Abstract
Oxidative damage in testicular DNA is associated with poor semen quality, reduced fertility and increased risk of stillbirths and birth defects. These DNA lesions are predominantly removed by base excision repair. Cellular extracts from human and rat testicular cells and three enriched populations of rat male germ cells (primary spermatocytes, round spermatids and elongating/elongated spermatids) all showed proficient excision/incision of 5-hydroxycytosine, thymine glycol and 2,6-diamino-4-hydroxy-5-formamidopyrimidine. DNA containing 8-oxo-7,8-dihydroguanine was excised poorly by human testicular cell extracts, although 8-oxoguanine-DNA glycosylase-1 (hOGG1) was present in human testicular cells, at levels that varied markedly between 13 individuals. This excision was as low as with human mononuclear blood cell extracts. The level of endonuclease III homologue-1 (NTH1), which excises oxidised pyrimidines, was higher in testicular than in somatic cells of both species. Cellular repair studies of lesions recognised by formamidopyrimidine-DNA glycosylase (Fpg) or endonuclease III (Nth) were assayed with alkaline elution and the Comet assay. Consistent with the enzymatic activities, human testicular cells showed poor removal of Fpg-sensitive lesions but efficient repair of Nth-sensitive lesions. Rat testicular cells efficiently repaired both Fpg- and Nth-sensitive lesions. In conclusion, human testicular cells have limited capacity to repair important oxidative DNA lesions, which could lead to impaired reproduction and de novo mutations.
INTRODUCTION
The integrity of the male germ cell genome is important for the successful production of offspring. Several biological systems have evolved that provide continual maintenance of the DNA molecule. Among these are the excision repair pathways that correct physical injuries to DNA as well as replication errors, thereby contributing to the quality control systems ensuring DNA fidelity. We recently showed efficient repair of alkylated bases, mainly via base excision repair (BER), in human and rat male germ cells (1) and the rather surprising results of suppressed or non-functional nucleotide excision repair (NER) in rat male germ cells (2,3). The present study focuses on the repair of oxidative DNA damage in human and rat male germ cells.
Reactive oxygen species (ROS) are formed in cells as by-products of normal cellular metabolism, or by external sources such as ionising radiation, near-UV light, redox-active drugs and sensitising dyes. ROS react with DNA to form genotoxic lesions such as 8-oxo-7,8-dihydroguanine (8-oxoG), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FaPy), thymine glycol (TG) and 5-hydroxycytosine (5-ohC). 8-oxoG residues are mutagenic, as they give rise to G:C→T:A transversions (4), and 5-ohC has a potential for pairing with A, resulting in C:G→T:A transitions, whereas FaPy and TG are cytotoxic. 8-oxoG residues were recently shown to block transcription by RNA polymerase II in vivo (5) whereas TG blocks both DNA and RNA polymerases in vitro (6,7). Both testicular cells and sperm contain oxidised DNA lesions (8–10) and the levels increase following exposure to a variety of drugs, irradiation and environmental factors, such as heavy metals (11,12).
Repair of oxidised bases by the BER pathway is initiated by DNA glycosylases that cleave the base–deoxyribose glycosylic bond of the damaged nucleotide residue. The resulting apurinic/apyrimidinic sites (AP sites) are incised by AP-endonucleases or AP-lyases, and the process proceeds with DNA synthesis and ligation. BER seems essential for the repair of endogenous DNA damage, as illustrated by the embryonic lethality of mice with gene knockout of proteins required for later steps in BER, including AP-endonuclease, DNA polymerase β (some survived to birth) and X-ray cross-complementing protein 1 (Xrcc1) (13). On the other hand, mice with gene knockout in early steps of BER, such as 8-oxoguanine-DNA glycosylase-1 (Ogg1) or uracil DNA glycosylase (Udg), are viable, suggesting the existence of back-up enzymes (14,15).
Mammalian OGG1 homologues have been cloned and characterised in humans (hOGG1) (16–21), mice (21,22) and rats (Ogg1) (23). OGG1 incises 8-oxoG as well as FaPy residues and has associated AP-lyase activity (18). The incision of 8-oxoG and the AP-lyase activity are most efficient when the base or AP site is located opposite a cytosine (18). OGG1 is essential for the repair of 8-oxoG in the non-transcribed strand of a gene whereas an OGG1-independent pathway for the removal of 8-oxoG in the transcribed strand seems to exist in vivo (24). It was recently shown that active hOGG1 bound to the nuclear matrix and mitotic chromatin is phosphorylated (25).
The human as well as the mouse genes for the DNA glycosylase endonuclease III homologue-1 (NTH1) have been cloned (26–28). The enzyme hNTH1 excises oxidised pyrimidines, such as 5-ohC and TG, as well as FaPy residues, and has associated AP-lyase activity (26,29,30). NTH1 is surprisingly ineffective compared to the bacterial enzyme, but it was recently shown that hNTH1 is stimulated by the presence of physiological concentrations of Mg2+ and that hNTH1 incises lesions positioned opposite G most efficiently (31). Furthermore, the activity of hNTH1 is stimulated by the XPG protein both in vivo and in vitro (14,32,33).
Recently several DNA glycosylases that belong to the formamidopyrimidine-DNA glycosylase (Fpg)/Nei family have been described (34–36). One of these (NEH1/hNEI1/hFPG1) efficiently excises FaPy, oxidised pyrimidines and, weakly, 8-oxoG, and its mRNA is moderately expressed in the testis. Another enzyme (NEH2/hNEI2/hFPG2), exclusively expressed in the testis and thymus, excises FaPy residues (36).
In this study the presence of activities like OGG1 and NTH1 in cellular extracts from human and rat male germ cells was studied, based on the activities for incising DNA lesions such as 8-oxoG, 5-ohC, TG and FaPy residues, and on western analysis using specific antibodies. Extracts were prepared from crude testicular cell suspensions as well as from enriched fractions of cells at different stages of spermatogenesis to detect possible cell-specific differences. Furthermore, cellular repair of DNA lesions removable by the Escherichia coli enzymes Fpg and endonuclease III (Nth) was studied using modified versions of the alkaline elution and the Comet assays.
MATERIALS AND METHODS
Cells
Human testis biopsies were obtained from organ transplant donors and rat testes from sexually mature male Wistar rats (MOL: WIST, 200–300 g; Møllegaard, Ejby, Denmark). The human tissues were obtained in accordance with the Norwegian law concerning transplantation, hospital autopsies and donation of corpses (revision of January 1, 1994). Human testis biopsy donors (n = 19; denoted by numbers) were 14–74 years old with spermatogenesis within the normal range for 18 of the 19 donors as judged by flow cytometric (37) and histological analyses, with one suffering from the ‘Sertoli cell-only’ syndrome (no. 14). Human and rat testicular cells (hTC and rTC, respectively) were isolated by enzymatic digestion with collagenase and trypsin and were kept in culture medium as previously described (1,3,38). Human and rat mononuclear blood cells (hMNC and rMNC, respectively) and rat primary hepatocytes (rHEP) were isolated as previously described (1,3). Lymphoblastoid cells (LC) (GM 00130; Coriell Cell Repositories, Camden, NJ) were cultivated in a humidified atmosphere in RPMI culture medium with 25 mM HEPES buffer, l-glutamine and 15% fetal calf serum. Primary normal human fibroblasts (VH25; a gift from Leon Mullenders) were cultivated as for GM 00130 cells, but in a 2.5% CO2 atmosphere. Viability of cells, assessed by trypan blue exclusion and for some samples also by propidium iodide exclusion, was >90%.
Enrichment and characterisation of male germ cells
Human and rat testicular cells were separated by centrifugal elutriation by two different procedures; human cells as described (39) and rat cells as described (1,40,41). Three cell fractions were collected containing predominantly primary spermatocytes (SC), round spermatids (RS) or elongating/elongated spermatids (ES). The rat ES were further enriched by metrizamide density gradient centrifugation (41). The composition of testicular cell suspensions was analysed with flow cytometric, immunocytochemical and microscopical methods as previously described (1).
The proportion of somatic cells in normal human crude testicular cell suspensions (hTC) averaged 17% (range 9–25%), as estimated by flow cytometry after immunostaining of the cytoskeletal protein vimentin (42) that is expressed in somatic cells only. Human testicular cells were fractionated into populations containing 50–70% primary spermatocytes (hSC), 50–70% round spermatids (hRS) or 70–85% elongating/elongated spermatids (hES). The three enriched rat male germ cell populations used contained 50 ± 15% spermatocytes (rSC), 93 ± 4% round spermatids (rRS) and 94 ± 2% elongating/elongated spermatids (rES).
