Abstract
hBRCA1 is involved in 20–45% of inherited breast cancer cases and is implicated in many mechanisms involved in response to DNA damage. To date, BRCA1 orthologs have been characterized in vertebrate genomes only. We have identified the ortholog of BRCA1 in Arabidopsis thaliana. AtBRCA1 is a 5.5 kb part of the locus At4g21070. The corresponding mRNA of 3.5 kb is composed of 14 exons and encodes a 941 amino acid protein (104 kDa). AtBRCA1, which has one N-terminal RING finger, two C-terminal BRCT and the p300/CBP interacting domain, shows a high similarity to hBRCA1 in these motifs and has the same characteristic molecular organization. We have also identified a putative ortholog in rice (OsBRCA1). With 941 and 968 amino acids, respectively, AtBRCA1 and OsBRCA1 are the shortest members of the BRCA1 family, and may represent a plant specificity. AtBRCA1 is expressed ubiquitously in plant tissues, at levels depending on organ type, with highest levels in flower buds and exponentially growing cell cultures. Increase of mRNA levels in all plant tissues 1 h after irradiation with the highest induction level of approximately 150 times for a 100 Gy dose is consistent with a putative role of AtBRCA1 in DNA repair and in cell-cycle control.
INTRODUCTION
DNA damage induced by gamma irradiation, or by other factors such as radio-mimetic drugs, leads to the enhancement of several pathways involved in DNA repair, cell-cycle control, transcriptional regulation and apoptosis. Two major mechanisms are responsible for the repair of double-strand breaks (DSB) generated by ionizing radiation (IR): homologous recombination (HR) and non-homologous end joining (NHEJ) (1–3). Several genes implicated in these pathways are remarkably conserved in different species and are characterized by specific domains (4,5). Genome sequencing of different organisms now allows us to find, by homology searches, new orthologs of genes described earlier.
Several Arabidopsis thaliana genes, identified previously in other organisms, are known to be involved in the repair of DSB. Among them, AtXrcc4 (6), AtLig4 (6), AtRad50 (7), AtKu70–80 (8) and AtMre11 (9) are mainly implicated in NHEJ, AtATM (10) and AtRAD51 (11) in HR. To date, orthologs of hBRCA1, implicated in either HR or NHEJ, and also in several other pathways, have been described only in vertebrates: in mammalian (12–14), Xenopus (15) and Gallus genomes (16).
hBRCA1 is located at 17q21 and is involved in 20–45% of inherited breast cancer cases and ∼80% of families predisposed to breast and ovarian cancer (17). hBRCA1 is a 220 kDa nuclear phosphoprotein with functional domains: an N-terminal RING finger domain, and two BRCA1 C-terminal (BRCT) domains with a transactivating activity in the C-terminus (18). hBRCA1 is phosphorylated by kinases like ATM (19), chk2 (20) or ATR (21) after DNA damage that leads to delocalization of hBRCA1 and enhancement of transcription-coupled DNA repair (22). Phosphorylation of the transcriptional repressor CtIP by ATM after DNA damage releases hBRCA1 from its interaction with CtIP and allows its transcriptional activity (23,24).
hBRCA1 is involved in transcription regulation, by transactivating transcription through direct protein interactions. hBRCA1 stimulates p53 transcriptional activity on the p21waf1/cip1, mdm2 and bax promoters (25,26), but also in a p53-independent manner for p21waf1/cip1 and c-myc promoters (27,28). Harkin et al. (29) have shown that enhanced hBRCA1 expression led to the modulation of the expression of several genes, such as GADD45. Moreover, hBRCA1 is a component of the RNA polymerase II holoenzyme (30,31) and interacts with proteins involved in the regulation or the transcription machinery, including CtIP (32), CREB binding protein (CBP)/p300 (33) and RNA helicase A (34). A role in chromatin remodeling has been attributed to hBRCA1 through its association with the histone deacytylases HDAC1, HDAC2 (35) and the BRG1 subunit of the SWI/SNF complex (36). Recently, it has been shown that hBRCA1 can also bind directly to DNA, a property which may be associated with its roles in transcription and in DNA repair (37).
hBRCA1 is also involved in maintaining genome integrity in several ways. First, it interacts in nuclear foci with several proteins involved in DNA repair such as hRad51 (38), BRCA2 (39,40), hRad50/hMre11/nibrin (41) and the BRCA1-associated genome surveillance complex (BASC) (42). Secondly, it is involved in controlling centrosome duplication by interaction with gamma-tubulin, a component essential for nucleation and mitotic spindle assembly (43). Xu et al. (44) have found that 30% of brca1Δ11/Δ11 cells (lacking exon 11) contained multiple centrosomes, leading to unequal chromosomal segregation and aneuploidy. Additional studies have linked hBRCA1 mRNA levels with sensitivity to microtubule-interfering agents which poison the mitotic spindle by inhibiting either the depolymerization or the polymerization of tubulin (45,46).
Recently, crystal structures of the RING and BRCT domains of hBRCA1 have been established, giving insight into the molecular properties of hBRCA1 (15,47). Brzovic et al. (47) determined the interaction structure between the RING domains of hBRCA1 and hBARD1 (BRCA1 associated ring domain 1), providing information on the structural effects of mutations in this domain and also on the ubiquitin ligase activity of this heterodimer. The crystal structure determination of the BRCT domains has revealed that it can homodimerize and this may be essential to BRCA1 function (48). Moreover, Xenopus orthologs of BRCA1 and BARD1 have been shown to also form functional heterodimers (15).
In an attempt to find Arabidopsis or plant genes implicated in IR response, with a strong implication in DNA repair and transcription regulation, and also because of the presence of DNA repair genes in the A.thaliana genome such as AtATM and AtRAD51, we have searched for an ortholog of BRCA1. Here, we report the characterization of AtBRCA1 and analysis of its mRNA expression.
MATERIALS AND METHODS
GenBank accession
Arabidopsis thaliana BRCA1 (AtBRCA1) had the GenBank accession no. AF515728.
