ∼5% of the total nuclear SAP130 is present in TFTC complexes. This may explain why no important decrease in splicing activity was observed in the TAFII30-depleted extracts (Figure 2B). In addition, we have repeated the splicing experiment using an S100 cytoplasmic extract (competent for splicing), that contained ∼50–100 times less SAP130 when compared with the NE, but even in this system the addition of the SAP130-containing TFTC complex had no effect on correct splicing (data not shown). Taken together, these results suggest that TFTC is not involved in splicing in vitro.
Fig. 2. The SAP130 contained in the TFTC is not involved in splicing. (A) A 500 µg aliquot of NE was immunodepleted with an anti-TAFII30 (ID TAFII30) antibody. Proteins were analysed by western blotting using the indicated antibodies. The efficiency of the anti-TAFII30 immunodepletion and that of the SAP130 depletion was calculated by using a Bio-Rad GS700 Imaging Densitometer. (B) The extracts tested in (A) were used for in vitro splicing assays with a short version of the adenovirus E1A gene, which is spliced only in the 13S mRNA. The pre-mRNA (Transcript) and the correctly spliced transcript (mRNA) are indicated. Where indicated, 100 ng of SR proteins and 150 ng of TFTC were added to the reactions. Res: control immunodepletion with protein G–Sepharose alone.
SAP130 as well as the TFTC are recruited preferentially on UV-damaged DNA
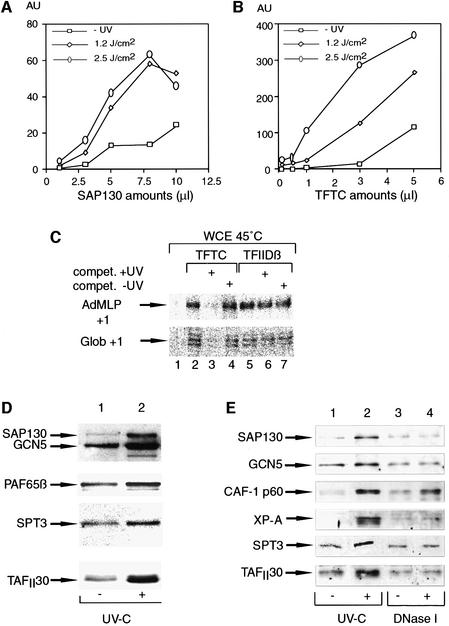
As SAP130 shows 50.7% similarity (24.5% identity) and a similar predicted structure (Neuwald and Poleksic, 2000) to the large subunit of the UV-damaged DNA-binding factor (p127 or DDB1), we tested whether the TFTC complex or the immunoprecipitated SAP130 is able to bind damaged DNA using a standard DNA filter binding assay (Materials and methods). Interestingly, SAP130, which is free of other TFTC and SF3b subunits tested (Figure 1A, lane 2), showed significant binding to the DNA fragment damaged by UV irradiation (Figure 3A). Similarly, the TFTC complex also bound preferentially to damaged DNA in a UV dose-dependent manner (Figure 3B). Surprisingly, at certain concentrations, TFTC bound ∼60- to 100-fold better to UV-damaged DNA (irradiated at 2.5 J/cm2) than to non-irradiated DNA (Figure 3B). Moreover, TFTC bound at least 10 times more efficiently to UV-damaged DNA than SAP130 alone, when equivalent amounts of SAP130 were used in both fractions (Figures 3A and B, and 1A, lanes 2 and 3). These results demonstrate that SAP130 free of other SF3b or TFTC subunits can bind to UV-damaged DNA as predicted from the sequence and the structure similarity between SAP130 and the p127 subunit of UV-DDB. Furthermore, they suggest that either the damaged DNA-binding capability of SAP130 is enhanced in the TFTC complex by additional factors, or another UV-damaged DNA-binding activity is present in the TFTC complex.
Fig. 3. SAP130 and TFTC preferentially bind to UV-damaged DNA. A 32P-labelled DNA fragment was UV irradiated at different doses as indicated, incubated with increasing amounts of either immunopurified SAP130 (A) or TFTC (B) and tested for retention on nitrocellulose filters. The amounts (µl) of SAP130 and TFTC used in this assay were previously normalized in Figure 1A. Graphs represent the amount of labelled DNA retained on the filters in arbitrary units (AU). AUs were obtained after phosphoimager scanning of the nitrocellulose filters and by deducting the buffer-only background from every TFTC- or SAP130-containing sample. Similar and comparable results (± 5%) were obtained in at least three independent experiments. (C) UV-irradiated DNA inhibits TFTC-mediated Pol II transcription in vitro. TFTC and TFIIDβ were pre-incubated for 10 min with 25 ng of templates, containing the AdMLP or the β-globin (Glob) promoter, in the absence or presence (100 ng) of either UV-irradiated (compet. +UV) or non-UV-irradiated (compet. –UV) competitor DNA fragments. Then the heat-treated HeLa whole-cell extract (WCE, 45°C) was added for 20 min and transcription initiated. The positions of the correctly initiated transcripts from the AdMLP (+1) and the Glob (+1) promoters, determined by quantitative S1 mapping, are indicated. (D) Nucleosomal arrays were assembled as in Figure 5A and the binding of TFTC (10 µl) to the non-irradiated (–) or UV-C-irradiated (2.5 J/cm2) templates was analysed by western blotting with the indicated antibodies. (E) Preferential recruitment of TFTC components and the repair damage recognition protein XP-A from a repair-competent HeLa NE onto bead-linked UV-damaged DNA (2.5 J/cm2) (UV-C), but not bead-linked DNA containing single-stranded breaks (DNase I). The different bead-linked DNA fragments were incubated with the repair-competent HeLa NE (50 µg), washed, and bound proteins were analysed by western blotting. Chromatin assembly factor-1 (CAF-1 p60) is recruited during the repair of both types of damage.
