Abstract
Ack/Ack1 is a nonreceptor protein tyrosine kinase that comprises a tyrosine kinase core, an SH3 domain, a Cdc42-binding region, a Ralt homology region, and a proline-rich region. Here we describe a detailed characterization of the Ack protein as well as the chromosomal localization of human Ack (chromosome 3q29) and the primary structure of murine Ack. We demonstrate that Ack is ubiquitously expressed, with highest expression seen in thymus, spleen, and brain. Activation of integrins by cell adhesion on fibronectin leads to strong tyrosine phosphorylation and activation of Ack. Upon cell stimulation with EGF or PDGF, Ack is tyrosine-phosphorylated and recruited to activated EGF or PDGF receptors, respectively. A pool of endogenous Ack molecules is constitutively tyrosine-phosphorylated, even in starved cells. Moreover, tyrosine-phosphorylated Ack forms a stable complex with the adapter protein Nck via its SH2 domain. Finally, we have characterized a membrane-targeting sterile α motif-like domain in the amino terminus of Ack. Using several Ack mutants, we show that the amino-terminal and CRIB domains are necessary for Ack autophosphorylation, whereas the SH3 domain appears to have an autoinhibitory role. These experiments suggest a functional role for Ack as an early transducer of multiple extracellular stimuli.
Keywords: cell signaling, tyrosine phosphorylation, surface receptors, growth factors, cell adhesion
A variety of extracellular signals are relayed intracellularly through activation of protein tyrosine kinases (PTKs). Unlike receptor tyrosine kinases, which are endowed with intrinsic PTK activity (1–3), lymphokine or antigen receptors transmit their signals into the cell by coupling to nonreceptor PTK such as Janus kinase, ZAP70 kinase, or Src kinase. Many nonreceptor tyrosine kinases contain protein modules, such as SH2 and SH3 domains, that are responsible for regulation of their enzymatic activity by an intramolecular mechanism. These modules are also responsible for mediating interactions with other proteins that regulate the kinase activity as well as the cellular localization of the PTK.
The nonreceptor PTK Ack was cloned by virtue of its binding to the GTP-bound form of the small G protein Cdc42 (4). In addition to its PTK domain, Ack contains an SH3 domain, a Cdc42 binding (CRIB) domain, and a proline-rich region. Earlier work reported that Ack1 specifically binds to activated GTP-bound Cdc42 but not to Rac or Rho via its CRIB domain (4). In vitro assays demonstrated that complex formation between human Ack and Cdc42 leads to inhibition of Cdc42 GTPase activity. However, these experiments were carried out with a mutant Ack protein (4).
Ack also contains a Ralt homology region, a region thought to mediate direct interactions with receptor tyrosine kinases (5–7). Because Ack has been shown to interact with clathrin and to localize in AP2-containing vesicles, it was proposed that Ack plays a role in receptor endocytosis (8). More recently, it was shown that Ack interacts with Nck, SNX9, and the lipid phosphatase synaptojanin, suggesting that Ack may indeed play a role in some aspects of control of vesicle dynamics (9–11). In addition, Ack was shown to be activated by agonists of the M3 muscarinic receptors (12) in response to EGF or bradykinin stimulation and after integrin-mediated cell adhesion (13–15). Ack1 and Cdc42 were also shown to be recruited by the melanoma chondroitin sulfate proteoglycan (16). Although Ack1 was shown to be activated by a variety of extracellular cues, the mechanism underlying Ack activation as well as the identity of its cellular substrates have not yet been revealed.
We initiated the work reported here by cloning the proline-rich region of murine Ack in an expression library screening using Grb2 SH3 domain as a probe for cloning of new targets of Grb2 (17). In this report we present a detailed biochemical characterization of endogenous murine Ack, including its stimulation by growth factors and cell adhesion, its interaction with the adaptor protein Nck, and the function of its individual domains. After the reported cloning of Ack2, recent nomenclature, including that adopted by the International Human Genome Sequencing Consortium (18), has referred to Ack as Ack1. However, we argue below that Ack2 is not coded by a separate gene. Therefore, in this article we refer to Ack/Ack1 as Ack, its original name, and we suggest the adoption of this nomenclature by the scientific community.
Results and Discussion
The adapter protein Grb2 was used as a probe for cloning target proteins from a mouse expression library. One positive clone coded for the carboxyl terminus of a protein rich in prolines that was named Pyk1 (for proline-rich tyrosine kinase 1). We have shown that Pyk1 is the murine homolog of Ack and demonstrated that a single gene codes for Ack1 and Ack2, and therefore we suggest that Ack1/Ack2 be referred to as Ack (4, 14) (see Supporting Text, which is published as supporting information on the PNAS web site). Human Ack gene was mapped to chromosome 3q29 (see Supporting Text), a region implicated in adenocortical carcinoma (19) and hematologic neoplasia (20) that is syntenic to mouse chromosome 16.
