Abstract
β cell failure is a common denominator of diabetes. Susceptibility to stress-induced apoptosis may underlie β cell failure and/or hamper islet transplantation therapy. The causal basis is not well understood. In efforts to identify important differences in gene expression in α vs. β cells, a gene termed HIMP1 (Hypoglycemia/hypoxia Inducible Mitochondrial Protein, or HIG1) has been cloned from an α cell cDNA library. It is a member of a well conserved eukaryote protein family. In mice, its two alternatively spliced products each form a transmembrane loop, having an Noutside–Coutside orientation and are expressed highly in the mitochondrial inner membrane in several tissues including heart and pancreatic α cells, but not in β cells. Ectopic expression of HIMP1 in MIN6 β cells protects the cells from apoptosis induced by several stimuli and prolongs their survival. These results suggest an important role for HIMP1 in stress protective programs in mitochondria.
Keywords: islet, apoptosis, inner membrane protein, stress
Diabetes, including two main types, is one of the most important health problems in the world today. In type 1 diabetes, β cells have been long-recognized to be especially sensitive to various noxious stimuli and also selectively prone to autoimmune destruction. In type 2 diabetes, accumulated findings illustrate that β cell mass is reduced (1–5). However, there is general agreement that an absolute or relative deficiency of insulin is a typical manifestation of this disease and is due to selective loss of β cell mass, often accompanied by elevated production and secretion of glucagon by an increased proportion of α cells. Functional disturbances of pancreatic β and/or α cells are thus central to the failure to maintain physiological glucose levels and the related metabolic concomitants of this disease.
In both types of diabetes, increasing β cell apoptosis is suggested to be mainly responsible for the reduction of β cell mass (6–7). The primary reason for the apoptotic susceptibility of β cells compared to other cell types remains unclear. Is it a cell type-specific property acquired inherently during differentiation due to the attenuation of some beneficial genes, and/or the turning on of detrimental genes? Or is it due to overloading the β cells under stressful conditions such as hyperglycemia or obesity and their concomitant toxic byproducts such as hydrogen peroxide? Albeit data supporting both views exist, increasing evidence tends to support the former notion because it has been observed that β cells are more sensitive to various apoptotic stimuli, such as the stress of low glucose (8, 9) or hypoxia (10), and β cell apoptosis increases during chronic hyperglycemic conditions (3, 11–14). The maturation of β cells has also been shown to link to increases in their sensitivity to the apoptotic stimuli of toxins and cytokines (15). In addition, low expression level of various protective genes against oxidative stress, such as catalase, in the β cells has been proposed to contribute to the apoptotic proneness of β cells (16, 17). On the other hand, defective or reduced expression of β cell-specific genes, e.g., partial PDX1 deficiency (18), has also been implicated in increased β cell loss.
Although mitochondria are believed to be the organelles that integrate cellular metabolism and apoptosis (19) and play a key role in β cell function (20), so far no mitochondrial, residential, or associated proteins linked to pro- or antiapoptosis have been found that differ profoundly between β cells and other cell types with high rates of glucose oxidation, such as cardiomyocytes. To assess differences in gene expression between α and β cells, we have analyzed profiles of gene expression by using mRNA differential display and microarrays. In the course of these studies, a low glucose and oxygen dually inducible gene, termed HIMP1, or HIG1, has been cloned from an α cell cDNA library. It is a member of a unique gene family that is conserved in eukaryotes. Various analyses demonstrate that two HIMP1 alternatively spliced gene products are mitochondrial inner membrane proteins, which are ubiquitously expressed, abundant in heart and pancreatic α, but not β cells, and are linked to cell survival. Transfection of HIMP1 into MIN6 cells enhanced their resistance to several apoptotic stimuli.
Results
Two Isoforms of a Unique Protein Family Conserved in Eukaryotes.
Two similar predicted full-length cDNAs were cloned from an αTC1.6 cell cDNA library using a probe of a cDNA fragment detected in αTC1.6, but not in MIN6 cells by mRNA differential display. Analysis of their sequences revealed ORFs encoding two similar protein products which we have designated as HIMP-a and -b (Fig. 6A, which is published as supporting information on the PNAS web site). One transcript of 1,930 bp (HIMP1-b) encodes a protein of 99 aa; whereas a shorter one (HIMP1-a) lacks 499 internal nucleotides of HIMP1-b, which leads to a frame shift at residue 79, generating a stop codon at residue 96, to yield a protein of 95 aa (Fig. 6B). A blast search revealed that HIMP1-a is identical with hypoxia-inducible gene 1 (HIG1), a cDNA deposited in GenBank that arises from a locus on mouse chromosome 9. Comparison of the cDNA sequences of HIMP1-a and -b with murine genomic DNA indicated that the two transcripts are alternatively spliced forms of a single copy HIMP1/HIG1 gene. The missing region in HIMP1-a is due to the excision of an intron-like DNA fragment in exon 3 of the HIMP1 gene. The corresponding two protein products share 84.2% amino acid sequence identity. The predicted molecular mass and isoelectric points are 10.6 kDa and 9.8 for HIMP1-a and 11.0 kDa and 11.2 for HIMP1-b, respectively.