Preparation of cellular extracts
Protein extracts were made by plasmolysis as described (1) or by homogenisation of tissue (testis, liver, kidney and spleen), and protein contents were determined by the method of Lowry using bovine serum albumin as a standard.
Preparation of DNA substrates
Duplex DNA substrate containing 8-oxoG was prepared as described (1,43). In short, single-stranded oligonucleotides (Eurogentech, Seraing, Belgium) containing the lesion were 5′-end-labelled with [γ-32P]ATP (5000 Ci/mmol; Amersham Pharmacia Biotech, Little Chalfont, UK) and T4 polynucleotide kinase (37°C for 30 min) and hybridised with complementary oligonucleotides. Duplex DNA was purified by separation by non-denaturing PAGE and electroelution. The DNA sequence of the oligonucleotide containing 8-oxoG was 5′-ATCACCGGC[8-oxoG]CCACACGAGCTG.
A duplex DNA substrate containing 5-ohC was made by annealing a 5′-32P-end-labelled 40mer (5′-AATTGCGATCT AGCTCGCCAG[5-ohC]AGCGACCTTATCTGATGA-3′) to a complementary oligonucleotide. Purification of the duplex DNA was by non-denaturing PAGE and electroelution.
A substrate containing TG was prepared as described (44,45). Four oligonucleotides were used (I, 5′-TTGACATT GCCCT; II, 5′-CGCGA[TG]ACGCC, a gift from Dr Philip Bolton; III, 5′-TAGACGAATTCCG; and a complementary 37mer oligonucleotide). Oligonucleotide I was 5′-end-labelled, the three oligonucleotides (I, II and III) were ligated (8 U T4 DNA ligase at 16°C for 3 h) and hybridised to the complementary 37mer oligonucleotide. The double-stranded substrate was treated with 2.5 ng E.coli endonuclease IV (Nfo) to remove oligonucleotides containing AP sites prior to purification by 15% denaturing PAGE. The full-length single-stranded oligonucleotide containing TG was extracted by electroelution and hybridised again to the complementary 37mer oligonucleotide.
A substrate containing 3H-labelled FaPy was prepared as described (46,47) with modifications. N-[3H]methyl-N′-nitrosourea ([3H]-MNU; Amersham Pharmacia Biotech) was used instead of [3H]dimethylsulfate. Poly(dG·dC) DNA was treated with [3H]-MNU (18.0 Ci/mmol) in 10 mM sodium cacodylate, pH 7.0, for 2 h at 37°C. DNA was precipitated twice with ethanol, washed and dissolved in 800 µl 10 mM Tris–HCl, pH 8, 1 mM EDTA (TE). A 270 µl aliquot of 50 mM Na2HPO4, pH 11.45, was added, followed by incubation at 25°C for 48 h and dialysis in TE for 16 h. The specific activity of the resulting substrate was 5000 d.p.m./µg DNA.
DNA glycosylase/AP-lyase assays
All enzyme reactions were carried out in 70 mM morpholinopropane sulfonic acid, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol and 5% glycerol, and incubated at 37°C, unless otherwise indicated. The oligonucleotide nick assay was carried out as described (1,43,45). Excision of the 8-oxoG-substrate (0.15 fmol) was performed in the presence of Nfo (20 ng/reaction) using 10 µg extract (13.7 µl total volume, 60 min). Incision of the 5-ohC:G substrate (1 fmol) was performed using 2 µg extract (5 µl total volume, 30 min). Incision of TG-DNA (10 fmol) was performed using 5 µg extract (10 µl total volume, 60 min). Incision of the 8-oxoG and 5-ohC substrates (100 fmol) by the 13 human testicular extracts (Fig. 2) was performed using 5 ng Saccharomyces cerevisiae AP-endonuclease 1 (Apn1) and 5 µg extract for each reaction and incubation for 30 min. Reactions were terminated by adding gel loading dye solution (95% formamide, 20 mM EDTA, pH 8, 0.05% bromophenol blue, 0.05% xylene cyanol) in 0.7 times the total volume of the reaction. The substrate and cleavage products were separated by 20% denaturing PAGE and visualised using a Molecular Dynamics PhosphorImager model 445 SI. The relative amount of cleaved substrate was calculated as the amount of cleaved substrate divided by the total amount of substrate. Excision of FaPy DNA residues was performed as described (43). Protein extracts were incubated with 0.24 µg [3H]FaPy DNA substrate in a total volume of 50 µl. Reactions were stopped by adding 75 µl solution A (2 mg calf thymus DNA, 80 mg BSA, 0.5 M NaAc, pH 5.5). DNA was ethanol precipitated, centrifuged and the amount of released [3H]FaPy residues in an aliquot of the supernatant was measured by liquid scintillation counting.
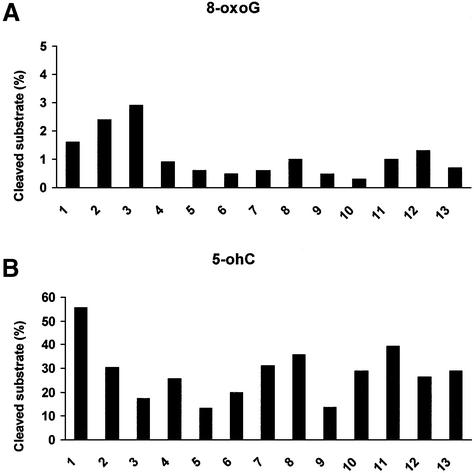
Figure 2.
Excision of 8-oxo-7,8-dihydroguanine (8-oxoG) and 5-hydroxycytosine (5-ohC) by human testicular extracts. Extracts (5 µg) from 13 testis biopsies (denoted by numbers) were assayed for their abilities to excise 8-oxoG:C (A) and 5-ohC:G (B). The amount of substrate was 100 fmol and 5 ng of S.cerevisiae AP-endonuclease (Apn1) was added to each reaction. The results are presented as per cent cleaved substrate (%) for each testis biopsy.
Western analyses
Standard western procedures were followed using 5–30 µg extract. Primary antibodies for the detection of OGG1 were a polyclonal rabbit anti-hOGG1 serum (1:3000 dilution; a gift from Dr Sancar Mitra) generated against a hOGG1 peptide, and monoclonal mouse anti-hOGG1 (1:100 dilution; IBL, Gunma, Japan). Primary antibody for the detection of NTH1 was rabbit anti-hNTH1 generated against the protein (1:1000 dilution; a gift from Dr Sancar Mitra). Secondary antibodies were donkey anti-rabbit–horseradish peroxidase (HRP) conjugate, donkey anti-mouse–HRP conjugate (Jackson ImmunoResearch, West Grove, PA) or mouse anti-rabbit– alkaline phosphatase conjugate (Promega, Madison, MI). Bands were visualised using enhanced chemiluminescence (ECL™; Amersham Pharmacia Biotech) followed by autoradiography or development of color (Protoblot®; Promega).
Cellular DNA repair assays
Treatment of cells. To analyse the removal of Fpg-sensitive DNA lesions, the compound ethyl 7-oxo-7H-thieno[2,3-A] quinolizine-8-carboxylate (Ro 12-9786; a gift from Dr Elmar Gocke) was used. Human and rat testicular cells (1.5–2.2 × 106 cells/ml) in phosphate-buffered saline on ice were treated with Ro 12-9786 (3–6 µM) in 0.5% dimethylsulphoxide combined with cold visible light (5 min, Schott KL 1500 electronic) at a distance of 10 cm from the fibre optic light guide. After exposure, the cells were centrifuged and resuspended in fresh culture medium. Repair of the induced DNA damage in testicular cells was allowed by incubation in a humidified atmosphere at 32°C, 5% CO2 for up to 8 h. Human normal primary fibroblasts were grown to near confluence, exposed to Ro 12-9786 (6 µM) and light (5 min, 15 cm distance). The cells were incubated for repair at 37°C and 2.5% CO2, and otherwise treated similarly to testicular cells. The incubation period was up to 4 h, after which the cells were harvested by scraping and transferred to the alkaline elution filters.
To analyse the repair of Nth-sensitive DNA lesions, oxidative DNA damage was introduced by irradiating cells (1.3 × 106/ml) on ice with X-rays (3 Gy; Phillips MG300 X-ray unit, operated at 10 mA and 260 kV). Cells were kept in a humidified atmosphere at 32°C, 5% CO2 for up to 2 h to allow repair.