Ack Is Expressed in Multiple Tissues and Cell Lines.
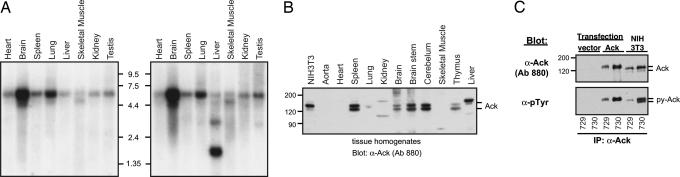
Northern blot analysis with two different probes demonstrated that Ack mRNA is ubiquitously expressed. A major ≈6-kb transcript is present in all tissues tested. Both a 5′ Ack probe (Fig. 1A Left) and a 3′ Ack probe (Fig. 1A Right) detected the ≈6-kb transcript in addition to a ≈5-kb transcript seen in skeletal muscle and brain. The 3′ probe additionally detected a 3.2-kb transcript and an abundant 1.8-kb transcript in the liver. The nature of the 1.8-kb transcript detected in liver tissue is unknown. We were not able to detect an Ack protein fragment in Ack immunoblots of liver lysates using antibodies that recognize this particular region (data not shown).
Fig. 1.
Expression of Ack mRNA and protein. (A) Tissue distribution of Ack mRNA expression. A CLONTECH blot containing 2 μg of polyA+ RNA per lane was probed with a 5′ (Left) or a 3′ (Right) probe derived from the mouse Ack cDNA. In both cases, blots were exposed to film for 14 h at −80°C. (B) Characterization of Ack protein with site-specific anti-Ack antibodies. Mouse tissue homogenates were obtained from a 3-month-old male, and ≈60 μg of tissue lysates together with ≈20 μg of lysates of NIH 3T3 cells were analyzed with anti-Ack antibodies (Ab 880; see Fig. 6). (C) Constitutive tyrosine phosphorylation of Ack. 293 cells were transfected with either vector alone or vector containing full-length Ack in pRK5, and the recombinant protein was immunoprecipitated with antibodies against the amino terminus (Ab 729) or carboxyl terminus (Ab 730) of Ack and visualized with anti-Ack antibodies (Ab 880). Endogenous Ack from NIH 3T3 lysates was also analyzed in the same way by immunoprecipitation followed by immunoblotting. The blot was stripped and probed with anti-pTyr antibodies.
To study the expression pattern of Ack protein we generated a panel of specific antibodies against Ack (Fig. 6C, which is published as supporting information on the PNAS web site). When whole-cell lysates from several mouse tissues were analyzed with anti-Ack antibodies, a protein doublet was identified (Fig. 1B): a faster migrating band of ≈120 kDa (predicted mass of unmodified Ack) and a slower migrating band of ≈150 kDa (with the same electrophoretic mobility as recombinant Ack). Both protein bands were visualized by anti-Ack antibodies that recognize either amino (Ab 729) or carboxyl (Ab730) regions of Ack (Fig. 1 B and C). In addition, immunoblotting of anti-Ack immunoprecipitates with anti-pTyr antibodies showed that Ack is constitutively tyrosine-phosphorylated. Both ectopically expressed and endogenous Ack proteins were found to be tyrosine-phosphorylated, even after prolonged cell starvation in serum-free medium (Fig. 1C).
The immunoblotting experiment presented in Fig. 1B shows that Ack is expressed in brain including brainstem, cerebellum, thymus, spleen, fat, heart, skin, testis, and liver tissues. Ack is also expressed in a variety of hematopoietic cell lines, including BaF3, CTLL, HL60, RBL, U937, and WEHI, and in embryonic stem cells (data not shown).
Activation of Ack by Fibronectin-Mediated Cell Adhesion.
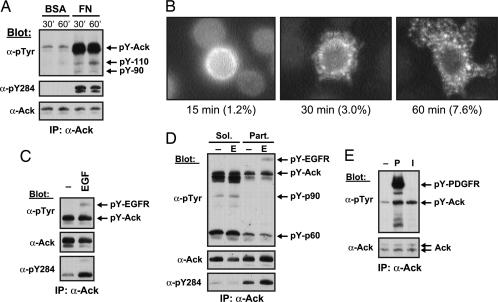
NIH 3T3 cells in suspension were lysed and subjected to immunoprecipitation with anti-Ack antibodies followed by immunoblotting with anti-pTyr antibodies. The experiment presented in Fig. 2A shows that tyrosine phosphorylation of endogenous Ack in NIH 3T3 cells in suspension is very low. However, when cells were plated onto fibronectin-coated dishes, tyrosine phosphorylation of Ack was greatly enhanced (Fig. 2A Top). When antibodies that specifically recognize the activated form of Ack (Ab pY284) were used to probe Ack immunoprecipitates, activation of Ack upon cell attachment to fibronectin was clearly detected. At least two other tyrosine-phosphorylated proteins that migrate in SDS gels with apparent molecular masses of 90 and 110 kDa were detected in complex with activated Ack, whose nature is currently unknown (Fig. 2A Top). Ack has been suggested to mediate tyrosine phosphorylation of p130Cas (16). However, we were not able to detect p130Cas or paxillin in complex with Ack by probing Ack immunoprecipitates with anti-p130Cas or anti-paxillin antibodies (data not shown).