Materials
Arabidopsis thaliana (var. Columbia, ecotype Col-0) seeds were grown in vitro for 4 or 20 days on modified Somerville medium in a chamber with a dark/light cycle of 10/14 h (49,50). Cell suspensions were cultivated at 25°C in an orbital shaker at 120 r.p.m. (Multitron II, Infors, Germany) under continuous light (60 µmol photons/m2/s) in MS medium as described earlier (51). Plants or cells were irradiated with a 60Co source at a dose rate of 22 Gy min–1, maintained for 1 h under respective standard growth conditions and then frozen in liquid nitrogen and ground for 5 min at 300 r.p.m. in the planetary ball mill PM400 (Retsch, Germany). Ground tissues harvested from independent experiments were pooled before RNA extraction.
Computational analysis
AtBRCA1 was identified using BLAST implemented on NCBI and TAIR databases with the hBRCA1 sequence, using the default parameter settings. Gene structure prediction was done on software implemented on the Softberry web page (http://www.softberry.com/), analysis of protein domains using the SMART database (http://smart.embl-heidelberg.de/), alignment of sequences using Alignp, and phylogenetic analysis using PHYlogeny Inference Package on Infobiogen web page (http://www.infobiogen.fr). BLAST research on rice genome (Oryza sativa) was performed on the Beijing Genomics Institute database (http://btn.genomics.org.cn/rice) (52).
RNA isolation and reverse transcription
Total RNA extraction was done using Trizol® (Invitrogen) on ground frozen tissues according to the manufacturer’s protocol. RNA was quantified spectrophotometrically at 260 nm. One microgram of RNA was reverse transcribed using the first-strand cDNA synthesis kit (Amersham Pharmacia) with random hexamers at 37°C for 1 h.
Poly(A)+ mRNAs were obtained using the Dynabeads mRNA direct kit (Dynal) following the instructions of the manufacturer. Briefly, 200 mg of ground frozen tissue was lysed using 1.5 ml of lysis/binding buffer. mRNA isolation was performed on centrifuged crude lysate using 500 µl of Dynabeads oligo (dT)25. cDNA synthesis was performed directly on the beads using oligo (dT)25 primers. Reverse transcription was performed for 1 h at 37°C using 500 U M-MLV reverse transcriptase in 10 mM DTT, 500 µM dNTP, 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2 (Invitrogen). The first-strand cDNA covalently linked to the beads was used for PCR amplification or 5′ rapid amplification of cDNA ends (RACE).
cDNA sequencing
To identify the 5′ end of AtBRCA1, 5′ RACE was done using poly(A)+ RNA as template and either poly(T) or a specific primer in the 3.5 kb predicted sequence for reverse transcription. A string of deoxyguanosine was added at the 3′ end of the cDNA by a DNA terminal transferase (10). PCR was done using (GA)12GCTCACTAGT5(C)14 as forward primer and AtB1q-R as reverse primer. PCR products were sequenced as described below.
Gene structure prediction allowed us to design oligonucleotides corresponding to the putative cDNA sequence. Amplification was performed in 25 µl containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 0.001% gelatin, 400 nM primers (MWG-biotech), 200 µM dNTP, 2.5 mM MgCl2, 1.5 U Taq polymerase (Sigma) and 5 ng of cDNA at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 10 min at 72°C. PCR products were purified on Qiaquick columns following the manufacturer’s instructions (Qiagen). Sequence reactions were done with the BigDye terminator kit (Applied Biosystems) on 30 ng of PCR product and analyzed on an ABI Prism 310 (Applied Biosystems).
Northern blot analysis
Poly(A)+ mRNA of cultured cells (5 µg/lane) were separated on gel under denaturing conditions and transferred to a nylon membrane (Hybond N+, Amersham). Gel was stained with ethidium bromide to ensure that equal amounts had been loaded. Hybridization was performed overnight at 50°C with digoxigenin (DIG)-labeled probes in buffer provided by the manufacturer (Roche). Labeling of probe (400 nt) was performed using the PCR DIG labeling kit (Roche) with two primers in the BRCA1 coding sequence (GAGAGATT CGAGGTGCTCCA and GGATGCTCATCCAAACGAAT). Washing and detection were done according to the manufacturer’s protocol. Filters were exposed for 1 h to chemiluminescent detection film (Amersham).
PCR amplification of cDNA coding sequence
Amplification of cDNA coding sequence was obtained by PCR in 25 µl of buffer containing 20 mM Tris–HCl (pH 8.3), 10 mM KCl, 2 mM MgSO4, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg/ml BSA, 400 nM primers (AtB1-5′3′-F ATCGAAAATGGCGGACACTA, AtB1-5′3′-R CAATTC TCCATAATCCTACAAACAA), 200 µM dNTP, 1 mM MgCl2, 1.5 U Pfu Turbo polymerase (Stratagene) and 1 µl of cDNA beads at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 3 min 30 s and 10 min at 72°C.
Real-time quantitative PCR
Quantification of transcript abundance was done by real-time PCR which took advantage of the affinity of SYBR Green (Molecular Probes) for double-stranded DNA. The measure of the fluorescence emitted by SYBR Green associated with PCR products is directly proportional to the number of amplicons produced during each cycle. The cycle threshold (Ct) is the number of cycles needed to reach the fluorescence detection threshold and depends on the number of DNA templates at the start of the PCR. 18S ribosomal RNA used as a reference gene provided a control for PCR quantification. Quantifications were done using the comparative Ct method. The ΔCt, which is the substraction of the average 18S Ct value from the average Ct value of the target gene, is sample-specific and can be compared to the ΔCt of a calibration sample (for example the unirradiated sample). The amount of target, determined by normalization to the endogenous reference (18S) and relative to the calibrator, is 2–ΔΔCt with ΔΔCt being the subtraction of the calibrator ΔCt from the ΔCt of the sample, assuming that the efficiency of both PCRs is close to one (53). Primers used were AtB1q-F CCATGTATTTTGCAATGCGTG, AtB1q-R TGTGGAGCACCTCGAATCTCT, AtRAD51q-F CGAGG AAGGATCTCTTGCAG, AtRAD51-R GCACTAGTGAAC CCCAGAGG, 18Sq-F CGGCTACCACATCCAAGGAA and 18Sq-R GCTGGAATTACCGCGGCT. Amplicon length was near 100 bp. Amplifications were done in triplicate and performed in 25 µl containing 12.5 µl of 2× platinum quantitative supermix-UDG (Invitrogen), 200 nM primers, 0.2× SYBR Green and 5 ng of cDNA (50°C, 2 min for uracil-N-glycosylase activation; 95°C for 10 min and 40 cycles at 95°C for 15 s; 60°C for 1 min).
