Abstract
Agrobacterium tumefaciens and related Agrobacterium species have been known as plant pathogens since the beginning of the 20th century. However, only in the past two decades has the ability of Agrobacterium to transfer DNA to plant cells been harnessed for the purposes of plant genetic engineering. Since the initial reports in the early 1980s using Agrobacterium to generate transgenic plants, scientists have attempted to improve this “natural genetic engineer” for biotechnology purposes. Some of these modifications have resulted in extending the host range of the bacterium to economically important crop species. However, in most instances, major improvements involved alterations in plant tissue culture transformation and regeneration conditions rather than manipulation of bacterial or host genes. Agrobacterium-mediated plant transformation is a highly complex and evolved process involving genetic determinants of both the bacterium and the host plant cell. In this article, I review some of the basic biology concerned with Agrobacterium-mediated genetic transformation. Knowledge of fundamental biological principles embracing both the host and the pathogen have been and will continue to be key to extending the utility of Agrobacterium for genetic engineering purposes.
INTRODUCTION
Twenty-five years ago, the concept of using Agrobacterium tumefaciens as a vector to create transgenic plants was viewed as a prospect and a “wish.” Today, many agronomically and horticulturally important species are routinely transformed using this bacterium, and the list of species that is susceptible to Agrobacterium-mediated transformation seems to grow daily. In some developed countries, a high percentage of the acreage of such economically important crops as corn, soybeans, cotton, canola, potatoes, and tomatoes is transgenic; an increasing number of these transgenic varieties are or will soon be generated by Agrobacterium-mediated, as opposed to particle bombardment-mediated transformation. There still remain, however, many challenges for genotype-independent transformation of many economically important crop species, as well as forest species used for lumber, paper, and pulp production. In addition, predictable and stable expression of transgenes remains problematic. Several excellent reviews have appeared recently that describe in detail various aspects of Agrobacterium biology (44, 73, 109, 325, 327, 328, 384, 385). In this review, I describe how scientists utilized knowledge of basic Agrobacterium biology to develop Agrobacterium as a “tool” for plant genetic engineering. I also explore how our increasing understanding of Agrobacterium biology may help extend the utility of Agrobacterium-mediated transformation. It is my belief that further improvements in transformation technology will necessarily involve the manipulation of these fundamental biological processes.
AGROBACTERIUM “SPECIES” AND HOST RANGE
The genus Agrobacterium has been divided into a number of species. However, this division has reflected, for the most part, disease symptomology and host range. Thus, A. radiobacter is an “avirulent” species, A. tumefaciens causes crown gall disease, A. rhizogenes causes hairy root disease, and A. rubi causes cane gall disease. More recently, a new species has been proposed, A. vitis, which causes galls on grape and a few other plant species (244). Although Bergey's Manual of Systematic Bacteriology still reflects this nomenclature, classification is complex and confusing; we now know that symptoms follow, for the most part, the type of tumorigenic plasmid contained within a particular strain. Curing a particular plasmid and replacing this plasmid with another type of tumorigenic plasmid can alter disease symptoms. For example, infection of plants with A. tumefaciens C58, containing the nopaline-type Ti plasmid pTiC58, results in the formation of crown gall teratomas. When this plasmid is cured, the strain becomes nonpathogenic. Introduction of Ri plasmids into the cured strain “converts” the bacterium into a rhizogenic strain (191, 358). Furthermore, one can introduce a Ti (tumor-inducing) plasmid from A. tumefaciens into A. rhizogenes; the resulting strain incites tumors of altered morphology on Kalanchoe plants (53). Thus, because A. tumefaciens can be “converted” into A. rhizogenes simply by substituting one type of oncogenic plasmid for another, the term “species” becomes meaningless. Perhaps a more meaningful classification system divides the genus Agrobacterium into “biovars” based on growth and metabolic characteristics (171). Using this system, most A. tumefaciens and A. rubi (316) strains belong to biovar I, A. rhizogenes strains fit into biovar II, and biovar III is represented by A. vitis strains. More recently, yet another taxonomic classification system for the genus Agrobacterium has been proposed (374). The recent completion of the DNA sequence of the entire A. tumefaciens C58 genome (which is composed of a linear and a circular chromosome, a Ti plasmid, and another large plasmid [114, 115, 363]) may provide a starting point for reclassification of Agrobacterium “strains” into true “species.”
Regardless of the current confusion in species classification, for the purposes of plant genetic engineering, the most important aspect may be the host range of different Agrobacterium strains. As a genus, Agrobacterium can transfer DNA to a remarkably broad group of organisms including numerous dicot and monocot angiosperm species (12, 68, 262, 341) and gymnosperms (198, 206, 215, 228, 307, 357, 371). In addition, Agrobacterium can transform fungi, including yeasts (32, 33, 260), ascomycetes (1, 71), and basidiomycetes (71). Recently, Agrobacterium was reported to transfer DNA to human cells (187).
The molecular and genetic basis for the host range of a given Agrobacterium strain remains unclear. Early work indicated that the Ti plasmid, rather than chromosomal genes, was the major genetic determinant of host range (207, 315). Several virulence (vir) loci on the Ti plasmid, including virC (367, 368) and virF (220, 267), were shown to determine the range of plant species that could be transformed to yield crown gall tumors. The virH (formerly called pinF) locus appeared to be involved in the ability of Agrobacterium to transform maize, as established by an assay in which symptoms of maize streak virus infection were determined following agroinoculation of maize plants (153). Other vir genes, including virG, contribute to the “hypervirulence” of particular strains (41, 146).
However, it is now clear that host range is a much more complex process, which is under the genetic control of multiple factors within both the bacterium and the plant host. The way one assays for transformation can affect the way one views host range. For example, many monocot plant species, including some cultivars of grasses such as maize (152), rice (39, 40, 85, 139, 265, 321), barley (317), and wheat (42), can now be genetically transformed by many Agrobacterium strains to the phenotype of antibiotic or herbicide resistance. However, these plant species do not support the growth of crown gall tumors. Host range may further result from an interaction of particular Ti plasmids with certain bacterial chromosomal backgrounds. For example, the Ti plasmid pTiBo542, when in its natural host strain A. tumefaciens Bo542, directs limited tumorigenic potential when assayed on many leguminous plant species. However, when placed in the C58 chromosomal background, pTiBo542 directs strong virulence toward soybeans and other legumes (143). Finally, susceptibility to crown gall disease has a genetic basis in cucurbits (292), peas (272), soybeans (15, 214, 246), and grapevines (312) and even among various ecotypes of Arabidopsis thaliana (231). The roles of both bacterial virulence genes and host genes in the transformation process, and the ways in which they may possibly be manipulated for genetic engineering purposes, are discussed below.
MOLECULAR BASIS OF AGROBACTERIUM-MEDIATED TRANSFORMATION
What Is T-DNA?
The molecular basis of genetic transformation of plant cells by Agrobacterium is transfer from the bacterium and integration into the plant nuclear genome of a region of a large tumor-inducing (Ti) or rhizogenic (Ri) plasmid resident in Agrobacterium (Fig. 1A). Ti plasmids are on the order of 200 to 800 kbp in size (81, 100, 111, 114, 145, 166, 175, 177, 245, 250, 251, 261, 311, 332, 342, 363). The transferred DNA (T-DNA) (Fig. 1B) is referred to as the T-region when located on the Ti or Ri plasmid. T-regions on native Ti and Ri plasmids are approximately 10 to 30 kbp in size (17, 34, 197, 311, 378). Thus, T-regions generally represent less than 10% of the Ti plasmid. Some Ti plasmids contain one T-region, whereas others contain multiple T-regions (17, 311). The processing of the T-DNA from the Ti plasmid and its subsequent export from the bacterium to the plant cell result in large part from the activity of virulence (vir) genes carried by the Ti plasmid (106, 147, 148, 174, 208, 303).
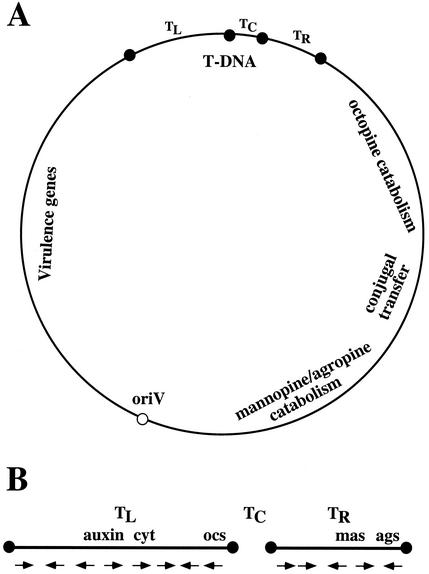
FIG. 1.
Schematic representation of a typical octopine-type Ti plasmid (A) and the T-DNA region of a typical octopine-type Ti plasmid (B). (A) The T-DNA is divided into three regions. TL (T-DNA left), TC (T-DNA center), and TR (T-DNA right). The black circles indicate T-DNA border repeat sequences. oriV, the vegetative origin of replication of the Ti plasmid, is indicated by a white circle. (B) The various T-DNA-encoded transcripts, and their direction of transcription, are indicated by arrows. Genes encoding functions involved in auxin synthesis (auxin), cytokinin synthesis (cyt), and the synthesis of the opines octopine (ocs), mannopine (mas), and agropine (ags) are indicated.
T-regions are defined by T-DNA border sequences. These borders are 25 bp in length and highly homologous in sequence (167, 366). They flank the T-region in a directly repeated orientation (257, 276, 335, 345, 352). In general, the T-DNA borders delimit the T-DNA (but see below for exceptions), because these sequences are the target of the VirD1/VirD2 border-specific endonuclease that processes the T-DNA from the Ti plasmid. There appears to be a polarity established among T-DNA borders: right borders initially appeared to be more important than left borders (136, 156, 286, 352, 353). We now know that this polarity may be caused by several factors. First, the border sequences not only serve as the target for the VirD1/VirD2 endonuclease but also serve as the covalent attachment site for VirD2 protein. Within the Ti or Ri plasmid (or T-DNA binary vectors [see below]), T-DNA borders are made up of double-stranded DNA. Cleavage of these double-stranded border sequences requires VirD1 and VirD2 proteins, both in vivo (82, 99, 155, 369) and in vitro (281). In vitro, however, VirD2 protein alone can cleave a single-stranded T-DNA border sequence (154, 249). Cleavage of the 25-bp T-DNA border results predominantly from the nicking of the T-DNA “lower strand,” as conventionally presented, between nucleotides 3 and 4 of the border sequence (301, 353). However, double-strand cleavage of the T-DNA border has also been noted (155, 305, 344). Nicking of the border is associated with the tight (probably covalent) linkage of the VirD2 protein, through tyrosine 29 (351), to the 5′ end of the resulting single-stranded T-DNA molecule termed the T-strand (91, 99, 137, 150, 355, 373). It is this T-strand, and not a double-stranded T-DNA molecule, that is transferred to the plant cell (318, 375). Thus, it is the VirD2 protein attached to the right border, and not the border sequence per se, that establishes polarity and the importance of right borders relative to left borders. It should be noted, however, that because left-border nicking is also associated with VirD2 attachment to the remaining molecule (the “non-T-DNA” portion of the Ti plasmid or “backbone” region of the T-DNA binary vector [91]), it may be possible to process T-strands from these regions of Ti and Ri plasmids and from T-DNA binary vectors (182, 264, 356). The problem of vector “backbone” sequence transfer to plants is discussed below.
Second, the presence of T-DNA “overdrive” sequences near many T-DNA right borders, but not left borders, may also help establish the functional polarity of right and left borders. Overdrive sequences enhance the transmission of T-strands to plants, although the molecular mechanism of how this occurs remains unknown (131, 156, 256, 291, 336, 337, 345). Early reports suggested that the VirC1 protein binds to the overdrive sequence and may enhance T-DNA border cleavage by the VirD1/VirD2 endonuclease (322). virC1 and virC2 functions are important for virulence; mutation of these genes results in loss of virulence on many plant species (299). However, several laboratories have noted that T-strand production in virC mutant Agrobacterium strains occurs at wild-type levels (301, 344). Thus, any effect of VirC must occur after T-DNA processing.
How Is T-DNA Transferred from Agrobacterium to Plant Cells?
As indicated above, many proteins encoded by vir genes play essential roles in the Agrobacterium-mediated transformation process. Some of these roles have been discussed in several excellent review articles (44, 109, 325, 327, 328, 384), and I shall therefore limit my description to the roles of Vir proteins that may serve as points of manipulation for the improvement of the transformation process.
VirA and VirG proteins function as members of a two-component sensory-signal transduction genetic regulatory system. VirA is a periplasmic antenna that senses the presence of particular plant phenolic compounds that are induced on wounding (3, 87, 162, 195, 303, 324, 359). In coordination with the monosaccharide transporter ChvE and in the presence of the appropriate phenolic and sugar molecules, VirA autophosphorylates and subsequently transphosphorylates the VirG protein (160, 161). VirG in the nonphosphorylated form is inactive; however, on phosphorylation, the protein helps activate or increase the level of transcription of the vir genes, most probably by interaction with vir-box sequences that form a component of vir gene promoters (59, 60, 252). Constitutively active VirA and VirG proteins that do not require phenolic inducers for activity, or VirG proteins that interact more productively with vir-box sequences to activate vir gene expression, may be useful to increase Agrobacterium transformation efficiency or host range. Experiments describing some attempts to manipulate VirA and/or VirG for these purposes are discussed below.
Together with the VirD4 protein, the 11 VirB proteins make up a type IV secretion system necessary for transfer of the T-DNA and several other Vir proteins, including VirE2 and VirF (44, 349). VirD4 may serve as a “linker” to promote the interaction of the processed T-DNA/VirD2 complex with the VirB-encoded secretion apparatus (126). Most VirB proteins either form the membrane channel or serve as ATPases to provide energy for channel assembly or export processes. Several proteins, including VirB2, VirB5, and possibly VirB7, make up the T-pilus (94, 163, 189, 190, 278, 283). VirB2, which is processed and cyclized, is the major pilin protein (94, 163, 189, 190). The function of the pilus in T-DNA transfer remains unclear; it may serve as the conduit for T-DNA and Vir protein transfer, or it may merely function as a “hook” to seize the recipient plant cell and bring the bacterium and plant into close proximity to effect molecular transfer. One aspect of pilus biology that may be important for transformation is its temperature lability. Although vir gene induction is maximal at approximately 25 to 27°C (8, 162, 323), the pilus of some, but not all, Agrobacterium strains is most stable at lower temperatures (approximately 18 to 20°C) (18, 105, 188). Early experiments by Riker indicated a temperature effect on transformation (268). Thus, one may consider cocultivating Agrobacterium with plant cells at lower temperatures during the initial few days of the transformation process.
The VirD2 and VirE2 proteins play essential and perhaps complementary roles in Agrobacterium-mediated transformation. These two proteins have been proposed to constitute, with the T-strand, a “T-complex” that is the transferred form of the T-DNA (149). Whether this complex assembles within the bacterium remains controversial. Citovsky et al. (50) showed that VirE2 could function in a plant cell: transgenic VirE2-expressing tobacco plants could “complement” infection by a virE2 mutant Agrobacterium strain. Several laboratories have shown that VirE2 can transfer to the plant cell in the absence of a T-strand (27, 193, 244, 309, 349), and it is possible that VirE2 complexes with the T-strand either in the bacterial export channel or within the plant cell. A recent report suggests perhaps another role for VirE2 early in the export process: Dumas et al. (90) showed that VirE2 could associate with artificial membranes in vitro and create a channel for the transport of DNA molecules. Thus, it is possible that one function of VirE2 is to form a pore in the plant cytoplasmic membrane to facilitate the passage of the T-strand.