Alkaline elution in combination with crude extract of Fpg. Alkaline elution was performed as previously described with modifications (48), using exogenous enzymes for removing modified bases, as described (49) with modifications. Following lysis of the cells, DNA was washed with 20 mM Na2EDTA, pH 9.6, for 35 min and then with BE1 buffer (20 mM Tris–HCl, 100 mM NaCl, 1 mM Na2EDTA, pH 7.5) for 1 h; these buffers were flushed through each filter at a flow rate of 0.25 ml/min. Subsequently, the DNA was treated with a crude Fpg extract (5 µg/ml, prepared from a Fpg overproducing E.coli strain in BE1 buffer with 0.01% BSA). The Fpg/BE1 mix was first flushed through each filter (0.25 ml/min) for 7 min at 22°C followed by 30 min at 0.034 ml/min at 37°C. The Fpg-treated DNA was washed with 20 mM Na2EDTA, pH 9.6, for 37 min at 22°C (0.25 ml/min). Finally, DNA was eluted at pH 12.25 ± 0.05 and quantified as previously described (48). The damage levels were calculated from elution profiles and are expressed as ‘normalised area above curve’ (NAAC) (2). Calibration of the system and calculation of the number of Fpg-sensitive DNA lesions was carried out using X-rays at the relevant elution pH, and assuming an induction of 90 × 10–9/nt/Gy [∼1000 single strand breaks (SSB)/diploid cell/Gy].
Comet assay in combination with purified Nth or crude extract of Fpg. The Comet assay was performed as described with modifications (1). After lysing cells embedded in agarose, slides were immersed in buffer A (40 mM HEPES, pH 8.0, 0.1 M KCl, 0.5 mM EDTA) twice for 10 min. Subsequently, 50 ng of purified Nth (a gift from Dr Serge Boiteux) in buffer A with 0.2 mg/ml BSA in a total volume of 50 µl was added to each slide, and the slides were incubated at 37°C for 45 min. To analyse the number of Fpg-sensitive DNA lesions, slides were incubated in buffer A with 0.2 mg/ml BSA and 0.7 µg/ml crude Fpg extract at 37°C for 30 min. The subsequent steps were performed as described previously (1). The tail moment was used as a quantitative measure of the DNA damage. The Comet assay was routinely calibrated with cells exposed to X-rays, inducing DNA lesions (SSB and alkali-labile DNA lesions) in a strictly linear fashion up to a tail moment of 40 (5 Gy), followed by a lower slope up to a tail moment of 60 (10 Gy). A comet with a tail moment of 25 represents a moderately damaged cell (∼3 Gy) with ∼35% tail DNA. The total fluorescence (proportional to the DNA content of each comet) was recorded to analyse cells of different ploidy (1C, spermatids; 2C, Sertoli cells, Leydig cells, secondary spermatocytes and leukocytes; 4C, primary spermatocytes).
Statistical analyses
Differences in enzymatic activities in cellular extracts were tested using Student’s t-test. Correlation coefficients, significance levels and power were calculated by linear regression tests using SigmaStat for Windows version 2.03.
RESULTS
Excision of oxidative lesions by human and rat male germ cell extracts
In mammalian cells 8-oxoG is mainly repaired via BER initiated by OGG1 (14). Excision of 8-oxoG was analysed by measuring the abilities of cellular extracts to cleave a DNA substrate, i.e. a linear duplex of two complementary oligonucleotides, one containing an 8-oxoG residue. Excess AP-endonuclease (Nfo) was added to each reaction to process abasic sites.
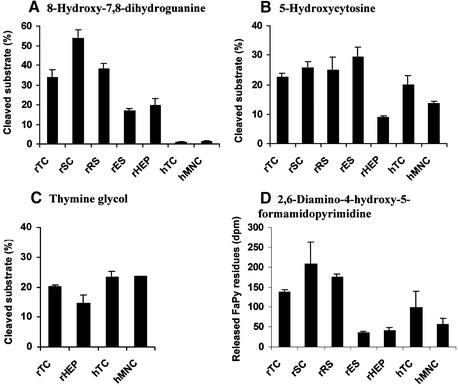
Human testicular extracts showed very limited excision of 8-oxoG when compared to rat testicular extracts, whereas similar low excision was observed in human mononuclear blood cell extracts (Fig. 1A). To establish optimal assay conditions a number of parameters were tested, such as different concentrations of extract, substrate, adding competitive DNA [poly(dA·dT)] or changing the total volume or the duration of the reaction (data not shown). In the rat there was a marked reduction in 8-oxoG removal along with the progression of spermatogenesis (Fig. 1A), with the early spermatocytes exhibiting the highest and the later elongating/elongated spermatids the lowest activity (3.7-fold lower than rSC; P < 0.01). Rat male germ cells removed 8-oxoG poorly when base paired with T, G or A, as opposed to efficient removal when base paired with C (data not shown). This is similar to the properties of OGG1 (18).
Figure 1.
Excision/incision of oxidative DNA substrates by human and rat cellular extracts. TC, testicular cells; SC, spermatocytes; RS, round spermatids; ES, elongating/elongated spermatids; HEP, primary hepatocytes; MNC, mononuclear blood cells. Rat or human samples are indicated with r or h, respectively. (A) Excision of 8-oxo-7,8-dihydroguanine (8-oxoG). Duplex oligonucleotides containing 8-oxoG:C (0.15 fmol), 10 µg extract and endonuclease IV (Nfo, 20 ng) were used in each reaction. The results are presented as mean cleaved substrate (%) ± SD of three to four independent extracts of each cell type (human testis extracts 8–10 and 13). (B) Excision of 5-hydroxycytosine (5-ohC). Duplex oligonucleotides containing 5-ohC:G (1 fmol) and 2 µg extracts were used. The results are presented as mean cleaved substrate (%) ± SD of three to four independent extracts of each cell type (human testis extracts 8–10 and 13). (C) Excision of thymine glycol (TG). Duplex oligonucleotides containing TG:A (10 fmol) and 5 µg extracts were used. The results are presented as mean cleaved substrate (%) ± SD of one to three independent extracts of each cell type (human testis extracts 1–3). (D) Release of 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FaPy) residues. The amount of [3H]FaPy substrate was 0.24 µg. Three independent extracts (25 µg) of each cell type (human testis extracts 8–10) were used and the results are expressed as mean released FaPy (fmol) ± SD.
Repair of the oxidised pyrimidines 5-ohC and TG in mammalian cells is initiated by enzymes such as NTH1 (26,29,30). The abilities of cellular extracts to remove these DNA lesions were measured as cleavage of linear duplex DNA containing one such damaged base per duplex. Different from the results with 8-oxoG, both human and rat male germ cells excise 5-ohC (Fig. 1B). Human male germ cells excised 5-ohC as efficiently as did rat male germ cells, and the various rat spermatogenic cell stages excised 5-ohC with similar efficiencies. Comparing somatic and male germ cells, human mononuclear blood cells showed 25% lower activity (P < 0.05) than human testicular cells, whereas rat hepatocytes exhibited 2.5- to 3-fold lower (P < 0.001) activity than rat testicular cells. TG was removed by both human and rat male germ cells (Fig. 1C), and the results were quantitatively similar to those for 5-ohC.
Several DNA glycosylases are able to remove FaPy residues, including hOGG1 and hNTH1, as well as the newly identified hFPG1 and hFPG2 (18,30,34–36). The removal of FaPy residues by cellular extracts was measured by quantifying the number of residues released from poly(dG·dC) DNA containing FaPy residues. Human testicular cell extracts did release FaPy residues (Fig. 1D), in contrast to their very low incision of 8-oxoG (Fig. 1A). Compared to rat male germ cells, the release of FaPy residues was lower (up to 5-fold) in human male germ cells, mononuclear blood cells and rat hepatocytes. Similar to the results with 8-oxoG (Fig. 1A), marked differences in the number of released FaPy residues were observed between rat male germ cells of different spermatogenic stages (Fig. 1D), with activity levels declining concurrently with the progression of spermatogenesis.
Excision of 8-oxoG and 5-oh-C by extracts from 13 different human testis biopsies
In order to identify possible differences in DNA glycosylase activities between individuals, several different human testicular extracts were tested for their abilities to excise 8-oxoG:C and 5-ohC:G (Fig. 2). Excess Apn1 was added to each reaction to ensure that all AP sites generated during the assay were cleaved. Thirteen different human testicular extracts showed low but significant excision of 8-oxoG (Fig. 2A), whereas 5-ohC was efficiently excised under similar conditions (Fig. 2B), and marked inter-individual variations were observed. The excision of 8-oxoG did not correlate with the excision of 5-ohC for any of the individuals.