RESULTS
Characterization and sequence of AtBRCA1 cDNA
BRCA1 orthologs have been defined by the presence of one RING and two BRCT domains, respectively, at the N- and C-termini (12–16). We looked for the Arabidopsis putative ortholog by first considering the typical hBRCA1 structural organization of these three domains. The human BRCA1 protein sequence matched on Arabidopsis chromosome 4 at the At4g21070 predicted locus, which encoded a putative protein fragment of 1495 amino acids with strong similarities at the N- and C-termini.
The gene structure of At4g21070 was determined with three gene structure prediction software packages (Softberry, GenScan, Grail). Two different types of results were given. One type predicted a single mRNA of >5.2 kb which encoded a protein with three PPR motifs, one RING domain and two BRCT domains. The other one predicted two distinct mRNA of 1.7 and 3.5 kb, respectively. The first was intron-less and encoded a protein containing a PPR domain which is never present in BRCA1 orthologs. The second has a predicted TATA box located 310 bp before the initiation codon and was made of 14 exons followed by a 3′ untranslated region (UTR) of 610 bp. The corresponding predicted protein has a RING and two BRCT domains at the N- and C-termini, respectively, the typical organization of the BRCA1 orthologs. To resolve this ambiguity in intron–exon prediction, we postulated the presence of two genes given by Softberry prediction software and performed northern blotting and 5′ RACE to characterize the structural organization of the At4g21070 locus. Northern hybridization of poly(A)+ mRNAs from cells with a probe corresponding to a part of the predicted 3.5 kb mRNA revealed a single band for a transcript of an overall size of 3.5 kb (Fig. 1A). The absence of signal in non-irradiated cells indicates a quite low abundance of the transcript in control conditions. Determination of 5′ UTR of the 3.5 kb transcript by 5′ RACE using either poly(T) or a specific primer for reverse transcription gave in each case a sequence of 50 bp before the translation initiation codon. We sequenced the cDNA of the BRCA1 domain-containing sequence of 3.5 kb that we named the putative AtBRCA1 gene. The AtBRCA1 mRNA is composed of 14 exons with a 2826 bp coding sequence (Table 1) located downstream of a 5′ UTR of 50 bp and upstream of a predicted 3′ UTR of 650 bp. It gave a mRNA length of 3.5 kb in accordance with the northern hybridization result (Fig. 1A). The corresponding AtBRCA1 genomic sequence from the TATA box to the polyadenylation signal is then 5.5 kb long. PCR amplification of the full-length coding sequence (Fig. 1B) gave a unique band as in northern blot analysis. This indicated the absence of splicing variants in the studied mRNA population. These results gave evidence of a wrong electronic annotation in this genomic area and confirmed the presence of two genes at the At4g21070 genomic locus. At this stage, they indicated that only one putative ortholog of BRCA1 was present in the Arabidopsis genome.
Figure 1.
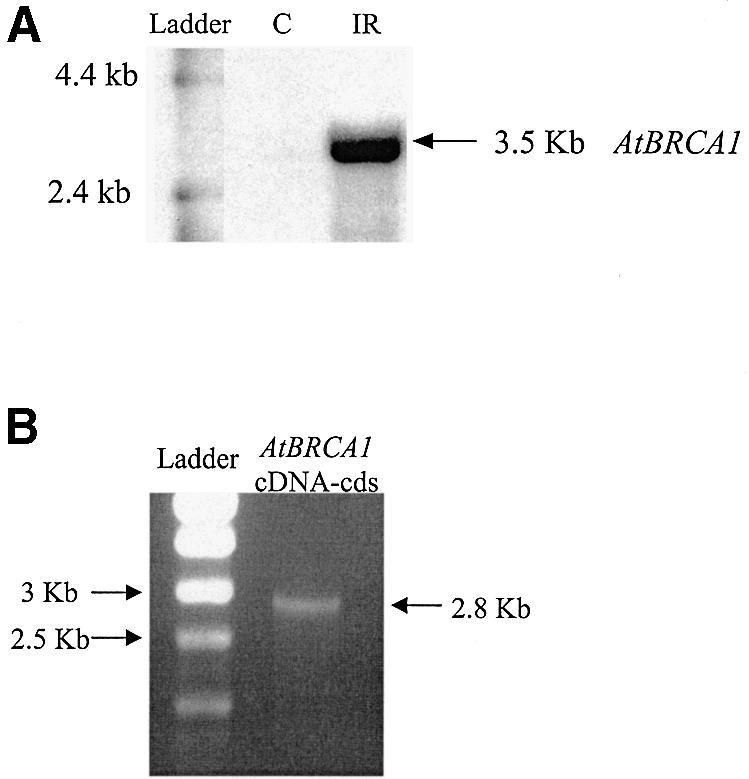
Analysis of AtBRCA1 expression. (A) Northern blot analysis of AtBRCA1 in control (C) and 25 Gy irradiated (IR) Arabidopsis cells. (B) Gel electrophoresis of AtBRCA1 coding sequence obtained by PCR amplification using cDNA population from cells.