UV-damaged DNA specifically blocks TFTC-mediated in vitro transcription
Mammalian cells respond to UV irradiation by a rapid, transient inhibition of overall transcription (Rockx et al., 2000 and references therein). TFTC was shown to nucleate Pol II transcription initiation (Wieczorek et al., 1998), thus, if the UV-damaged DNA-binding activity of TFTC has a functional relevance, UV-irradiated DNA should inhibit TFTC-mediated Pol II transcription. To test this hypothesis, TFTC and, as a control, TFIIDβ, which does not contain SAP130, were first pre-incubated with the promoter-containing DNA templates [the adenovirus major late (AdMLP) and the rabbit β-globin (Glob) promoters] in the presence of UV-irradiated or non-irradiated competitor DNA fragments, lacking functional promoters (Figure 3C). Then, as a source of additional transcription factors, a heat-inactivated HeLa cell whole-cell extract (Nakajima et al., 1988; Wieczorek et al., 1998) was added to the reactions for 30 min at 25°C and transcription started. In good agreement with the specific UV-damaged DNA-binding activity of TFTC, an excess of the UV-irradiated competitor DNA completely abolished the TFTC-dependent transcription initiation (Figure 3C, lane 3), whereas the same amount of non-irradiated DNA had no effect on the TFTC-mediated transcription (lane 4). Moreover, neither the UV-irradiated nor the non-irradiated competitor DNA had an effect on the TFIID-dependent transcription initiation (Figure 3C, lanes 5–7), indicating that TFTC has a much higher affinity for UV-irradiated DNA than has TFIID (Vichi et al., 1997). These results together demonstrate the specificity of the UV-damaged DNA-binding activity of TFTC in a functional in vitro assay and suggest that TFTC can be recruited rapidly to UV-induced lesions in the cells, leading to the observed transcription inhibition after UV irradiation.
TFTC binds preferentially to nucleosomes assembled on UV-damaged DNA
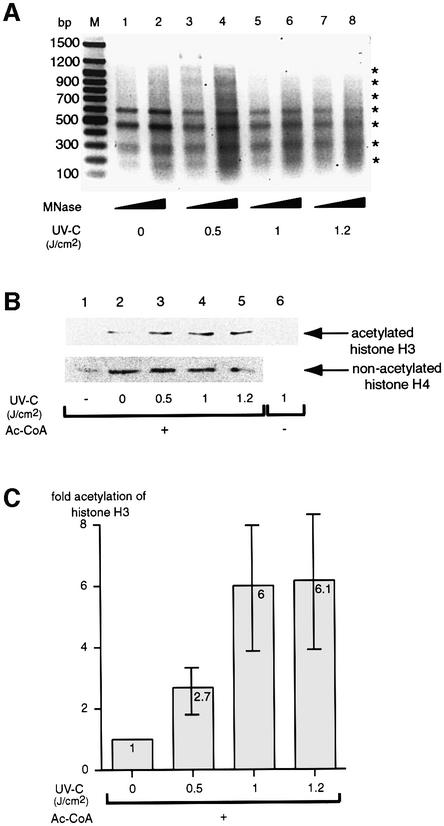
In order to examine whether TFTC could bind to UV-damaged DNA assembled with nucleosomes, we used a 1.2 kb DNA fragment linked to paramagnetic beads. After UV irradiation, this fragment was assembled in a regularly spaced nucleosomal array containing 5–6 nucleosomes (Materials and methods). The efficiency of assembly was tested by micrococcal nuclease (MNase) digestion (Figure 5A; and see below). Nucleosomal arrays reconstituted with UV-damaged DNA were then incubated with the purified TFTC complex and, after several washes, bound proteins were eluted and analysed by western blotting (Figure 3D). On these nucleosomal array- containing templates, TFTC also bound preferentially to the template that was assembled on UV-irradiated DNA (Figure 3D, compare lane 1 with lane 2). Thus, nucleosomes do not prevent the preferential binding of TFTC to the UV-damaged DNA.
Fig. 5. TFTC preferentially acetylates nucleosomal arrays assembled on UV-damaged DNA templates. (A) A bead-linked DNA fragment, with five copies of the 208 bp 5S rDNA nucleosome positioning sequence that allows the formation of regular nucleosomal arrays, was irradiated with increasing UV-C doses (as indicated) and reconstituted with purified human histone octamers. Bead-linked nucleosomal arrays were either digested with MNase, separated on an agarose gel and stained with ethidium bromide (asterisks indicate nucleosome boundaries), or incubated with 200 ng of TFTC in the presence (+) or absence (–) of acetyl-CoA and washed. (B) Acetylated histones were analysed by western blotting using antibodies recognizing acetylated histones. Nucleosomes assembled on the different DNA fragments were verified with antibodies recognizing non-acetylated histone H4 or acetylated histone H3. Lane 1, magnetic only beads. (C) The graph represents the acetylation of histone H3 in four independent experiments after normalization for DNA and histone content in the UV-irradiated samples. The standard deviation is given by error bars.