Because of its attachment to the 5′ end of the T-strand, VirD2 may serve as a pilot protein to guide the T-strand to and through the type IV export apparatus. Once in the plant cell, VirD2 may function in additional steps of the transformation process. VirD2 contains nuclear localization signal (NLS) sequences that may help direct it and the attached T-DNA to the plant nucleus. The NLS of VirD2 can direct fused reporter proteins and in vitro-assembled T-complexes to the nuclei of plant, animal, and yeast cells (48, 119, 138, 151, 185, 186, 229, 319, 326, 381, 382). Furthermore, VirD2 can associate with a number of Arabidopsis importin-α proteins in an NLS-dependent manner, both in yeast and in vitro (16; S. Bhattacharjee and S. B. Gelvin, unpublished data). Importin-α is a component of one of the protein nuclear transport pathways found in eukaryotes. Recent data, however, suggest that VirD2 may not be sufficient to direct T-strands to the nucleus. Ziemienowicz et al. (382) showed that in permeabilized cells, VirD2 could effect the nuclear targeting of small linked oligonucleotides generated in vitro but could not direct the nuclear transport of larger linked molecules. To achieve nuclear targeting of these larger molecules, VirE2 additionally had to be associated with the T-strands. Finally, VirD2 may play a role in integration of the T-DNA into the plant genome. Various mutations in VirD2 can affect either the efficiency (229) or the “precision” (320) of T-DNA integration.
The role of VirE2 in T-DNA nuclear transport also remains controversial. VirE2 is a non-sequence-specific single-stranded DNA binding protein (45, 46, 49, 112, 286). In Agrobacterium cells, VirE2 probably interacts with the VirE1 molecular chaperone and may therefore not be available to bind T-strands (77, 84, 310, 380). However, when bound to single-stranded DNA (perhaps in the plant cell?), VirE2 can alter the DNA from a random-coil conformation to a shape that resembles a coiled telephone cord (47). This elongated shape may help direct the T-strand through the nuclear pore. VirE2 also contains NLS sequences that can direct fused reporter proteins to plant nuclei (48, 50, 326, 383). As with VirD2, VirE2 interacts in yeast with Arabidopsis importin-α proteins in an NLS-dependent manner (Bhattacharjee and Gelvin, unpublished). One report indicates that VirE2 bound to single-stranded DNA and microinjected into plant cells can direct the DNA to the nucleus (381). However, other reports demonstrate that VirE2 cannot direct bound single-stranded DNA to the nuclei of either plant or animal cells that are permeabilized in order to effect DNA uptake (380). The cause of these contradictory results remains unclear but may reflect differences in the cell types and DNA delivery systems used by the two groups. When T-DNA is delivered to plant cells from Agrobacterium strains that encode a mutant form of VirD2 containing a precise deletion of the NLS, there is at most only a 40% decrease in transformation efficiency (229, 233, 290). Transgenic plants expressing VirE2 can complement a double-mutant Agrobacterium strain that lacks virE2 and contains a deletion in the NLS-encoding region of virD2 (110). These results suggest that in the absence of NLS sequences in VirD2, some other nuclear targeting mechanism (perhaps involving VirE2) may take place.
When bound to DNA, the NLS motifs of VirE2 may be occluded and inactive. This is because the NLS and DNA binding domains of VirE2 overlap (50). Hohn's group has hypothesized that the primary role of VirE2 in nuclear transport is NLS independent and that VirE2 merely shapes the T-strand so that it can snake through the nuclear pores (274).
Further controversy involves the ability of VirE2 protein to localize to the nuclei of animal cells. Ziemienowicz et al. (381) showed that in permeabilized HeLa cells, octopine-type VirE2 could target to the nucleus, whereas in microinjected Drosophila and Xenopus cells, the NLS sequence of nopaline-type VirE2 had to be changed in order to effect nuclear localization of the altered protein (50). Although the reason for this discrepancy is not known, it is not likely that it results from the use of octopine- versus nopaline-type VirE2 by the two groups (326).
Finally, VirE2 may protect T-strands from nucleolytic degradation that can occur both in the plant cytoplasm and perhaps in the nucleus (274, 374).
The existence of a T-complex composed of a single molecule of VirD2 covalently attached to the 5′ end of the T-strand, which in turn is coated by VirE2 molecules, has generally been accepted by the Agrobacterium research community (149). However, the reader should be aware that such a complex has not yet been identified in either Agrobacterium or plant cells. It is possible that other proteins, such as importins (16), VIP1 (329), and even VirF (285), may additionally interact, either directly or indirectly, with the T-strand to form larger T-complexes in the plant cell.
Although the role of Ti plasmid-encoded vir genes has often been considered of primary importance for transformation, many Agrobacterium chromosomal genes are also essential for this process. The role of chromosomal genes was first established by random insertional mutagenesis of the entire Agrobacterium genome (106). Further research defined the roles of many of these genes. Included among these functions are exopolysaccharide production, modification, and secretion (pscA/exoC, chvA, and chvB [36, 37, 88, 89, 313]) and other roles in bacterial attachment to plant cells (att genes [212, 213]), sugar transporters involved in coinduction of vir genes (chvE [86, 172, 289]), regulation of vir gene induction (chvD [204]), and T-DNA transport (acvB [168, 248, 360, 361, 362]). Other genes, such as miaA (116), may also play a more minor role in the transformation process. The recent elucidation of the entire A. tumefaciens C58 sequence (114, 363) will surely provide fertile ground for the discovery of additional genes involved in Agrobacterium-mediated transformation.
MANIPULATION OF AGROBACTERIUM FOR GENETIC ENGINEERING PURPOSES
Introduction of Genes into Plants by Using Agrobacterium
Years before scientists elucidated the molecular mechanism of Agrobacterium-mediated transformation of plants, Armin Braun proposed the concept of a “tumor-inducing principle” that was stably transferred to and propagated in the plant genome (30). Research in the 1970s resulted in the identification of large plasmids in virulent Agrobacterium strains (376), although we now know that many strains contain plasmids unrelated to virulence. Genetic experiments indicated that a particular class of plasmids, the Ti (and later Ri) plasmids, were responsible for tumorigenesis (339) and that a portion of these plasmids, the T-DNA, was transferred to plant cells and incorporated into the plant genome (43). It was thus obvious to propose that Ti plasmids be used as a vector to introduce foreign genes into plant cells.
However, Ti plasmids are very large and T-DNA regions do not generally contain unique restriction endonuclease sites not found elsewhere on the Ti plasmid. Therefore, one cannot simply clone a gene of interest into the T-region. Scientists therefore developed a number of strategies to introduce foreign genes into the T-DNA. These strategies involved two different approaches: cloning the gene, by indirect means, into the Ti plasmid such that the new gene was in cis with the virulence genes on the same plasmid, or cloning the gene into a T-region that was on a separate replicon from the vir genes (T-DNA binary vectors).
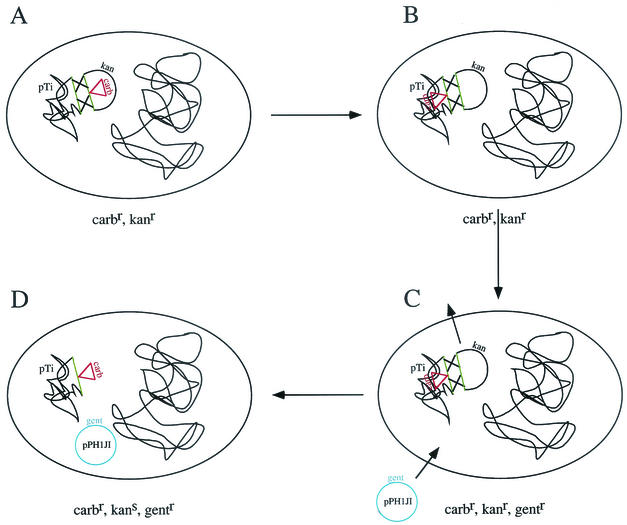
Two methods were used for cloning foreign DNA into the Ti plasmid. The first method was based on a strategy developed by Ruvkin and Ausubel (277) (Fig. 2). A region of DNA (either the T-region or any region of DNA targeted for disruption) containing unique restriction endonuclease sites is cloned into a broad-host-range plasmid, such as an IncPα-based vector. These plasmids can replicate both in Escherichia coli, in which the initial cloning is performed, and in Agrobacterium. The exogenous gene of interest, along with an antibiotic resistance marker, is next cloned into a unique restriction endonuclease site within the target region of DNA. Alternatively, an antibiotic resistance gene can be introduced into the DNA fragment of interest by transposition (107, 297). The resulting plasmid is introduced into Agrobacterium by conjugation or transformation. The presence of this plasmid in Agrobacterium is confirmed by selection for resistance to antibiotics encoded by both the plasmid vector backbone and the resistance marker near the gene of interest. Next, another plasmid of the same incompatibility group as the first plasmid, but harboring yet another antibiotic resistance marker, is introduced into the Agrobacterium strain containing the first plasmid. The resulting bacteria are plated on medium containing antibiotics to select for the second (eviction) plasmid and the resistance marker next to the gene of interest. Because plasmids of the same incompatibility group cannot usually coreside within the same bacterial cell, the bacteria can become resistant to both these antibiotics only if either (i) the first plasmid cointegrates into the Ti plasmid and uses the oriV of the Ti plasmid to replicate or (ii) an exchange of DNA on the first plasmid and the Ti plasmid occurs by double homologous recombination (homogenotization) using homologous sequences on the Ti plasmid flanking both sides of the gene of interest plus the resistance marker. In the first case (cointegration of the entire plasmid with the Ti plasmid), the resistance marker of the plasmid backbone would be expressed; these bacteria are screened for and discarded. In the second instance (homogenotization), the resistance marker encoded by the plasmid backbone is lost. Double homologous recombination can be confirmed by DNA blot analysis of total DNA from the resulting strain (107). A variant of this procedure utilizes a sacB gene on the plasmid backbone of the first plasmid. Only elimination of the plasmid backbone after homogenotization renders the bacterium resistant to growth on sucrose (194).
FIG. 2.
Schematic representation of the steps involved in gene replacement by double homologous recombination (homogenotization [107, 277]). The green lines represent regions targeted for disruption. (A) An antibiotic resistance gene (in this case, encoding a β-lactamase that confers resistance to carbenicillin) has been inserted into the targeted gene that has been cloned into an IncPα plasmid (containing a kanamycin resistance gene [kan] in its backbone) and introduced into Agrobacterium. Double homologous recombination is allowed to take place. (B) Following double homologous recombination, the disrupted gene is exchanged onto the Ti plasmid (pTi). (C) A plasmid of the same incompatibility group as the first plasmid is introduced into Agrobacterium. An example is the IncPα plasmid pPH1JI, containing a gentamicin resistance gene (gent). (D) Because plasmids of the same incompatibility group (in this case IncPα) cannot replicate independently in the cell at the same time, selection for gentamicin resistance results in eviction of the other IncPα plasmid, onto which has been exchanged the wild-type gene.
Another method to introduce foreign DNA into the T-region of the Ti plasmid involves first introducing a ColE1 replicon, such as pBR322, into the T-region of a Ti plasmid. DNA to be integrated into this T-region is cloned into a separate pBR322-derived molecule containing a second antibiotic resistance marker. This plasmid is introduced into the altered Agrobacterium strain, and the resulting strain is selected for resistance to the second antibiotic. Because ColE1 replicons cannot function in Agrobacterium, the pBR322-based plasmid would have to cointegrate into the pBR322 segment of the altered T-region for the stable expression of the plasmid-encoded resistance gene (379). A modification of this procedure was used to develop the “split-end vector” system. Using this system, a gene of interest is cloned into a pBR322-based vector that contains a T-DNA right border, a nos-nptII chimaeric gene for selection of transgenic plants, a spectinomycin-streptomycin resistance marker to select for the presence of the plasmid in Agrobacterium, and a region of homology with a nononcogenic portion of an octopine-type T-region. Cointegration of this plasmid with a Ti-plasmid lacking a right border but containing the T-DNA homologous region restores border activity and localizes the gene of interest and the plant selectable marker within the reconstituted T-region (101).
Each of these cis-integration methods has advantages and disadvantages. The first strategy can target the foreign gene to any part of the T-region (or other region in the Ti plasmid). However, it is cumbersome to perform and involves somewhat sophisticated microbial genetic procedures that many laboratories shunned. The second method is technically easier but allows cointegration of the foreign gene only into Ti-plasmid locations where pBR322 had previously been placed. However, a modification of this procedure allows cointegration of a pBR322-based plasmid into any region of the Ti plasmid (338, 377). An advantage of both of these systems is that they maintain the foreign gene at the same low copy number as that of the Ti plasmid in Agrobacterium.
Because of the complexity of introducing foreign genes directly into the T-region of a Ti plasmid, several laboratories developed an alternative strategy to use Agrobacterium to deliver foreign genes to plants. This strategy was based on seminal findings of Hoekema et al. (140) and de Frammond et al. (70). These authors determined that the T-region and the vir genes could be separated into two different replicons. When these replicons were within the same Agrobacterium cell, products of the vir genes could act in trans on the T-region to effect T-DNA processing and transfer to a plant cell. Hoekema et al. called this a binary-vector system; the replicon harboring the T-region constituted the binary vector, whereas the replicon containing the vir genes became known as the vir helper. The vir helper plasmid generally contained a complete or partial deletion of the T-region, rendering strains containing this plasmid unable to incite tumors. A number of Agrobacterium strains containing nononcogenic vir helper plasmids have been developed, including LBA4404 (242), GV3101 MP90 (181), AGL0 (192), EHA101 and its derivative strain EHA105 (144, 146), and NT1 (pKPSF2) (247).
T-DNA binary vectors revolutionized the use of Agrobacterium to introduce genes into plants. Scientists without specialized training in microbial genetics could now easily manipulate Agrobacterium to create transgenic plants. These plasmids are small and easy to manipulate in both E. coli and Agrobacterium and generally contain multiple unique restriction endonuclease sites within the T-region into which genes of interest could be cloned. Many vectors were designed for specialized purposes, containing different plant selectable markers, promoters, and poly(A) addition signals between which genes of interest could be inserted, translational enhancers to boost the expression of transgenes, and protein-targeting signals to direct the transgene-encoded protein to particular locations within the plant cell (some representative T-DNA binary vector systems are described in references 10, 20, 25, 26, 62, 113, 120, 216, 364, and 386 and at http://www.cambia.org). Hellens et al. (134) provide a summary of many A. tumefaciens strains and vectors commonly used for plant genetic engineering.
Although the term “binary vector system” is usually used to describe two constituents (a T-DNA component and a vir helper component), each located on a separate plasmid, the original definition placed the two modules only on different replicons. These replicons do not necessarily have to be plasmids. Several groups have shown that T-DNA, when located in the Agrobacterium chromosome, can be mobilized to plant cells by a vir helper plasmid (141, 224).
How Much DNA Can Be Transferred from Agrobacterium to Plants?
The T-regions of natural Ti and Ri plasmids can be large enough to encode tens of genes. For example, the T-region of pTiC58 is approximately 23 kbp in size. In addition, some Ti and Ri plasmids contain multiple T-regions, each of which can be transferred to plants individually or in combination (34, 314). For purposes of plant genetic engineering, scientists may wish to introduce into plants large T-DNAs with the capacity to encode multiple gene products in a biosynthetic pathway. Alternatively, the reintroduction of large regions of a plant genome into a mutant plant may be useful to identify, by genetic complementation, genes responsible for a particular phenotype. How large a T-region can be transferred to plants?