Table 1. Structure organization of AtBRCA1 mRNA.
| Position on the mRNA | Length (bp) | |
|---|---|---|
| 5′ UTR | –50 | 50 |
| Exon 1 | 1–83 | 83 |
| Exon 2 | 84–137 | 54 |
| Exon 3 | 138–208 | 71 |
| Exon 4 | 209–316 | 108 |
| Exon 5 | 317–777 | 461 |
| Exon 6 | 778–888 | 111 |
| Exon 7 | 889–1749 | 861 |
| Exon 8 | 1750–1850 | 101 |
| Exon 9 | 1851–2037 | 187 |
| Exon 10 | 2038–2135 | 98 |
| Exon 11 | 2136–2262 | 127 |
| Exon 12 | 2263–2428 | 166 |
| Exon 13 | 2429–2547 | 119 |
| Exon 14 | 2548–2854 | 306 |
| Predicted 3′ UTR | 2855–3474 | 619 |
| Overall size | 3.5 kb |
AtBRCA1: protein motifs and homologies with BRCA1 orthologs
The putative AtBRCA1 cDNA encoded a protein of 941 amino acids with a predicted molecular weight of 104 kDa. Analysis of domains confirmed the presence of one RING domain between amino acids 16 and 53 (E-value 9.74e–08) and two BRCT domains between amino acids 727 and 809, and 842 and 935 (E-values 2.91e–15 and 6.95e–13, respectively) (see Supplementary Material, Figure S1A and B). Alignment of AtBRCA1 and hBRCA1 showed 34% identity–61% similarity in the RING domain, and 28% identity–61% similarity in the BRCT region (see Figure S1C and D). In the SMART database, four protein sequences contained both RING and BRCT domains in the A.thaliana proteome. One of them carried a single BRCT domain in N-terminal and one RING in its central part. The wrong number and location of the domains does not correspond to the organization of BRCA1 orthologs and this candidate gene was discarded in our study. Among the three other sequences predicted from three sequencing projects of the At4g21070 locus only one (ID no. Q8RXD4) corresponded to the protein translated from the putative AtBRCA1 cDNA we have sequenced. Thus, only one protein in the A.thaliana proteome, at the At4g21070 locus, carried a RING and two BRCT domains located at the N- and C-termini, respectively, as with other BRCA1 orthologs. We did not find Arabidopsis sequences containing specific ankyrin repeats together with RING and BRCT domains (54–56), characteristic of BARD1 orthologs. Altogether, these data showed that only one protein in the Arabidopsis genome carries the typical domain organization of BRCA1 orthologs.
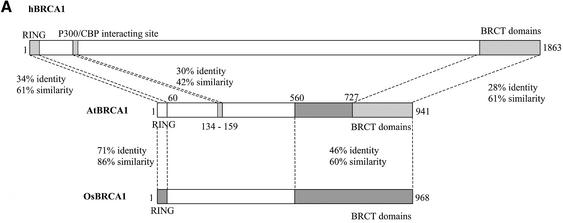
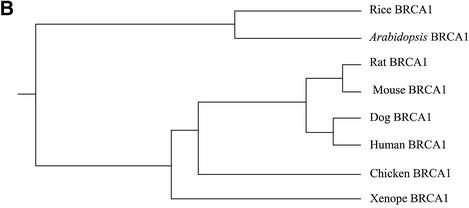
Alignment of BRCA1 orthologs using Clustal W (57) confirmed the preceding results but also revealed another homology region between amino acids 134 and 159 of AtBRCA1 and amino acids 181 and 206 for hBRCA1 (30% identity–42% similarity) (Fig. 2A), which correspond to the p300/CBP interacting site. On the overall sequence, identity between AtBRCA1 and hBRCA1 was 15.7 and 21% with the splicing variant hBRCA1Δ11. AtBRCA1 lacks the major part of the region between RING and BRCT domains, corresponding to exon 11 in hBRCA1, giving a coding region twice as short as other orthologs. In order to see if this shorter length could be plant-specific, we looked for a putative BRCA1 ortholog in the recently sequenced rice (O.sativa) genome. Only one candidate gene encoding a predicted protein (Scaffold6799_3, see Materials and Methods) of 968 amino acids contained a RING and two BRCT domains at the N- and C-termini, respectively, and was also twice as short as hBRCA1. Alignment of AtBRCA1 with this putative OsBRCA1 showed 35% identity–49% similarity on the overall sequence, 71% identity–86% similarity in the RING domain and 46% identity–60% similarity between amino acids 560 and 941 of AtBRCA1 (Fig. 2A). Phylogenetic analysis of these plant BRCA1 orthologs using PHYlogeny Inference Package with kitsch-Fitch-Margoliash and least squares methods with evolutionary clock (58) clearly showed the distribution of sequences between vertebrates versus plants and in vertebrates, mammalian versus non-mammalian (Fig. 2B).
Figure 2.
(A) Schematic representation of similarities between human, A.thaliana and rice BRCA1 orthologs. (B) Phylogenetic analysis of BRCA1 orthologs using PHYlogeny Inference Package with kitsch-Fitch-Margoliash and least squares methods with evolutionary clock.
With all these elements, we proposed that the 3.5 kb mRNA, which is transcribed at the locus At4g21070, is the Arabidopsis ortholog of BRCA1, and suggested that plant orthologs might always be twice as short as vertebrate ones.