Fig. 2.
Integrin-mediated and growth factor-induced Ack activation. (A) Ack is activated upon cell attachment to fibronectin. NIH 3T3 cells were starved in serum-free medium (DMEM) for 20 h, and the cells were trypsinized and replated on fibronectin (FN)-coated 10-cm dishes. Cells were lysed, and extracts (0.5 mg) were immunoprecipitated with a combination of anti-Ack antibodies (Ab 729 and 730). Upon SDS/PAGE and transfer to nitrocellulose, immunoprecipitates were analyzed with anti-pTyr antibodies, then stripped and probed with anti-Ack antibodies (Ab 880). The blot was stripped again and probed with affinity-purified antibodies that specifically recognize tyrosine-phosphorylated Y284, an autophosphorylation site in the Ack kinase domain. (B) Integrin-mediated cellular redistribution of Ack. HeLa cells were transfected with an expression vector for Ack in pRK5. After 48 h, the cells were trypsinized and replated onto fibronectin-coated coverslips for the indicated time. At those times, nonattached cells were washed off, and attached cells were fixed with 4% paraformaldehyde, permeabilized, and stained with antibodies against pY284 at 1 μg/ml. The numbers below the pictures represent the percentage of stained cells on the coverslips. (C) Ack is activated and recruited by activated EGFR. Starved Her14 cells were stimulated with EGF for 5 min. Ack was immunoprecipitated from precleared lysates by using a combination of anti-Ack antibodies (Ab 729 and 730). Upon SDS/PAGE and transfer to nitrocellulose, the blot was probed with anti-pTyr antibodies. The blot was stripped and probed with anti-Ack antibodies (Ab 880) to visualize Ack and stripped again and probed with antibodies that recognize activated Ack (pY284). (D) Subcellular fractionation of Her14 cell lysates. Her14 cells were mock-stimulated (−) or EGF-stimulated (E) as in A and lysed in hypotonic lysis buffer (34). Upon separation of both soluble (Sol.) and particulate (Part.) fractions, equal amounts of protein (1 mg) were incubated with anti-Ack antibodies (Ab 730). Upon SDS/PAGE and transfer to nitrocellulose, immunocomplexes were analyzed with anti-pTyr antibodies. The blot was stripped and probed with anti-Ack antibodies (Ab 880). The blot was stripped again and probed with antibodies against pY284. (E) Ack is activated after PDGF or insulin stimulation. Starved L6 myoblasts were either mock-stimulated or stimulated with PDGF (P) or insulin (I) for 5 min. Ack was immunoprecipitated from the precleared lysates (≈0.3 mg) by using Ab 730, and immunocomplexes were analyzed with anti-pTyr antibodies. The blot was stripped and probed with anti-Ack (Ab 880) antibodies.
Antibodies against active Ack (Ab pY284) were used to visualize the cellular distribution of the active kinase in HeLa cells after cell attachment on fibronectin. The fluorescence micrograph presented in Fig. 2B shows punctate distribution of activated Ack in the cell cytoplasm and perinuclear regions. In addition, the fraction of cells containing activated Ack was increased with time of attachment to fibronectin-coated coverslips. The punctate distribution appears to represent intracellular organelles or vesicles containing Ack.
Endogenous Ack Protein Is Activated upon EGF Stimulation and Is Recruited to Activated EGF Receptor (EGFR).
Her14 cells were either mock- or EGF-stimulated, and, after cell lysis, equal amounts of protein lysate were immunoprecipitated with anti-Ack antibodies. Immunoprecipitates were first analyzed by immunoblotting with anti-pTyr antibodies. Because of the high degree of tyrosine phosphorylation of Ack in starved cells, experiments in which anti-Ack immunoprecipitates were analyzed by immunoblotting with anti-pTyr antibodies usually revealed weak stimulation of tyrosine phosphorylation of Ack in response to EGF stimulation (Fig. 2C). However, when antibodies that specifically recognize activated Ack were used for immunoblotting (Ab pY284), clear activation of Ack was detected upon EGF stimulation (Fig. 2C Bottom).