A search for HIMP1 homologues yielded >70 hits arising mainly from 12 species of eukaryotes ranging from fungi to man. As shown in Fig. 6B, a cluster analysis of 11 HIMP1-related members, this family is evolutionarily well conserved at the amino acid level, exhibiting >40% identity to HIMP1-a. These findings imply a conserved function in a basic cellular pathway in eukaryotes generally. Using a prediction program (21), HIMP1 and its homologues are predicted to have two transmembrane helices (TMHs) linked by a small loop. The predicted TMH and loop regions represent the most highly conserved regions in these proteins. The murine 2310056K19Rik gene encodes a homologue that shows 44.8% identity with HIMP1-a (Fig. 6B) and can be traced to its locus on chromosome 11.
HIMP1 Is Expressed Predominantly in α, not β, Cells Within the Pancreas and at Highest Levels in Heart Among Other Tissues Examined.
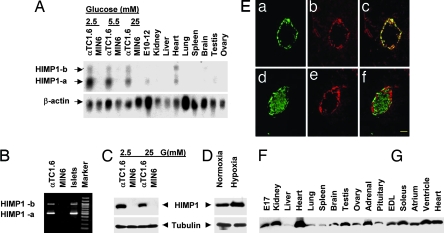
Coincident with the findings using mRNA differential display, HIMP1 transcripts were detected at high levels in αTC1.6, but not in MIN6 cells by Northern blotting (Fig. 1A). The two detected bands (≈1.5 and ≈2.0 kb) correspond to the predicted sizes of HIMP1-a and -b transcripts, respectively. HIMP1-a mRNA is the major transcript detected in αTC1.6 cells, E10–12 embryo, adult organs including heart, testis, and kidney, and is also detected in various other tissues upon longer exposure of Northern blots. In brain, the HIMP1-b signal is slightly stronger than HIMP1-a. Interestingly, both forms are detected at similar high levels in the heart among major organs.
Fig. 1.
Contrasting tissue distribution and induction of expression by low glucose or O2 of the two HIMP1 gene products. (A) Northern blot. E10–12, 10- to 12-day embryos. (B) RT-PCR. The primer set (5′-GGGTTGTACAAGCTGAAGAG-3′, 5′-TGACACCCTCCCAAAGCATG-3′) is derived from the upstream and downstream regions beyond the 499 internal nucleotides of the HIMP1-b cDNA. HIMP1-a (500 bp) and HIMP1-b (999 bp) are shown. (C and D) Immunoblot analysis of HIMP1 level inducible at low glucose (C) or 5% O2 hypoxia (D) for 24 h; 50 μg of protein per lane. (E) Double immunofluorescent detection of HIMP1 (b and e) with glucagon (a) or C-peptide (d) in mouse pancreatic islets. (c and f) Merged images of a and b and d and e, respectively. (Scale bar, 50 μm.) (F and G) Immunoblot analysis of HIMP1 level in various tissues; 50 μg of protein per lane. E17, 17-day embryo. EDL, extensor digitorum longus muscle.
The expression of the two HIMP1 transcripts in islets was further analyzed by RT-PCR (Fig. 1B). Two PCR products corresponding to the expected sizes of the two cDNA fragments were amplified from αTC1.6 cells and islets, but were absent in MIN6 cells similar to the pattern obtained by Northern blotting. The contrasting expression pattern between α and β cells was further examined by immunoblotting using a polyclonal anti-HIMP1 serum generated against the shared N-terminal region of the two isoforms (Fig. 1C). A ≈12-kDa band, which corresponds with the calculated molecular mass of the HIMP1 proteins, was identified only in αTC1.6, but not in MIN6 cells. No signal was detected by using the preimmune serum (data not shown). As HIMP1-a and -b differ in size by only 4 aa, they apparently comigrated on SDS/PAGE immunoblots (Fig. 1D). To further verify the distribution of HIMP1 in pancreas, double immunostaining of pancreas sections was performed by using antibodies against HIMP1 and glucagon or C-peptide. The results revealed that HIMP1 immunoreactivity is predominant in α cells, but not in β cells (Fig. 1E). The analysis indicates that HIMP1 is normally expressed predominantly in α, but not in β, cells within islets. The expression of the HIMP1-related 2310056K19Rik gene (see Fig. 1) in islets was also examined by RT-PCR. No detectable expression was found in islets, αTC1.6 or MIN6 cell lines, but was detected at high levels in heart (data not shown). The tissue distribution of the HIMP1 proteins was further examined by immunoblotting (Fig. 1 F and G). HIMP1 was found to be expressed at highest levels in heart and ubiquitously detected in various organs and some endocrine tissues, consistent with the Northern blot results.
HIMP1 Expression Is Induced by Low Glucose and Oxygen.
When we examined the effects of glucose on HIMP1 levels in αTCI.6 cells by Northern blot and immunoblotting, we found that low (2.5 mM) glucose substantially increased levels, whereas high (25 mM) glucose decreased them (Fig. 1 A and C). This finding was of interest in view of the observation by Denko et al. (26) that the HIMP1 homologue in human tumor cells is inducible by hypoxia. When we examined the response of HIMP1 to hypoxia at 1, 2, 6, and 24 h in αTCI.6 cells, we found a significant response (Fig. 1D). In contrast, no detectable induction was observed in MIN6 cells. Similarly, induction of HIMP1 levels by 1 mM glucose over a 24-h time course occurred in αTC1.6, but not in MIN6 cells.