Expression of AtBRCA1 mRNA in plant organs and after gamma irradiation
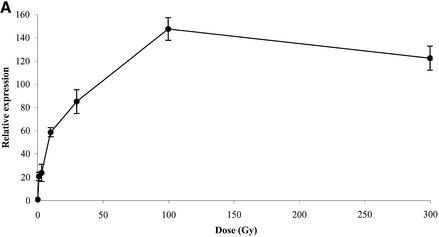
First, AtBRCA1 mRNAs were quantified in whole plantlets after gamma irradiation with doses ranging from 1 to 300 Gy (Fig. 3A). Real-time quantitative PCR was used due to the hard detection conditions by northern analysis without genotoxic stress. A maximal induction of 140 times was observed for a dose close to 100 Gy, but a more than 20-fold induction was already reached at the low doses of 1–3 Gy. Expression levels of transcripts in plantlet leaves versus roots were hardly different within the control tissues (Table 2A) but were both enhanced 160-fold by IR. In adult flowering plants, a gradient of expression of AtBRCA1 transcript was observed from the bottom to the top of the control plants, with the highest value in flower buds, being 10 times higher than in rosette leaves (Table 2B). As in plantlets, AtBRCA1 was strongly induced in all tissues after IR and expression levels reached the same maximal value whatever the basal levels, except for cauline leaves. We can underline the highest induction of AtBRCA1 mRNA by a factor of more than 800 times in rosette leaves. In siliques, the relative proportion of 18S rRNA levels was too low compared to other tissues to allow a meaningful comparison between the tissues. Even the siliques presumably showed a reduced translational activity compared to the highly dividing tissues, the induction of AtBRCA1 was still more than 10 times (Table 2C).
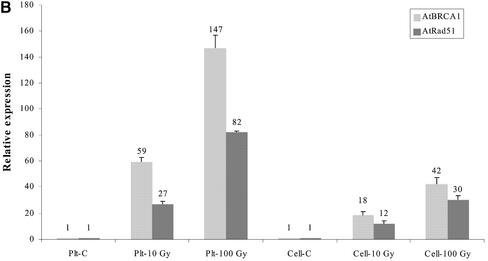
Figure 3.
Quantification by real-time quantitative PCR of: (A) AtBRCA1 mRNA level determined 1 h after irradiation of plantlets with different doses; (B) AtBRCA1 and AtRAD51 mRNA levels in control (C) and irradiated samples (dose-Gy). Samples were 4-day plantlets (Plt) and cells suspensions (cell). All results are normalized to the unirradiated sample. Each quantification was done on two independent RNA extractions and reverse transcription reactions.
Table 2. Relative expression of AtBRCA1 mRNA in control Arabidopsis tissues and 1 h after 100 Gy irradiation (IR) determined by real-time quantitative PCR.
| Tissues | Control | IR | |
|---|---|---|---|
| (A) | Leaves | 1.00 | 163.14 ± 0.53 |
| Roots | 1.52 ± 0.02 | 166.53 ± 32.28 | |
| (B) | Flower buds | 1.00 | 92.67 ± 18.85 |
| Cauline leaves | 0.50 ± 0.05 | 40.21 ± 4.52 | |
| Stem | 0.23 ± 0.01 | 105.69 ± 10.36 | |
| Rosette leaves | 0.12 ± 0.01 | 99.26 ± 8.71 | |
| (C) | Siliques | 1.00 | 16.33 ± 3.51 |
All results were normalized using the comparative Ct method (A) to the leaves control sample value, (B) to the flower bud control sample value and (C) to the silique control sample value. Each quantification was done on two independent RNA extractions and reverse transcription reactions.
In human cell cultures, hBRCA1 has been found to interact with the DSB repair protein hRAD51. So, we looked for the co-induction of the Arabidopsis transcripts of these two genes after IR in cell suspensions and in plant organs. Basal levels of AtRAD51 and AtBRCA1 mRNA were 10-fold higher in exponentially growing cell cultures than in entire plants (data not shown). Interestingly, transcripts were co-induced in the same relative proportion in Arabidopsis cell cultures as well as in entire plantlets (Fig. 3B). The fold induction value of AtRAD51 mRNA was between 10 and 80 times and was, in all cases, 50–70% of the AtBRCA1 induction factor. But since AtRAD51 was expressed twice more than AtBRCA1 in both models without IR (data not shown), the same final stoichiometry was finally reached. Taken together, these results demonstrated the ubiquity of AtBRCA1 transcripts in all Arabidopsis tissues and its strong transcriptional induction after DNA damage by IR in the same way as AtRAD51.
DISCUSSION
Identification of AtBRCA1 was done by similarity search and the gene predicted in databases differed from that experimentally isolated. Errors in some exon/intron boundaries were found and some programs merged the sequence of the AtBRCA1 gene with a second gene. During this study, this fusion gene, which does not exist, containing three PPR motifs, one RING and two BRCT domains, was given as a potential Arabidopsis BRCA1/BARD1 ortholog in an article on BRCA1 and BARD1 identification in the Xenopus laevis genome (15). Thus, we have characterized the Arabidopsis ortholog of BRCA1, which was located on chromosome 4 and gave a 3.5 kb transcript that encoded a 941 amino acid protein (104 kDa) with one N-terminal RING and two C-terminal BRCT domains as hBRCA1. In the RIKEN Arabidopsis full-length complementary (RAFL) cDNA collection (59), we have found the clone corresponding to AtBRCA1 cDNA sequence (RAFL09-61-E20), which confirmed our results. This showed the difficulty of precise gene prediction, and also that orthologs can still be identified between distantly related species. To date, AtBRCA1 is the first non-vertebrate BRCA1 ortholog.
The known functions of hBRCA1 are nearly all linked to RING and BRCT domains with an E3 ubiquitin autoligase activity and interaction with several proteins in the RING domain, while the BRCT domains are involved in transcriptional transactivation and are also required for interaction with several proteins. This BRCT domain is a signature of proteins involved in DNA repair and is present in numerous species, including vertebrates, yeast and plants, suggesting a conserved role for this domain. As shown by Joukov et al. (15), the Xenopus structural ortholog of BRCA1 was also reported as a functional ortholog. So, conserved domains strongly indicated that this structural organization is the signature of BRCA1 orthologs, and are linked to conserved activities. In addition to these two kinds of functional domains, AtBRCA1 shared an additional similarity between amino acids 181 and 206 of hBRCA1, a region of interacting domains with c-myc (60) and p300/CBP (61), proteins which are endowed with acetyltransferase activity and interact with transcription factors. Since such orthologs have been described in Arabidopsis (62), it may be possible that these two Arabidopsis proteins also interact.