Subcellular localization of Ack was next examined in Her14 cells before and upon EGF stimulation. Soluble and particulate fractions were separated by ultracentrifugation, and Ack was immunoprecipitated from the lysates obtained from both fractions. Ack is present in both soluble and particulate fractions before and after EGF stimulation (Fig. 2D). Upon EGF stimulation, the amount of Ack found in the soluble fraction was decreased, with concomitant increase in the particulate fraction. This finding indicates that a pool of Ack molecules is translocated from the soluble to the particulate fraction, possibly because of the translocation of Ack to the plasma membrane as a consequence of association with activated EGFR. Activated Ack, detected with antibodies against pY284, was present predominantly in the particulate fraction and at higher levels upon EGF stimulation. In addition, at least two uncharacterized tyrosine-phosphorylated proteins that migrate in SDS gels with apparent molecular masses of ≈60 and 90 kDa were found in complex with soluble Ack. Similar results were obtained in cells stimulated with PDGF. Lysates from mock- or PDGF-treated L6 myoblasts were immunoprecipitated with anti-Ack antibodies and analyzed with anti-pTyr antibodies. Enhanced tyrosine phosphorylation of Ack and recruitment to activated PDGFR were detected upon PDGF stimulation (Fig. 2E). In addition, insulin stimulation of L6 cells resulted in enhanced tyrosine phosphorylation of Ack (Fig. 2E).
Ack Forms a Complex with the Adapter Protein Nck.
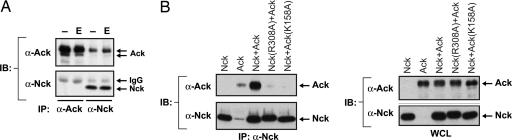
We previously reported the existence of a constitutively tyrosine-phosphorylated protein of 140 kDa present in Nck immunoprecipitates of several cell types, in both starved and growth factor-stimulated cells (21). Indeed, immunoblotting of Nck immunoprecipitates with anti-Ack antibodies identified the 140-kDa tyrosine-phosphorylated protein as Ack (Fig. 3A). A stable complex between Nck and Ack was also detected in lysates from mouse embryonic fibroblasts and PC12, Swiss-3T3, NIH 3T3, and L6 cells, but not in T cells.
Fig. 3.
Endogenous Ack forms a stable complex with the adaptor protein Nck. (A) Her14 cells were starved overnight in DMEM with 0.1% calf serum and then either mock-stimulated or stimulated with EGF (E) for 5 min. Precleared lysates (≈2 mg) were incubated with anti-Ack (Ab 729) or anti-Nck antibodies previously crosslinked to protein A Sepharose beads. Immunocomplexes were analyzed with anti-Ack (Ab 730) or anti-Nck antibodies. (B) 293 cells were transfected with expression vectors for Nck and wild-type or mutant Ack as shown. (Left) Precleared lysates (250 μg) were immunoprecipitated with anti-Nck antibodies and analyzed by immunoblotting with anti-Ack (Ab 880) or anti-Nck antibodies. (Right) Whole-cell lysates (WCL; 30 μg) were directly analyzed by immunoblotting with anti-Ack (Ab 880) or anti-Nck antibodies.
The interaction between Ack and Nck was analyzed in detail. Tyrosine phosphorylation of Ack was enhanced when serum-starved L6 cells were treated with the protein phosphatase inhibitor pervanadate, which increased the extent of complex formation between Ack and Nck (data not shown). This result suggests that the association between these two proteins is mediated by binding of the Nck SH2 domain to tyrosine phosphorylation site(s) on Ack. To test this possibility, we generated a point mutation in the FLVR motif of the Nck SH2 domain (R308A), a conserved residue involved in pTyr binding, and examined whether the Nck mutant was able to associate with Ack in transfected 293 cells. As shown in Fig. 3B, wild-type Nck formed a complex with Ack, whereas Nck (R308A) did not. Moreover, an Ack mutant (K158A) that has lost the ability to become autophosphorylated no longer formed a complex with Nck. These experiments demonstrate that the association between Ack and Nck is mediated by binding of the Nck SH2 domain to tyrosine-phosphorylated Ack.
Amino Terminus of Ack Contains a Membrane-Targeting Sterile α Motif (SAM) Domain.