Miranda et al. (224) showed that by reversing the orientation of a T-DNA right border, they could mobilize an entire Ti plasmid, approximately 200 kbp, into plants. Although the event was rare, this study showed that very large DNA molecules could be introduced into plants using Agrobacterium-mediated transformation. Hamilton et al. (124) first demonstrated the directed transfer of large DNA molecules from Agrobacterium to plants by the development of a binary BAC (BIBAC) system. These authors showed that a 150-kbp cloned insert of human DNA could be introduced into plant cells by using this system. However, the efficient transfer of such a large DNA segment required the overexpression of either virG or both virG and virE. VirE2 encodes a single-stranded DNA binding protein that protects the T-DNA from degradation in the plant cell (275). Because virG is a transcriptional activator of the vir operons (303), expression of additional copies of this regulatory vir gene was thought to enhance the expression of VirE2 and other Vir proteins involved in T-DNA transfer. Overexpression of virE formed part of the BIBAC system that was used to transform large (30- to 150-kbp) DNA fragments into tobacco and the more recalcitrant tomato and Brassica (56, 103, 123, 125). However, the transfer of different-size T-DNAs from various Agrobacterium strains had different requirements for overexpression of virG and virE (103). Liu et al. (203, 204) developed a transformation-competent artificial chromosome vector system based on a P1 origin of replication and used this system to generate libraries of large (40- to 120-kbp) Arabidopsis and wheat DNA molecules. This system did not require overexpression of virG or virE to effect the accurate transfer of large fragments to Arabidopsis.
What DNA Is Transferred from Agrobacterium to Plants?
T-DNA was initially defined as the portion (the T-region) of the Ti plasmid that was transferred from Agrobacterium to plant cells to form crown gall tumors. T-DNA border repeat sequences defined the T-region (366), and regions of the Ti plasmid outside these borders were not initially found in tumor cells (43). However, the transfer of Ti-plasmid sequences outside the conventional T-region may at first have been missed because of a lack of known selectable (e.g., tumorigenesis) or screenable (e.g., opine production) markers. Ooms et al. (241) observed the incorporation into plant DNA of regions of the Ti plasmid later shown to be outside the classical T-DNA borders. Ramanathan and Veluthambi (264) also showed that a nos-nptII cassette, placed outside the T-DNA left border, could be transferred to and confer kanamycin resistance on infected tobacco cells.
The use of relatively small T-DNA binary vectors made it easier for scientists to evaluate the transfer of “non-T-DNA” regions to plants. Martineau et al. (211) first reported the transfer of binary vector backbone sequences into transgenic plant DNA and questioned the definition of T-DNA. Wenck et al. (356) found that the entire binary vector, including backbone sequences as well as T-DNA sequences, could frequently be transferred to Nicotiana plumbaginifolia and Arabidopsis thaliana cells. Kononov et al. (182) carefully examined the structure of binary vector backbone sequences that could be found in up to 75% of transgenic tobacco plants and concluded that such transfer could result from either skipping the left T-DNA border when T-DNA was processed from the binary vector or initiation of T-DNA transfer from the left border to bring vector backbone sequences into plant cells. Considering the previous observation by Durrenberger et al. (91) that VirD2 protein could covalently attach to the 5′ end of the non-T-DNA strand, Kononov et al. suggested that transfer of vector backbone sequences to plants was a natural consequence of the mechanism of VirD2 function. Thus, the definition of T-DNA and vector backbone constitutes a semantic argument. It would thus appear that the transfer of non-T-DNA sequences to plants may be an unavoidable, but frequently unobserved and untested, result of transformation. Indeed, Frary and Hamilton (103) observed incorporation of BIBAC plasmid sequences into 9 to 38% of tested tomato transformants.
Although the transfer of plasmid backbone sequences may be an unavoidable consequence of the mechanism of Agrobacterium-mediated transformation, it may be possible to select against transgenic plants containing this unwanted DNA. Hanson et al. (132) showed that the incorporation of a toxic “killer” gene into the binary vector backbone sequences could severely reduce the percentage of transgenic plants containing such extra sequences. Remarkably, the transformation frequency of tobacco, tomato, and grape plants infected using this modified binary vector did not substantially differ from that of plants infected using a binary vector lacking the killer gene. Because the presence of uncharacterized DNA in transgenic plants is important for regulatory concerns, such an approach may be useful in the future for the production of plants (especially difficult to transform species) with a more highly defined transgenic composition.
Transfer of Multiple T-DNAs into the Same Plant Cell, and Generation of “Marker-Free” Transgenic Plants
Because of concerns regarding the spread of antibiotic resistance genes in nature or the escape of herbicide resistance genes to wild weedy species, scientists have developed several methods to generate marker-free transgenic plants. These plants would initially be selected for resistance to an antibiotic or herbicide, but the selection marker would be removed on subsequent manipulation and plant growth. Several methods have been proposed to eliminate the selection marker from the primary transformant. These include use of a site-specific recombination system, such as Cre-lox or Flp-Frt (2, 19, 57, 209, 235, 347, 348) to remove the marker, transposon-based movement of the selection marker from the initial site of insertion from the plant genome entirely or to another unlinked site from which it can be segregated in subsequent generations (93), or the use of multiple T-DNAs which can insert into unlinked sites for future segregation (reviewed in references 142 and 372). Each of these systems has advantages and disadvantages. For example, excision of marker genes using a site-specific recombination system requires introduction of the site-specific recombinase into plants, either by transformation or by genetic crossing. Segregation of markers can occur only in progeny following the generation of the initial transgenic plant and is limited to species naturally propagated through seed and not those propagated vegetatively.
Early research that characterized the integration pattern of T-DNAs in crown gall tumors indicated that each of the two T-DNAs encoded by an octopine-type Ti plasmid could independently integrate into the plant genome, sometimes in multiple copies (43, 63, 314). The molecular analyses suggested that these T-DNAs could be integrated into unlinked sites. These results suggested that cotransformation could be performed to integrate transgenes carried by two different T-DNAs and that perhaps these T-DNAs would segregate in subsequent generations. Three approaches were subsequently used for cotransformation: (i) the introduction of two T-DNAs, each from a different bacterium; (ii) the introduction of two T-DNAs carried by different replicons within the same bacterium; and (iii) the introduction of two T-DNAs located on the same replicon within a bacterium.
Early experiments using these various approaches indicated that cotransformation could be a frequent event. An et al. (9) showed that tobacco cells could be cotransformed to two different phenotypes by a single Agrobacterium strain containing both a Ti plasmid (phytohormone-independent growth) and a T-DNA binary vector (kanamycin-resistant growth). This experiment represents a “one-strain, two-replicon” approach to cotransformation. When the cells were first selected for kanamycin resistance, 10 to 20% of them also displayed phytohormone-independent growth; when the cells were first selected for phytohormone-independent growth, 60% of the resulting calli were also kanamycin resistant. The authors credited these differing frequencies to the higher copy number (5 to 10) of the binary vector in the bacterium relative to the single copy Ti plasmid.
These experiments were extended by de Frammond et al. (69), who showed that fertile transgenic plants could be regenerated from cloned tobacco tissue that was cotransformed by T-DNA from a Ti plasmid and from a micro-Ti (the one-strain, two-replicon approach). The two T-DNAs segregated in progeny plants, indicating that the T-DNAs had integrated into genetically separable loci. Other groups have used the one-strain, two-replicon approach to generate transgenic plants which initially expressed both T-DNA markers but could subsequently segregate the markers from each other (58).
Depicker et al. (80) performed a similar experiment in which the selection markers were phytohormone-independent growth and nopaline synthesis (encoded by a Ti plasmid) and kanamycin-resistant growth (encoded by a T-DNA binary vector). They performed the experiment in two ways: either the two T-DNAs were delivered by two different Agrobacterium strains (the two-strains, two-replicons approach), or both T-DNAs were delivered from a single replicon in one strain (the one-strain, one-replicon approach). The results of these experiments indicated that cotransfer of T-DNAs from the same plasmid in one strain was considerably more efficient than was transfer from two different strains. The use of a single Agrobacterium strain to cotransform plants with two T-DNAs from the same replicon, followed by segregation of the selection gene to generate marker-free transgenic plants, has been described by Komari et al. (178) and Xing et al. (365). In each of these studies, the authors were able to generate marker-free transgenic plants at high frequency.
The use of two Agrobacterium strains to deliver different T-DNAs to the same plant cells was studied by a number of groups (65, 66, 67, 76, 217). Although cotransfer of T-DNAs to genetically unlinked sites was reported, some authors also reported close linkage of the two different T-DNAs in many instances. It thus remains unclear which of the three cotransformation protocols will be reproducibly best for the generation of marker-free transgenic plants.
Virulence Gene Expression and Plant Transformation
The processing and transfer of T-DNA from Agrobacterium to plant cells is regulated by the activity of the vir genes. Virulence gene activity is induced by plant wound-induced phenolic compounds such as acetosyringone and related molecules (28, 74, 75, 92, 228, 293, 295, 298, 300). However, there may be instances in which scientists would like to induce vir genes to levels higher than that accomplished by plant extracts. Several groups have therefore identified virA and virG mutants that function constitutively, in the absence of phenolic inducers. Constitutive virA mutants were characterized by several groups (13, 218, 253). However, more emphasis has been placed on inducer-independent virG mutants, possibly because virG functions downstream of virA.
Extensive genetic studies resulted in the identification of a number of mutations that render the VirG protein active in the absence of phenolic inducing compounds (127, 254). These altered proteins contain mutations that converted either asparagine-54 to aspartic acid (virGN54D) or isoleucine-106 to leucine (virGI106L). Both of these mutant proteins stimulated a high level of vir gene expression, especially when expressed from a high-copy-number plasmid (118). When tested in transient tobacco and maize transformation assays, strains containing the virGN54D mutant effected a higher level of transformation than did strains encoding the wild-type virG gene (130). An even greater effect was seen when the virGN54D allele was harbored on a high-copy-number plasmid; the presence of this mutant gene in Agrobacterium increased the transient transformation of rice and soybean two- to sevenfold (170).
Several laboratories have determined the effect of additional copies of wild-type virG genes on vir gene induction and plant transformation. Rogowsky et al. (273) showed that additional copies of nopaline-type virG resulted in increased vir gene expression. Liu et al. (200) showed that multiple copies of virG altered the pH response profile for vir gene induction. Normally, vir gene induction is very poor at neutral or alkaline pH or in rich medium; additional copies of virG permitted a substantial degree of induction in rich medium even at pH 8.5. Additional copies of virG also increased the transient transformation frequency of rice, celery, and carrot tissues (199).
Given these results in toto, one may conclude that increasing the copy number of virA or virG or decreasing the dependence of the encoded proteins on phenolic inducers would generally increase the transformation efficiency of the resulting strains. However, the situation is likely to be more complex. Belanger et al. (23) showed that individual virA genes may be particularly suited to function in certain genetic backgrounds, and Krishnamohan et al. (183) recently demonstrated that Ti plasmids may have evolved to optimize specific combinations of virA, virG, and vir boxes. As noted above, the Ti-plasmid pTiBo542 in the C58 chromosomal background is hypervirulent on certain legume species, possibly because of the associated virG gene (41, 146, 159), but not in its native Bo542 chromosomal background (143). Recent results from my laboratory indicate that vir gene induction and T-strand production by and transformation efficiency of particular Agrobacterium strains may not correlate well. A. tumefaciens A277, containing the Ti plasmid pTiBo542 within the C58 chromosomal background, is considerably more virulent than are strains A348 and A208, containing the Ti plasmids pTiA6 and pTiT37, respectively, in the same chromosomal background. However, vir gene induction by plant exudates and T-strand production are highest in A. tumefaciens A208 (L.-Y. Lee and S. B. Gelvin, unpublished data). These data further suggest that increased vir gene induction and T-strand production may not necessarily be reliable predictors of transformation efficiency.
T-DNA Integration and Transgene Expression
Plant transformation does not always result in efficient transgene expression. The literature is replete with examples of variable expression levels of transgenes, which frequently did not correlate with transgene copy number (see, for example, reference 255). This lack of correspondence was initially attributed to position effects, i.e., the position within the genome into which the T-DNA integrated was credited with the ability of transgenes to express. T-DNA could integrate near to or far from transcriptional activating elements or enhancers, resulting in the activation (or lack thereof) of T-DNA-carried transgenes (22, 35, 296, 308). T-DNA could also integrate into transcriptionally competent or transcriptionally silent regions of the plant genome. The high percentage (approximately 30%) of T-DNA integration events that resulted in activation of a promoterless reporter transgene positioned near a T-DNA border suggested that T-DNA may preferentially integrate into transcriptionally active regions of the genome. Only integration events that would link the promoterless transgene with an active promoter would result in reporter activity (180). However, a drawback to some of these experiments was that transgenic events may have been biased by the selection of antibiotic resistant plants expressing an antibiotic marker gene carried by the T-DNA. It is not clear whether T-DNA insertions into transcriptionally inert regions of the genome would have gone unnoticed because of lack of expression of the antibiotic resistance marker gene.
An obvious way to circumvent the presumed problems of position effect is to integrate T-DNA into known transcriptionally active regions of the plant genome. However, gene targeting in plants by homologous recombination has been at best extremely inefficient (72, 173, 223, 237, 238, 269, 270). An alternative system for gene targeting is the use of site-specific integration systems such as Cre-lox. However, single-copy transgenes introduced into a lox site in the same position of the plant genome also showed variable levels of expression in independent transformants. Transgene silencing in these instances may have resulted from transgene DNA methylation (61). Such methylation-associated silencing was reported earlier for naturally occurring T-DNA genes (135, 340). Thus, transcriptional silencing may result from integration of transgenes into regions of the plant genome susceptible to DNA methylation and may be a natural consequence of the process of plant transformation.
We now know not only that transgene silencing results from “transcriptional” mechanisms, usually associated with methylation of the transgene promoter (222), but also that transgene silencing is often “posttranscriptional”; i.e., the transgene is transcribed, but the resulting RNA is unstable (219). Such posttranscriptional gene silencing is frequently associated with multiple transgene copies within a cell. Transgenic plants generated by direct DNA transfer methods (e.g., polyethylene glycol- or liposome-mediated transformation, electroporation, or particle bombardment) often integrate a large number of copies of the transgene in tandem or inverted repeat arrays, in either multiple or single loci (176). Although Agrobacterium-mediated transformation usually results in a lower copy number of integrated transgenes, it is common to find tandem copies of a few T-DNAs integrated at a single locus (165). Transgene silencing can occur in plants harboring a single integrated T-DNA (95). However, integration of T-DNA repeats, especially "head-to-head' inverted repeats around the T-DNA right border, frequently results in transgene silencing (51, 164, 304). Thus, a procedure or Agrobacterium strain that could be used to generate transgenic plants with a single integrated T-DNA would be a boon to the agricultural biotechnology industry and to plant molecular biology in general. Grevelding et al. (117) noted that transgenic Arabidopsis plants derived from a root transformation procedure tended to have fewer T-DNA insertions than did plants derived from leaf disks. However, it is not clear if this observation can be generally applicable to other plant species. Anecdotal information from several laboratories suggests that Agrobacterium strains that are less efficient in delivering T-DNA may be more efficient in producing single-copy T-DNA insertions. However, these findings need to be tested rigorously; it is possible that T-DNA copy number may also correlate with the growth state of the bacterium or the plants to be transformed.
Use of Matrix Attachment Regions To Ameliorate Transgene Silencing
At present, the generation of single-copy transgenic plants is still somewhat hit and miss. Scientists usually produce a relatively large number of independent transformants and screen them for plants containing a single-copy T-DNA insertion. At best, this can be a time-consuming nuisance. However, for agronomically important species, elite cultivars, or lines that are recalcitrant to transformation, it can become a rate-limiting step. An alternative to this approach may be to generate transgenic plants containing a few copies of T-DNA that are insulated from each other. One proposed mechanism to accomplish this is to flank transgenes within the T-DNA with matrix attachment regions (MARs). MARs are DNA sequences that either are associated with chromosome “matrices” as isolated or can associate with these matrices in vitro (121, 122, 294, 350). Among other properties, they have been ascribed the role of insulating genes within a looped chromatin domain from transcription-activating or -silencing effects of neighboring domains. In animal cells, such insulating effects may render transgene expression proportional to transgene copy number (306). However, some of the MARs initially used in animal experiments may also have contained enhancer elements, confounding the interpretation of the original experiments (29, 259).