HIMP1 Proteins Localize to the Mitochondrial Inner Membrane and Cristae.
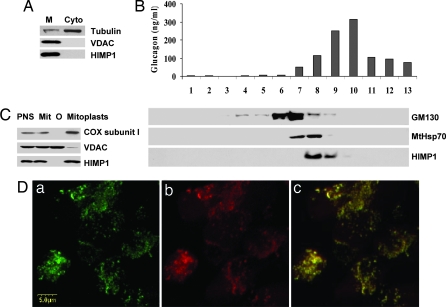
The subcellular localization of HIMP1 was analyzed by using αTC1.6 cells. Initial analyses showed that HIMP1 proteins were detected only in the membrane fraction by immunoblotting (Fig. 2A). This finding, along with the predicted structure, suggests that HIMP1 proteins are integral membrane proteins. To determine which organelle contains HIMP1 proteins, fractions of αTC1.6 cells were separated on sucrose gradients (22) and subjected to immunoblotting and RIA for several markers. As shown in Fig. 2B, the peak of HIMP1 immunoreactivity was detected in fraction 9, which also contained mitochondrial heat shock protein MtHsp70, whereas peaks of Golgi components identified by Golgi matrix protein GM130 and glucagon-containing secretory granules were enriched at fractions 8 and 10, respectively. Consistent with these analyses, HIMP1 immunoreactivity was observed to colocalize with mitochondria along with cytochrome c oxidase (Cox) subunit I by confocal immunofluorescence staining of αTC1.6 cells (Fig. 2D). To study the localization of HIMP1 proteins in detail, two fractions of mitochondrial outer and inner membranes were separated by centrifugation after digitonin treatment (23) and examined by immunoblotting. The results show that the HIMP1 proteins colocalize with inner membrane (mitoplast, containing matrix) markers such as Cox subunit I, but not with an outer membrane marker, the voltage-dependent anion channel protein (VDAC) (Fig. 2C). These data strongly suggest that the HIMP1 proteins are located in the mitochondrial inner membrane of αTC1.6 cells.
Fig. 2.
Analyses of HIMP1 subcellular localization. (A) Immunoblot. M, membrane; Cyto, cytosol; 50 μg of protein per lane. (B) RIA of glucagon and immunoblot analyses of proteins (GM130, MtHsp70, and HIMP1) in 13 fractions of αTC1.6 cells separated by sucrose gradient centrifugation. (C) Immunoblot. PNS, postnuclear supernatant; Mit, crude mitochondrial fraction; O, outer membrane fraction; mitoplast, inner membrane and matrix. (D) Confocal immunofluorescence staining. (a) HIMP1. (b) Cox subunit I. (c) Merged image of a and b.
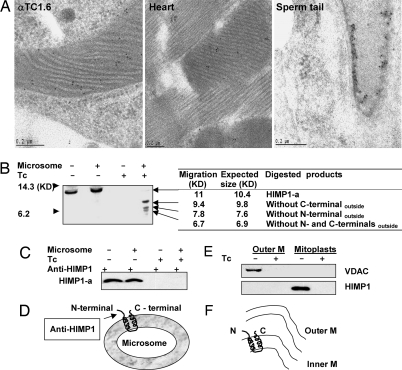
To validate these findings, immunoelectron microscopy studies were performed comparing immunogold labeling of mitochondria in αTC1.6 cells, heart, or testis samples (Fig. 3A). The specific HIMP1 immunoreactivity was clearly identifiable along the inner mitochondrial membrane and its enfolded cristae in αTC1.6 cells, heart, and also in the helix-shaped chondriosomes fused with mitochondria and wound around the flagellum of a sperm tail in the testis (Fig. 3A). No comparable labeling of mitochondria in MIN6 cells is seen (data not shown). These findings clearly indicate that HIMP1 proteins are localized in the inner membrane of mitochondria in α cells, cardiomyocytes, and mature sperm cells in testis.
Fig. 3.
Subcellular localization and membrane topology of HIMP1. (A) Examination of HIMP1 in αTC1.6 cells, cardiomyocytes, and mature sperm by immunoelectron microscopy. (B) In vitro synthesized [35S]HIMP1-a protein with/without microsomes was subjected to digestion with Tc and separation by SDS/PAGE gel for autoradiography. (C) Immunoprecipitants with anti-HIMP1 serum from aliquots treated with/without Tc of translation products in B were subjected to SDS/PAGE for autoradiography. (D) Membrane topology sketch of HIMP1-a, as determined in the in vitro system. (E) Immunoblot analysis of HIMP1in mitochondrial outer and inner membrane fractions treated with or without Tc. Outer M, outer membrane fraction; mitoplast, inner membrane and matrix. (F) Membrane orientation sketch of HIMP1 determined in mitochondria from αTC1.6 cells. N, N-terminal; C, C-terminal; M, membrane.