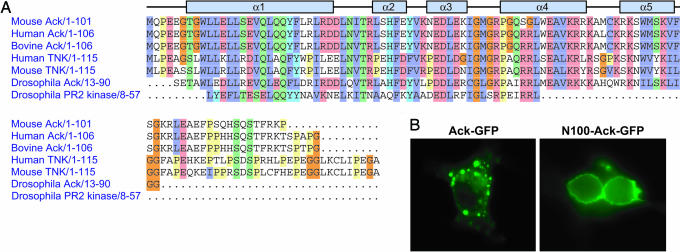
Sequence analysis using the integrated program interpro (protein accession no. O54967) and structure-fold based programs fugue, mgenthreader, and ffas revealed that the amino terminus of Ack belongs to the SAM domain superfamily. Although SAM domains adopt similar folds, their amino acid sequences are quite diverse, and their binding properties are rather versatile. Alignment of murine Ack amino terminus 85 aa with 96 SAM domains listed in the SMART database revealed that key consensus sites are conserved in murine Ack (Fig. 7A, which is published as supporting information on the PNAS web site). Among these SAM domains, highest homology is observed between the amino terminus of murine Ack and the SAM domain of Odin (ankyrin repeat and SAM domain-containing protein 1), which shares 25.5% identity and 37.2% similarity with murine Ack based on pairwise alignment using emboss-align. In addition to the clear sequence homology, the predicted secondary structure of the Ack amino terminus region consists of five to six α-helixes (Fig. 4A). This feature is compatible with all known SAM domain structures, which are composed of a four- to five-helix bundle of two orthogonally packed α-hairpins (22). Based on this analysis, we have concluded that Ack has a SAM-like domain at its amino terminus. The SAM-like domain of Ack appears conserved across species and, in addition, appears to be present in the nonreceptor tyrosine kinase Tnk1 (Fig. 4A).
Fig. 4.
Ack contains a SAM domain-like membrane-targeting region. (A) Alignment of the amino terminus of Ack (residues 1–85) to a panel of SAM domains in closely related protein kinases by using the program pfam. Secondary structure elements of this region consisting of five α-helixes were predicted by using psi-pred. (B) Cos1 cells were transfected with expression vectors for Ack-GFP (Left) or for SAM-like (N100) domain-GFP (Right). Cellular localization of fluorescently labeled Ack or SAM-like domain were visualized with an Axioskop2 microscope (Zeiss).
Unlike other functional domains, SAM domains display exceptional diversity with respect to their binding partners’ characteristics and the subcellular location of their host proteins (22). To investigate the function of SAM-like domain in Ack, cellular localization of this domain alone and full-length Ack were examined by fluorescence microscopy using GFP fusion proteins. Cos1 (Fig. 4B) or HeLa (data not shown) cells were transfected with either SAM–GFP or full-length Ack–GFP expression vectors. The experiments presented in Fig. 4B show that the SAM domain of Ack is primarily localized at the plasma membrane in both serum-starved (Fig. 4B Right) and EGF-stimulated (data not shown) cells. This result shows that the SAM-like domain of Ack may function as a membrane-targeting signal. In contrast, GFP-tagged full-length Ack displays a punctuate distribution pattern in what appears to be the membrane of large, tubular intracellular structures. Moreover, the cellular distribution of Ack, when visualized by staining with anti-Ack antibodies, is similar to the localization of activated Ack, visualized by anti-pY284 antibodies, except that the activated form of Ack appears to be more abundant in the perinuclear region (Fig. 2B). Thus, the presence of additional sequences in the context of the full-length protein and/or its interactions with other proteins are likely to be responsible for determining the subcellular localization of Ack.
Altered Activity of Ack Mutants.
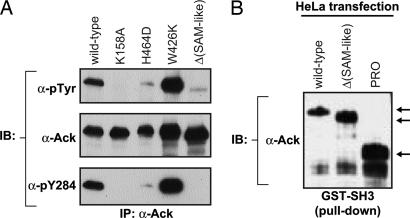
To determine the regions of Ack that are important for Ack activation, several Ack mutants were generated. These include a kinase-negative mutant (K158A), a CRIB domain mutant deficient in binding to Cdc42 (H464D), a deletion mutant lacking the SAM-like domain, and a mutant with a functionally defective SH3 domain (W424K). Expression vectors for wild-type and mutant proteins were transfected into HeLa cells, and antibodies that recognize wild-type and mutant proteins (Ab 730) were used for immunoprecipitation. Analysis of Ack immunoprecipitates with anti-pTyr antibodies revealed that the kinase-negative mutant is indeed inactive (Fig. 5A). Moreover, the CRIB domain and deletion mutant lacking the SAM-like domain were severely impaired in their ability to undergo tyrosine autophosphorylation. In contrast, a point mutation in the Ack SH3 domain resulted in enhanced tyrosine autophosphorylation, suggesting that the SH3 domain of Ack has an autoinhibitory role.
Fig. 5.
Role of Ack domains in regulation of Ack kinase activity. (A) HeLa cells were transfected with the indicated Ack wild type or mutants in the pRK5 expression vector. Forty-eight hours after transfection, cells were lysed and subjected to immunoprecipitation with anti-Ack antibodies that recognize wild type and Ack mutants (Ab 730). Tyrosine phosphorylation was detected by immunoblotting with anti-pTyr antibodies. The blot was stripped and probed with anti-Ack antibodies (Ab 880) to visualize the amount of Ack protein per lane. The blot was stripped again and probed with antibodies directed against the activated form of Ack (anti-pY284). (B) Cell extracts (≈0.4 mg) were incubated with 15 μg of purified GST–Ack SH3 fusion protein. After SDS/PAGE and transfer to nitrocellulose, bound proteins were analyzed by immunoblotting with anti-Ack (Ab 880) antibodies.