Expression of OGG1
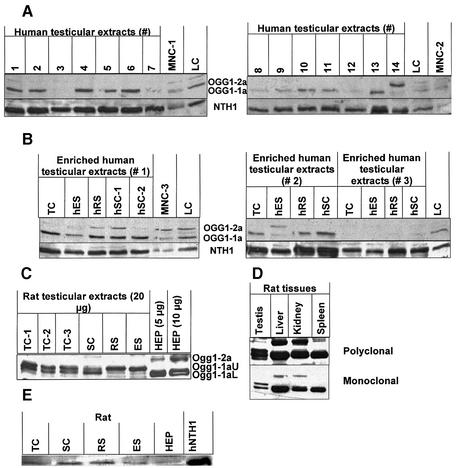
Two antibodies were used for the detection and analysis of OGG1. One monoclonal antibody identifies both the nuclear hOGG1-1a and the mitochondrial hOGG1-1b (50). The ability to visualise purified hOGG1-1a was confirmed (data not shown). A polyclonal antibody also recognised both hOGG1 species, and gave a much stronger staining signal than the monoclonal antibody. The polyclonal antibody was used subsequent to the identification of specific bands by means of the monoclonal antibody (Fig. 3A–D). As a gel-to-gel standard, equal amounts of a LC extract were applied in one lane of each gel (Fig. 3A and B). Both human and rat male germ cells expressed OGG1 (Fig. 3A, upper panel, and B–D). Human testicular cells from 13 different biopsies generally expressed higher levels of hOGG1 than mononuclear blood cells and lymphoblastoid cells (Fig. 3A, upper panel), but lower than primary human fibroblasts (data not shown). Furthermore the expression of hOGG1 in testicular extracts varied markedly between individuals. The predominant version of hOGG1 expressed by human testicular cells was the nuclear hOGG1-1a (∼36 kDa), similar to somatic cells. Somatic cells also expressed significant amounts of the mitochondrial hOGG1-2a (∼40 kDa). Interestingly, among the 13 human testis biopsies showing normal spermatogenesis, three exhibited almost non-detectable levels of hOGG1 (Fig. 3A, upper panel), indicating significant inter-individual differences in hOGG1 expression. The same three biopsies did however express normal levels of hNTH1, using the same western membrane (Fig. 3A, lower panel). Cell populations enriched in human testicular cells of specific stages expressed hOGG1 non-uniformly (Fig. 3B, upper panel), with spermatocytes expressing the highest and elongating/elongated spermatids the lowest amounts of the nuclear hOGG1-1a, respectively. One particular exception is enriched cell populations from testis biopsy 3 that showed very low expression of hOGG1. This extract was however able to incise 8-oxoG (Fig. 2A), indicating the presence of alternative DNA glycosylases for the excision of 8-oxoG. Interestingly, one testis biopsy (no. 14) from a donor with Sertoli cell-only syndrome (no male germ cells) did not express the nuclear hOGG1-1a but significant amounts of the mitochondrial hOGG1-2a (Fig. 3A, upper panel).
Figure 3.
Western analyses of 8-oxoguanine DNA glycosylase-1 (OGG1) and endonuclease III homologue-1 (NTH1) in human and rat extracts. (A) hOGG1 and hNTH1 in human testicular extracts. Testicular extracts (30 µg/well) were from 14 different testis biopsies, indicated with numbers. Somatic controls were three independent mononuclear blood cell extracts (MNC1-3) and one human lymphoblastoid cell (LC) extract. Upper panels show nuclear hOGG1-1a (lower band) and mitochondrial hOGG1-2a (upper band) using polyclonal anti-hOGG1; lower panels show hNTH1 using polyclonal anti-hNTH1. (B) hOGG1 and hNTH1 in human testicular extracts from enriched spermatogenic cell populations. Cells from three testicular biopsies (nos 1–3) were examined. The spermatocytes (SC) from biopsy 1 were in two different fractions after centrifugal elutriation (hSC-1 and hSC-2). Conditions as in (A). (C) Ogg1 in rat extracts. Extracts from crude and enriched populations of rat male germ cells (20 µg) and from rat primary hepatocytes (5 and 10 µg) were analysed using polyclonal anti-OGG1. Three species are recognised: upper band, Ogg1-2a; middle band, Ogg1-1aU; lower band, Ogg1-1aL. (D) Ogg1 in different rat tissues. Expression of rat Ogg1 in testis, liver, kidney and spleen extracts (20 µg each) analysed with both the polyclonal (upper panel) and the monoclonal (lower panel) OGG1 antibodies. (E) Nth1 in rat extracts. Expression of Nth1 in rat extracts (20 µg each) from different enriched male germ cell populations as well as primary hepatocytes. Abbreviations as in Figure 1.
The expression of Ogg1 in the rat was different from that in human male germ cells. Rat germ cells expressed two bands of ∼35–37 kDa (Fig. 3C), and Ogg1 was present in markedly smaller amounts in rat male germ cells compared to rat somatic tissues (Fig. 3D). Furthermore, the expression of the two smaller molecular weight Ogg1 species varied in the different spermatogenic cell types (Fig. 3C).
Expression of NTH1
Using the same western membranes as those for detecting OGG1, we found that human testicular cells consistently expressed substantially higher amounts of hNTH1 compared to mononuclear blood cells and lymphoblastoid cells (Fig. 3A, lower panel), and the expression varied between the biopsies. Furthermore, the expression of hNTH1 was higher in spermatocytes and round spermatids compared to elongating/elongated spermatids (Fig. 3B, lower panel). Similar to human extracts, rat testicular cells expressed higher amounts of Nth1 than somatic cells (hepatocytes), and the expression varied similarly between the male germ cell stages (Fig. 3E).
Cellular repair of Fpg-sensitive DNA lesions
Repair of Fpg-sensitive DNA lesions, i.e. predominantly 8-oxoG residues, was measured in human and rat male germ cells by the alkaline elution or Comet assay, both in combination with excess amounts of a crude Fpg extract. Oxidative DNA lesions were introduced by exposing cells to the photoactive substance Ro 12-9786 that mediates oxidation of bases when activated by visible light. Unlike most other oxidative agents, Ro 12-9786 induces very low numbers of SSB relative to the number of Fpg-sensitive base lesions, and this compound is hence very useful for repair studies of oxidative base damage. The level of Fpg-sensitive DNA lesions increased linearly with the concentration of Ro 12-9786; visible light or Ro 12-9786 alone induced very low levels of DNA lesions (data not shown).
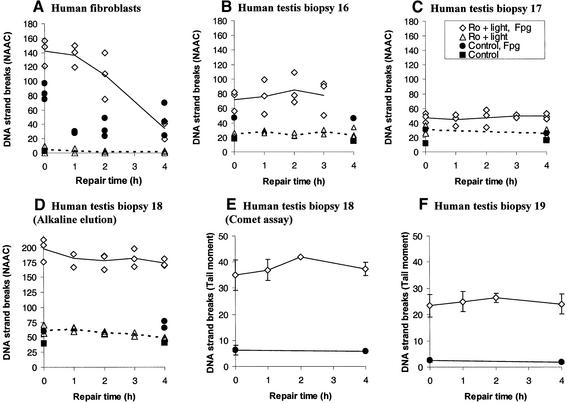
Testicular cells from four different humans showed very poor repair of Fpg-sensitive DNA lesions (Fig. 4B–F), as opposed to normal human fibroblasts exhibiting efficient repair with completion within 4 h (Fig. 4A). The two cellular repair assays were used to measure repair in populations of cells (alkaline elution) and in single cells (Comet assay). The results were consistent with the enzymatic activities for excision of 8-oxoG (Figs 1A and 2A). Limited repair efficiency by human testicular cells was observed following exposure to two different concentrations of Ro 12-9786 (Fig. 4B–F).
Figure 4.
Cellular repair of Fpg-sensitive DNA lesions by human cells. The number of Fpg-sensitive DNA lesions is measured as SSB in a modified alkaline elution assay (NAAC) or Comet assay (tail moment). In these assays genomic DNA is treated with an excess amount of Fpg crude extract to transform Fpg-sensitive DNA lesions into SSB. (A) Repair of Fpg-sensitive DNA lesions by normal primary human fibroblasts. Open diamonds and triangles are cell samples treated with Ro 12-9786 (6 µM) plus light (5 min) and with or without Fpg, respectively. Solid and broken lines show their mean values. Filled circles and squares represent control cell samples treated with or without Fpg, respectively. (B–D) Repair of Fpg-sensitive DNA lesions by human testicular cells from three individual testis biopsies (nos 16–18). Cells in (B) and (C) were exposed to 3 µM Ro 12-9786, whereas the cells in (D) were exposed to 6 µM Ro 12-9786 and analysed by alkaline elution. For symbols, see (A) and box in (C). (E–F) Repair of Fpg-sensitive DNA lesions by human testicular cells from two individual testis biopsies (nos 18 and 19). Cells were exposed to 3 µM Ro 12-9786 plus light (5 min) and analysed in the Comet assay. For symbols, see (A) and box in (C).