To date, with 941 and 968 amino acids, respectively, AtBRCA1 and OsBRCA1 are the shortest members of the BRCA1 family, except the alternative splicing variant hBRCA1Δ11, which still contains the two kinds of functional domains and has a molecular weight of 110 kDa. Analysis of other orthologs showed that sequence conservation is particularly important in the RING and BRCT domains but less significant between them. This may suggest that the BRCA1 central region is species-specific. We also notice the reduced length in the predicted sequences of the Arabidopsis and rice putative orthologs of BRCA2 (63), an interacting partner of hBRCA1 and hRAD51 in human cells. If we looked at orthologs of other genes implicated in DNA repair between Arabidopsis and human, some of them are different in length and/or in structural composition. For instance, in AtLig IV (1219 amino acids), there is only one BRCT domain and two in hLig IV (911 amino acids). AtATM (3856 amino acids) is longer than human ortholog (3056 amino acids) and presents additional domains, while AtRAD51 is closely related to the human ortholog which shares the same domains and length (342 versus 339 amino acids for hRAD51). So, there are slight differences between Arabidopsis and human orthologs, but domains linked to protein activities are always conserved within the same structural organization.
AtBRCA1 is expressed ubiquitously but seemed to be elevated in proliferating tissues like in flower buds and in cell cultures. This may be linked to a higher rate of division and suggests a similar role in cell-cycle control of AtBRCA1 as shown for hBRCA1. AtBRCA1 mRNA levels are highly increased after gamma irradiation; this could be due to an enhanced transcription and/or a higher mRNA stability. We observed maximal induction near 100 Gy, a non-lethal dose generally used to screen mutants hypersensitive to gamma irradiation, but AtBRCA1 was also significantly up-regulated at doses between 1 and 20 Gy. Then, this increase seems to be dose-related from 1 to 100 Gy suggesting a DNA lesion amount dependency. Basal AtBRCA1 expression in different plant tissues varied, but was quantitatively the same after gamma irradiation, showing a higher induction factor in tissues with low basal levels and a plateau response level of AtBRCA1 mRNA. More strikingly, a lower induction observed in cell suspensions compared to plantlets whatever the irradiation dose, may be linked to a higher basal mRNA level. This could be related to homogeneity of cultured cells growing in exponential phase compared to heterogeneity of plantlets, which are a mosaic of meristems and differentiated tissues. The strong correlation between fold induction and dose suggests a highly sensitive and precise regulation system linked to DNA repair and/or to cell-cycle control, which is consistent with the role of hBRCA1. However, there are some discrepancies in the regulation of mRNA levels after DNA damage among species. Indeed, hBRCA1 mRNA is induced by irradiation but much less than AtBRCA1 (64), and hRAD51 is not induced after IR, whereas levels of AtRAD51 mRNA were found to be increased by irradiation, confirming a previous study (11). In our study, fold induction of both genes seemed to be correlated whatever the tissues or irradiation doses, suggesting a common regulation of transcript amount.
Considering molecular organization, transcript behaviour after IR of AtBRCA1 and the large conservation of DNA repair functions and associated proteins among eucaryotes strongly argue in favor of the fact that AtBRCA1 is indeed the ortholog of hBRCA1. The presence of RING, BRCT and p300/CBP interacting domains in AtBRCA1 may guide us in determining the function of this plant ortholog, using our knowledge of the human gene. As several partners of hBRCA1 are present in Arabidopsis, including AtRAD51, AtATM, AtBRCA2 and p300/CBP, there may be conserved functional interactions between them. But we have also to find out its functions specific to plant physiology. For instance, absence of Arabidopsis orthologs of genes such as BARD1 or p53, which are key elements in the DNA damage response of human cells and partners of hBRCA1, may help in answering the question of species specificity of conserved functions.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elena Marin-Nussaume, V. Garcia, A. Tissier and C. Triantaphylides for support and advice, and N. Uhrhammer for critically reading the manuscript.
DDBJ/EMBL/GenBank accession no. AF515728
REFERENCES
- 1.Khanna K.K. and Jackson,S.P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet., 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 2.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 3.van Gent D.C., Hoeijmakers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 4.Bennett C.B., Lewis,L.K., Karthikeyan,G., Lobachev,K.S., Jin,Y.H., Sterling,J.F., Snipe,J.R. and Resnick,M.A. (2001) Genes required for ionizing radiation resistance in yeast. Nature Genet., 29, 426–434. [DOI] [PubMed] [Google Scholar]
- 5.Boulton S.J., Gartner,A., Reboul,J., Vaglio,P., Dyson,N., Hill,D.E. and Vidal,M. (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science, 295, 127–131. [DOI] [PubMed] [Google Scholar]
- 6.West C.E., Waterworth,W.M., Jiang,Q. and Bray,C.M. (2000) Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J., 24, 67–78. [DOI] [PubMed] [Google Scholar]
- 7.Gallego M.E., Jeanneau,M., Granier,F., Bouchez,D., Bechtold,N. and White,C.I. (2001) Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J., 25, 31–41. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K., Adachi,Y., Chiba,K., Oguchi,K. and Takahashi,H. (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J., 29, 771–781. [DOI] [PubMed] [Google Scholar]