A GST–SH3 fusion protein was generated to examine whether the Ack SH3 domain is capable of binding to the Ack proline-rich region to mediate intramolecular or intermolecular interactions that could potentially control PTK activity. Fig. 5B shows that the Ack SH3 domain is able to form a complex with both full-length Ack and the deletion mutant devoid of the SAM-like domain (Fig. 5B). Moreover, the Ack SH3 domain also bound to the isolated proline-rich region of the protein. Taken together, these results demonstrate that the different domains of Ack mediate complex interactions that control its PTK activity and its interactions with other cellular components.
Conclusion
In this report we describe the biochemical characterization of the nonreceptor tyrosine kinase Ack. Around the same time Ack was cloned in our laboratory as a Grb2 binding partner >10 years ago (17), the same protein was cloned by others by means of its in vitro binding to activated Cdc42 (4). The corrected primary structure of Ack contains 56 fewer amino acids in the amino terminus of the protein, and the correct sequence reveals a novel SAM domain, which appears to function as a membrane-targeting signal. The central portion of the nonreceptor tyrosine kinase Tnk1 is homologous to Ack (23). When we used the corrected Ack sequence for this analysis, we noticed that, in addition to the central portion, the Tnk1 amino terminus is also homologous to that of Ack, and we propose that it may serve as a membrane-targeting domain as well. We have identified two isoforms of Ack that differ by an insertion of 15 aa (amino acids 515–529 in murine Ack) in the beginning of the proline-rich region. Analysis of the murine Ack genomic DNA suggests that these two isoforms represent splicing variants (M.L.G. and J.S., unpublished results). Not surprisingly, we have also found this exon in the Ack gene in the draft sequence of the human genome, and it is flanked by the standard GT/AG dinucleotides at the splice sites (18).
Using antibodies against phosphotyrosine for immunoblotting, we have demonstrated that Ack is tyrosine-phosphorylated in response to cell adhesion, EGF, PDGF, or insulin stimulation. Moreover, analysis of Ack stimulation by EGF using antibodies that recognize the activated form of Ack (anti-pY284) has shown EGF-induced complex formation between activated Ack and activated EGFR molecules. Recruitment of Ack by activated EGFR may occur directly through the Ralt homology region (6) and/or indirectly through an adapter protein. Nck is not likely to be responsible for mediating association with EGFR, because the Nck SH2 domain is engaged in binding to Ack. Another potential candidate is Grb2, which can associate directly with the proline-rich region of Ack in an in vitro binding assay. We have detected a constitutive association between endogenous Grb2 and Ack in cells, but the amount of Ack found in Grb2 immunoprecipitates appears to be low (M.L.G. and J.S., unpublished results). This finding may suggest that the interaction between these two proteins is indirect or that the Grb2/Ack complexes are not very abundant in the cell. Genetic experiments in Caenorhabditis elegans have shown that a putative C. elegans homologue of Ack (ARK-1) plays a negative role in signaling via Let-23, the C. elegans homologue of EGFR (24). It has been proposed that the negative signal exerted by ARK-1 may be mediated by Sem-5, the C. elegans homologue of Grb2 (24). However, the mechanism underlying the negative effect of ARK-1 on signaling via Let-23 is not understood (25). Moreover, it is not clear whether Ark1 is a true homolog of Ack.
We have shown that endogenous Ack became highly tyrosine-phosphorylated and activated upon cell attachment to fibronectin, suggesting that Ack is involved in integrin-mediated signaling pathways. The cellular distribution of activated Ack, tracked by immunofluorescence microscopy of permeabilized cells with anti-pY284 antibodies, exhibited punctate structures in the cytoplasm, particularly in the perinuclear region.
Analysis of the activity of Ack mutants has shown that tyrosine phosphorylation of Ack is severely reduced in a mutant that lacks the amino terminus, as well as in a mutant with a nonfunctional CRIB domain. However, a point mutation in the Ack SH3 domain that impedes this domain from binding to its ligands resulted in enhanced Ack tyrosine autophosphorylation. Moreover, the isolated Ack SH3 domain is capable of binding to intact Ack or to the isolated proline-rich region of the protein. These results suggest that the Ack SH3 domain may play an autoinhibitory role, perhaps by binding to the proline-rich region of the enzyme by an intramolecular mechanism similar to the autoinhibition conferred by the Src SH3 domain (26–30).
The experiments presented in this article demonstrate that Ack is activated in response to multiple extracellular stimuli. Experiments described here and in earlier studies demonstrate that Ack can interact with adapter proteins (i.e., Nck and Grb2) and with the small G protein Cdc42, suggesting that Ack is an intracellular mediator of a variety of extracellular signals.