Because several potential trypsin and chymotrypsin (Tc) cleavage sites exist beyond the predicted TMH regions of HIMP1, we chose the method described in ref. 24 using in vitro translated HIMP1-a protein (Fig. 3 B and C) and separated outer and inner membrane fractions of mitochondria from αTC1.6 cells (Fig. 3E). We initially tested whether in vitro synthesized HIMP1-a can insert into canine pancreatic microsomal membranes, despite the lack of evidence of localization normally to the endoplasmic reticulum (ER) in vivo. As shown in Fig. 3B, [35S]HIMP1-a protein synthesized with microsomes was partially protected during Tc treatment compared to the control without microsomes (lanes 4 vs. 3). The estimated sizes of the three observed cleavage products on 16.5% tricine SDS/PAGE gel are 9.4, 7.8, and 6.7 kDa, based on the migration of molecular markers. These correspond respectively to the three predicted cleavage products generated by removing the C-terminal, the N-terminal, or both N- and C-terminal regions that lie beyond the predicted TMH and loop regions of HIMP1-a (Fig. 6B). Furthermore, no evidence was found for cleavage in the loop region, supporting its luminal localization. These results indicate that in vitro translated HIMP1-a can insert into microsomal membranes and has a membrane topology with Noutside–Coutside and loopinside orientations (Fig. 3D). The possibility that the microsomal membranes were contaminated with mitochondria was ruled out because HIMP1 in a crude mitochondrial fraction of αTC1.6 cells could not be digested by Tc treatment as shown by immunoblots using the same procedure described for Fig. 3B (data not shown); this may be due to a protective effect of the intact mitochondrial outer membrane. The reason for the insertion into microsomal membranes of in vitro synthesized HIMP1-a is currently unclear. Full-length HIMP1-a, synthesized in vitro with or without microsomes (Fig. 3B, lanes 1 and 2), is not cleaved by endogenous proteinases whether or not it inserts into microsomal membranes, whereas the signal peptide of the control protein, Escherichia coli β-lactamase, was removed when synthesized with microsomes in this system (data not shown). To confirm the finding of Noutside orientation, aliquots of the same translation mixtures described in Fig. 3B were subjected to digestion with or without Tc treatment, immunoprecipitated with anti-HIMP1 serum, separated by SDS/PAGE, and examined by autoradiography (Fig. 3C). The results showed that intact HIMP1-a was immunoprecipitated from the products without Tc treatment in the presence or absence of microsomes (lanes 1 and 2); but was absent after Tc treatment (lanes 3 and 4). This result clearly indicates that HIMP1-a inserted into microsomal membranes in vitro in Noutside orientation.
Using a similar procedure, outer and inner membrane fractions of mitochondria from αTC1.6 cells were subjected to digestion with or without Tc, then examined by immunoblotting with the N-terminally directed anti-HIMP1 serum. As shown in Fig. 3E, HIMP1 proteins were detected only in the inner membrane fraction without Tc digestion, but were not detectable after Tc digestion. VDAC protein, an outer membrane marker, which was recognized by an antibody against one of its cytosolic loops, also disappeared with Tc treatment (Fig. 3E). The size of HIMP1 was identical to that detected in postnuclear or crude mitochondrial fractions (data not shown), and no HIMP1 cleavage products corresponding to the reverse orientation, with Ninside–Cinside and loopoutside, were observed. All of these data indicate that HIMP1 proteins are normally oriented with Noutside–Coutside and loopinside within the mitochondrial inner membrane (Fig. 3F). In addition, no evidence of cleavage during trafficking and localization of HIMP1 proteins in the mitochondrial inner membrane have been observed.
Ectopic Expression of HIMP1-a in β Cells Protects Cells from Apoptosis and Extends Cell Survival Under Conditions of Hypoxia or Low Glucose Levels.
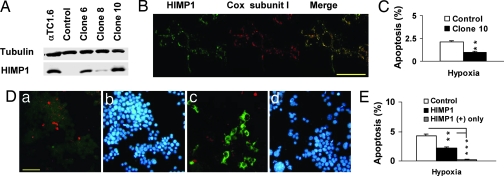
One of the mitochondrial roles is the linkage to cellular apoptosis. Because HIMP1 is a mitochondrial protein that is up-regulated by low glucose or low O2, we were interested in its possible effects in β cells. To examine this question, we have analyzed the effects of ectopic expression of HIMP1-a in two widely used β cell models, MIN6 and βTC3 cells, under these conditions. From several stable MIN6HIMP1-a clones, clone 10 cells were selected for experiments because they have a similar expression level (Fig. 4 A) and mitochondrial localization (Fig. 4B) of HIMP1-a protein compared to αTC1.6 cells. As shown in Fig. 4C, under hypoxic (5%) conditions for 24 h, the clone 10 cells exhibited a significantly reduced percentage of apoptotic cells compared to vector control MIN6 cells (0.95 ± 0.14% vs. 2.13 ± 0.13%, P = 0.02). In βTC3 cells transiently transfected with either vector or HIMP1-a cDNA, triple staining of TUNEL, HIMP1-a, and DAPI was performed (Fig. 4D). The results, summarized in Fig. 4E, show that the percent apoptosis of the HIMP1-a transfected βTC3 cells (2.53 ± 0.13%, P = 0.028), or of the HIMP1-positive βTC3 cells only (0.17 ± 0.12%, P = 0.0003), is significantly lower compared to the nontransfected control cells (4.06 ± 0.36%).
Fig. 4.