When they flank transgenes delivered to plants via Agrobacterium-mediated transformation, MARs appear to have only a small effect on transgene expression (128, 198, 201, 225, 226, 227, 334). Larger increases in transgene expression have been observed using particle bombardment-mediated transformation (5, 6, 236). However, this increase is generally associated with expression of transgenes in plant cells rather than in whole plants (330, 333). It is possible that the higher transgene expression effects of MARs using particle bombardment reflects the higher integrated transgene copy number resulting from this technique as opposed to the relatively lower copy number of integrated T-DNAs delivered by Agrobacterium (7). As such, it is not clear whether MARs will be, on their own, highly useful for decreasing the silencing of transgenes delivered to plants by Agrobacterium-mediated transformation.
Use of Viral Suppressors of Gene Silencing To Increase Transgene Expression
Recent data from a number of laboratories indicates that some plant viruses, both DNA and RNA viruses, contain genes that suppress gene silencing (4, 11, 21, 31, 38, 55, 169, 210). Several investigators have speculated that viral antisilencing has evolved as a mechanism for viruses to evade a plant's defense through viral gene silencing (55, 266). Regardless of the reason for and mechanism of antisilencing, viral suppressors of silencing may be useful to mitigate the silencing of transgenes.
As indicated in some of the references cited above, viral suppressors of gene silencing can activate a previously silenced stable transgene. One would then wonder whether such silencing suppressors could prevent the silencing of transgenes stably introduced into plants by Agrobacterium-mediated transformation. Although this hypothesis has not yet been tested and possible negative consequences (such as increased viral susceptibility) may ensue from the stable incorporation of antisilencing genes into a plant genome, experiments in which viral silencing suppressors have been used to increase the levels of transient expression of Agrobacterium-introduced transgenes appear promising. O. Voinnet and colleagues (unpublished data) have recently demonstrated that when cotransformed with various transgenes encoding green fluorescent protein, the potato virus Y Nia protein, or tomato Cf-9 and Cf-4 disease resistance proteins, various viral silencing suppressors dramatically increased the expression of these other proteins. Expression levels up to 50-fold higher than those achieved in control transformations (lacking the viral silencing suppressor genes) were obtained. Several different viral silencing suppressors, including the p25 protein of PVX, the P1-HcPro protein of tobacco etch virus, and the p19 protein of tomato bushy stunt virus, were able to enhance transient transgene expression from both the cauliflower mosaic virus 35S promoter and from native transgene promoters. Of these, the p19 protein was most effective in both increasing transient transgene expression and decreasing the levels of small (21- to 25-bp) RNA molecules associated with posttranscriptional gene silencing. The authors concluded that viral suppressors of gene silencing could be useful for the production of large amounts of proteins in plants.
When Transgene Expression Is Not Forever
Experiments to express transgenes in plants initially used elements, such as the cauliflower mosaic virus 35S and 19S promoters or opine synthase promoters, that would express the transgene in a relatively constitutive manner (24, 133, 179, 196, 234, 280, 343). However, as plant genetic engineering experiments became more refined, scientists turned to regulated promoters that would express a transgene in a particular developmental, environmental, or tissue-specific pattern. Systems were also developed that would allow scientists to induce transgene expression at will, allowing for the overexpression of a particular product or expression of a product that may be toxic during certain stages of plant development. Such inducible systems included those regulated by tetracycline (108), alcohol (279), copper (221), heat shock (284), and steroid hormones (14, 282) (see reference 158 for a recent review of chemically inducible gene induction systems). Many of these systems were leaky, permitting the expression of transgenes under noninduced conditions.
There may be instances, however, when one would not want a transgene or its product to be present after the initial few hours or days following transformation. Such traits include those that would aid in the transformation process itself or would effect plant DNA rearrangements desired only during the initial transformation event (e.g., gene targeting using site-specific recombinase systems). Two strategies are currently being developed to permit only transient expression of gene products in plants. These are the use of “nonintegrating” T-DNA systems and the transfer of proteins, rather than DNA molecules, from Agrobacterium to plant cells.
Nonintegrating T-DNA systems include the use of mutant Agrobacterium strains and/or plant cells that are proficient in T-DNA nuclear transfer but deficient in T-DNA integration. During a search for domains of VirD2 necessary for nuclear targeting of the T-DNA, Shurvinton et al. (290) defined a C-terminal domain, termed ω, that showed high amino acid sequence homology among virD2 genes. Although this domain was not required for either VirD2 endonuclease activity or nuclear targeting of the T-DNA, replacement of four conserved amino acids by two serine residues resulted in a mutant protein that rendered the encoding Agrobacterium strain highly attenuated in virulence. Narasimhulu et al. (233) and Mysore et al. (229) further showed that Agrobacterium strains harboring this VirD2 ω substitution were highly deficient in stable transformation (with 2% of the efficiency of wild-type strains) but were still able to transform plant cells transiently at 20% of the efficiency of wild-type strains. Thus, this mutation rendered Agrobacterium strains highly deficient in T-DNA integration but still relatively proficient in delivering T-DNA to the plant nucleus. This mutant VirD2 protein can therefore be used to target T-strands to the nucleus, where they can transiently express but not efficiently integrate.
Nam et al. (231) used a root assay to screen almost 40 A. thaliana ecotypes for their ability to be transformed by Agrobacterium. Among these ecotypes, UE-1 was easily transiently transformed but poorly stably transformed. Genetic and molecular characterization of this ecotype indicated that the block in transformation occurred at the T-DNA integration step. Nam et al. (232) further identified a large number of Arabidopsis T-DNA insertion mutants that were resistant to Agrobacterium transformation (rat mutants). Of the initial 21 mutants, 5 were efficiently transiently transformed but were highly recalcitrant to stable transformation, a phenotype associated with a deficiency in T-DNA integration. Mysore et al. (230) characterized one of these mutants, the rat5 mutant, in greater detail. This mutant was generated by the insertion of T-DNA into the 3′ untranslated region of a histone H2A gene (HTA1). Biochemical and molecular data indicated that this mutant could be transiently transformed efficiently but that T-DNA integration was disrupted. Although the precise role of the HTA1 gene in T-DNA integration has yet to be elucidated, root transformation of this mutant (and perhaps other T-DNA integration-deficient mutants) can be used for the transient delivery of T-DNA without efficient subsequent T-DNA integration. The use of the HTA1 gene to improve the transformation efficiency of wild-type plants is discussed below.
Vergunst et al. (349) recently described a novel procedure to transfer proteins directly from Agrobacterium to plant cells. This system relies on the ability of the type IV protein secretion system encoded by the Agrobacterium virB and virD4 genes to transfer certain Vir proteins to plant cells. VirD2, VirE2, and VirF are the three proteins identified to date that can be transferred by this system. These authors showed that translational fusions of the Cre recombinase protein to the N terminus of either VirE2 or VirF could be transferred to plant cells and effect recombination at lox sites. They further showed that the C-terminal 37 amino acids of VirF were sufficient to transfer the fusion protein. These experiments lead to the possibility of using Vir proteins as carriers to introduce other proteins transiently into plant cells.
MANIPULATION OF PLANT GENES TO IMPROVE TRANSFORMATION
Plant Response to Agrobacterium Infection
Although great advances have been made over the past decade to increase the number of plant species that can be transformed and regenerated using Agrobacterium, many important species or inbred lines remain highly recalcitrant to Agrobacterium-mediated transformation. The question has often arisen, “Who has the problem with transformation, Agrobacterium or the researcher?” The very wide host range of Agrobacterium, including gymnosperms and perhaps lower plant phyla, a variety of fungi, and even animal cells, suggests that T-DNA transfer to the recipient (i.e., entry exclusion) may not be the problem. That Agrobacterium can transiently transform a number of these species efficiently, including agronomically important species such as maize and soybean (170, 271, 288), suggests that in many instances T-DNA integration may remain the limiting step. Alteration of the tissue culture conditions, for example by the use of antioxidants during the transformation of grape, rice, maize, and soybean, has increased the probability of stably transforming cell types that can be regenerated (96, 102, 239, 240, 258). However, such manipulations of the transformation conditions may have limitations.
Agrobacterium infection of plant tissues may in some instances result in plant tissue necrosis. Several groups have described a slow, spreading necrosis in grape infected by particular Agrobacterium strains (79, 263). More recently, Hansen (129) described an apoptotic response of maize to Agrobacterium infection. The response included both rapid tissue necrosis and cleavage of nuclear DNA into oligonucleosome-sized fragments by endogenous nucleases and is characteristic of a caspase-protease cascade. The expression of two baculovirus cell death suppressor genes, p35 and iap, greatly inhibited both tissue necrosis and endogenous DNA cleavage. Manipulation of these genes during the Agrobacterium-mediated transformation process may thus be useful to increase both plant cell viability and transformation efficiency in plant species with an apoptotic response to Agrobacterium.
Several groups have recently begun to identify plant genes and protein products involved in the transformation process. One of the rationales for these experiments is the hope that identification of these genes may eventually result in their manipulation either to improve transformation or to make plants resistant to crown gall disease. A number of approaches have been employed to identify these plant genes, including (i) use of yeast two-hybrid systems to identify plant proteins that may interact with virulence proteins, (ii) direct “forward genetic” screening to identify plant mutants that cannot be transformed, (iii) “reverse genetic” screening to test whether particular genes of interest may be involved in transformation, and (iv) various genomics approaches to identify plant genes that may be induced or repressed soon after infection by Agrobacterium.
Identification of Plant Genes Encoding Proteins That Interact with Agrobacterium Virulence Proteins
Several Agrobacterium virulence proteins would be expected to interact with plant proteins. These include the processed form of VirB2, the major component of the T-pilus that is required for transformation; VirD2, the protein that caps the 5′ end of the transferred T-strand; VirE2, the single-stranded DNA binding protein that presumably coats the T-strand; and VirF, which is transferred to plant cells but whose function remains unknown. Several other Vir proteins that are on the bacterial cell surface, such as VirB5 and VirB7 (minor components of the T-pilus), and VirB1∗ (a processed product of VirB1 that can be found in the extracellular medium), may also interact with proteins on the surface of plant cells.
Early work (16) utilized VirD2 as the bait protein in a yeast two-hybrid system to identify an A. thaliana importin-α (AtKAP, now known as importin-α1) as an interacting partner. Importin-α proteins are involved in the nuclear translocation of many proteins harboring NLS sequences, and Arabidopsis encodes at least nine of these proteins (Bhattacharjee and Gelvin, unpublished). Ballas and Citovsky (16) showed that interaction of VirD2 with importin-α AtKAP was NLS dependent both in yeast and in vitro. The importance of importin-α proteins in the Agrobacterium transformation process has recently been suggested by demonstrating that a T-DNA insertion into the importin-α7 gene, or antisense inhibition of expression of the importin-α1 (AtKAP) gene, results in a highly attenuated transformation phenotype (Bhattacharjee and Gelvin, unpublished).
VirD2 also interacts with at least two other plant proteins by using a yeast two-hybrid system. Deng et al. (78) identified three VirD2- and two VirE2-interacting proteins. They characterized more fully one of the VirD2 interactors, an Arabidopsis cyclophilin. This protein, as a glutathione S-transferase fusion, interacted strongly with VirD2 in vitro. The authors further showed that the interaction domain of VirD2 was in the central portion of the protein, a region to which no previous function had been ascribed. Cyclosporin A, an inhibitor of cyclophilins, inhibited Agrobacterium-mediated transformation of Arabidopsis roots and tobacco suspension cell cultures. The authors suggested that this plant protein may serve as a chaperone to help in T-complex trafficking within the plant cell. Experiments in my laboratory identified a tomato type 2C protein phosphatase as an interacting partner with VirD2. This phosphatase may be involved in the phosphorylation and dephosphorylation of a serine residue near the C-terminal NLS motif in VirD2. Overexpression of this phosphatase in transfected tobacco BY-2 cells resulted in decreased nuclear targeting of a GUS-VirD2-NLS fusion protein, suggesting that phosphorylation of the VirD2 NLS region may be involved in nuclear targeting of the VirD2/T-strand complex (Y. Tao, P. Rao, and S. B. Gelvin, submitted for publication).
Using VirE2 as the bait protein in a yeast two-hybrid system, Tzfira et al. (329) identified two interacting proteins from Arabidopsis, VIP1 and VIP2. VIP1 may be involved in nuclear targeting of the T-complex because antisense inhibition of VIP1 expression resulted in a deficiency in nuclear targeting of VirE2. Tobacco VIP1 antisense plants were also highly recalcitrant to Agrobacterium infection. Recent results further suggest a use for VIP1 in improving plant transformation: transgenic plants that overexpress VIP1 are hypersusceptible to Agrobacterium transformation (327, 330). VirE2 also interacts in yeast with several of the Arabidopsis importin-α proteins, suggesting that VirD2 and VirE2 may have a common mechanism of nuclear import (Bhattacharjee and Gelvin, unpublished).
Schrammeijer et al. (285) recently identified an Arabidopsis Skp1 protein as an interactor with the F-box domain of VirF. Skp1 proteins may be involved in targeting proteins such as cyclins to the 26S proteosome, suggesting that VirF may function in setting the plant cell cycle to effect better transformation. The interaction of VirF with VIP1, but not VirE2, may also suggest a mechanism for Vir protein turnover: if VirF is targeted to the proteosome, it may help target other Vir proteins for proteolysis.
The T-pilus is essential for Agrobacterium-mediated transformation. Mutations in various VirB proteins disrupt transformation but not T-DNA processing (301, 344). As mentioned above, the major T-pilus component is the processed and cyclized VirB2 protein (189), although other virulence proteins, including VirB5 and possibly VirB7, also are minor T-pilus constituents (278, 283). Although the precise role of the T-pilus remains controversial, it is expected that the T-pilus would interact with the plant cell wall or membrane. Experiments in my laboratory have begun to address possible plant-interacting partners with T-pilus components. Using a yeast two-hybrid system and the processed, but not cyclized, form of VirB2 as a bait protein, we have identified two classes of Arabidopsis proteins that strongly and specifically interact with this major T-pilus constituent (H.-H. Hwang and S. B. Gelvin, unpublished data). One of these classes of plant proteins is encoded by a three-member gene family. Although the identity of these three related proteins is not currently known, their hydropathy profiles suggest that they contain membrane-spanning domains. The other interacting protein is a Rab-type GTPase. Each of these four plant proteins interacts in yeast with itself and with each other but not with other tested virulence proteins, including VirB1, VirB1∗, VirB5, VirD2, VirE2, and VirF. In vivo data indicate that each of these proteins is involved in Agrobacterium-mediated transformation. Antisense or RNAi inhibition of expression of the genes encoding these proteins results in a transformation-deficient phenotype. In addition, an Arabidopsis mutant line containing a T-DNA insertion into the promoter region of one of the “unknown protein” genes also is highly recalcitrant to Agrobacterium-mediated transformation.
Forward Genetic Screening To Identify Plant Genes Involved in Agrobacterium-Mediated Transformation
Scientists have shown a genetic basis for susceptibility to crown gall disease in some plant species (15, 214, 231, 246, 272, 292, 312). In an effort to identify plant genes involved in Agrobacterium-mediated transformation, my laboratory embarked on a major project to identify Arabidopsis T-DNA insertion mutants that are resistant to Agrobacterium transformation (rat mutants [232]). These studies have resulted in the identification of more than 70 such mutants to date. The roles of many of the mutated genes in the transformation process have been revealed by various assays. Thus, rat1 (encoding an arabinogalactan protein) and rat3 (probably encoding a plant cell wall protein) are involved in bacterial attachment to roots (23). Other rat genes that may affect cell wall structure include a xylan synthase (rat4 [232]) and a β-expansin (A. Kaiser, A. Kopecki, Y. Zhu, and S. B. Gelvin, unpublished data). Because bacterial attachment to the roots of the rat4 mutant appears nearly normal (A. Matthysse, unpublished data), RAT4 may be involved in T-DNA transfer to the cytoplasm.