Methods
Antibodies and Plasmids.
Anti-Nck (Ab 66) and anti-phosphotyrosine (Ab 72) antibodies were described previously (31, 32). Details regarding anti-Ack antibodies Ab729, Ab731, Ab880, and phosphospecific anti-pY284 are described in Supporting Text. Wild-type human Nck in pRK5 vector has been described (31). Single point mutations in Nck (R308A) and Ack (K158A, H464D, and W264K) were generated by site-directed mutagenesis according to the manufacturer’s protocol (Stratagene). A detailed description of the generation of these Ack mutants is presented in Supporting Text.
Northern Blotting.
A mouse multitissue CLONTECH blot containing 2 μg of mRNA per lane was probed with two set of probes. The 5′ probe includes 331 bp around the initiation ATG (nucleotides 239–570) from mouse Ack, and the 3′ probe contains 774 bp, from a region corresponding to the proline-rich region (nucleotides 2036–2810). Both probes were generated by 32P[dCTP] labeling by using Ready-To-Go DNA labeling beads (Amersham Pharmacia Biotech). Northern blots were performed with ExpressHyb hybridization solution (CLONTECH) at 68°C for 1 h followed by several washes, as recommended by the manufacturer.
Cell Lysis, Immunoprecipitation, and Immunoblotting.
After overnight starvation, cells were either mock-stimulated or stimulated with 100 ng/ml EGF (Intergen), 40 ng/ml PDGF (Intergen), or 100 nM insulin (Sigma) for 5 min at 37°C. Cell solubilization, immunoprecipitation, and immunoblotting with different antibodies were performed as previously described (33). Procedures for subcellular fractionation of Her14 cells were described in ref. 34.
Cell Adhesion Experiments.
For cell adhesion experiments, tissue culture dishes were incubated with fibronectin (8 μg/ml) in PBS at 4°C overnight and blocked with BSA (0.5%) in PBS at 37°C for 1.5–2 h. Control dishes were incubated with the BSA solution only. Cells were trypsinized, resuspended in soybean trypsin solution, and collected by centrifugation. Cell pellets were then washed, resuspended in serum-free DMEM, and plated onto the BSA control or fibronectin-coated dishes for the indicated time.
Generation of GST Fusion Protein and GST Pull-Down Assay.
To generate the GST–Ack SH3 fusion protein, the DNA fragment coding for amino acids 395–445 of mouse Ack was amplified by PCR and subcloned into the BamHI/EcoRI sites in pGEX-2T (Amersham Pharmacia). Fifteen micrograms of purified GST–Ack SH3 fusion protein on glutathione Sepharose beads was incubated with ≈0.4 mg of lysates of transfected HeLa cells for 3 h, washed as described, and then subjected to Western blot analysis.
Fluorescence Microscopy.
HeLa and Cos1 cells were transfected with the (SAM-like)–GFP or Ack–GFP constructs by lipofectamine (GIBCO/BRL) according to the manufacturer’s recommendations. Transfected cells were grown on coverslips, fixed in 4% PFA in PBS for 20 min, and permeabilized with 0.2% Triton X-100 in PBS for 20 min. After several washes in PBS, cells were mounted by using Fluorostab embedding medium (ICN) and directly visualized with an Axioskop2 microscope (Zeiss).
For indirect immunofluorescent microscopy, transfected cells were incubated with affinity-purified Ab 880 to visualize Ack and with affinity-purified anti-pY284 antibodies to visualize activated Ack. Cells were then washed with PBS and incubated in Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes). Coverslips were mounted and visualized as above.
Supplementary Material
Acknowledgments
We thank Avner Schlessinger for help with the analysis of the SAM domain of Ack. J.S. is supported by National Institutes of Health Grants R01-AR 051448 and R01-AR 051886 and funds from the Ludwig Institute for Cancer Research.