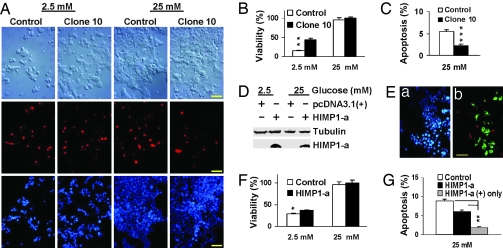
Ectopic expression of HIMP1-a in MIN6 and βTC3 β cells protects cells from apoptosis and extends cell survival under hypoxia (5% O2) for 24 h. (A-C) Analysis in stable MIN6HIMP1-a clonal cells. (A) Immunoblot, 50 μg of protein per lane. (B) Confocal immunofluorescent staining in MIN6HIMP1-a clone 10 cells. (Scale bar, 50 μm.) (C) Protective effects of HIMP1-a on MIN6 cell survival at hypoxia for 24 h. (D and E) Transient transfection assay in βTC3 cell. (D) Images of triple staining of TUNEL (red), HIMP1-a (green), and DAPI (blue) in βTC3 cells transfected with either vector (a and b) or HIMP1-a (c and d) under hypoxic conditions for 24 h. (Scale bar, 50 μm.) (E) Protective effects of HIMP1-a on βTC3 cell survival at hypoxia for 24 h.
To reveal whether this action of HIMP1-a is sustained with another stress, we examined the effects of high (25 mM) or low (2.5 mM) glucose levels using a similar protocol. As shown in Fig. 5 A, clone 10 and control cells were subjected to double staining with TUNEL/DAPI and morphological observation using differential interface contrast (DIC) microscopy at various time points after seeding equal numbers of cells for a 48-h preculture under normal conditions. The percents of viability and apoptosis observed at the 3-day time point are shown in Fig. 5 B and C, respectively. At low glucose, the percent viability of clone 10 cells is significantly higher than the vector control (44 ± 3.8% vs.15 ± 2.1%, P = 0.001) and the corresponding percent apoptosis is significantly lower. At high glucose, the number of apoptotic clone 10 cells is also significantly lower than in control cultures (2.3 ± 1.2% vs.5.5 ± 1.9%, P = 0.0002), but no significant difference in percent viability between them was observed. To further validate these findings, similar experiments were performed in βTC3 cells transiently transfected with either vector or HIMP1-a cDNA (Fig. 5 D–G). Ectopic HIMP1-a expression in βTC3 cells was confirmed by immunoblotting (Fig. 5D). Triple staining of DAPI, TUNEL, and HIMP1 in βTC3 cells transfected with HIMP1-a (Fig. 5E) or vector cDNA was carried out by using the same procedure described for Fig. 4D. Analyses of the stained cells (Fig. 5F) show that, at low glucose, the viability of βTC3 cells transfected with HIMP1-a is significantly higher than that of the control (37.2 ± 1.6% vs.29.3 ± 1.2%, P = 0.02), whereas the corresponding percentage of apoptotic cells is reversed. At high glucose (Fig. 5G), the percent apoptosis (5.9 ± 0.6%) for all βTC3 cells transfected with HIMP1-a was lower than the control (8.8 ± 0.5%, P = 0.064), whereas for the HIMP1-positive staining βTC3 cells, it was significantly lower (1.8 ± 0.3%) than the control (8.8 ± 0.5%, P = 0.0078). However, no significant difference in viability was evident at this glucose level. These data show that HIMP1 proteins can increase β cell survival under the stress of either hypoxic or hypoglycemic conditions.
Fig. 5.
Ectopic expression of HIMP1-a in MIN6 and βTC3 β cells protects cells from apoptosis and extends β cell survival at high (25 mM) or low (2.5 mM) glucose levels after exposure for 3 days. (A–C) Analysis in stable MIN6HIMP1-a clonal cells. (A) Images of DIC and double staining of TUNEL (red) and DAPI (blue) in MIN6HIMP1-a clone 10 and control cells at low and high glucose. (Scale bar, 50 μm.) (B) Protective effects of HIMP1-a on MIN6 cell survival at low glucose. (C) Protective effects of HIMP1-a on MIN6 cell survival at high glucose. (D–G) Transient transfection assay in βTC3 cells. (D) Immunoblot analysis of HIMP1-a expression in βTC3 cells. (E) An image sample of triple staining of TUNEL (red image of B), HIMP1-a (green image of B), and DAPI (blue image of A) in βTC3 cells transfected with HIMP1-a for 3 days at high glucose. (Scale bar, 50 μm.) (F) Protective effects of HIMP1-a on βTC3 cell survival at low glucose. (G) Protective effects of HIMP1-a on MIN6 cell survival at high glucose.
Because the mitochondria are the main source of cellular energy and link to insulin secretion (20), it was of interest to assess whether ATP production and insulin secretion are affected by the ectopic expression of HIMP1-a in β cells. No significant changes in these parameters were observed in MIN6HIMP1-a clone10 cells under normal growth conditions (data not shown). Thus, the protective effects of HIMP1 on β cell survival appear unlikely to be mediated by direct affects on ATP production.