Using this forward-genetics approach, we have identified a number of other rat mutants in later stages of the transformation process. T-DNA insertions into genes encoding α- and β-importins are probably blocked in the T-DNA nuclear targeting process (S. Bhattacharjee, H. Cao, J. Humara, Y. Zhu, and S. B. Gelvin, unpublished data). Other mutants, including the rat5 (a histone H2A mutant [230, 232]), rat17, rat18, rat20, and rat22 mutants, are probably involved in T-DNA integration (232). A T-DNA insertion between two closely spaced replacement histone H3 genes (histone H3-4 and H3-5) also results in the rat phenotype (J. Li, Y. Zhu, and S. B. Gelvin, unpublished data).
The finding that the histone H2A-1 gene affects T-DNA integration has led to a more extensive characterization of this gene. The Arabidopsis histone H2A gene family includes 13 members. We have initiated a study of the expression pattern of each of these genes and an examination of the role that each of these genes may play in Agrobacterium-mediated transformation (370; H. Yi, T. Fujinuma, and S. B. Gelvin, unpublished data). The histone H2A-1 gene, encoded by RAT5, is expressed in numerous cell types, including cells that are not undergoing rapid division. This expression pattern is characteristic of a “replacement” histone gene. In roots, the gene is expressed in lateral root primordia, the meristem region, and the elongation zone. Interestingly, the root elongation zone is the region most highly susceptible to Agrobacterium-mediated transformation (370). Other experiments indicate that histone H2A-1 gene expression and susceptibility to Agrobacterium-mediated transformation are highly correlated. Thus, expression of this gene may be predictive of cell types that are most sensitive to transformation. Knowledge of plant cell transformation competency may be important for the genetic engineering of recalcitrant plant species and cultivars.
Because mutation of the histone H2A-1 gene resulted in decreased Arabidopsis root transformation, we examined whether overexpression of this gene would increase the efficiency of Agrobacterium-mediated transformation. Transgenic Arabidopsis plants containing additional genomic (230) or cDNA (H. Yi and S. B. Gelvin, unpublished data) H2A-1 copies are two- to sixfold more transformation competent than are plants containing the normal histone H2A-1 gene complement. In addition, transient expression of the histone H2A-1 gene from an incoming T-DNA both complements the rat5 mutant (230) and increases the transformation efficiency of normally susceptible and highly recalcitrant Arabidopsis ecotypes (L.-Y. Lee and S. B. Gelvin, unpublished data). Finally, overexpression of the RAT5 histone H2A-1 gene in various rat mutants (other than the rat5 mutant) also restores transformation competency (L.-Y. Lee, S. Davis, X. Sui, and S. B. Gelvin, unpublished data). Expression of the RAT5 gene is therefore epistatic over the rat phenotype of other rat mutants and thus may sensitize plant cells to Agrobacterium-mediated transformation. We suggest that overexpression of the RAT5 histone H2A-1 gene may improve the transformation efficiency of recalcitrant plants.
Reverse Genetic Screening for Plant Genes Involved in Agrobacterium-Mediated Transformation
Plant genes encoding several proteins that interact with virulence proteins have been identified using a yeast two-hybrid system. Such interactions are at best suggestive of a role for these genes in plant transformation. Their roles must be shown directly. One way to accomplish this is to inhibit gene expression in planta using techniques such as antisense RNA, RNAi, or mutagenesis. I have discussed above the use of antisense RNA and RNAi to show that VIP1 (a VirE2 interactor), a Rab GTPase, and several proteins of unidentified function (VirB2 interactors) are involved in Agrobacterium-mediated transformation. Suppression of expression of these genes may be one method to generate plants resistant to crown gall disease.
Another method to test the role of a particular gene in transformation would be to mutate that gene and then assay the plant for transformation susceptibility. However, at present site-directed mutagenesis is not an efficient method for use in plants. An alternative reverse genetic approach is to identify mutant plants containing transposon or T-DNA insertions in genes of interest. Several PCR-based strategies have been described to identify such knockout mutations in Arabidopsis (98, 104, 184), tomato (52), and rice (157). Using one such strategy, my laboratory has identified Arabidopsis mutant lines containing disruptions in various importin-α and importin-β genes (putatively involved in nuclear transport of the T-complex) and various genes involved in plant chromatin structure (putatively involved in T-DNA integration into the plant genome). Some of these mutants are either moderately or highly resistant to Agrobacterium-mediated transformation (S. Bhattacharjee, H. Cao, H. Humara, A. Kaiser, A. Kopecki, J. Li, X. Zhao, and S. B. Gelvin, unpublished data), and contain T-DNA insertions in or near genes encoding importin-α7 or importin-β3, various histones (including histones H2A1, H2A3, H2B5, H2B6, H3-4, H3-5, and H4-1), histone acetyltransferases (including HAC4, HAC6, HAC9, HAC10, and HAC11), and a histone deacetylase (HDA1). We have not yet, however, established the precise roles of these plant genes in the Agrobacterium-mediated transformation process.
Escobar et al. (97) have recently described a novel reverse genetic strategy to produce crown gall-resistant plants. They generated transgenic Arabidopsis and tomato plants expressing double-stranded RNA constructions targeted to T-DNA-encoded auxin and cytokinin biosynthetic oncogenes. These genes are highly homologous among a large variety of different Agrobacterium strains. Many transgenic plants expressing these RNAi constructions were highly resistant to crown gall disease directed by a broad range of oncogenic strains, although they were not generally resistant to Agrobacterium-mediated transformation per se. A similar approach has been used to generate crown gall disease-resistant tobacco and apple plants (W. Ream, unpublished data).
Genomics Approaches To Identify Plant Genes That Respond to Agrobacterium Infection
As described above (129), plants may respond to infection by Agrobacterium, and this response may involve differential plant gene expression. Genes that are induced or repressed during the early stages of Agrobacterium-mediated transformation may provide targets for manipulation of the host to improve the efficiency of transformation of recalcitrant plant species. Several laboratories have consequently begun investigations to identify these differentially expressed plant genes.
Ditt et al. (83) recently investigated the response of Ageratum conyzoides suspension cell cultures to infection by a nontumorigenic supervirulent A. tumefaciens strain (233). Using cDNA-amplification fragment length polymorphism (AFLP) to amplify 16,000 fragments, they identified 251 bands that were differentially regulated 48 h after infection. Reverse transcription-PCR analysis of some of these genes confirmed the results of the cDNA-AFLP analysis. Some of these bands were also induced or repressed 24 h after inoculation. Whereas most of the bands investigated (encoding, e.g., an RNase, a putative recpetor kinase, a peroxidase, and a pathogenesis-related protein) were also differentially regulated following incubation of plant cells with E. coli, four genes, including one encoding a nodulin-like protein, responded specifically to Agrobacterium infection. The authors speculated that this nodulin gene may respond to signals from the bacterium to regulate plant cell division or differentiation.
Our laboratory has conducted a similar study, using tobacco BY-2 suspension cell cultures inoculated with five different non-tumorigenic Agrobacterium strains (Veena, H. Jiang, R. W. Doerge, and S. B. Gelvin, submitted for publication). One strain could transfer T-DNA but not VirE2 protein, one could transfer virulence proteins but not T-DNA, one could transfer neither, and two could transfer both. Using suppressive subtractive hybridization followed by DNA and RNA macroarray analyses of RNA samples from eight different time points following inoculation (from 0 to 36 h), we identified more than 400 genes that were differentially regulated after various periods of infection. Most of these genes showed a general differential response to Agrobacterium inoculation; however, some genes responded specifically to a T-DNA and Vir protein transfer-competent strain. A few genes responded specifically to T-DNA or Vir protein transfer only. Among the genes that were induced (or whose expression was maintained at a high level) were those encoding histones and ribosomal proteins. The activity of several plant defense and stress response genes was repressed by Agrobacterium infection. Because of the importance of histones in the T-DNA integration process (230), we propose that Agrobacterium infection induces the expression of plant genes necessary for transformation while simultaneously repressing the host defense response. Further analysis of these differentially expressed genes will indicate whether they play a direct or indirect role in Agrobacterium-mediated plant transformation.
PROSPECTS
In less than 20 years, the use of Agrobacterium to genetically transform plants has advanced from a dream to a reality. Modern agricultural biotechnology is heavily dependent on using Agrobacterium to create transgenic plants, and it is difficult to think of an area of plant science research that has not benefited from this technology. However, there remain many challenges. Many economically important plant species, or elite varieties of particular species, remain highly recalcitrant to Agrobacterium-mediated transformation, and the day has not yet arrived when flowers will be the only things seen coming from the barrels of gene guns. However, I feel that such a day is not too far in the distant future. I also feel that Agrobacterium evolved millions of years ago to genetically transform a very wide range of organisms; it is now up to the scientist to harness the natural ability of this bacterium. In addition to extending the host range and transformation efficiency of plants by Agrobacterium, some of the remaining challenges to the scientific biotechnology community are summarized below.
(i) The first is the use of Agrobacterium for homologous or site-directed recombination. Many scientists consider homologous recombination to be one of the remaining “holy grails” of plant molecular biology. The ability to perform gene replacement experiments has become a staple of bacterial, fungal, and even animal cell and molecular biology research. However, homologous recombination in plants generally occurs at 10−5 the frequency of illegitimate recombination (71, 173, 223, 237, 238, 269, 270). We need an Agrobacterium-mediated transformation system that delivers T-DNA to the plant nucleus efficiently, but is deficient in random T-DNA integration.
(ii) The second involves stable and predictable transgene expression in plants. Too often, the level of transgene expression in plants is highly variable. Often, lines of transgenic plants that are “good expressers” lose this characteristic after several generations of growth under field conditions. We need to understand the roles of position effects, chromatin effects, and T-DNA integration patterns in transcriptional and posttranscriptional gene silencing in order to develop strategies to enhance the extent and stability of transgene expression.
(iii) The third is manipulation of the Agrobacterium genome. The availability of the complete A. tumefaciens C58 genomic sequence (114, 363) presents us with an unparalleled opportunity to investigate Agrobacterium gene expression patterns and the ways in which they may be altered during cocultivation of the bacterium with various plant species. Such information may provide clues to methods to further manipulate Agrobacterium in order to effect higher levels of transformation of recalcitrant plant species.
(iv) The fourth is plastid genetic transformation by Agrobacterium. Although a few scattered references to chloroplast transformation by Agrobacterium exist in the literature (see, e.g., references 64 and 346), these reports have not been confirmed by the scientific community. The existence of NLS sequences in VirD2 and VirE2 proteins may ensure T-DNA targeting to the nucleus. Even if these NLS sequences could be removed without altering other essential functions of these proteins, the recent finding that the plant actin cytoskeleton is involved in Agrobacterium-mediated transformation (P. Rao and S. B. Gelvin, unpublished data) may preclude redirection of the T-DNA from the nucleus to plastids.
(v) The fifth is genetic transformation of animal and plant pathogenic fungi. Many medically or agronomically important pathogenic fungi remain highly recalcitrant to genetic transformation. Recent reports of Agrobacterium-mediated transformation of several filamentous fungal species (1, 71) suggest that Agrobacterium may be a useful “gene-jockeying tool” for more than just plant species.
(vi) The final challenge involves genetic transformation of human and animal cells. The recent report of Agrobacterium-mediated genetic transformation of human cells (187) suggests the exciting possibility of using Agrobacterium, or Agrobacterium-like processes, for human and animal gene therapy.
Acknowledgments
I thank Paula Olhoft, Walt Ream, Olivier Voinnet, and Kan Wang for sharing data prior to publication.
Work in my laboratory is funded by the National Science Foundation, the U.S. Department of Agriculture, the Biotechnology Research and Development Corporation, the Corporation for Plant Biotechnology Research, and Pioneer Hi-bred.