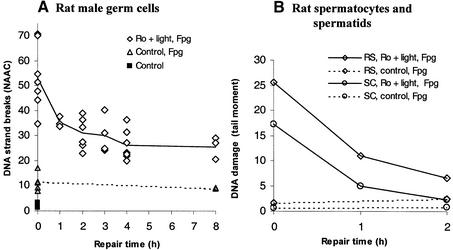
Unlike human male germ cells, rat male germ cells efficiently repaired the Fpg-sensitive DNA lesions (Fig. 5A). Using the alkaline elution technique, repair was found to be rapid during the first 2 h of incubation, when about 40% of the initial DNA lesions were eliminated, after which the number of Fpg-sensitive DNA lesions reached a steady state. To obtain information about different spermatogenic cell types, the Comet assay was used since it allows separate analyses of primary spermatocytes (4C) and round spermatids (1C) on the basis of their amount of DNA. The results clearly show that both primary spermatocytes and round spermatids repaired Fpg-sensitive DNA lesions efficiently, and to completion within 2 h (Fig. 5B). Elongating/elongated spermatids were not included in these analyses due to their distinctive morphology and tight packaging of DNA, which prevent tail formation under the conditions used.
Figure 5.
Cellular repair of Fpg-sensitive DNA lesions by rat cells. The experiments were conducted similarly to as described in Figure 4. (A) Repair of Fpg-sensitive DNA lesions in rat male germ cells. The cells were exposed to Ro 12-9786 (3 µM) plus visible light (5 min) and repair was analysed by alkaline elution. Open diamonds and triangles represent single cell samples with or without exposure to Ro 12-9786 plus light, respectively, and treatment with Fpg. Solid and broken lines show their mean values. Filled squares represent untreated cells. (B) Repair of Fpg-sensitive DNA lesions in rat germ cells at different stages of spermatogenesis. Spermatocytes (rSC; open circles) and round spermatids (rRS; open diamonds) were treated similarly as in (A) and analysed in the modified Comet assay. Solid and broken lines are results of cells treated with and without Ro 12-9786 plus light, respectively.
Cellular repair of Nth-sensitive DNA lesions
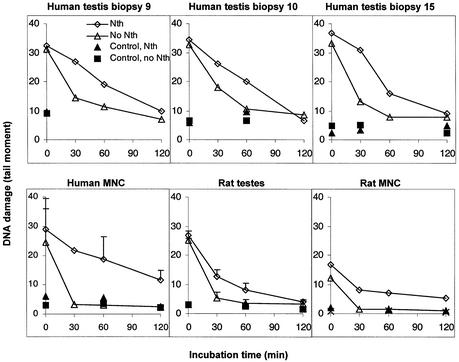
Cellular repair of oxidised pyrimidines was analysed following exposure of the cells to X-rays, allowing repair by incubation, and analysis of the number of Nth-sensitive DNA lesions (mainly hydrated pyrimidines) in each cell using the Comet assay in combination with purified Nth. Both human and rat male germ cells exhibited efficient and complete repair of the Nth-sensitive DNA lesions within 2 h (Fig. 6). Furthermore, both round spermatids and primary spermatocytes repaired Nth-sensitive DNA lesions at approximately similar rates (data not shown). Compared to male germ cells, mononuclear blood cells from both humans and rats removed Nth-sensitive DNA lesions slowly (Fig. 6). Furthermore, rat male germ cells exposed to 30 or 50 µM hydrogen peroxide for 5 min revealed similar efficient repair of the Nth-sensitive DNA lesions (data not shown).
Figure 6.
Cellular repair of Nth-sensitive DNA lesions by human and rat cells. Human and rat male germ cells and mononuclear blood cells exposed to X-rays (3 Gy) were allowed to repair for different time periods. Nth-sensitive DNA lesions were measured as SSB (tail moment) in the Comet assay after treating nuclear DNA with purified Nth to transform Nth-sensitive DNA lesions into SSB. Human testicular cells from three individual testis biopsies (nos 9, 10 and 15) are presented. The results from rat male germ cells and human mononuclear blood cells (mean ± SD) are from three independent experiments, whereas the results with rat mononuclear blood cells are from one representative experiment. Open diamonds and triangles represent exposed cells treated with or without Nth, respectively, whereas filled triangles and squares represent unexposed cells treated with or without Nth, respectively.
DISCUSSION
There are several lines of evidence indicating harmful effects of oxidative DNA damage in male germ cells. The presence of oxidative DNA damage in sperm is associated with male infertility, reduced sperm number and function (51,52), and the number of 8-oxoG residues in the DNA correlates with lifestyle factors such as smoking (53). Sperm exposed to even high doses of X-rays retain their ability to fertilise an oocyte (54), whereas extensive DNA damage affects embryonic development and leads to a high rate of early pregnancy loss (55). Mutations in DNA repair genes leading to meiotic defects have been proposed as one reason why mutations are common in infertile men with meiotic arrest (56).
The blood–testis barrier partly protects male germ cells from exogenous genotoxic agents. The requirement for removal of bulky lesions via NER may thus be limited, and indeed NER appears to be low or non-functional in rat male germ cells (2,3). However, spontaneously induced DNA lesions such as oxidative DNA damage do arise in all cells including male germ cells, and they are likely to be repaired. Accordingly, we have shown that methylated DNA is efficiently repaired by human and rat male germ cells (1). Intano et al. have shown high activity in vitro for BER of uracil in extracts of a mixture of mouse male germ cells (57) and in extracts of cells of different stages of spermatogenesis (58).
To our knowledge, this is the first study of repair of oxidative DNA base lesions by BER in male germ cells of humans and rats. The results show distinctive differences in repair capacity of various oxidative DNA lesions between human and rat male germ cells. Rat male germ cells efficiently repaired both Fpg- and Nth-sensitive DNA lesions, whereas human male germ cells efficiently repaired Nth-sensitive DNA lesions only. The presence and amounts of DNA glycosylases such as OGG1 and NTH1 in human and rat male germ cells, measured in extracts by western analyses and indirectly by enzymatic analyses, were largely consistent with the observed cellular repair.
OGG1 in human and rat male germ cells
In contrast to the rodent male germ cell results, human testicular extracts excised 8-oxoG poorly. A number of assay conditions were tested, since 8-oxoG excision by human extracts has been difficult to detect (59). Previous studies have shown that 8-oxoG is poorly repaired by normal human or rodent somatic extracts compared with other lesions such as uracil and abasic sites (60). Despite a low 8-oxoG DNA glycosylase activity in human testicular cells, western analyses show that the hOGG1 protein is present. The functionality of the detected protein species should therefore be questioned, or it could be that hOGG1 has a low turnover. The apparent lack of consistency between 8-oxoG excision and the amount of hOGG1 in protein extracts (Figs 2A and 3A) suggests the presence of back-up activities, and also that the alternative activities may be up-regulated when the amount of OGG1 is low. Statistical analyses indeed suggest a negative correlation between hOGG1 and 8-oxoG incision, although non-significant (r = –0.495, P = 0.085). One descriptive example is the extract from human testis biopsy 3 that showed very low expression of hOGG1 by western analyses, even in enriched cell fractions (Fig. 3), whereas the extract exhibited the highest amount of 8-oxoG excision activity among the 13 testicular extracts (Fig. 2A).
The efficient incision of 8-oxoG observed in rat male germ cell extracts is consistent with previous results from mice (14). In rodent male germ cells, the major 8-oxoG-excising activity measured in vitro most likely reflects that of Ogg1, since testicular extracts from Ogg1 null mice showed no residual incision of 8-oxoG using a similar DNA glycosylase assay (14). The declining amount of Ogg1 in rats with the progression of spermatogenesis is probably related to differences in transcriptional and translational activity accompanying the changes in chromatin structure. The high expression of OGG1 in spermatocytes could suggest a role for OGG1 in recombination. Heteroduplexes formed between DNA strands derived from non-identical homologous chromosomes are intermediates in meiotic crossing over, and they contain base mismatches that need to be corrected. Both the yeast Ogg1 and the E.coli Fpg have very high affinity for C:C mismatches, which are poorly repaired by the mismatch repair machinery (61), and OGG1 bound to such mismatches may thus act as a signal for initiating repair or preventing recombination.
The elevated rate of metabolism in the rat compared to the human most likely requires a higher capacity for the removal of oxidative DNA damage. Very low levels of Ogg1 mRNA were detected in many mouse tissues except the testis (22). The two bands of Ogg1 in the western analyses of rat testicular extracts (Fig. 3C and D) may represent alternatively spliced variants of Ogg1 or post-translationally modified versions of Ogg1. One of the bands may represent phosphorylated Ogg1, since hOGG1 associated with the nuclear matrix was recently shown to be phosphorylated (25).