Abbreviations
- PTK
protein tyrosine kinase
- SAM
sterile αmotif
- EGFR
EGF receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Schlessinger J. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T., Scott J. D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 4.Manser E., Leung T., Salihuddin H., Tan L., Lim L. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 5.Makkinje A., Quinn D. A., Chen A., Cadilla C. L., Force T., Bonventre J. V., Kyriakis J. M. J. Biol. Chem. 2000;275:17838–17847. doi: 10.1074/jbc.M909735199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiorentino L., Pertica C., Fiorini M., Talora C., Crescenzi M., Castellani L., Alema S., Benedetti P., Segatto O. Mol. Cell. Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anastasi S., Fiorentino L., Fiorini M., Fraioli R., Sala G., Castellani L., Alema S., Alimandi M., Segatto O. Oncogene. 2003;22:4221–4234. doi: 10.1038/sj.onc.1206516. [DOI] [PubMed] [Google Scholar]
- 8.Teo M., Tan L., Lim L., Manser E. J. Biol. Chem. 2001;276:18392–18398. doi: 10.1074/jbc.M008795200. [DOI] [PubMed] [Google Scholar]
- 9.Worby C. A., Simonson-Leff N., Clemens J. C., Huddler D., Jr., Muda M., Dixon J. E. J. Biol. Chem. 2002;277:9422–9428. doi: 10.1074/jbc.M110172200. [DOI] [PubMed] [Google Scholar]
- 10.Lin Q., Lo C. G., Cerione R. A., Yang W. J. Biol. Chem. 2002;277:10134–10138. doi: 10.1074/jbc.M110329200. [DOI] [PubMed] [Google Scholar]
- 11.Yeow-Fong L., Lim L., Manser E. FEBS Lett. 2005;579:5040–5048. doi: 10.1016/j.febslet.2005.07.093. [DOI] [PubMed] [Google Scholar]
- 12.Linseman D. A., Heidenreich K. A., Fisher S. K. J. Biol. Chem. 2001;276:5622–5628. doi: 10.1074/jbc.M006812200. [DOI] [PubMed] [Google Scholar]
- 13.Yang W., Cerione R. A. J. Biol. Chem. 1997;272:24819–24824. doi: 10.1074/jbc.272.40.24819. [DOI] [PubMed] [Google Scholar]
- 14.Yang W., Lin Q., Guan J. L., Cerione R. A. J. Biol. Chem. 1999;274:8524–8530. doi: 10.1074/jbc.274.13.8524. [DOI] [PubMed] [Google Scholar]
- 15.van der Horst E. H., Degenhardt Y. Y., Strelow A., Slavin A., Chinn L., Orf J., Rong M., Li S., See L. H., Nguyen K. Q., et al. Proc. Natl. Acad. Sci. USA. 2005;102:15901–15906. doi: 10.1073/pnas.0508014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenmann K. M., McCarthy J. B., Simpson M. A., Keely P. J., Guan J. L., Tachibana K., Lim L., Manser E., Furcht L. T., Iida J. Nat. Cell Biol. 1999;1:507–513. doi: 10.1038/70302. [DOI] [PubMed] [Google Scholar]
- 17.Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J. M., Plowman G. D., Rudy B., Schlessinger J. Nature. 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- 18.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Dohna M., Reincke M., Mincheva A., Allolio B., Solinas-Toldo S., Lichter P. Genes Chromosomes Cancer. 2000;28:145–152. [PubMed] [Google Scholar]
- 20.Slavutsky I., de Vinuesa M. L., Larripa I., Dupont J., de Salum S. B. Cancer Genet. Cytogenet. 1986;21:335–342. doi: 10.1016/0165-4608(86)90214-1. [DOI] [PubMed] [Google Scholar]
- 21.Galisteo M. L., Chernoff J., Su Y. C., Skolnik E. Y., Schlessinger J. J. Biol. Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 22.Qiao F., Bowie J. U. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 23.Hoehn G. T., Stokland T., Amin S., Ramirez M., Hawkins A. L., Griffin C. A., Small D., Civin C. I. Oncogene. 1996;12:903–913. [PubMed] [Google Scholar]
- 24.Hopper N. A., Lee J., Sternberg P. W. Mol. Cell. 2000;6:65–75. [PubMed] [Google Scholar]
- 25.Worby C., Margolis B. Sci. STKE. 2000;2000:PE2. doi: 10.1126/stke.2000.63.pe2. [DOI] [PubMed] [Google Scholar]
- 26.Xu W., Harrison S. C., Eck M. J. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 27.Sicheri F., Moarefi I., Kuriyan J. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 28.Moarefi I., LaFevre-Bernt M., Sicheri F., Huse M., Lee C. H., Kuriyan J., Miller W. T. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 29.Barila D., Superti-Furga G. Nat. Genet. 1998;18:280–282. doi: 10.1038/ng0398-280. [DOI] [PubMed] [Google Scholar]
- 30.Andreotti A. H., Bunnell S. C., Feng S., Berg L. J., Schreiber S. L. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 31.Li W., Hu P., Skolnik E. Y., Ullrich A., Schlessinger J. Mol. Cell. Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batzer A. G., Rotin D., Urena J. M., Skolnik E. Y., Schlessinger J. Mol. Cell. Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolis B., Rhee S. G., Felder S., Mervic M., Lyall R., Levitzki A., Ullrich A., Zilberstein A., Schlessinger J. Cell. 1989;57:1101–1107. doi: 10.1016/0092-8674(89)90047-0. [DOI] [PubMed] [Google Scholar]
- 34.Sieh M., Batzer A., Schlessinger J., Weiss A. Mol. Cell. Biol. 1994;14:4435–4442. doi: 10.1128/mcb.14.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.