Parallel recruitment of TFTC subunits and the nucleotide excision repair damage recognition protein XP-A to UV-damaged DNA
The observation that purified TFTC complex could bind to UV-irradiated DNA both in a free and a nucleosomal context led us to examine whether this complex could also be recruited to UV-induced DNA photoproducts in a cell-free system competent for DNA repair. Linearized plasmid DNA was linked to paramagnetic beads and either UV photoproducts were introduced using a UV-C lamp or single strand breaks were generated using DNase I. Bead-linked DNA substrates containing either no damage, UV-C photoproducts or single strand breaks were incubated in a protein extract competent for DNA repair and bound proteins were eluted and analysed by western blotting (Moggs et al., 2000) (Figure 3E). Different TFTC components including SAP130, GCN5, SPT3 and TAFII30 bound preferentially to UV-damaged DNA (lanes 1 and 2). Consistent with previous observations, this damage- specific binding was paralleled by that of the NER damage recognition protein XP-A (Jones and Wood, 1993). Both TFTC and XP-A exhibit a specificity for UV photoproducts since no preferential binding was observed for either factor on DNA containing single strand breaks (Figure 3E, lanes 3 and 4). In contrast, the histone chaperone chromatin assembly factor-1 (CAF-1 p60) was recruited preferentially during the repair of both types of damage (lanes 1–4) (Moggs et al., 2000). In this extract, the preferential binding of several TFTC components (i.e. SAP130, GCN5, TAFII30 and SPT3) to UV-irradiated DNA increased in proportion to the UV fluence and in parallel with that observed for XP-A (data not shown). These results show that TFTC preferentially binds to UV-irradiated DNA in a cell-free system competent for DNA repair. Thus, we have shown that TFTC can bind preferentially to UV-damaged DNA in three independent systems: DNA free of histones, nucleosomes and in the context of a cellular extract.
TFTC preferentially acetylates nucleosomes assembled on UV-damaged DNA templates
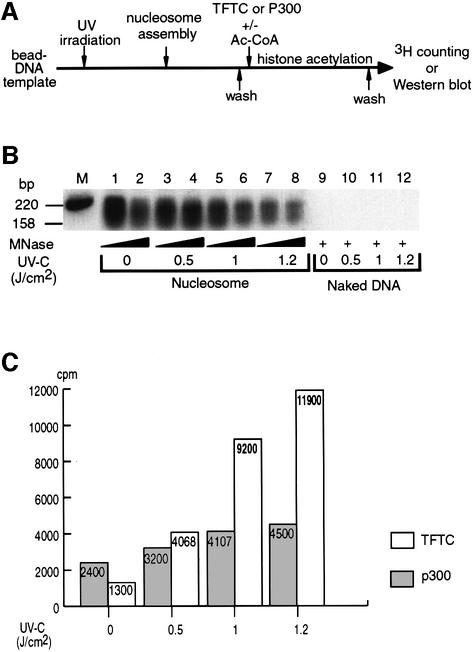
We have shown previously that TFTC is able to acetylate histone H3 in a nucleosomal context (Brand et al., 1999b). We next wanted to examine whether this property is modified when the nucleosomes are assembled on UV-damaged DNA. Thus, to study the possible involvement of the UV-damaged DNA-binding capacity of TFTC at the chromatin level, we developed two assays in which nucleosome acetylation can be monitored on UV-damaged DNA templates (Figures 4 and 5). In the first assay, a 220 bp DNA fragment was linked to paramagnetic beads and UV irradiated. Mononucleosomes were assembled using these fragments and the efficiency of assembly was assessed by either MNase digestion followed by Southern blot analysis (Figure 4B) or silver staining of histones (data not shown). After irradiation, we observed a UV dose-dependent loss of DNA fragments from the beads, probably due to double-stranded breaks within the template (data not shown). Moreover, it has been reported that the assembly of mononucleosomes on UV-damaged DNA is less efficient than on non-irradiated DNA (Matsumoto et al., 1995) (see also Figure 4B, lanes 1–8). After the assembly, TFTC or p300, a transcriptional coactivator with HAT activity (Ogryzko et al., 1996), was added to the nucleosome-containing beads in the presence of [3H]acetyl-CoA, and then incubated for 30 min. The excess [3H]acetyl-CoA was washed away and the radioactivity incorporated into the mononucleosomes was counted (Figure 4A). Interestingly, not only was TFTC able to acetylate histones in a mononucleosomal context containing UV-damaged DNA, but the acetylation was significantly increased (9-fold) with mononucleosomes assembled on damaged DNA (Figure 4C). In contrast, p300-mediated acetylation on mononucleosomes assembled on UV-irradiated DNA was only slightly increased when compared with non-treated nucleosomes (1.8-fold; Figure 4C). Consistent with our previous results, the TFTC-dependent acetylation was specific to histone H3 (data not shown; and see Brand et al., 1999b).