REFERENCES
- 1.Abuodeh, R. O., M. J. Orbach, M. A. Mandel, A. Das, and J. N. Galgiani. 2000. Genetic transformation of Coccidioides immitis facilitated by Agrobacterium tumefaciens. J. Infect. Dis. 181:2106-2110. [DOI] [PubMed] [Google Scholar]
- 2.Albert, H., E. C. Dale, E. Lee, and D. W. Ow. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649-659. [DOI] [PubMed] [Google Scholar]
- 3.Albright, L. M., E. Huala, and F. M. Ausubel. 1989. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu. Rev. Genet. 23:311-336. [DOI] [PubMed] [Google Scholar]
- 4.Al-Kaff, N. S., and S. N. Covey. 1996. Unusual accumulations of cauliflower mosaic virus in local lesions, dark green leaf tissue and roots of infected plants. Mol. Plant-Microbe Interact. 9:357-363. [Google Scholar]
- 5.Allen, G. C., G. E. Hall, L. C. Childs, A. K. Weissinger, S. Spiker, and W. F. Thompson. 1993. Scaffold attachment regions increase reporter gene expression in stably transformed plant cells. Plant Cell 5:603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen, G. C., G. E. Hall, S. Michalowski, W. Newman, S. Spiker, A. K. Weissinger, and W. F. Thompson. 1996. High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell 8:899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen, G. C., S. Spiker, and W. F. Thompson. 2000. Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol. Biol. 43:361-376. [DOI] [PubMed] [Google Scholar]
- 8.Alt-Moerbe, J., P. Neddermann, J. von Lintig, and J. Schroder. 1988. Temperature-sensitive step in Ti plasmid vir-region induction and correlation with cytokinin secretion by Agrobacteria. Mol. Gen. Genet. 213:1-8. [Google Scholar]
- 9.An, G. 1985. High efficiency transformation of cultured tobacco cells. Plant Physiol. 79:568-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An, G., B. D. Watson, S. Stachel, M. P. Gordon, and E. W. Nester. 1985. New cloning vehicles for transformation of higher plants. EMBO J. 4:277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson, A., and L. Moore. 1979. Host specificity in the genus Agrobacterium. Phytopathology 69:320-323. [Google Scholar]
- 13.Ankenbauer, R. G., E. A. Best, C. A. Palanca, and E. W. Nester. 1991. Mutants of the Agrobacterium tumefaciens virA gene exhibiting acetosyringone-independent expression of the vir region. Mol. Plant-Microbe Interact. 4:400-406. [DOI] [PubMed] [Google Scholar]
- 14.Aoyama, T. A., and N.-H. Chua. 1997. A glucocorticoid-mediated transcriptional induction system for transgenic plants. Plant J. 11:605-612. [DOI] [PubMed] [Google Scholar]
- 15.Bailey, M. A., H. R. Boerma, and W. A. Parrott. 1994. Inheritance of Agrobacterium tumefaciens-induced tumorigenesis of soybean. Crop Sci. 34:514-519. [Google Scholar]
- 16.Ballas, N., and V. Citovsky. 1997. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA 94:10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker, R. F., K. B. Idler, D. V. Thompson, and J. D. Kemp. 1983. Nucleotide sequence of the T-DNA region from the Agrobacterium tumefaciens octopine Ti-plasmid pTi15955. Plant Mol. Biol. 2:335-350. [DOI] [PubMed] [Google Scholar]
- 18.Baron, C., N. Domke, M. Beinhofer, and S. Hapfelmeier. 2001. Elevated temperature differentially affects virulence, VirB protein accumlation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 183:6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayley, C. C., M. Morgan, E. C. Dale, and D. W. Ow. 1992. Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol. Biol. 18:353-361. [DOI] [PubMed] [Google Scholar]
- 20.Becker, D. 1990. Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18:203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beclin, C., R. Berthome, J. C. Palaqui, M. Tepfer, and H. Vaucheret. 1998. Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252:313-317. [DOI] [PubMed] [Google Scholar]
- 22.Beilmann, A., K. Albrecht, S. Shultze, G. Wanner, and U. M. Pfitzner. 1992. Activation of a truncated PR-1 promoter by endogenous enhancers in transgenic plants. Plant Mol. Biol. 18:65-78. [DOI] [PubMed] [Google Scholar]
- 23.Belanger, C., M. L. Canfield, L. W. Moore, and P. Dion. 1995. Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors. J. Bacteriol. 177:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benfey, P. N., L. Ren, and N.-H. Chua. 1989. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8:2195-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bevan, M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12:8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharyya, M., B. A. Stermer, and R. A. Dixon. 1994. Reduced variation in transgene expression from a binary vector with selectable markers at the right and left T-DNA borders. Plant J. 6:957-968. [Google Scholar]
- 27.Binns, A. N., C. E. Beaupre, and E. M. Dale. 1995. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J. Bacteriol. 177:4890-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolton, G. W., E. W. Nester, and M. P. Gordon. 1986. Plant phenolic compounds induce expression of the Agrobacterium tumefaciens loci needed for virulence. Science 232:983-985. [DOI] [PubMed] [Google Scholar]
- 29.Bonifer, C., N. Yannoutosos, G. Kruger, F. Grosveld, and A. E. Sippel. 1994. Dissection of the locus control function located on the chicken lysozyme gene domain in transgenic mice. Nucleic Acids Res. 22:4202-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun, A. C. 1947. Thermal studies on the factors responsible for tumor initiation in crown gall. Am. J. Bot. 34:234-240. [PubMed] [Google Scholar]
- 31.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Bundock, P., A. den Dulk-Ras, A. Beijersbergen, and P. J. J. Hooykaas. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14:3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bundock, P., and P. J. J. Hooykaas. 1996. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl. Acad. Sci. USA 93:15272-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne, M. C., J. Koplow, C. David, J. Tempe, and M.-D. Chilton. 1983. Structure of T-DNA in roots transformed by Agrobacterium rhizogenes. J. Mol. Appl. Genet. 2:201-209. [PubMed] [Google Scholar]
- 35.Campisi, L., Y. Z. Yang, Y. Yi, E. Heilig, B. Herman, A. J. Cassissta, D. W. Allen, H. J. Xiang, and T. Jack. 1999. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17:699-707. [DOI] [PubMed] [Google Scholar]
- 36.Cangelosi, G. A., L. Hung, V. Puvanesarajah, G. Stacey, D. A. Ozga, J. A. Leigh, and E. W. Nester. 1987. Common loci for Agrobacterium tumefaciens and Rhizobium meliloti exopolysaccharide synthesis and their roles in plant interactions. J. Bacteriol. 169:2086-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cangelosi, G. A., G. Martinetti, J. A. Leigh, C. C. Lee, C. Theines, and E. W. Nester. 1989. Role of Agrobacterium tumefaciens chvA protein in export of β-1,2-glucan. J. Bacteriol. 171:1609-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrington, J. C., and S. A. Whitham. 1998. Viral invasion and host defense: Strategies and counter-strategies. Curr. Opin. Plant Biol. 1:336-341. [DOI] [PubMed] [Google Scholar]
- 39.Chan, M.-T., H.-H. Chang, S.-L. Ho, W.-F. Tong, and S.-M. Yu. 1993. Agrobacterium-mediated production of transgenic rice plants expressing a chimeric alpha-amylase promoter/beta-glucuronidase gene. Plant Mol. Biol. 22:491-506. [DOI] [PubMed] [Google Scholar]
- 40.Chan, M.-T., T.-M. Lee, and H.-H. Chang. 1992. Transformation of Indica rice (Oryza sativa L.) mediated by Agrobacterium tumefaciens. Plant Cell Physiol. 33:577. [Google Scholar]
- 41.Chen, C.-Y., L. Wang, and S. C. Winans. 1991. Characterization of the supervirulent virG gene of the Agrobacterium tumefaciens plasmid pTiBo542. Mol. Gen. Genet. 230:302-309. [DOI] [PubMed] [Google Scholar]
- 42.Cheng, M., J. E. Fry, S. Pang, H. Zhou, C. M. Hironaka, D. R. Duncan, T. W. Conner, and Y. Wan. 1997. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chilton, M.-D., M. H. Drummond, D. J. Merlo, D. Sciaky, A. L. Montoya, M. P. Gordon, and E. W. Nester. 1977. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11:263-271. [DOI] [PubMed] [Google Scholar]
- 44.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christie, P. J., J. E. Ward, S. C. Winans, and E. W. Nester. 1988. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J. Bacteriol. 170:2659-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Citovsky, V., G. De Vos, and P. Zambryski. 1988. Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science 240:501-504. [DOI] [PubMed] [Google Scholar]
- 47.Citovsky, V., B. Guralnick, M. N. Simon, and J. S. Wall. 1997. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J. Mol. Biol. 271:718-727. [DOI] [PubMed] [Google Scholar]
- 48.Citovsky, V., D. Warnick, and P. Zambryski. 1994. Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl. Acad. Sci. USA 91:3210-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Citovsky, V., M. L. Wong, and P. Zambryski. 1989. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. USA 86:1193-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Citovsky, V., J. Zupan, D. Warnick, and P. Zambryski. 1992. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256:1802-1805. [DOI] [PubMed] [Google Scholar]
- 51.Cluster, P. D., M. O'Dell, M. Metzlaff, and R. B. Flavell. 1996. Details of T-DNA structural organization from a transgenic Petunia population exhibiting co-suppression. Plant Mol. Biol. 32:1197-1203. [DOI] [PubMed] [Google Scholar]
- 52.Cooley, M. B., A. P. Goldsbrough, D. W. Still, and J. I. Yoder. 1996. Site-selected insertional mutagenesis of tomato with maize Ac and Ds elements. Mol. Gen. Genet. 252:184-194. [PubMed] [Google Scholar]
- 53.Costantino, P., P. J. J. Hooykaas, H. den Dulk-Ras, and R. A. Schilperoort. 1980. Tumor formation and rhizogenicity of Agrobacterium rhizogenes carrying Ti plasmids. Gene 11:79-87. [DOI] [PubMed] [Google Scholar]
- 54.Covey, S. N., and N. S. Al-Kaff. 2000. Plant DNA viruses and gene silencing. Plant Mol. Biol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 55.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner. 1997. Plants combat infection by gene silencing. Nature 38:781-182. [Google Scholar]
- 56.Cui, Y., Y.-M. Bi, N. Brugiere, M. Arnoldo, and S. J. Sothstein. 2000. The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc. Natl. Acad. Sci. USA 97:373-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dale, E. C., and D. W. Ow. 1990. Intra and inter-molecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene 91:79-85. [DOI] [PubMed] [Google Scholar]
- 58.Daley, M., V. C. Knauf, K. R. Summerfelt, and J. C. Turner. 1998. Co-transformation with one Agrobacterium tumefaciens strain containing two binary plasmids as a method for producing marker-free transgenic plants. Plant Cell Rep. 17:489-496. [DOI] [PubMed] [Google Scholar]
- 59.Das, A., and G. J. Pazour. 1989. Delineation of the regulatory region sequences of Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 17:4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das, A., S. Stachel, P. Ebert, P. Allenza, A. Montoya, and E. Nester. 1986. Promoters of Agrobacterium tumefaciens Ti-plasmid virulence genes. Nucleic Acids Res. 14:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Day, C. D., E. Lee, T. Kobayashi, L. D. Holappa, H. Albert, and D. W. Ow. 2000. Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14:2869-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day, M. J. D., J. L. Ashurst, and R. A. Dixon. 1994. Plant expression cassettes for enhanced translational efficiency. Plant Mol. Biol. Rep. 12:347-357. [Google Scholar]
- 63.DeBeuckeleer, M., M. Lemmers, G. DeVos, L. Willmitzer, M. Van Montagu, and J. Schell. 1981. Further insight on the transferred-DNA of octopine crown gall. Mol. Gen. Genet. 183:283-288. [DOI] [PubMed] [Google Scholar]
- 64.DeBlock, M., J. Schell, and M. Van Montagu. 1985. Chloroplast transformation by Agrobacterium tumefaciens. EMBO J. 4:1367-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Buck, S., C. De Wilde, M. Van Montagu, and A. Depicker. 2000. Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol. Plant-Microbe Interact. 13:658-665. [DOI] [PubMed] [Google Scholar]
- 66.De Buck, S., A. Jacobs, M. Van Montagu, and A. Depicker. 1998. Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol. Plant-Microbe Interact. 11:449-457. [DOI] [PubMed] [Google Scholar]
- 67.De Buck, S., A. Jacobs, M. Van Montagu, and A. Depicker. 1999. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 20:295-304. [DOI] [PubMed] [Google Scholar]
- 68.DeCleene, M., and J. DeLey. 1976. The host range of crown gall. Bot. Rev. 42:389-466. [Google Scholar]
- 69.de Frammond, A. J., E. W. Back, W. S. Chilton, L. Kayes, and M.-D. Chilton. 1986. Two unlinked T-DNAs can transform the same tobacco plant cell and segregate in the F1 generation. Mol. Gen. Genet. 202:125-131. [Google Scholar]
- 70.de Frammond, A. J., K. A. Barton, and M.-D. Chilton. 1983. Mini-Ti-: a new vector strategy for plant genetic engineering. Bio/Technology 5:262-269.
- 71.de Groot, M. J. A., P. Bundock, P. J. J. Hooykaas, and A. G. M. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 72.de Groot, M. J. A., R. Offringa, J. Groet, M. P. Does, P. J. J. Hooykaas, and P. J. M. van den Elzen. 1994. Non-recombinant background in gene targeting: Illegitimate recombination between a hpt gene and a defective 5′ deleted nptII gene can restore a Kmr phenotype in tobacco. Plant Mol. Biol. 25:721-733.8061322 [Google Scholar]
- 73.de la Riva, G. A., J. Gonzalez-Cabrera, R. Vazquez-Padron, and C. Ayra-Pardo. 1998. Agrobacterium tumefaciens: a natural tool for plant transformation. Electron. J. Biotechnol. 1:1-16. [Google Scholar]
- 74.Delay, D., J. Cizeau, and F. Delmotte. 1992. Synthesis of aryl glycosides as vir gene inducers of Agrobacterium tumefaciens. Carbohydr. Res. 225:179. [Google Scholar]
- 75.Delmotte, F. M., D. Delay, J. Cizeau, B. Guerin, and J.-C. Leple. 1991. Agrobacterium vir-inducing activities of glycosylated acetosyringone, acetovanillone, syringaldehyde and syringic acid derivatives. Phytochemistry 30:3549-3552. [Google Scholar]
- 76.De Neve, M., S. De Buck, A. Jacobs, M. Van Montagu, and A. Depicker. 1997. T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J. 11:15-29. [DOI] [PubMed] [Google Scholar]
- 77.Deng, W., L. Chen, W.-T. Peng, X. Liang, S. Sekiguchi, M. P. Gordon, L. Comai, and E. W. Nester. 1999. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 31:1795-1807. [DOI] [PubMed] [Google Scholar]
- 78.Deng, W., L. Chen, D. W. Wood, T. Metcalfe, X. Liang, M. P. Gordon, L. Comai, and E. W. Nester. 1998. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl. Acad. Sci. USA 95:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng, W., X.-A. Pu, R. N. Goodman, M. P. Gordon, and E. W. Nester. 1995. T-DNA genes responsible for inducing a necrotic response on grape vines. Mol. Plant-Microbe Interact. 8:538-548. [DOI] [PubMed] [Google Scholar]
- 80.Depicker, A., L. Herman, A. Jacobs, J. Schell, and M. Van Montagu. 1985. Frequencies of simultaneous transformation with different T-DNAs and their relevance to the Agrobacterium/plant cell interaction. Mol. Gen. Genet. 201:477-484. [Google Scholar]
- 81.De Vos, G., M. DeBeuckeleer, M. Van Montagu, and J. Schell. 1981. Restriction endonuclease mapping of the octopine tumor-inducing plasmid pTiAch5 of Agrobacterium tumefaciens—addendum. Plasmid 6:249-253. [DOI] [PubMed] [Google Scholar]
- 82.De Vos, G., and P. Zambryski. 1989. Expression of Agrobacterium nopaline-specific VirD1, VirD2, and VirC1 proteins and their requirement for T-strand production in E. coli. Mol. Plant-Microbe Interact. 2:43-52. [DOI] [PubMed] [Google Scholar]
- 83.Ditt, R. F., E. W. Nester, and L. Comai. 2001. Plant gene expression response to Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 98:10954-10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dombek, P., and W. Ream. 1997. Functional domains of Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J. Bacteriol. 179:1165-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong, J., W. Teng, W. G. Buchholz, and T. C. Hall. 1996. Agrobacterium-mediated transformation of Javanica rice. Mol. Breeding 2:267-276. [Google Scholar]