Discussion
In the course of studies to identify differentially expressed proteins in α vs. β cells (25), we have identified a small protein that is present in α cells, heart, and several other tissues, but is absent or very low in β cells. In the meantime, this protein has been found to be expressed in other cell types and its homologues have been demonstrated to exist in other species by several groups (26–31). It is well conserved among eukaryotes, including lower vertebrates and Drosophila, suggesting an important function. In mice, a single gene encodes this protein, which we have called HIMP1, giving rise through alternative splicing to two isoforms differing only at their C termini. The predicted structure of this protein suggests that it is an integral membrane protein consisting of two hydrophobic helices, 21–23 residues in length that might tend to form a hairpin-like loop across the bilayer. Indeed, when expressed in a cell-free system, the protein was found to be inserted into microsomes with an orientation of both N and C termini located externally and with a central small loop on the luminal side (Fig. 3 B–D). However, fractionation of αTC1.6 cell homogenates on sucrose gradients did not support an ER/Golgi localization, but rather pointed toward the mitochondria as a major subcellular site. The mitochondrial inner membrane location of HIMP1 is well demonstrated. Its possible location in secretory granules or ER in addition to mitochondria was investigated because some HMIP1 immunoreactivity was seen in the glucagon granule fraction (Fig. 2B). Observations in αTC1.6 cells with immunoelectron microscopy and double staining with antibodies against HIMP1 and ER or granule markers did not support these possible localizations (data not shown). Comparison of HIMP1 with the N-terminal signal sequence of the P4501A1 protein, which exhibits dual targeting to ER and mitochondria (32), did not indicate much primary sequence similarity other than some shared positively charged residues suggested to be critical for mitochondrial targeting (33). The reason for the in vitro insertion of HIMP1-a into microsomal membranes is an issue to be investigated. Further immunocytochemical analysis confirmed the mitochondrial localization and indicated a localization mainly in the cristae. Subsequent fractionation experiments confirmed the inner mitochondrial membrane to be the major site of this protein in α cell, heart, and sperm tail mitochondria (Fig. 3). The topology here again appears to be with N and C termini outside and central loop inside (Fig. 3). Moreover, the high degree of species conservation includes especially the transmembrane domains, possibly indicating that these interact either with themselves to form an oligomeric structure such as a channel or with some other component(s) of the mitochondrial inner membrane. Thus, HIMP1 proteins have a membrane topology similar to that of some potassium channel proteins, and also have a key structural requirement (Tyr–Gly) of a K+ channel pore (34), but not the conserved signature sequence (Gly–Tyr–Gly) (35). Because heart and sperm mitochondria are known to be highly active metabolically, a possible regulatory role for this protein seems likely.
Further studies (Fig. 1 A and C) revealed that HIMP1 is up-regulated in αTC1.6 cells when incubated at low glucose levels. Also, like homologues reported (26, 27) HIMP1 levels were also elevated by exposure of αTC1.6 cells to hypoxia, consistent with its induction by hypoxia found in several other cell types (28, 29). These findings suggest a role in metabolic or oxidative stress. To pursue this possibility, we tried expressing HIMP1 stably in MIN6 cells. Several positive clones were obtained, and one having levels of expression similar to that in αTC1.6 cells was exposed to high or low glucose conditions for several days. We were surprised to find that the presence of HIMP1 significantly increased the viability of the cells at low glucose, but not appreciably at high glucose. This effect on survival was accompanied by a highly significant reduction in rates of apoptotic cell death at both glucose concentrations. Intrigued by these findings, we compared the expression of cellular proteins known to be associated with apoptosis and/or oxidative stress in α vs. β cells. Surprisingly there were only minor differences in all instances (data not shown).
As an important cellular organelle, mitochondria exist in most eukaryotic cells in varying numbers (36). A great deal of evidence has implicated the mitochondria as important regulators of apoptosis. Thus, in addition to their role as cellular components equipped with various oxidative and biosynthetic pathways and serving as a power plant for cellular ATP supply, they are the site of integration of cellular metabolism and apoptosis (19). Although the importance of mitochondria in β cells for insulin secretion has been long recognized (20, 37) their link with apoptotic proneness of β cells is less clear.
The data presented here support the conclusion that mitochondria in β cells lack both HIMP1 and a known homologue, albeit other molecules having potential complementary functions may exist. Probing the role(s) of HIMP1 in the mitochondria should thus provide insights into mechanisms linked to mitochondrial apoptotic susceptibility in β cells. The high level of expression of HIMP1 in cardiomyocytes suggests an important role, perhaps in adapting these cells to their vigorous work role and/or their highly efficient utilization of metabolic fuels. The conservation of this gene in many eukaryotic organisms, including both vertebrates and invertebrates, is also consistent with an evolutionarily conserved essential function. Further insight into the interactions of HIMP1 may provide strategies for treating diabetes and/or preventing other diseases resulting from mitochondrial dysfunction such as heart failure (36, 38).
Materials and Methods
For further detail, please see Supporting Text, which is published as supporting information on the PNAS web site.
Construction of a αTC1.6 cDNA Library and Molecular Cloning of HIMP1 cDNAs.
Characterized cDNA sequences for the HIMP1 have been deposited in GenBank (accession numbers AY028386 and AY028387).
Cell Culture, Islet Isolation, RNA Preparation, RT-PCR, Northern Blot, and in Vitro Synthesis.