Fig. 4. TFTC preferentially acetylates mononucleosomes assembled on UV-damaged DNA templates. (A) A bead-linked 220 bp DNA fragment irradiated with increasing UV-C doses was reconstituted either with (nucleosome) or without (naked DNA) human histone octamers. Bead-linked nucleosomes (or naked DNA) were then either (B) digested with MNase, transfered to a nylon membrane and hybridized with a 32P-labelled probe, or (C) incubated with 200 ng of TFTC or 5 ng of p300 in the presence of [3H]acetylCoA, washed and counted for incorporation of radioactivity in histones. The graph represents the average of two independent experiments performed in duplicate and is normalized for the loss of DNA due to UV irradiation. Grey column, p300; white column, TFTC.
In the second assay, a DNA fragment, with five copies of the 208 bp 5S rDNA nucleosome positioning sequence, which generates regularly spaced nucleosomal arrays (Steger and Workman, 1999), was immobilized on magnetic beads and UV irradiated. The immobilized array fragments were then reconstituted with core histones by salt dilution (Steger and Workman, 1999), and the formation of the nucleosomal array was assessed by MNase digestion (Figure 5A). After incubation of the nucleosomal array-containing templates with TFTC in the presence of acetyl-CoA, the acetylation of nucleosomes was assessed by western blot analysis using an mAb that specifically recognizes the acetylated form of histone H3 (Figure 5B). This experiment demonstrated that histone H3 acetylation by TFTC was stimulated in nucleosomal arrays that were assembled on UV-irradiated DNA in a UV dose-dependent manner (Figure 5B). An average of four independent experiments (each in duplicate) is represented by the graph in Figure 5C. These data demonstrate that in accordance with the preferential recruitment of TFTC to the UV-damaged nucleosomal arrays (Figure 3D), TFTC preferentially acetylates nucleosomes on UV-damaged DNA templates in vitro.
Following UV irradiation, histone acetylation is increased in HeLa cells in vivo
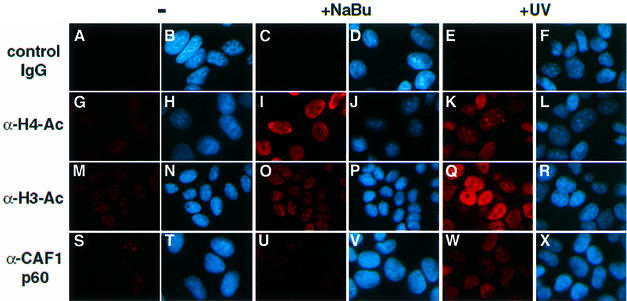
To test the validity of the above-described in vitro observations, we have investigated histone acetylation in vivo after UV irradiation. HeLa cells were UV irradiated and after 2 h histone acetylation was tested by using immunofluorescence labelling. DNA-containing structures were visualized by Hoechst staining and endogenous proteins were detected by using specific antibodies, recognizing either acetylated histone H3 (α-H3-Ac) or histone H4 (α-H4-Ac), and a Cy3-fluorochrome conjugated secondary antibody (Figure 6). As a positive control for acetylation, cells were treated with sodium butyrate (NaBu), an inhibitor of histone deacetylation (Perry et al., 1993; Boggs et al., 1996). As expected, in the NaBu-treated cells the antibodies efficiently recognized acetylated histone H3 and histone H4 (Figure 6I and O). In good agreement with our in vitro results, after UV irradiation HeLa cells were labelled efficiently with the α-H3-Ac antibodies when compared with the non-irradiated cells (Figure 6, compare M with Q), whereas cells stained with the α-H4-Ac antibodies exhibited a significant, but weaker and a more speckled distribution throughout the nucleoplasm than that seen with α-H3-Ac antibodies (Figure 6, compare K with Q). Moreover, histone H3 and H4 acetylation was paralleled by the increased binding of CAF-1 p60 or XP-A to the chromatin of UV-irradiated cells (Figure 6W; and data not shown), consistent with previous studies (Otrin et al., 1997; Martini et al., 1998). Thus, the preferential histone H3 acetylation after UV treatment in vivo, together with the above in vitro results, further suggest that TFTC may acetylate nucleosomes on UV-damaged DNA templates in vivo.
Fig. 6. Histone H3 and H4 acetylation is increased in HeLa cells after UV irradiation. HeLa cells were either untreated (–), UV irradiated (+UV) or treated with sodium butyrate (+NaBu). Cells were then fixed and subjected to immunofluorescence detection. In each column, the right-hand panels show the Hoechst DNA staining (blue), and the left-hand panels correspond to immunodetection with specific antibodies (red), as indicated. A mouse IgG fraction was used as a control in (A), (C) and (E).
Discussion
In this study, we have shown that TFTC is able to bind UV-damaged DNA in vitro in both a free and a nucleosomal context and that this binding is paralleled by the recruitment of the repair factor XP-A. Moreover, we demonstrate that as a consequence of this increased binding, TFTC preferentially acetylates histone H3 in nucleosomes assembled on UV-irradiated DNA. In accordance, we show that after UV irradiation, histone H3 acetylation is also increased in vivo.