- 86.Doty, S. L., M. Chang, and E. W. Nester. 1993. The chromosomal virulence gene, chvE, of Agrobacterium tumefaciens is regulated by a LysR family member. J. Bacteriol. 175:7880-7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doty, S. L., M. C. Yu, J. I. Lundin, J. D. Heath, and E. W. Nester. 1996. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J. Bacteriol. 178:961-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Douglas, C., W. Halperin, M. Gordon, and E. Nester. 1985. Specific attachment of Agrobacterium tumefaciens to bamboo cells in suspension cultures. J. Bacteriol. 161:764-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Douglas, C. J., W. Halperin, and E. W. Nester. 1982. Agrobacterium tumefaciens mutants affected in attachment to plant cells. J. Bacteriol. 152:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dumas, F., M. Duckely, P. Pelczar, P. Van Gelder, and B. Hohn. 2001. An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl. Acad. Sci. USA 98:485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Durrenberger, F., A. Crameri, B. Hohn, and Z. Koukolikova-Nicola. 1989. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl. Acad. Sci. USA 86:9154-9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dye, F., K. Berthelot, B. Griffon, D. Delay, and F. M. Delmotte. 1997. Alkylsyringamides, new inducers of Agrobacterium tumefaciens virulence genes. Biochimie 79:3-6. [DOI] [PubMed] [Google Scholar]
- 93.Ebinuma, H., K. Sugita, E. Matsunaga, and M. Yamakado. 1997. Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc. Natl. Acad. Sci. USA 94:2117-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eisenbrandt, R., M. Kalkum, E.-M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 95.Elmayan, T., and H. Vaucheret. 1996. Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J 9:787-797. [Google Scholar]
- 96.Enriquez-Obregon G. A., D. L. Prieto-Samsonov, G. A. de la Riva, M. Perez, G. Selman-Housein, and R. I. Vazquez-Padron. 1999. Agrobacterium-mediated Japonica rice transformation: a procedure assisted by an anti-necrotic treatment. Plant Cell Tissue Organ Cult. 59:159-168. [Google Scholar]
- 97.Escobar, M. A., E. L. Civerolo, K. R. Summerfelt, and A. M. Dandekar. 2001. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 98:13437-13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feldmann, K. A. 1991. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1:71-82. [Google Scholar]
- 99.Filichkin, S. A., and S. B. Gelvin. 1993. Formation of a putative relaxation intermediate during T-DNA processing directed by the Agrobacterium tumefaciens VirD1,D2 endonuclease. Mol. Microbiol. 8:915-926. [DOI] [PubMed] [Google Scholar]
- 100.Fortin, C., C. Marquis, E. W. Nester, and P. Dion. 1993. Dynamic structure of Agrobacterium tumefaciens Ti plasmids. J. Bacteriol. 175:4790-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fraley, R. T., S. G. Rogers, R. B. Horsch, D. A. Eichholtz, J. S. Flick, C. L. Fink, N. L. Hoffmann, and P. R. Sanders. 1985. The SEV system: a new disarmed Ti plasmid vector system for plant transformation. Bio/Technology 3:629-635. [Google Scholar]
- 102.Frame, B., H. Shou, R. Chikwamba, Z. Zhang, C. Xiang, T. Fonger, S. E. Pegg, B. Li, D. Nettleton, D. Pei, and K. Wang. 2002. Agrobacterium-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frary, A., and C. M. Hamilton. 2001. Efficiency and stability of high molecular weight DNA transformation: an analysis in tomato. Transgenic Res. 10:121-132. [DOI] [PubMed] [Google Scholar]
- 104.Frey, M., C. Stettner, and A. Gierl. 1998. A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J. 13:717-721. [Google Scholar]
- 105.Fullner, K. J., and E. W. Nester. 1996. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J. Bacteriol. 178:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garfinkel, D. J., and E. W. Nester. 1980. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J. Bacteriol. 144:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 108.Gatz, C., and P. H. Quail. 1988. Tn10-encoded tet repressor can regulate an operator-containing plant promoter. Proc. Natl. Acad. Sci. USA 85:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gelvin, S. B. 2000. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:223-256. [DOI] [PubMed]
- 110.Gelvin, S. B. 1998. Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol. 180:4300-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gerard, J.-C., J. Canaday, E. Szegedi, H. de la Salle, and L. Otten. 1992. Physical map of the vitopine Ti plasmid pTiS4. Plasmid 28:146-156. [DOI] [PubMed] [Google Scholar]
- 112.Gietl, C., Z. Koukolikova-Nicola, and B. Hohn. 1987. Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc. Natl. Acad. Sci. USA 84:9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gleave, A. P. 1992. A versitile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20:1203-1207. [DOI] [PubMed] [Google Scholar]
- 114.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 115.Goodner, B. W., B. P. Markelz, M. C. Flanagan, C. B. Crowell, J. L. Racette, A. Schilling, L. M. Halfon, J. S. Mellors, and G. Grabowski. 1999. Combined genetic and physical map of the complex genome of Agrobacterium tumefaciens. J. Bacteriol. 181:5160-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gray, J., J. Wang, and S. B. Gelvin. 1992. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J. Bacteriol. 174:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grevelding, C., V. Fantes, E. Kemper, J. Schell, and R. Masterson. 1993. Single-copy T-DNA insertions in Arabidopsis are the predominant form of integration in root-derived transgenics, whereas multiple insertions are found in leaf discs. Plant Mol. Biol. 23:847-860. [DOI] [PubMed] [Google Scholar]
- 118.Gubba, S., Y.-H. Xie, and A. Das. 1995. Regulation of Agrobacterium tumefaciens virulence gene expression: isolation of a mutation that restores virGD52E function. Mol. Plant-Microbe Interact. 8:788-791. [DOI] [PubMed] [Google Scholar]
- 119.Guralnick, B., G. Thomsen, and V. Citovsky. 1996. Transport of DNA into the nuclei of Xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell 8:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hajdukiewicz, P., Z. Svab, and P. Maliga. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25:989-994. [DOI] [PubMed] [Google Scholar]
- 121.Hall, G. E., G. C. Allen, D. S. Loer, W. F. Thompson, and S. Spiker. 1991. Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc. Natl. Acad. Sci. USA 88:932-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hall, G. E., and S. Spiker. 1994. Isolation and characterization of nuclear scaffolds, p. 1-12. In S. B. Gelvin and R. A. Schilperoort (ed.), Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 123.Hamilton, C. M. 1997. A binary-BAC system for plant transformation with high-molecular weight DNA. Gene 200:107-116. [DOI] [PubMed] [Google Scholar]
- 124.Hamilton, C. M., A. Frary, C. Lewis, and S. D. Tanksley. 1996. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. USA 93:9975-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamilton, C. M., A. Frary, Y. Xu, S. D. Tanksley, and H.-B. Zhang. 1999. Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant J. 18:223-229. [Google Scholar]
- 126.Hamilton, C. M., H. Lee, P.-L. Li, D. M. Cook, K. R. Piper, S. D. von Bodman, E. Lanka, W. Ream, and S. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Han, D. C., C.-Y. Chen, Y.-F. Chen, and S. C. Winans. 1992. Altered-function mutations of the transcriptional regulatory gene virG of Agrobacterium tumefaciens. J. Bacteriol. 174:7040-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Han, K. H., C. P. Ma, and S. H. Strauss. 1997. Matrix attachment regions (MARs) enhance transformation frequency and transgene expression in poplar. Transgenic Res. 6:415-420. [Google Scholar]
- 129.Hansen, G. 2000. Evidence for Agrobacterium-induced apoptosis in maize cells. Mol. Plant-Microbe Interact. 13:649-657. [DOI] [PubMed] [Google Scholar]
- 130.Hansen, G., A. Das, and M.-D. Chilton. 1994. Constitutive expression of the virulence genes improves the efficiency of plant transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA 91:7603-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hansen, G., J. Tempe, and J. Brevet. 1992. A T-DNA transfer stimulator sequence in the vicinity of the right border of pRi8196. Plant Mol. Biol. 20:113-122. [DOI] [PubMed] [Google Scholar]
- 132.Hanson, B., D. Engler, Y. Moy, B. Newman, E. Ralston, and N. Gutterson. 1999. A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. Plant J. 19:727-734. [DOI] [PubMed] [Google Scholar]
- 133.Harpster, M. H., J. A. Townsend, J. D. G. Jones, J. Bedbrook, and P. Dunsmuir. 1988. Relative strengths of the 35S cauliflower mosaic virus, 1′, 2′, and nopaline synthase promoters in transformed tobacco, sugarbeet, and oilseed rape callus tissue. Mol. Gen. Genet. 212:182-190. [DOI] [PubMed] [Google Scholar]
- 134.Hellens, R., P. Mullineaux, and H. Klee. 2000. A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5:446-451. [DOI] [PubMed] [Google Scholar]
- 135.Hepburn, A. G., L. E. Clarke, L. Pearson, and J. White. 1983. The role of cytosine methylation in the control of nopaline synthase gene expression in a plant tumor. J. Mol. Appl. Genet. 2:315-329. [PubMed] [Google Scholar]
- 136.Hepburn, A. G., and J. White. 1985. The effect of right terminal repeat deletion on the oncogenicity of the T-region of pTiT37. Plant Mol. Biol. 5:3-11. [DOI] [PubMed] [Google Scholar]
- 137.Herrera-Estrella, A., Z.-M. Chen, M. Van Montagu, and K. Wang. 1988. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 7:4055-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Herrera-Estrella, A., M. Van Montagu, and K. Wang. 1990. A bacterial peptide acting as a plant nuclear targeting signal: The amino-terminal portion of Agrobacterium VirD2 protein directs a β-galactosidase fusion protein into tobacco nuclei. Proc. Natl. Acad. Sci. USA 87:9534-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hiei, Y., S. Ohta, T. Komari, and T. Kumashiro. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6:271-282. [DOI] [PubMed] [Google Scholar]
- 140.Hoekema, A., P. R. Hirsh, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 141.Hoekema, A., P. W. Roelvink, P. J. J. Hooykaas, and R. A. Schilperoort. 1984. Delivery of T-DNA from the Agrobacterium tumefaciens chromosome into plant cells. EMBO J. 3:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hohn, B., A. A. Levy, and H. Puchta. 2001. Elimination of selection markers from transgenic plants. Curr. Opin. Plant Biotechnol. 12:139-143. [DOI] [PubMed] [Google Scholar]
- 143.Hood, E. E., R. T. Fraley, and M.-D. Chilton. 1987. Virulence of Agrobacterium tumefaciens strain A281 on legumes. Plant Physiol. 83:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hood, E. E., S. B. Gelvin, L. S. Melchers, and A. Hoekema. 1993. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2:208-218. [Google Scholar]
- 145.Hood, E. E., G. Jen, L. Kayes, J. Kramer, R. T. Fraley, and M.-D. Chilton. 1984. Restriction endonuclease map of pTiBo542, a potential Ti-plasmid vector for genetic engineering of plants. Bio/Technology 2:702-709. [Google Scholar]
- 146.Hood, E. E., G. L. Helmer, R. T. Fraley, and M.-D. Chilton. 1986. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 168:1291-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hooykaas, P. J. J., M. Hofker, H. den Dulk-Ras, and R. A. Schilperoort. 1984. A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid 11:195-205. [DOI] [PubMed] [Google Scholar]
- 148.Horsch, R. B., H. J. Klee, S. Stachel, S. C. Winans, E. W. Nester, S. G. Rogers, and R. T. Fraley. 1986. Analysis of Agrobacterium tumefaciens virulence mutants in leaf disks. Proc. Natl. Acad. Sci. USA 83:2571-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Howard, E., and V. Citovsky. 1990. The emerging structure of the Agrobacterium T-DNA transfer complex. Bioessays 12:103-108. [Google Scholar]
- 150.Howard, E. A., B. A. Winsor, G. De Vos, and P. Zambryski. 1989. Activation of the T-DNA transfer process in Agrobacterium results in the generation of a T-strand-protein complex: tight association of VirD2 with the 5′ ends of T-strands. Proc. Natl. Acad. Sci. USA 86:4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Howard, E. A., J. R. Zupan, V. Citovsky, and P. C. Zambryski. 1992. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68:109-118. [DOI] [PubMed] [Google Scholar]
- 152.Ishida, Y., H. Saito, S. Ohta, Y. Hiei, T. Komari, and T. Kumashiro. 1996. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14:745-750. [DOI] [PubMed] [Google Scholar]
- 153.Jarchow, E., N. H. Grimsley, and B. Hohn. 1991. virF, the host-range-determining virulence gene of Agrobacterium tumefaciens, affects T-DNA transfer to Zea mays. Proc. Natl. Acad. Sci. USA 88:10426-10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jasper, F., C. Koncz, J. Schell, and H.-H. Steinbiss. 1994. Agrobacterium T-strand production in vitro: sequence-specific cleavage and 5′ protection of single-stranded DNA templates by purified VirD2 protein. Proc. Natl. Acad. Sci. USA 91:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jayaswal, R. K., K. Veluthambi, S. B. Gelvin, and J. L. Slightom. 1987. Double-stranded cleavage of T-DNA and generation of single-stranded T-DNA molecules in Escherichia coli by a virD-encoded border-specific endonuclease from Agrobacterium tumefaciens. J. Bacteriol. 169:5035-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jen, G. C., and M.-D. Chilton. 1986. The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Proc. Natl. Acad. Sci. USA 83:3895-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jeon, J.-S., S. Lee, K.-H. Jung, S.-H. Jun, D.-H. Jeong, J. Lee, C. Kim, S. Jang, S. Lee, K. Yang, J. Nam, K. An, M.-H. Han, R.-J. Sung, H.-S. Choi, J.-H. Yu, J.-H. Choi, S.-S. Cha, S.-I. Kim, and G. An. 2000. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22:561-570. [DOI] [PubMed] [Google Scholar]
- 158.Jepson, I., A. Martinez, and J. P. Sweetman. 1998. Chemical-inducible gene expression systems for plants—a review. Pestic. Sci. 54:360-367. [Google Scholar]
- 159.Jin, S., T. Komari, M. P. Gordon, and E. W. Nester. 1987. Genes responsible for the supervirulence phenotype of Agrobacterium tumefaciens A281. J. Bacteriol. 169:4417-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jin, S., R. K. Prusti, T. Roitsch, R. G. Ankenbauer, and E. W. Nester. 1990. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J. Bacteriol. 172:4945-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jin, S., T. Roitsch, R. G. Ankenbauer, M. P. Gordon, and E. W. Nester. 1990. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J. Bacteriol. 172:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jin, S., Y.-N. Song, W.-Y. Deng, M. P. Gordon, and E. W. Nester. 1993. The regulatory VirA protein of Agrobacterium tumefaciens does not function at elevated temperatures. J. Bacteriol. 175:6830-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jones, A. L., E.-M. Lai, K. Shirasu, and C. I. Kado. 1996. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jorgensen, R. A., P. D. Cluster, J. English, Q. Que, and C. A. Napoli. 1996. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 31:957-973. [DOI] [PubMed] [Google Scholar]
- 165.Jorgensen, R., C. Snyder, and J. D. G. Jones. 1987. T-DNA is organized predominantly in inverted repeat structures in plants transformed with Agrobacterium tumefaciens C58 derivatives. Mol. Gen. Genet. 207:471-477. [Google Scholar]
- 166.Jouanin, L. 1984. Restriction map of an agropine-type Ri-plasmid and its homologies with Ti-plasmids. Plasmid 12:91-102. [DOI] [PubMed] [Google Scholar]
- 167.Jouanin, L., D. Bouchez, R. F. Drong, D. Tepfer, and J. L. Slightom. 1989. Analysis of TR-DNA/plant junctions in the genome of a Convolvulus arvensis clone transformed by Agrobacterum rhizogenes strain A4. Plant Mol. Biol. 12:75-85. [DOI] [PubMed] [Google Scholar]
- 168.Kalogeraki, V. S., and S. C. Winans. 1995. The octopine-type Ti plasmid pTiA6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J. Bacteriol. 177:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 170.Ke, J., R. Khan, T. Johnson, D. A. Somers, and A. Das. 2001. High-efficiency gene transfer to recalcitrant plants by Agrobacterium tumefaciens. Plant Cell Rep. 20:150-156. [DOI] [PubMed] [Google Scholar]
- 171.Keane, P. J., A. Kerr, and R. B. New. 1970. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23:585-595. [Google Scholar]
- 172.Kemner, J. M., X. Liang, and E. W. Nester. 1997. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 179:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kempin, S. A., S. J. Liljegren, L. M. Block, S. D. Roundsley, M. F. Yanofsky, and E. Lam. 1997. Targeted disruption in Arabidopsis. Nature 389:802-803. [DOI] [PubMed] [Google Scholar]
- 174.Klee, H. J., F. F. White, V. N. Iyer, M. P. Gordon, and E. W. Nester. 1983. Mutational analysis of the virulence region of an Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 153:878-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Knauf, V., M. Yanofsky, A. Montoya, and E. Nester. 1984. Physical and functional map of an Agrobacterium tumefaciens tumor inducing plasmid that confers a narrow host range. J. Bacteriol. 160:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kohli, A., D. Gahakwa, P. Vain, D. A. Laurie, and P. Christou. 1999. Transgene expression in rice engineered through particle bombardment: molecular factors controlling stable expression and transgene silencing. Planta 208:88-97. [Google Scholar]