- 9.Hartung F. and Puchta,H. (1999) Isolation of the complete cDNA of the Mre11 homologue of Arabidopsis. Plant Physiol., 121, 312. [Google Scholar]
- 10.Garcia V., Salanoubat,M., Choisne,N. and Tissier,A. (2000) An ATM homologue from Arabidopsis thaliana: complete genomic organisation and expression analysis. Nucleic Acids Res., 28, 1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doutriaux M.P., Couteau,F., Bergounioux,C. and White,C. (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet., 257, 283–291. [DOI] [PubMed] [Google Scholar]
- 12.Bennett L.M., Haugen-Strano,A., Cochran,C., Brownlee,H.A., Fiedorek,F.T.,Jr and Wiseman,R.W. (1995) Isolation of the mouse homologue of BRCA1 and genetic mapping to mouse chromosome 11. Genomics, 29, 576–581. [DOI] [PubMed] [Google Scholar]
- 13.Bennett L.M., Brownlee,H.A., Hagavik,S. and Wiseman,R.W. (1999) Sequence analysis of the rat Brca1 homolog and its promoter region. Mamm. Genome, 10, 19–25. [DOI] [PubMed] [Google Scholar]
- 14.Szabo C.I., Wagner,L.A., Francisco,L.V., Roach,J.C., Argonza,R., King,M.C. and Ostrander,E.A. (1996) Human, canine and murine BRCA1 genes: sequence comparison among species. Hum. Mol. Genet., 5, 1289–1298. [DOI] [PubMed] [Google Scholar]
- 15.Joukov V., Chen,J., Fox,E.A., Green,J.B. and Livingston,D.M. (2001) Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl Acad. Sci. USA, 98, 12078–12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orelli B.J., Logsdon,J.M.,Jr and Bishop,D.K. (2001) Nine novel conserved motifs in BRCA1 identified by the chicken orthologue. Oncogene, 20, 4433–4438. [DOI] [PubMed] [Google Scholar]
- 17.Miki Y., Swensen,J., Shattuck-Eidens,D., Futreal,P.A., Harshman,K., Tavtigian,S., Liu,Q., Cochran,C., Bennett,L.M., Ding,W. et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science, 266, 66–71. [DOI] [PubMed] [Google Scholar]
- 18.Deng C.X. and Brodie,S.G. (2000) Roles of BRCA1 and its interacting proteins. Bioessays, 22, 728–737. [DOI] [PubMed] [Google Scholar]
- 19.Cortez D., Wang,Y., Qin,J. and Elledge,S.J. (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science, 286, 1162–1166. [DOI] [PubMed] [Google Scholar]
- 20.Lee J.S., Collins,K.M., Brown,A.L., Lee,C.H. and Chung,J.H. (2000) hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature, 404, 201–204. [DOI] [PubMed] [Google Scholar]
- 21.Chen J. (2000) Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res., 60, 5037–5039. [PubMed] [Google Scholar]
- 22.Scully R., Chen,J., Ochs,R.L., Keegan,K., Hoekstra,M., Feunteun,J. and Livingston,D.M. (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell, 90, 425–435. [DOI] [PubMed] [Google Scholar]
- 23.Wu-Baer F. and Baer,R. (2001) Effect of DNA damage on a BRCA1 complex. Nature, 414, 36. [DOI] [PubMed] [Google Scholar]
- 24.Li S., Ting,N.S., Zheng,L., Chen,P.L., Ziv,Y., Shiloh,Y., Lee,E.Y. and Lee,W.H. (2000) Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature, 406, 210–215. [DOI] [PubMed] [Google Scholar]
- 25.Ouchi T., Monteiro,A.N., August,A., Aaronson,S.A. and Hanafusa,H. (1998) BRCA1 regulates p53-dependent gene expression. Proc. Natl Acad. Sci. USA, 95, 2302–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Somasundaram,K., Peng,Y., Tian,H., Bi,D., Weber,B.L. and El-Deiry,W.S. (1998) BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene, 16, 1713–1721. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q., Zhang,H., Kajino,K. and Greene,M.I. (1998) BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene, 17, 1939–1948. [DOI] [PubMed] [Google Scholar]
- 28.Somasundaram K., Zhang,H., Zeng,Y.X., Houvras,Y., Peng,Y., Wu,G.S., Licht,J.D., Weber,B.L. and El-Deiry,W.S. (1997) Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature, 389, 187–190. [DOI] [PubMed] [Google Scholar]
- 29.Harkin D.P., Bean,J.M., Miklos,D., Song,Y.H., Truong,V.B., Englert,C., Christians,F.C., Ellisen,L.W., Maheswaran,S., Oliner,J.D. and Haber,D.A. (1999) Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell, 97, 575–586. [DOI] [PubMed] [Google Scholar]
- 30.Schlegel B.P., Green,V.J., Ladias,J.A. and Parvin,J.D. (2000) BRCA1 interaction with RNA polymerase II reveals a role for hRPB2 and hRPB10alpha in activated transcription. Proc. Natl Acad. Sci. USA, 97, 3148–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scully R., Anderson,S.F., Chao,D.M., Wei,W., Ye,L., Young,R.A., Livingston,D.M. and Parvin,J.D. (1997) BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 94, 5605–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S., Chen,P.L., Subramanian,T., Chinnadurai,G., Tomlinson,G., Osborne,C.K., Sharp,Z.D. and Lee,W.H. (1999) Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J. Biol. Chem., 274, 11334–11338. [DOI] [PubMed] [Google Scholar]
- 33.Pao G.M., Janknecht,R., Ruffner,H., Hunter,T. and Verma,I.M. (2000) CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc. Natl Acad. Sci. USA, 97, 1020–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson S.F., Schlegel,B.P., Nakajima,T., Wolpin,E.S. and Parvin,J.D. (1998) BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nature Genet., 19, 254–256. [DOI] [PubMed] [Google Scholar]
- 35.Yarden R.I. and Brody,L.C. (1999) BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 4983–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochar D.A., Wang,L., Beniya,H., Kinev,A., Xue,Y., Lane,W.S., Wang,W., Kashanchi,F. and Shiekhattar,R. (2000) BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell, 102, 257–265. [DOI] [PubMed] [Google Scholar]