What is the ‘real’ function of TFTC, mediating initiation of transcription or facilitation of DNA repair by nucleosome acetylation?
The results of this study imply that TFTC could have two functions: one in transcription initiation and one in facilitating the DNA repair. These two functions of TFTC seem to be closely related (using the same enzymatic activity) and can very probably be performed by the same complex because they are founded on the same principle: (i) recognition of the transcription start site by certain TAFIIs and acetylation of nucleosomes around the promoter in the absence of UV irradiation and (ii) recognition of a 6–4 photoproduct by SAP130 and acetylation of histones in the presence of a UV damage. More importantly, initiation of transcription and DNA repair appear to be closely related as they have been shown to be interdependent processes that can implicate the same transcription factors (e.g. TFIIH or TBP). Indeed, among the diverse biological responses of human cells to UV irradiation, a rapid, transient inhibition of overall transcription is observed. This phenomenon has been explained partly by the observation that TBP/TFIID can bind directly to UV-damaged DNA, potentially making these factors unavailable for transcription (Vichi et al., 1997). Interestingly, it has been demonstrated that the recruitment, by the NER machinery, of some factors essential for transcription initiation is at least partially responsible for the UV-induced inhibition of transcription (Bootsma and Hoeijmakers, 1993; Rockx et al., 2000). In good agreement with the identification of the UV-damaged DNA-binding capability of TFTC, previously shown to play a role in transcription initiation (Wieczorek et al., 1998), here we clearly demonstrate that TFTC can be sequestered by DNA lesions in vitro and thus contributes to the general reduction of Pol II transcription observed in UV-irradiated human cells. In the case of TFTC, the affinity for UV lesions may deliver HAT activity to lesion sites within chromatin, thus facilitating the rapid access of the core NER machinery. Thus, our experiments have revealed a possible link between chromatin modifications and DNA repair.
Similarities between SAP130 and UV-DDB p127 and their relationship to chromatin
Although the most abundant lesions formed by UV irradiation are cyclobutane pyrimidine dimers, TFTC (SAP130) is most likely to be recognizing 6–4 photoproducts since the UV fluences we have used are above the range in which cyclobutane pyrimidine dimer formation and reversal have reached photochemical equilibrium, whereas the formation of 6–4 photoproducts continues to increase with UV fluences above 2 kJ/m2 (Lippke et al., 1981). Consistent with this notion, the affinity of XP-A for UV-irradiated DNA has been shown to be due mainly to 6–4 photoproducts (Jones and Wood, 1993). Interestingly, the same type of UV lesion is also recognized specifically by UV-DDB (Otrin et al., 1997). It is thus possible that the UV-DDB-127-type repeats-containing proteins (i.e. UV-DDB p127, SAP130 and CPSF160), which possess a conserved β-propeller structure (Neuwald and Poleksic, 2000), may be involved in specific DNA and/or RNA recognition events. It is also noteworthy that many protein complexes involved in chromatin assembly, modification and/or transcriptional silencing contain subunits with either UV-DDB-127- or WD40-type repeats that form β-propeller structures (Neuwald and Poleksic, 2000).
Moreover, the similarity between SAP130 and the large subunit of UV-DDB is intriguing as the UV-DDB complex may also play a specific role in the repair of chromosomal DNA in a nuclear environment (Otrin et al., 1997; Rapic Otrin et al., 1998). A tight association with chromatinized DNA was found for the p127 component of DDB as well as for other NER proteins, i.e. XP-A, RP-A and PCNA, immediately after UV irradiation (Otrin et al., 1997). The high affinity of UV-DDB, SAP130 and possibly additional damage recognition proteins for DNA lesions may be required for the recognition of DNA damage within specific chromatin structures. The fact that TFTC can recognize damaged DNA and subsequently acetylate histones more efficiently than on undamaged templates raises the possibility that the TFTC complex plays a role in making the DNA damage accessible for the repair machinery in the context of chromatin. Consistent with this hypothesis, pre-treatment of human cells with n-butyrate (producing a global increase in cellular acetylation levels) prior to UV irradiation increased subsequent NER activity (Smerdon et al., 1982; Williams and Friedberg, 1982; Ramanathan and Smerdon, 1989). Together, our data point towards a mechanism in which the access of DNA repair machinery to lesions within chromatin may be achieved via chromatin modifications similar to those associated with the activation of transcription (Meijer and Smerdon, 1999; Thoma, 1999).
Materials and methods
Microsequencing of TFTC subunits
TFTC was prepared as described in Wieczorek et al. (1998) and ∼100 µg of TFTC were separated on a preparative 8% SDS–polyacrylamide gel. The proteins were stained with Coomassie brilliant blue, excised and digested with trypsin. The eluted peptides were fractionated by reverse-phase HPLC and microsequenced as described (Brou et al., 1993b). The following microsequences were obtained: 102-IHQETFGK-109, 1160-DHLSFR-1165 and 1166-SYYFPVK-1171 (numbers indicate the corresponding amino acid positions in the human KIAA0017 protein; DDBJ/EMBL/GenBank accession No. D87686).