Recent publications indicate that the most important role for Ogg1 in mice could be repair of mitochondrial rather than nuclear 8-oxoG (62). A somatic mutation leading to impaired mitochondrial targeting in hOGG1 found in kidney cancer was recently identified (63). In our analyses, the expression of mitochondrial OGG1 in humans and rat male germ cells is limited, and the mitochondrial repair capacity of oxidative DNA lesions in male germ cells remains to be investigated.
Repair of Fpg-sensitive DNA lesions
Treatment of cells with Ro 12-9786 plus visible light induced high numbers of Fpg-sensitive DNA lesions relative to the number of SSB. Similar methods have proven useful as tools for the study of the repair of oxidative DNA lesions in somatic cells (14,49). In these studies the induced DNA lesions were identified as almost exclusively 8-oxoG (49,64). In the present study, the spontaneous number of Fpg-sensitive DNA lesions is estimated to be 0.3–0.4 per 106 bp in human testicular cells compared to 0.14 per 106 bp in rat testicular cells. Using a similar technique Pflaum et al. (49) reported 0.07–0.3 Fpg-sensitive DNA lesions per 106 bp in various human cell types. The higher number of spontaneous Fpg-sensitive DNA lesions in human testicular cells corresponds with the low rate of repair of such lesions. The amount of 8-oxoG in nuclear liver DNA of Ogg1 null mice is approximately 2-fold higher than in the wild type (14).
The poor repair of Fpg-sensitive DNA lesions in human testicular cells is in contrast to the efficient repair in rat testicular cells (Fig. 5) and in human primary fibroblasts (Fig. 4). Furthermore, in correspondence with the results with hMNC extracts, others have previously shown that human lymphocytes poorly repair both Fpg- and Nth-sensitive DNA lesions (65,66). The repair of 8-oxoG has previously been reported to be approximately similar in rodent and human somatic cells (67). The low repair of this specific type of oxidative DNA lesion in human male germ cells is contrasted by efficient BER of other types of lesions in human testicular cells prepared similarly, such as Nth-sensitive sites (Fig. 6) and methylated bases (1), indicating that the primary cells are proficient in repairing at least some types of lesions via BER. Four consecutive human biopsies consistently showed limited cellular removal of Fpg-sensitive DNA lesions, and the probability that these represent individuals with no expression of DNA glycosylases removing 8-oxoG is very low. The hOGG1 levels were not recorded in the four individuals; however, in the extended set of 13 donor biopsies the expression of hOGG1 varied markedly and three individuals; showed non-detectable levels of hOGG1 (Fig. 3). Also, the activities for excision of 8-oxoG varied markedly between individuals (Fig. 2A) and the amount of hOGG1 was negatively, although non-significantly, correlated with the 8-oxoG-excising activity.
The human testicular cell populations studied here contain a high proportion of cells at late stages of spermatogenesis (spermatids), and we find it plausible that earlier stages may retain the ability to remove these biologically important DNA lesions, as was observed in the rat. On the other hand, male germ cells may not accumulate very high numbers of oxidative DNA lesions, since the testicle exhibits low blood flow and hence low partial O2 pressure, and some of the cells use lactate as their energy source. However, this does not explain the difference between humans and rats. Consistent with our results, a large study concluded that spermatogenesis in man is 3.1 times more sensitive to ionising radiation than in the mouse (68). In general, an alternative to repairing Fpg-sensitive DNA lesions is to eliminate the cells that contain such DNA lesions via apoptosis, which is dependent on the level of DNA damage (69).
The proficient repair of Fpg-sensitive DNA lesions in rat testicular cells is consistent with induced unscheduled DNA synthesis in rodent male germ cells following exposure to oxidising agents (70). While the repair of Fpg-sensitive DNA lesions in rat male germ cells using alkaline elution was initially rapid, a substantial level of non-repaired lesions remained even at extended (8 h) repair periods (Fig. 5A). This apparent bimodal repair of Fpg-sensitive DNA lesions could reflect heterogeneous repair within the cell or among male germ cells at different spermatogenic stages, since mixed cell samples were analysed (1). The non-repaired fraction of DNA observed with alkaline elution, in which repair in the whole population of cells is measured, compared with the complete repair observed in both primary spermatocytes and round spermatids in the Comet assay, suggest that elongating/elongated spermatids may exhibit limited or no repair of Fpg-sensitive DNA lesions. The higher excision of 8-oxoG by extracts from rat spermatocytes compared to round spermatids (Fig. 1A) could have indicated more efficient cellular repair in the former cell type, and the data in Figure 5B do not exclude this possibility. Recently it was shown that overexpression of hOGG1 conferred increased repair rates of Fpg-sensitive DNA lesions in Chinese hamster ovary cells (71). On the other hand, Cappelli et al. (72) showed that the repair rate in vitro is not solely dependent on the amount of DNA glycosylase.
NTH1 and the repair of Nth-sensitive DNA lesions
Unlike the repair of Fpg-sensitive DNA lesions, Nth-sensitive DNA lesions were efficiently repaired in both human and rat testicular cells. This corresponds with the presence of NTH1 protein in human and rat male germ cells (Fig. 3) and with the activity levels against 5-ohC:G (Fig. 1B). The contribution of the recently characterised DNA glycosylases of the Nei/Fpg family may be significant for the efficient repair observed, especially since one of the genes was exclusively expressed in the testis and thymus (36). Furthermore, humans showed marked inter-individual differences in their abilities to incise 5-ohC as well as 8-oxoG, and the abilities to excise 5-ohC and 8-oxoG were not correlated in each human testis biopsy.
The cellular repair of Nth-sensitive DNA lesions in the various spermatogenic cell types was similar and correlated well with the corresponding incision activities of the extracts (Fig. 1B) and the amount of NTH1 (Fig. 3B and E). Similar cell type-specific correlations were recently observed with other BER repair functions (Mpg, Ung and Apex) in rat male germ cells (1). Consistent with our findings, a transcript of hNTH1 was expressed in the human testis at amounts that were not different from those of many other tissues (26,73).
The apparently low initial numbers of Nth-sensitive DNA lesions in the Comet assay have also been observed by others in somatic cells (65,74). It is not due to saturation of the Comet assay and may reflect an initial binding of purified bifunctional Nth to high numbers of SSB or AP sites induced by X-rays, in addition to oxidised pyrimidines. This is reflected in the lack of initial Fpg-sensitive DNA lesions measured in the Comet assay in cells exposed to X-rays compared to cells exposed to Ro 12-9786 plus light (unpublished results). The latter treatment induces mostly base damage, giving rise to high initial levels of Fpg-sensitive DNA lesions. Supporting our observations of cells exposed to X-rays, the clustered DNA damage generated by X-rays has been shown to have an inhibitory effect on the formation of SSB at sites of base damage (75).
Since FaPy residues are excised by both OGG1 and NTH1, as well as by one of the newly characterised Nei/Fpg homologues (36), their release may represent the activity of several enzymes (18,30). The relative excision of FaPy residues in the various cell types (Fig. 1D) correlates with the removal of 5-ohC:G in humans (Fig. 1B) and with the incision of 8-oxoG:C in rats (Fig. 1A).
CONCLUSIONS
Human and rat testicular cells differ in their ability to repair certain oxidative DNA lesions. Cells from both species repair Nth-sensitive DNA lesions efficiently. Human testicular cells, at least in the late stages of spermatogenesis (spermatids), poorly repair Fpg-sensitive DNA lesions such as 8-oxoG, whereas rat testicular cells repair these lesions efficiently. Human male germ cells may thus be particularly sensitive to some environmental agents that induce oxidative DNA lesions, with negative effects on male reproduction and de novo germline mutations as likely consequences.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the assistance of the transplantation team at the National Hospital, Oslo, Norway, for providing the human testicular biopsies. Furthermore, thanks to Dr Philip Bolton (Department of Chemistry, Wesleyan University, Middletown, CO) for the oligonucleotide containing TG, Dr Serge Boiteux (CEA, Département de Radiobiologie et Radiopathologie, Fontenay aux Roses, France) for purified endonuclease III, Dr Sancar Mitra (Sealy Center for Molecular Science and Department of Human Biological Chemistry and Genetics, University of Texas, TX) for the hOGG1 and hNTH1 sera, Dr Peter de Boer (Department of Genetics, Wageningen University, The Netherlands) for the anti-mouse SCP3 antibody and Dr Leon Mullenders (Department of Radiation Genetics and Chemical Carcinogenesis, University of Leiden, The Netherlands) for the primary normal human fibroblasts. Thanks to Dr Elmar Gocke (F. Hoffmann-La Roche Ltd, Basel, Switzerland) for the kind gift of Ro 12-9786. E.C.S. and M.B. acknowledge support from the Norwegian Cancer Society and Anders Jahres Medical Foundation. The study was supported by the EU-Environment and Climate program (contract no. ENV4-CT95-0204) and the Norwegian Research Council (grant no. 129614/310).