- 37.Paull T.T., Cortez,D., Bowers,B., Elledge,S.J. and Gellert,M. (2001) Direct DNA binding by Brca1. Proc. Natl Acad. Sci. USA, 98, 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scully R., Chen,J., Plug,A., Xiao,Y., Weaver,D., Feunteun,J., Ashley,T. and Livingston,D.M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88, 265–275. [DOI] [PubMed] [Google Scholar]
- 39.Chen J., Silver,D.P., Walpita,D., Cantor,S.B., Gazdar,A.F., Tomlinson,G., Couch,F.J., Weber,B.L., Ashley,T., Livingston,D.M. and Scully,R. (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell, 2, 317–328. [DOI] [PubMed] [Google Scholar]
- 40.Chen J.J., Silver,D., Cantor,S., Livingston,D.M. and Scully,R. (1999) BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res., 59, 1752s–1756s. [PubMed] [Google Scholar]
- 41.Zhong Q., Chen,C.F., Li,S., Chen,Y., Wang,C.C., Xiao,J., Chen,P.L., Sharp,Z.D. and Lee,W.H. (1999) Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science, 285, 747–750. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Cortez,D., Yazdi,P., Neff,N., Elledge,S.J. and Qin,J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev., 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu L.C. and White,R.L. (1998) BRCA1 is associated with the centrosome during mitosis. Proc. Natl Acad. Sci. USA, 95, 12983–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X., Weaver,Z., Linke,S.P., Li,C., Gotay,J., Wang,X.W., Harris,C.C., Ried,T. and Deng,C.X. (1999) Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell, 3, 389–395. [DOI] [PubMed] [Google Scholar]
- 45.Lafarge S., Sylvain,V., Ferrara,M. and Bignon,Y.J. (2001) Inhibition of BRCA1 leads to increased chemoresistance to microtubule-interfering agents, an effect that involves the JNK pathway. Oncogene, 20, 6597–6606. [DOI] [PubMed] [Google Scholar]
- 46.Mullan P.B., Quinn,J.E., Gilmore,P.M., McWilliams,S., Andrews,H., Gervin,C., McCabe,N., McKenna,S., White,P., Song,Y.H., Maheswaran,S., Liu,E., Haber,D.A., Johnston,P.G. and Harkin,D.P. (2001) BRCA1 and GADD45 mediated G2/M cell cycle arrest in response to antimicrotubule agents. Oncogene, 20, 6123–6131. [DOI] [PubMed] [Google Scholar]
- 47.Brzovic P.S., Rajagopal,P., Hoyt,D.W., King,M.C. and Klevit,R.E. (2001) Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nature Struct. Biol., 8, 833–837. [DOI] [PubMed] [Google Scholar]
- 48.Williams R.S., Green,R. and Glover,J.N. (2001) Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nature Struct. Biol., 8, 838–842. [DOI] [PubMed] [Google Scholar]
- 49.Estelle M.A. and Somerville,C. (1967) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet., 206, 200–206. [Google Scholar]
- 50.Santoni V., Bellini,C. and Caboche,M. (1994) Use of two-dimensional protein-pattern analysis for the characterisation of Arabidopsis mutants. Planta, 192, 557–566. [Google Scholar]
- 51.Banzet N., Richaud,C., Deveaux,Y., Kazmaier,M., Gagnon,J. and Triantaphylides,C. (1998) Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J., 13, 519–527. [DOI] [PubMed] [Google Scholar]
- 52.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 53.Favy D.A., Lafarge,S., Rio,P., Vissac,C., Bignon,Y.J. and Bernard-Gallon,D. (2000) Real-time PCR quantification of full-length and exon 11 spliced BRCA1 transcripts in human breast cancer cell lines. Biochem. Biophys. Res. Commun., 274, 73–78. [DOI] [PubMed] [Google Scholar]
- 54.Gautier F., Irminger-Finger,I., Gregoire,M., Meflah,K. and Harb,J. (2000) Identification of an apoptotic cleavage product of BARD1 as an autoantigen: a potential factor in the antitumoral response mediated by apoptotic bodies. Cancer Res., 60, 6895–6900. [PubMed] [Google Scholar]
- 55.Ayi T.C., Tsan,J.T., Hwang,L.Y., Bowcock,A.M. and Baer,R. (1998) Conservation of function and primary structure in the BRCA1-associated RING domain (BARD1) protein. Oncogene, 17, 2143–2148. [DOI] [PubMed] [Google Scholar]
- 56.Wu L.C., Wang,Z.W., Tsan,J.T., Spillman,M.A., Phung,A., Xu,X.L., Yang,M.C., Hwang,L.Y., Bowcock,A.M. and Baer,R. (1996) Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genet., 14, 430–440. [DOI] [PubMed] [Google Scholar]
- 57.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsenstein J. (1989) Phylogeny Inference Package (Version 3.2). Cladistics, 5, 164–166. [Google Scholar]
- 59.Seki M., Narusaka,M., Kamiya,A., Ishida,J., Satou,M., Sakurai,T., Nakajima,M., Enju,A., Akiyama,K., Oono,Y., Muramatsu,M., Hayashizaki,Y., Kawai,J., Carninci,P., Itoh,M., Ishii,Y., Arakawa,T., Shibata,K., Shinagawa,A. and Shinozaki,K. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science, 296, 141–145. [DOI] [PubMed] [Google Scholar]
- 60.Wang Q., Zhang,H., Kajino,K. and Greene,M.I. (1998) BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene, 17, 1939–1948. [DOI] [PubMed] [Google Scholar]
- 61.Pao G.M., Janknecht,R., Ruffner,H., Hunter,T. and Verma,I.M. (2000) CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc. Natl Acad. Sci. USA, 97, 1020–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bordoli L., Netsch,M., Luthi,U., Lutz,W. and Eckner,R. (2001) Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res., 29, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H., Jeffrey,P.D., Miller,J., Kinnucan,E., Sun,Y., Thoma,N.H., Zheng N., Chen,P.L., Lee,W.H. and Pavletich,N.P. (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science, 297, 1837–1848. [DOI] [PubMed] [Google Scholar]
- 64.Aunoble B., Bernard-Gallon,D. and Bignon,Y.J. (2001) Regulation of BRCA1 and BRCA2 transcript in response to cisplatin, adriamycin, taxol and ionising radiation is correlated to p53 functional status in ovarian cancer cell lines. Oncol. Rep., 8, 663–668. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.