Immunization, antibody production and in vivo immunofluorescence detection
To generate the anti-SAP130, anti-GCN5 and anti-SPT3 monoclonal antibodies, peptides corresponding to amino acids 1057–1076 of hSAP130 (RLPPNTNDEVDEDPTGNKAL), amino acids 118–131 of hGCN5 (CKCNGWKNPKPPTA) and amino acids 10–27 of hSPT3 (STATSSSGRSTGKSISFA) were synthesized, coupled to ovalbumin as a carrier protein and used for immunization of mice. Immunization and monoclonal antibody production were essentially as described by Brou et al. (1993a). For immunofluorescence, HeLa cells were grown on Leighton tubes (Costar, Cambridge, MA), left untreated, irradiated with UV light (0.8 J/cm2) and left to recover for 2 h, or treated with sodium butyrate (10 mM) for 90 min. The cells were then fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 4 min at room temperature and permeabilized twice for 10 min with PBS containing 0.1% Triton X-100 (PBS-Tx). After a PBS wash, the cells were incubated overnight at room temperature in an humidified chamber with mouse mAb 3HH4 2C2, raised against an acetylated histone H4 peptide (Dilworth et al., 2000), or with polyclonal antibodies raised against acetylated histone H3 tails (kind gift from D.Allis) or CAF1 p60 (Martini et al., 1998). Then, the cells were washed with PBS-Tx and incubated with either a goat Cy3-anti-mouse IgG secondary antibody or with a goat Cy3-anti-rabbit secondary antibody (Jackson Immunoresearch Laboratories), diluted 1/500 in PBS-Tx for 1 h at room temperature. The cells were washed with PBS-Tx, rinsed with PBS and counterstained with Hoechst 33258 DNA dye (5 mg/ml bis-benzimide, Sigma) for 20 s. The cells were covered with mounting medium and analysed by fluorescence microscopy. Control was performed with a mouse (Figure 6) or rabbit (not shown) polyclonal IgG fraction (Jackson Immunoresearch Laboratories).
Immunoprecipitation and western blot analysis
Routinely, 100–500 µl of the indicated protein fractions were immuno precipitated (IP) with 50 µl of protein G–Sepharose (Pharmacia) and ∼2–5 µg of the different antibodies (as indicated in the figures). Antibody–protein G–Sepharose-bound protein complexes were washed three times with IP buffer [25 mM Tris–HCl pH 7.9, 10% (v/v) glycerol, 0.1% NP-40, 0.5 mM dithiothreitol (DTT), 5 mM MgCl2] containing 0.5 M KCl, and twice with IP buffer containing 100 mM KCl. After washing, proteins were eluted either by an excess of the corresponding epitope peptide (in the case of the anti-SAP130 IP) or under acidic conditions (in the case of the anti-TAFII135 IP). Proteins were boiled in SDS sample buffer, separated by SDS–PAGE, transferred to nitrocellulose membrane and probed with the indicated primary antibodies. Chemiluminescence detection was performed according to the manufacturer’s instructions (Amersham). The anti-SAP145 and SAP155 antibodies (Das et al., 1999), the anti-TRRAP antisera (McMahon et al., 1998), the anti-SPT3 antibody (Martinez et al., 1998), the anti-TAFII30, anti-TAFII100 and anti-TAFII135 mAbs (Wieczorek et al., 1998), the antibodies raised against ADA3 and PAF65β (Ogryzko et al., 1998), the antibodies against CAF-1 (p60) (Marheineke and Krude, 1998), XP-A (kind gift from R.Wood) and the anti-acetylated histone 3HH4 3H10 mAb (Dilworth et al., 2000) were described previously.
Filter binding assay
A 32P-labelled 220 bp DNA fragment was synthesized by PCR using the oligonucleotide primers 5′-CCTTCGGTCCTCCGATCGTTG-3′ and 5′-AGTTCTGCTATGTGGCGCGGT-3′ on the pBSK+ template. The labelled DNA fragment was exposed to different doses (0–2.5 J/cm2) of UV-C (254 nm) light. Each sample (TFTC or SAP130) was incubated for 30 min at 30°C with 2.5 ng of the DNA fragment (5000 c.p.m.) in 20 µl of RG10 buffer (50 mM Tris–HCl pH 7.9, 100 mM KCl, 10% glycerol, 1 mM EDTA, 0.5 mM DTT). Nitrocellulose membranes were pre-incubated in 0.4 mM KOH, washed in water and equilibrated in RG10 buffer prior to use. Each sample was then applied in duplicate under vacuum to a nitrocellulose membrane using a dot-blot apparatus (Bio-Rad). The samples were washed once with 400 µl of RG10 buffer and three times with 400 µl of RG10 buffer containing 50 mM KCl. The membrane was then removed, dried and exposed. Quantitative phosphoimager analysis was performed on a Fujix BAS 2000 apparatus. Similar and comparable results (± 5%) were obtained in at least three independent experiments.
Assay for protein factors recruited to UV-damaged DNA
The assay for detecting the recruitment of DNA repair and chromatin assembly factors to damaged DNA was described in Moggs et al. (2000).