REFERENCES
- 1.Olsen A.K., Bjortuft,H., Wiger,R., Holme,J., Seeberg,E., Bjoras,M. and Brunborg,G. (2001) Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res., 29, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunborg G., Holme,J.A. and Hongslo,J.K. (1995) Inhibitory effects of paracetamol on DNA repair in mammalian cells. Mutat. Res., 342, 157–170. [DOI] [PubMed] [Google Scholar]
- 3.Jansen J., Olsen,A.K., Wiger,R., Naegeli,H., de Boer,P., van Der,H.F., Holme,J.A., Brunborg,G. and Mullenders,L. (2001) Nucleotide excision repair in rat male germ cells: low level of repair in intact cells contrasts with high dual incision activity in vitro. Nucleic Acids Res., 29, 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 5.Le Page F., Kwoh,E.E., Avrutskaya,A., Gentil,A., Leadon,S.A., Sarasin,A. and Cooper,P.K. (2000) Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH and CSB and implications for Cockayne syndrome. Cell, 101, 159–171. [DOI] [PubMed] [Google Scholar]
- 6.Evans J., Maccabee,M., Hatahet,Z., Courcelle,J., Bockrath,R., Ide,H. and Wallace,S. (1993) Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat. Res., 299, 147–156. [DOI] [PubMed] [Google Scholar]
- 7.Hatahet Z., Purmal,A.A. and Wallace,S.S. (1994) Oxidative DNA lesions as blocks to in vitro transcription by phage T7 RNA polymerase. Ann. N. Y. Acad. Sci., 726, 346–348. [DOI] [PubMed] [Google Scholar]
- 8.Loft S., Deng,X.S., Tuo,J., Wellejus,A., Sorensen,M. and Poulsen,H.E. (1998) Experimental study of oxidative DNA damage. Free Radic. Res., 29, 525–539. [DOI] [PubMed] [Google Scholar]
- 9.Wellejus A., Poulsen,H.E. and Loft,S. (2000) Iron-induced oxidative DNA damage in rat sperm cells in vivo and in vitro. Free Radic. Res., 32, 75–83. [DOI] [PubMed] [Google Scholar]
- 10.Shen H. and Ong,C. (2000) Detection of oxidative DNA damage in human sperm and its association with sperm function and male infertility. Free Radic. Biol. Med., 28, 529–536. [DOI] [PubMed] [Google Scholar]
- 11.Sakkas D., Mariethoz,E., Manicardi,G., Bizzaro,D., Bianchi,P.G. and Bianchi,U. (1999) Origin of DNA damage in ejaculated human spermatozoa. Rev. Reprod., 4, 31–37. [DOI] [PubMed] [Google Scholar]
- 12.Bialkowski K., Bialkowska,A. and Kasprzak,K.S. (1999) Cadmium(II), unlike nickel(II), inhibits 8-oxo-dGTPase activity and increases 8-oxo-dG level in DNA of the rat testis, a target organ for cadmium(II) carcinogenesis. Carcinogenesis, 20, 1621–1624. [DOI] [PubMed] [Google Scholar]
- 13.Wilson D.M. III and Thompson,L.H. (1997) Life without DNA repair. Proc. Natl Acad. Sci. USA, 94, 12754–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klungland A., Rosewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindahl,T. and Barnes,D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsen H., Rosewell,I., Robins,P., Skjelbred,C.F., Andersen,S., Slupphaug,G., Daly,G., Krokan,H.E., Lindahl,T. and Barnes,D.E. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 16.Aburatani H., Hippo,Y., Ishida,T., Takashima,R., Matsuba,C., Kodama,T., Takao,M., Yasui,A., Yamamoto,K. and Asano,M. (1997) Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res., 57, 2151–2156. [PubMed] [Google Scholar]
- 17.Arai K., Morishita,K., Shinmura,K., Kohno,T., Kim,S.R., Nohmi,T., Taniwaki,M., Ohwada,S. and Yokota,J. (1997) Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene, 14, 2857–2861. [DOI] [PubMed] [Google Scholar]
- 18.Bjoras M., Luna,L., Johnsen,B., Hoff,E., Haug,T., Rognes,T. and Seeberg,E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J., 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radicella J.P., Dherin,C., Desmaze,C., Fox,M.S. and Boiteux,S. (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roldan-Arjona T., Wei,Y.F., Carter,K.C., Klungland,A., Anselmino,C., Wang,R.P., Augustus,M. and Lindahl,T. (1997) Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 8016–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu R., Nash,H.M. and Verdine,G.L. (1997) A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol., 7, 397–407. [DOI] [PubMed] [Google Scholar]
- 22.Rosenquist T.A., Zharkov,D.O. and Grollman,A.P. (1997) Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto Alamo M.J., Jurado,J., Francastel,E. and Laval,F. (1998) Rat 7,8-dihydro-8-oxoguanine DNA glycosylase: substrate specificity, kinetics and cleavage mechanism at an apurinic site. Nucleic Acids Res., 26, 5199–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Page F., Klungland,A., Barnes,D.E., Sarasin,A. and Boiteux,S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl Acad. Sci. USA, 97, 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dantzer F., Luna,L., Bjoras,M. and Seeberg,E. (2002) Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res., 30, 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aspinwall R., Rothwell,D.G., Roldan-Arjona,T., Anselmino,C., Ward,C.J., Cheadle,J.P., Sampson,J.R., Lindahl,T., Harris,P.C. and Hickson,I.D. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilbert T.P., Chaung,W., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (1997) Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J. Biol. Chem., 272, 6733–6740. [DOI] [PubMed] [Google Scholar]
- 28.Sarker A.H., Ikeda,S., Nakano,H., Terato,H., Ide,H., Imai,K., Akiyama,K., Tsutsui,K., Bo,Z., Kubo,K. et al. (1998) Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol., 282, 761–774. [DOI] [PubMed] [Google Scholar]
- 29.Dizdaroglu M., Karahalil,B., Senturker,S., Buckley,T.J. and Roldan-Arjona,T. (1999) Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry, 38, 243–246. [DOI] [PubMed] [Google Scholar]
- 30.Luna L., Bjoras,M., Hoff,E., Rognes,T. and Seeberg,E. (2000) Cell-cycle regulation, intracellular sorting and induced overexpression of the human NTH1 DNA glycosylase involved in removal of formamidopyrimidine residues from DNA. Mutat. Res., 460, 95–104. [DOI] [PubMed] [Google Scholar]
- 31.Eide L., Luna,L., Gustad,E.C., Henderson,P.T., Essigmann,J.M., Demple,B. and Seeberg,E. (2001) Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry, 40, 6653–6659. [DOI] [PubMed] [Google Scholar]
- 32.Cooper P.K., Nouspikel,T., Clarkson,S.G. and Leadon,S.A. (1997) Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science, 275, 990–993. [DOI] [PubMed] [Google Scholar]
- 33.Bessho T. (1999) Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res., 27, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazra T.K., Izumi,T., Boldogh,I., Imhoff,B., Kow,Y.W., Jaruga,P., Dizdaroglu,M. and Mitra,S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA, 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandaru V., Sunkara,S., Wallace,S.S. and Bond,J.P. (2002) A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair, 1, 517–529. [DOI] [PubMed] [Google Scholar]
- 36.Morland I., Rolseth,V., Luna,L., Rognes,T., Bjoras,M. and Seeberg,E. (2002) Human DNA glycosylases of the bacterial Fpg/MutM superfamily: as alternaitve pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res., 30, 4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman D.G. and Nagler,H.M. (1987) Aspiration flow cytometry of the testes in the evaluation of spermatogenesis in the infertile male. Fertil. Steril., 48, 287–291. [DOI] [PubMed] [Google Scholar]
- 38.Bjorge C., Wiger,R., Holme,J.A., Brunborg,G., Scholz,T., Dybing,E. and Soderlund,E.J. (1996) DNA strand breaks in testicular cells from humans and rats following in vitro exposure to 1,2-dibromo-3-chloropropane (DBCP). Reprod. Toxicol., 10, 51–59. [DOI] [PubMed] [Google Scholar]