The procedures have been described (25, 39). The HIMP1-a cDNA was cloned into the pcDNA3.1(+) vector for in vitro synthesis and transfection.
Antibodies, Immunoblot, Immunoprecipitation, and Histology.
Antiserum against the N-terminal 1–17 residues of HIMP1 was raised in rabbits. Antibodies against mtHsp70, VDAC, COX subunit I, GM130, glucagon, C-peptide, and tubulin were purchased commercially.
Immunoblot, immunoprecipitation, and immunofluorescent staining were performed similarly as described (25, 39). TUNEL staining was carried out by using DeadEnd system (Promega) with modifications. For immunoelectron microscopy, sectioned samples were mounted on grids and examined after staining with gold-labeled anti-HIMP1 serum.
Subcellular Fractionation and Topology Studies.
Separation of cellular membrane and cytosol fractions or sucrose gradient fractionation was performed as described (22, 40). Separated fractions were analyzed by immunoblotting and/or assayed for glucagon by RIA. The outer and inner membrane fractions of mitochondria were separated with digitonin treatment (23). The topology studies were performed with the method of Blobel and Dobberstein (24) using in vitro synthesized HIMP1-a protein and various membrane subfractions of mitochondria with or without Tc treatment.
Expression of HIMP1-a Stably in MIN6 Cells, and Transiently in βTC3 Cells.
The HIMP1-a or vector plasmid DNA was transfected into MIN6 or βTC3 β cells. Clones of MIN6 cell expressing HIMP1-a permanently, termed MIN6HIMP1-a, were selected with G418-resistant and immunoblots with anti-HIMP1 serum. βTC3 cells were used for transient transfection studies.
Apoptosis and Viability Examination of β Cells with Ectopic Expression of HIMP1-a Under Different O2 or Glucose Conditions.
Equal numbers of clone 10 MIN6HIMP1-a and vector control cells were subjected to hypoxia (5% O2) and normal culture condition (95% air/5% CO2) for 24 h, or subjected to either low (2.5 mM) or high (25 mM) glucose DMEM culture in 95% air/5% CO2 incubator for 3 days. In βTC3 cells, at 48 h after transfection, cells were subjected to the same treatments as for the MIN6HIMP1-a cells. The determination, count, and calculation methods for the numbers and percent of total, apoptotic, and viable cells are provided in supporting information.
Data are presented as the means ± SEM. Statistical significance was assessed by the Student t test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗,P < 0.0005.
Supplementary Material
Acknowledgments
We thank Raymond Carroll, Jeff Stein, Vytas Bindokas, Yingli Duan, and William Chutkow (University of Chicago) for technical assistance; Rosie Ricks for expert assistance in preparing this manuscript; and Graeme Bell, Louis Philipson, Manami Hara, Gene Webb (University of Chicago), and Paul Schumacker (Northwestern University, Chicago) for helpful discussions. This work was supported by National Institutes of Health Grants DK-13914 and DK-20595 and by the Howard Hughes Medical Institute.
Abbreviations
- TMH
transmembrane helice
- Tc
trypsin and chymotrypsin.
Footnotes
References
- 1.Kloppel G., Lohr M., Habich K., Oberholzer M., Heitz P. U. Surv. Synth. Pathol. Res. 1985;4:110–125. doi: 10.1159/000156969. [DOI] [PubMed] [Google Scholar]
- 2.Clark A., Wells C. A., Buley I. D., Cruickshank J. K., Vanhegan R. I., Matthews D. R., Cooper G. J., Holman R. R., Turner R. C. Diabetes Res. 1988;9:151–159. [PubMed] [Google Scholar]
- 3.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 4.Yoon K. H., Ko S. H., Cho J. H., Lee J. M., Ahn Y. B., Song K. H., Yoo S. J., Kang M. I., Cha B. Y., Lee K. W., et al. J. Clin. Endocrinol. Metab. 2003;88:2300–2308. doi: 10.1210/jc.2002-020735. [DOI] [PubMed] [Google Scholar]
- 5.Marchetti P., Del Guerra S., Marselli L., Lupi R., Masini M., Pollera M., Bugliani M., Boggi U., Vistoli F., Mosca F., Del Prato S. J. Clin. Endocrinol. Metab. 2004;89:5535–5541. doi: 10.1210/jc.2004-0150. [DOI] [PubMed] [Google Scholar]
- 6.Mathis D., Vence L., Benoist C. Nature. 2001;414:792–798. doi: 10.1038/414792a. [DOI] [PubMed] [Google Scholar]
- 7.Rhodes C. J. Science. 2005;307:380–384. doi: 10.1126/science.1104345. [DOI] [PubMed] [Google Scholar]