Mononucleosome and nucleosome array assembly
For mononucleosome assembly, the DNA fragment used in the filter binding experiment was generated by PCR using the same primers described above except that the 5′ primer was biotinylated. This fragment was bound to streptavidin-linked paramagnetic dynabeads (Dynal) following the manufacturer’s instructions. The linked DNA fragments were exposed to UV-C light, as described in Moggs et al. (2000), and incubated with HeLa core histones (1:1 molar ratio) in a buffer containing 2 M KCl. The mononucleosomes were assembled by salt dilution (Steger and Workman, 1999), and washed extensively in buffer B [10 mM HEPES pH 7.6, 50 mM KCl, 5% glycerol, 10 mM sodium butyrate, 5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.25 mg/ml bovine serum albumin (BSA)]. For the nucleosome array assembly, the pIC-2085S/G5-E4R plasmid (Steger and Workman, 1999) was digested with HaeIII and Asp718 to generate a 1241 bp fragment with a 5′ overhang end. This fragment was coupled enzymatically to 14dATP biotin (Gibco-BRL) following the manufacturer’s instructions (Dynal). The free biotin and a 17 bp biotinylated fragment were removed by Sephadex G-50 gel filtration. The 1241 bp biotinylated fragment was then coupled to paramagnetic streptavidin-bound Dynabeads using the kilobases Binder Kit (Dynal), and the non-biotinylated DNA fragments were washed away. The beads were then resuspended in water and irradiated at different doses with a UV-C lamp. Fragments were then assembled in chromatin using the salt dilution method (Steger and Workman, 1999). Nucleosomal assembly was confirmed by MNase digestion. The bead-linked chromatin template contained on average 5–6 nucleosomes.
Histone acetyltransferase assay
TFTC or recombinant p300 (Kraus and Kadonaga, 1998) was incubated with nucleosomes bound to the beads in buffer B in the presence of either [3H]acetyl CoA (ICN) or cold acetyl-CoA (1.7 µM final) for 30 min at 30°C. The beads were then washed three times with 2 ml of the same buffer and either the incorporation of 3H into nucleosomes was counted or nucleosome acetylation was analysed by western blot.
For further details on Materials and methods, see Supplementary data available at The EMBO Journal Online.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to A.Staub for microsequencing, to T.Nagase and the Kazusa DNA Research Institute (Japan) for the KIAA0017 cDNA clone, to D.Allis, I.Davidson, R.Reed, Y.Nakatani, E.Martinez, K.Mareinheke, S.McMahon, C.J.Jones, R.D.Wood, Y.Lutz and S.Iwai for antibodies and reagents, to F.Coin and P.Becker for advice, to W.L.Kraus and J.T.Kadonaga for the recombinant p300 baculovirus, to D.Steger and J.L.Workman for the pIC-2085S/G5-E4R plasmid and advice, to D.Roche for technical assistance, to J.P.Quivy for his help, and to J.H.J.Hoeijmaker for helpful discussions. We also thank P.Eberling for peptide synthesis, and the cell culture group for providing cells. M.B. and F.L were supported by a fellowship from the Ministère de l’Education Nationale, de l’Enseignement Supérieur, de la Recherche et de la Technologie. J.G.M. was supported by a TMR fellowship from the EU. This work was supported by funds from INSERM, CNRS, Hôpital Universitaire de Strasbourg, Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, Ligue Nationale contre le Cancer and Human Frontier Science Foundation (to L.T.), and Institut Curie and EU TMR (to G.A).
References
- Andel F., Ladurner,A.G., Inouye,C., Tjian,R. and Nogales,E. (1999) Three-dimensional structure of the human TFIID-IIA-IIB complex. Science, 286, 2153–2156. [DOI] [PubMed] [Google Scholar]
- Bell B. and Tora,L. (1999) Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res., 246, 11–19. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Connors,B., Sobel,R.E., Chinault,A.C. and Allis,C.D. (1996) Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma, 105, 303–309. [DOI] [PubMed] [Google Scholar]
- Bootsma D. and Hoeijmakers,J.H. (1993) DNA repair. Engagement with transcription. Nature, 363, 114–115. [DOI] [PubMed] [Google Scholar]
- Brand M., Leurent,C., Mallouh,V., Tora,L. and Schultz,P. (1999a) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science, 286, 2151–2153. [DOI] [PubMed] [Google Scholar]
- Brand M., Yamamoto,K., Staub,A. and Tora,L. (1999b) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem., 274, 18285–18289. [DOI] [PubMed] [Google Scholar]
- Brosi R., Groning,K., Behrens,S.E., Luhrmann,R. and Kramer,A. (1993a) Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science, 262, 102–105. [DOI] [PubMed] [Google Scholar]
- Brosi R., Hauri,H.P. and Kramer,A. (1993b) Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem., 268, 17640–17646. [PubMed] [Google Scholar]
- Brou C., Chaudhary,S., Davidson,I., Lutz,Y., Wu,J., Egly,J.M., Tora,L. and Chambon,P. (1993a) Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J., 12, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C., Kuhn,A., Staub,A., Chaudhary,S., Grummt,I., Davidson,I. and Tora,L. (1993b) Sequence-specific transactivators counteract topo isomerase II-mediated inhibition of in vitro transcription by RNA polymerase I and II. Nucleic Acids Res., 21, 4011–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Lechner,I., Howe,I. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Caspary F., Shevchenko,A., Wilm,M. and Seraphin,B. (1999) Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J., 18, 3463–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B.K., Xia,L., Palandjian,L., Gozani,O., Chyung,Y. and Reed,R. (1999) Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145 and 155. Mol. Cell. Biol., 19, 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth F.J., Fromental-Ramain,C., Yamamoto,K. and Chambon,P. (2000) ATP-driven chromatin remodelling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell, 6, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates, J.R. and Workman,J.L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Jones C.J. and Wood,R.D. (1993) Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry, 32, 12096–12104. [DOI] [PubMed] [Google Scholar]