- 39.Blanchard Y., Lavault,M.T., Quernee,D., Le Lannou,D., Lobel,B. and Lescoat,D. (1991) Preparation of spermatogenic cell populations at specific stages of differentiation in the human. Mol. Reprod. Dev., 30, 275–282. [DOI] [PubMed] [Google Scholar]
- 40.Meistrich M.L., Longtin,J., Brock,W.A., Grimes,S.R.,Jr and Mace,M.L. (1981) Purification of rat spermatogenic cells and preliminary biochemical analysis of these cells. Biol. Reprod., 25, 1065–1077. [DOI] [PubMed] [Google Scholar]
- 41.Bjorge C., Wiger,R., Holme,J.A., Brunborg,G., Andersen,R., Dybing,E. and Soderlund,E.J. (1995) In vitro toxicity of 1,2-dibromo-3-chloropropane (DBCP) in different testicular cell types from rats. Reprod. Toxicol., 9, 461–473. [DOI] [PubMed] [Google Scholar]
- 42.Suter L., Koch,E., Bechter,R. and Bobadilla,M. (1997) Three-parameter flow cytometric analysis of rat spermatogenesis. Cytometry, 27, 161–168. [DOI] [PubMed] [Google Scholar]
- 43.Eide L., Bjoras,M., Pirovano,M., Alseth,I., Berdal,K.G. and Seeberg,E. (1996) Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klungland A., Hoss,M., Gunz,D., Constantinou,A., Clarkson,S.G., Doetsch,P.W., Bolton,P.H., Wood,R.D. and Lindahl,T. (1999) Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell, 3, 33–42. [DOI] [PubMed] [Google Scholar]
- 45.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riazuddin S. and Lindahl,T. (1978) Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry, 17, 2110–2118. [DOI] [PubMed] [Google Scholar]
- 47.Boiteux S., Gajewski,E., Laval,J. and Dizdaroglu,M. (1992) Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry, 31, 106–110. [DOI] [PubMed] [Google Scholar]
- 48.Brunborg G., Holme,J.A., Soderlund,E.J., Omichinski,J.G. and Dybing,E. (1988) An automated alkaline elution system: DNA damage induced by 1,2-dibromo-3-chloropropane in vivo and in vitro. Anal. Biochem., 174, 522–536. [DOI] [PubMed] [Google Scholar]
- 49.Pflaum M., Will,O. and Epe,B. (1997) Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis, 18, 2225–2231. [DOI] [PubMed] [Google Scholar]
- 50.Shinmura K., Kohno,T., Takeuchi-Sasaki,M., Maeda,M., Segawa,T., Kamo,T., Sugimura,H. and Yokota,J. (2000) Expression of the OGG1-type 1a (nuclear form) protein in cancerous and non-cancerous human cells. Int. J. Oncol., 16, 701–707. [DOI] [PubMed] [Google Scholar]
- 51.Ni Z.Y., Liu,Y.Q., Shen,H.M., Chia,S.E. and Ong,C.N. (1997) Does the increase of 8-hydroxydeoxyguanosine lead to poor sperm quality? Mutat. Res., 381, 77–82. [DOI] [PubMed] [Google Scholar]
- 52.Shen H.M., Chia,S.E. and Ong,C.N. (1999) Evaluation of oxidative DNA damage in human sperm and its association with male infertility. J. Androl., 20, 718–723. [PubMed] [Google Scholar]
- 53.Shen H.M., Chia,S.E., Ni,Z.Y., New,A.L., Lee,B.L. and Ong,C.N. (1997) Detection of oxidative DNA damage in human sperm and the association with cigarette smoking. Reprod. Toxicol., 11, 675–680. [DOI] [PubMed] [Google Scholar]
- 54.Twigg J.P., Irvine,D.S. and Aitken,R.J. (1998) Oxidative damage to DNA in human spermatozoa does not preclude pronucleus formation at intracytoplasmic sperm injection. Hum. Reprod., 13, 1864–1871. [DOI] [PubMed] [Google Scholar]
- 55.Ahmadi A. and Ng,S.C. (1999) Fertilizing ability of DNA-damaged spermatozoa. J. Exp. Zool., 284, 696–704. [DOI] [PubMed] [Google Scholar]
- 56.Nudell D., Castillo,M., Turek,P.J. and Pera,R.R. (2000) Increased frequency of mutations in DNA from infertile men with meiotic arrest. Hum. Reprod., 15, 1289–1294. [DOI] [PubMed] [Google Scholar]
- 57.Intano G.W., McMahan,C.A., Walter,R.B., McCarrey,J.R. and Walter,C.A. (2001) Mixed spermatogenic germ cell nuclear extracts exhibit high base excision repair activity. Nucleic Acids Res., 29, 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Intano G.W., McMahan,C.A., McCarrey,J.R., Walter,R.B., McKenna,A.E., Matsumoto,Y., MacInnes,M.A., Chen,D.J. and Walter,C.A. (2002) Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol. Cell. Biol., 22, 2410–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazra T.K., Izumi,T., Maidt,L., Floyd,R.A. and Mitra,S. (1998) The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res., 26, 5116–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cappelli E., Degan,P. and Frosina,G. (2000) Comparative repair of the endogenous lesions 8-oxo-7,8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis, 21, 1135–1141. [PubMed] [Google Scholar]
- 61.Nakahara T., Zhang,Q.M., Hashiguchi,K. and Yonei,S. (2000) Identification of proteins of Escherichia coli and Saccharomyces cerevisiae that specifically bind to C/C mismatches in DNA. Nucleic Acids Res., 28, 2551–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Souza-Pinto N.C., Eide,L., Hogue,B.A., Thybo,T., Stevnsner,T., Seeberg,E., Klungland,A. and Bohr,V.A. (2001) Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res., 61, 5378–5381. [PubMed] [Google Scholar]
- 63.Audebert M., Chevillard,S., Levalois,C., Gyapay,G., Vieillefond,A., Klijanienko,J., Vielh,P., El Naggar,A.K., Oudard,S., Boiteux,S. et al. (2000) Alterations of the DNA repair gene OGG1 in human clear cell carcinomas of the kidney. Cancer Res., 60, 4740–4744. [PubMed] [Google Scholar]
- 64.Will O., Gocke,E., Eckert,I., Schulz,I., Pflaum,M., Mahler,H.C. and Epe,B. (1999) Oxidative DNA damage and mutations induced by a polar photosensitizer, Ro19-8022. Mutat. Res., 435, 89–101. [DOI] [PubMed] [Google Scholar]
- 65.Collins A.R., Ma,A.G. and Duthie,S.J. (1995) The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat. Res., 336, 69–77. [DOI] [PubMed] [Google Scholar]
- 66.Collins A.R., Dobson,V.L., Dusinska,M., Kennedy,G. and Stetina,R. (1997) The comet assay: what can it really tell us? Mutat. Res., 375, 183–193. [DOI] [PubMed] [Google Scholar]
- 67.Cappelli E., Degan,P., Thompson,L.H. and Frosina,G. (2000) Efficient repair of 8-oxo-7,8-dihydrodeoxyguanosine in human and hamster xeroderma pigmentosum D cells. Biochemistry, 39, 10408–10412. [DOI] [PubMed] [Google Scholar]
- 68.Clifton D.K. and Bremner,W.J. (1983) The effect of testicular X-irradiation on spermatogenesis in man. A comparison with the mouse. J. Androl., 4, 387–392. [DOI] [PubMed] [Google Scholar]
- 69.Oldereid N.B., Angelis,P.D., Wiger,R. and Clausen,O.P. (2001) Expression of Bcl-2 family proteins and spontaneous apoptosis in normal human testis. Mol. Hum. Reprod., 7, 403–408. [DOI] [PubMed] [Google Scholar]
- 70.Sotomayor R.E. and Sega,G.A. (2000) Unscheduled DNA synthesis assay in mammalian spermatogenic cells: an update. Environ. Mol. Mutagen., 36, 255–265. [PubMed] [Google Scholar]
- 71.Hollenbach S., Dhenaut,A., Eckert,I., Radicella,J.P. and Epe,B. (1999) Overexpression of Ogg1 in mammalian cells: effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis, 20, 1863–1868. [DOI] [PubMed] [Google Scholar]
- 72.Cappelli E., Hazra,T., Hill,J.W., Slupphaug,G., Bogliolo,M. and Frosina,G. (2001) Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis, 22, 387–393. [DOI] [PubMed] [Google Scholar]
- 73.Imai K., Sarker,A.H., Akiyama,K., Ikeda,S., Yao,M., Tsutsui,K., Shohmori,T. and Seki,S. (1998) Genomic structure and sequence of a human homologue (NTHL1/NTH1) of Escherichia coli endonuclease III with those of the adjacent parts of TSC2 and SLC9A3R2 genes. Gene, 222, 287–295. [DOI] [PubMed] [Google Scholar]
- 74.Banath J.P., Wallace,S.S., Thompson,J. and Olive,P.L. (1999) Radiation-induced DNA base damage detected in individual aerobic and hypoxic cells with endonuclease III and formamidopyrimidine-glycosylase. Radiat. Res., 151, 550–558. [PubMed] [Google Scholar]
- 75.David-Cordonnier M.H., Laval,J. and O’Neill,P. (2001) Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry, 40, 5738–5746. [DOI] [PubMed] [Google Scholar]