- 8.Hoorens A., Van de Casteele M., Kloppel G., Pipeleers D. J. Clin. Invest. 1996;98:1568–1574. doi: 10.1172/JCI118950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Casteele M., Kefas B. A., Cai Y., Heimberg H., Scott D. K., Henquin J. C., Pipeleers D., Jonas J. C. Biochem. Biophys. Res. Commun. 2003;312:937–944. doi: 10.1016/j.bbrc.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Moritz W., Meier F., Stroka D. M., Giuliani M., Kugelmeier P., Nett P. C., Lehmann R., Candinas D., Gassmann M., Weber M. FASEB J. 2002;16:745–747. doi: 10.1096/fj.01-0403fje. [DOI] [PubMed] [Google Scholar]
- 11.Pick A., Clark J., Kubstrup C., Levisetti M., Pugh W., Bonner-Weir S., Polonsky K. S. Diabetes. 1998;47:358–364. doi: 10.2337/diabetes.47.3.358. [DOI] [PubMed] [Google Scholar]
- 12.Donath M. Y., Gross D. J., Cerasi E., Kaiser N. Diabetes. 1999;48:738–744. doi: 10.2337/diabetes.48.4.738. [DOI] [PubMed] [Google Scholar]
- 13.Federici M., Hribal M., Perego L., Ranalli M., Caradonna Z., Perego C., Usellini L., Nano R., Bonini P., Bertuzzi F., et al. Diabetes. 2001;50:1290–1301. doi: 10.2337/diabetes.50.6.1290. [DOI] [PubMed] [Google Scholar]
- 14.Unger R. H., Orci L. Biochim. Biophys. Acta. 2002;1585:202–212. doi: 10.1016/s1388-1981(02)00342-6. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen K., Karlsen A. E., Deckert M., Madsen O. D., Serup P., Mandrup-Poulsen T., Nerup J. Diabetes. 1999;48:2324–2332. doi: 10.2337/diabetes.48.12.2324. [DOI] [PubMed] [Google Scholar]
- 16.Welsh N., Margulis B., Borg L. A., Wiklund H. J., Saldeen J., Flodstrom M., Mello M. A., Andersson A., Pipeleers D. G., Hellerstrom C., et al. Mol. Med. 1995;1:806–820. [PMC free article] [PubMed] [Google Scholar]
- 17.Tiedge M., Lortz S., Drinkgern J., Lenzen S. Diabetes. 1997;46:1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J. D., Ahmed N. T., Luciani D. S., Han Z., Tran H., Fujita J., Misler S., Edlund H., Polonsky K. S. J. Clin. Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danial N. N., Korsmeyer S. J. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 20.Wollheim C. B., Maechler P. Diabetes. 2002;51(Suppl. 1):S37–S42. doi: 10.2337/diabetes.51.2007.s37. [DOI] [PubMed] [Google Scholar]
- 21.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 22.Wendland B., Scheller R. H. Mol. Endocrinol. 1994;8:1070–1082. doi: 10.1210/mend.8.8.7997233. [DOI] [PubMed] [Google Scholar]
- 23.Schnaitman C., Erwin V. G., Greenawalt J. W. J. Cell Biol. 1967;32:719–735. doi: 10.1083/jcb.32.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blobel G., Dobberstein B. J. Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Webb G., Cao Y., Steiner D. F. Proc. Natl. Acad. Sci. USA. 2003;100:12660–12665. doi: 10.1073/pnas.1735286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denko N., Schindler C., Koong A., Laderoute K., Green C., Giaccia A. Clin. Cancer Res. 2000;6:480–487. [PubMed] [Google Scholar]
- 27.Gracey A. Y., Troll J. V., Somero G. N. Proc. Natl. Acad. Sci. USA. 2001;98:1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin K., Mao X. O., Eshoo M. W., del Rio G., Rao R., Chen D., Simon R. P., Greenberg D. A. Neurochem. Res. 2002;27:1105–1112. doi: 10.1023/a:1020913123054. [DOI] [PubMed] [Google Scholar]
- 29.Salnikow K., Davidson T., Zhang Q., Chen L. C., Su W., Costa M. Cancer Res. 2003;63:3524–3530. [PubMed] [Google Scholar]
- 30.Bedo G., Vargas M., Ferreiro M. J., Chalar C., Agrati D. Int. J. Dev. Biol. 2005;49:431–436. doi: 10.1387/ijdb.041901gb. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Fink T., Zhang X. Y., Ebbesen P., Zachar V. Differentiation. 2005;73:350–363. doi: 10.1111/j.1432-0436.2005.00038.x. [DOI] [PubMed] [Google Scholar]
- 32.Bhagwat S. V., Biswas G., Anandatheerthavarada H. K., Addya S., Pandak W., Avadhani N. G. J. Biol. Chem. 1999;274:24014–24022. doi: 10.1074/jbc.274.34.24014. [DOI] [PubMed] [Google Scholar]
- 33.Neupert W. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura R. L., Anderson J. A., Gaber R. F. J. Biol. Chem. 1997;272:1011–1018. doi: 10.1074/jbc.272.2.1011. [DOI] [PubMed] [Google Scholar]
- 35.Heginbotham L., Lu Z., Abramson T., MacKinnon R. Biophys. J. 1994;66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace D. C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 37.Maechler P., Wollheim C. B. Nature. 2001;414:807–812. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 38.Duchen M. R. Diabetes. 2004;53(Suppl. 1):S96–S102. doi: 10.2337/diabetes.53.2007.s96. [DOI] [PubMed] [Google Scholar]
- 39.Wang J., Cao Y., Steiner D. F. J. Biol. Chem. 2003;278:32899–32904. doi: 10.1074/jbc.M305456200. [DOI] [PubMed] [Google Scholar]
- 40.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.