- Keeney S., Eker,A.P., Brody,T., Vermeulen,W., Bootsma,D., Hoeijmakers,J.H. and Linn,S. (1994) Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl Acad. Sci. USA, 91, 4053–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev., 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Lippke J.A., Gordon,L.K., Brash,D.E. and Haseltine,W.A. (1981) Distribution of UV light-induced damage in a defined sequence of human DNA: detection of alkaline-sensitive lesions at pyrimidine nucleoside-cytidine sequences. Proc. Natl Acad. Sci. USA, 78, 3388–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marheineke K. and Krude,T. (1998) Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem., 273, 15279–15286. [DOI] [PubMed] [Google Scholar]
- Martinez E., Kundu,T.K., Fu,J. and Roeder,R.G. (1998) A human SPT3–TAFII31–GCN5-L acetylase complex distinct from tran scription factor IID. J. Biol. Chem., 273, 23781–23785. [DOI] [PubMed] [Google Scholar]
- Martini E., Roche,D.M., Marheineke,K., Verreault,A. and Almouzni,G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol., 143, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H., Takakusu,A., Mori,T., Ihara,M., Todo,T. and Ohnishi,T. (1995) Preferential inhibition of nucleosome assembly by ultraviolet-induced (6–4) photoproducts. Photochem. Photobiol., 61, 459–462. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk,H.A., Dugan,K.A., Copeland,T.D. and Cole,M.D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 94, 363–374. [DOI] [PubMed] [Google Scholar]
- Meijer M. and Smerdon,M.J. (1999) Accessing DNA damage in chromatin: insights from transcription. BioEssays, 21, 596–603. [DOI] [PubMed] [Google Scholar]
- Mintz P.J., Patterson,S.D., Neuwald,A.F., Spahr,C.S. and Spector,D.L. (1999) Purification and biochemical characterization of interchromatin granule clusters. EMBO J., 18, 4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs J.G., Grandi,P., Quivy,J.P., Jonsson,Z.O., Hubscher,U., Becker, P.B. and Almouzni,G. (2000) A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol., 20, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N., Horikoshi,M. and Roeder,R.G. (1988) Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity and TATA box–promoter interactions of TFIID. Mol. Cell. Biol., 8, 4028–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature, 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F. and Poleksic,A. (2000) PSI-BLAST searches using hidden markov models of structural repeats: prediction of an unusual sliding DNA clamp and of β-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res., 28, 3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N. et al. (1994) Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001–KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. Suppl., 1, 47–56. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani, Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schlitz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Otrin V.R., McLenigan,M., Takao,M., Levine,A.S. and Protic,M. (1997) Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J. Cell Sci., 110, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Perry C.A., Dadd,C.A., Allis,C.D. and Annunziato,A.T. (1993) Analysis of nucleosome assembly and histone exchange using antibodies specific for acetylated H4. Biochemistry, 32, 13605–13614. [DOI] [PubMed] [Google Scholar]
- Ramanathan B. and Smerdon,M.J. (1989) Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J. Biol. Chem., 264, 11026–11034. [PubMed] [Google Scholar]
- Rapic Otrin V., Kuraoka,I., Nardo,T., McLenigan,M., Eker,A.P., Stefanini,M., Levine,A.S. and Wood,R.D. (1998) Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol., 18, 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx D.A., Mason,R., van Hoffen,A., Barton,M.C., Citterio,E., Bregman,D.B., van Zeeland,A.A., Vrieling,H. and Mullenders,L.H. (2000) UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl Acad. Sci. USA, 97, 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon M.J., Lan,S.Y., Calza,R.E. and Reeves,R. (1982) Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem., 257, 13441–13447. [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Steger D.J. and Workman,J.L. (1999) Transcriptional analysis of purified histone acetyltransferase complexes. Methods, 19, 410–416. [DOI] [PubMed] [Google Scholar]
- Tansey W.P. and Herr,W. (1997) TAFs: guilt by association? Cell, 88, 729–732. [DOI] [PubMed] [Google Scholar]
- Thoma F. (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel N-CoR complex contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- Vichi P., Coin,F., Renaud,J.P., Vermeulen,W., Hoeijmakers,J.H., Moras,D. and Egly,J.M. (1997) Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J., 16, 7444–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E., Brand,M., Jacq,X. and Tora,L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Williams J.I. and Friedberg,E.C. (1982) Increased levels of unscheduled DNA synthesis in UV-irradiated human fibroblasts pretreated with sodium butyrate. Photochem. Photobiol., 36, 423–427. [DOI] [PubMed] [Google Scholar]