- 177.Komari, T., W. Halperin, and E. W. Nester. 1986. Physical and functional map of supervirulent Agrobacterium tumefaciens tumor-inducing plasmid pTiBo542. J. Bacteriol. 166:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Komari, T., Y. Hiei, Y. Saito, N. Murai, and T. Kumasiashiro. 1996. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10:165-174. [DOI] [PubMed] [Google Scholar]
- 179.Koncz, C., H. DeGreve, D. Andre, F. Deboeck, M. Van Montagu, and J. Schell. 1983. The opine synthase genes carried by Ti plasmids contain all signals necessary for expression in plants. EMBO J. 2:1597-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koncz, C., N. Martini, R. Mayerhofer, Z. Koncz-Kalman, H. Korber, G. P. Redei, and J. Schell. 1989. High-frequency T-DNA-mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86:8467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koncz, C., and J. Schell. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204:383-396. [Google Scholar]
- 182.Kononov, M. E., B. Bassuner, and S. B. Gelvin. 1997. Integration of T-DNA binary vector “backbone” sequences into the tobacco genome: evidence for multiple complex pattrns of integration. Plant J. 11:945-957. [DOI] [PubMed] [Google Scholar]
- 183.Krishnamohan, A., V. Balaji, and K. Veluthambi. 2001. Efficient vir gene induction in Agrobacterium tumefaciens requires virA, virG, and vir box from the same Ti plasmid. J. Bacteriol. 183:4079-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Krysan, W., P. J., J. C. Young, and M. R. Sussman. 1999. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11:2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koukolikova-Nicola, Z., and B. Hohn. 1993. How does the T-DNA of Agrobacterium tumefaciens find its way into the plant cell nucleus? Biochimie 75:635-638. [DOI] [PubMed] [Google Scholar]
- 186.Kukolikova-Nicola, Z., D. Raineri, K. Stephens, C. Ramos, B. Tinland, E. Nester, and B. Hohn. 1993. Genetic analysis of the virD operon of Agrobacterium tumefaciens: a search for functions involved in transport of T-DNA into the plant cell nucleus and in T-DNA integration. J. Bacteriol. 175:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kunik, T., T. Tzfira, Y. Kapulnik, Y. Gafni, C. Dingwall, and V. Citovsky. 2001. Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA 98:1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lai, E.-M., O. Chesnokova, L. M. Banta, and C. I. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lai, E.-M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lai, E.-M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 191.Lam, S., B. Lam, L. Harrison, and G. Strobel. 1984. Genetic information of the Ri plasmid of Agrobacterium rhizogenes determines host specificity. Plant Sci. Lett. 34:345-352. [Google Scholar]
- 192.Lazo, G. R., P. A. Stein, and R. A. Ludwig. 1991. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology 9:963-967. [DOI] [PubMed] [Google Scholar]
- 193.Lee, L.-Y., S. B. Gelvin, and C. I. Kado. 1999. pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export. J. Bacteriol. 181:186-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee, L.-Y., J. M. Humara, and S. Gelvin. 2001. Novel constructions to enable the integration of genes into the Agrobacterium tumefaciens C58 chromosome. Mol. Plant-Microbe Interact. 14:577-579. [DOI] [PubMed] [Google Scholar]
- 195.Lee, Y.-W., S. Jin, W.-S. Sim, and E. W. Nester. 1995. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 92:12245-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Leisner, S. M., and S. B. Gelvin. 1989. Multiple domains exist within the upstream activator sequence of the octopine synthase gene. Plant Cell 1:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lemmers, M., M. DeBeuckeleer, M. Holsters, P. Zambryski, A. Depicker, J. P. Hernalsteens, M. Van Montagu, and J. Schell. 1980. Internal organization, boundaries and integration of Ti-plasmid DNA in nopaline crown gall tumours. J. Mol. Biol. 144:353-376. [DOI] [PubMed] [Google Scholar]
- 198.Levee, V., E. Garin, K. Klimaszewska, and A. Seguin. 1999. Stable genetic transformation of white pine (Pinus strobus L.) after cocultivation of embryogenic tisues with Agrobacterium tumefaciens. Mol. Breeding 5:429-440. [Google Scholar]
- 199.Liu, C.-N., X.-Q. Li, and S. B. Gelvin. 1992. Multiple copies of virG enhance the transient transformation of celery, carrot, and rice tissues by Agrobacterium tumefaciens. Plant Mol. Biol. 20:1071-1087. [DOI] [PubMed] [Google Scholar]
- 200.Liu, C.-N., T. R. Steck, L. L. Habeck, J. A. Meyer, and S. B. Gelvin. 1993. Multiple copies of virG allow induction of Agrobacterium tumefaciens vir genes and T-DNA processing at alkaline pH. Mol. Plant-Microbe Interact. 6:144-156. [Google Scholar]
- 201.Liu, J. W., and L. M. Tabe. 1998. The influences of two plant nuclear matrix attachment regions (MARs) on gene expression in transgenic plants. Plant Cell Physiol. 39:115-123. [DOI] [PubMed] [Google Scholar]
- 202.Liu, Y.-G., N. Mitsukawa, T. Oosumi, and R. F. Whittier. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8:457-463. [DOI] [PubMed] [Google Scholar]
- 203.Liu, Y.-G., K. Nagaki, M. Fujita, K. Kawaura, M. Uozumi, and Y. Ohigara. 2000. Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in a transformation-competent artificial chromosome (TAC) vector. Plant J. 23:687-695. [DOI] [PubMed] [Google Scholar]
- 204.Liu, Y.-G., Y. Shirano, H. Fukaki, Y. Yanai, M. Tasaka, S. Tabata, and D. Shibata. 1999. Complementation of plant mutants with large genomic DNA fagments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96:6535-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu, Z., M. Jacobs, D. A. Schaff, C. A. McCullen, and A. N. Binns. 2001. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 183:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Loopstra, C. A., A.-M. Stomp, and R. R. Sederoff. 1990. Agrobacterium-mediated DNA transfer in sugar pine. Plant Mol. Biol. 15:1-9. [DOI] [PubMed] [Google Scholar]
- 207.Loper, J. E., and C. I. Kado. 1979. Host range conferred by the virulence-specifying plasmid of Agrobacterium tumefaciens. J. Bacteriol. 139:591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lundquist, R. C., T. J. Close, and C. I. Kado. 1984. Genetic complementation of Agrobacterium tumefaciens Ti plasmid mutants in the virulence region. Mol. Gen. Genet. 193:1-7. [DOI] [PubMed] [Google Scholar]
- 209.Lyznik, L. A., K. V. Rao, and T. K. Hodges. 1996. FLP-mediated recombination of FRT sites in the maize genome. Nucleic Acids Res. 24:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Marathe, R., R. Anandalakshmi, T. H. Smith, G. J. Pruss, and V. B. Vance. 2000. RNA viruses as inducers, suppressors and targets of post-transcriptional gene silencing. Plant Mol. Biol. 43:295-306. [DOI] [PubMed] [Google Scholar]
- 211.Martineau, B., T. A. Voelker, and R. A. Sanders. 1994. On defining T-DNA. Plant Cell 6:1032-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Matthysse, A. G. 1987. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J. Bacteriol. 169:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mauro, A. O., T. W. Pfeiffer, and G. B. Collins. 1995. Inheritance of soybean susceptibility to Agrobacterium tumefaciens and its relationship to transformation. Crop Sci. 35:1152-1156. [Google Scholar]
- 215.McAfee, B. J., E. E. White, L. E. Pelcher, and M. S. Lapp. 1993. Root induction in pine (Pinus) and larch (Larix) spp. using Agrobacterium rhizogenes. Plant Cell Tissue Organ Cult. 34:53-62. [Google Scholar]
- 216.McBride, K. E., and K. R. Summerfelt. 1990. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14:269-276. [DOI] [PubMed] [Google Scholar]
- 217.McKnight, T. D., M. T. Lillis, and R. B. Simpson. 1987. Segregation of genes transferred to one plant cell from two separate Agrobacterium strains. Plant Mol. Biol. 8:439-445. [DOI] [PubMed] [Google Scholar]
- 218.McLean, B. G., E. A. Greene, and P. C. Zambryski. 1994. Mutants of Agrobacterium VirA that activate the vir gene expression in the absence of the inducer acetosyringone. J. Biol. Chem. 269:2645-2651. [PubMed] [Google Scholar]
- 219.Meins, F. 2000. RNA degradation and models for post-transcriptional gene silencing. Plant Mol. Biol. 43:261-273. [DOI] [PubMed] [Google Scholar]
- 220.Melchers, L. S., M. J. Maroney, A. den Dulk-Ras, D. V. Thompson, H. A. J. van Vuuren, R. A. Schilperoort, and P. J. J. Hooykaas. 1990. Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence: molecular characterization of the virF locus. Plant Mol. Biol. 14:249-259. [DOI] [PubMed] [Google Scholar]
- 221.Mett, V. L., L. P. Lochhead, and P. H. S. Reynolds. 1991. Copper-controllable gene expression system for whole plants. Proc. Natl. Acad. Sci. USA 90:4567-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Meyer, P. 2000. Transcriptional transgene silencing and chromatin components. Plant Mol. Biol. 43:221-234. [DOI] [PubMed] [Google Scholar]
- 223.Miao, Z.-H., and E. Lam. 1995. Targeted disruption of the TGA3 locus in Arabidopsis thaliana. Plant J. 7:359-365. [DOI] [PubMed] [Google Scholar]
- 224.Miranda, A., G. Janssen, L. Hodges, E. G. Peralta, and W. Ream. 1992. Agrobacterium tumefaciens transfers extremely long T-DNAs by a unidirectional mechanism. J. Bacteriol. 174:2288-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mlynarova, L., R. C. Jansen, A. J. Conner, W. J. Stiekema, and J.-P. Nap. 1995. The MAR-mediated reduction in position effect can be uncoupled from copy number-dependent expression in transgenic plants. Plant Cell 7:599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mlynarova, L., L. C. P. Keizer, W. J. Stiekema, and J.-P. Nap. 1996. Approaching the lower limits of transgene variability. Plant Cell 8:1589-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mlynarova, L., A. Loonen, J. Heldens, R. C. Jansen, P. Keizer, W. J. Stiekema, and J.-P. Nap. 1994. Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 6:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Morris, J. W., and R. O. Morris. 1990. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc. Natl. Acad. Sci. USA 87:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mysore, K. S., B. Bassuner, X.-B. Deng, N. S. Darbinian, A. Motchoulski, W. Ream, and S. B. Gelvin. 1998. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant-Microbe Interact. 11:668-683. [DOI] [PubMed] [Google Scholar]
- 230.Mysore, K. S., J. Nam, and S. B. Gelvin. 2000. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 97:948-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Nam, J., A. G. Matthysse, and S. B. Gelvin. 1997. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9:317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nam, J., K. S. Mysore, C. Zheng, M. K. Knue, A. G. Matthysse, and S. B. Gelvin. 1999. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261:429-438. [DOI] [PubMed] [Google Scholar]
- 233.Narasimhulu, S. B., X.-B. Deng, R. Sarria, and S. B. Gelvin. 1996. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8:873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ni, M., D. Cui, J. Einstein, S. Narasimhulu, C. E. Vergara, and S. B. Gelvin. 1995. Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7:661-676. [Google Scholar]
- 235.Odell, J., P. Caimi, B. Sauer, and S. Russell. 1990. Site-directed recombination in the genome of transgenic tobacco. Mol. Gen. Genet. 223:369-378. [DOI] [PubMed] [Google Scholar]
- 236.Odell, J. T., and E. Krebbers. 1998. Enhanced transgene expression in a population of monocot cells employing scaffold attachment regions. World Peanut Office.
- 237.Offringa, R., M. J. A. de Groot, H. J. Haagsman, M. P. Does, P. J. M. van den Elzen, and P. J. J. Hooykaas. 1990. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 9:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Offringa, R., P. J. M. van den Elzen, and P. J. J. Hooykaas. 1992. Gene targeting in plants using the Agrobacterium vector system. Transgenic Res. 1:114-123. [Google Scholar]
- 239.Olhoft, P. M., K. Lin, J. Galbraith, N. C. Nielsen, and D. A. Somers. 2001.. The role of thiol compounds in increasing Agrobacterium-mediated transformation of soybean cotyledonary-node cells. Plant Cell Rep 20:731-737.
- 240.Olhoft, P. M., and D. A. Somers. 2001. L-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary-node cells. Plant Cell Rep. 20:706-711. [Google Scholar]
- 241.Ooms, G., A. Bakker, L. Molendijk, G. J. Wullems, M. P. Gordon, E. W. Nester, and R. A. Schilperoort. 1982. T-DNA organization in homogeneous and heterogeneous octopine-type crown gall tissues of Nicotiana tabacum. Cell 30:589-597. [DOI] [PubMed] [Google Scholar]
- 242.Ooms, G., P. J. J. Hooykaas, G. Moolenaar, and R. A. Schilperoort. 1981. Crown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene 14:33-50. [DOI] [PubMed] [Google Scholar]
- 243.Ophel, K., and A. Kerr. 1990. Agrobacterium vitis sp. nov. for strains of Agrobacterium biovar 3 from grapevines. Int. J. Syst. Bacteriol. 40:236-241. [Google Scholar]
- 244.Otten, L., H. DeGreve, J. Leemans, R. Hain, P. Hooykaas, and J. Schell. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195:159-163. [Google Scholar]
- 245.Otten, L., P. de Ruffray, E. A. Momol, M. T. Momol, and T. J. Burr. 1996. Phylogenetic relationships between Agrobacterium vitis isolates and their Ti plasmids. Mol. Plant-Microbe Interact. 9:782-786. [Google Scholar]
- 246.Owens, L. D., and D. E. Cress. 1984. Genotypic variability of soybean response to Agrobacterium strains harboring the Ti or Ri plasmids. Plant Physiol. 77:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Palanichelvam, K., P. Oger, S. J. Clough, C. Cha, A. F. Bent, and S. K. Farrand. 2000. A second T-region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol. Plant-Microbe Interact. 13:1081-1091. [DOI] [PubMed] [Google Scholar]
- 248.Pan, S. Q., S. Jin, M. I. Boulton, M. Hawes, M. P. Gordon, and E. W. Nester. 1995. An Agrobacterium virulence factor encoded by a Ti plasmid gene or a chromosomal gene is required for T-DNA transfer into plants. Mol. Microbiol. 17:259-269. [DOI] [PubMed] [Google Scholar]
- 249.Pansegrau, W., F. Schoumacher, B. Hohn, and E. Lanka. 1993. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc. Natl. Acad. Sci. USA 90:11538-11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Paulus, F., J. Canaday, and L. Otten. 1991. Limited host range Ti plasmids: Recent origin from wide host range Ti plasmids and involvement of a novel IS element, IS868. Mol. Plant-Microbe Interact. 4:190-197. [DOI] [PubMed] [Google Scholar]
- 251.Paulus, F., B. Huss, G. Bonnard, M. Ride, E. Szegedi, J. Tempe, A. Petit, and L. Otten. 1989. Molecular systematics of biotype III Ti plasmids of Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 2:64-74. [Google Scholar]
- 252.Pazour, G. J., and A. Das. 1990. Characterization of the VirG binding site of Agrobacterium tumefaciens. Nucleic Acids Res. 18:6909-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pazour, G. J., C. N. Ta, and A. Das. 1991. Mutants of Agrobacterium tumefaciens with elevated vir gene expression. Proc. Natl. Acad. Sci. USA 88:6941-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pazour, G. J., C. N. Ta, and A. Das. 1992. Constitutive mutations of Agrobacterium tumefaciens transcriptional activator virG. J. Bacteriol. 174:4169-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Peach, C., and J. Velten. 1991. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol. Biol. 17:49-60. [DOI] [PubMed] [Google Scholar]
- 256.Peralta, E. G., R. Hellmiss, and W. Ream. 1986. Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J. 5:1137-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Peralta, E. G., and L. W. Ream. 1985. T-DNA border sequences required for crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 82:5112-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Perl, A., O. Lotan, M. Abu-Abied, and D. Holland. 1996. Establishment of an Agrobacterium-mediated transformation system for grape (Vitis vinifera L.): the role of antioxidants during grape-Agrobacterium interactions. Nat. Biotechnol. 14:624-628. [DOI] [PubMed] [Google Scholar]
- 259.Phi-Van, L., and W. H. Stratling. 1996. Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry 35:10735-10742. [DOI] [PubMed] [Google Scholar]
- 260.Piers, K. L., J. D. Heath, X. Liang, K. M. Stephens, and E. W. Nester. 1996. Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. USA 93:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pomponi, M., L. Spano, M. G. Sabbadini, and P. Costantino. 1983. Restriction endonuclease mapping of the root-inducing plasmid of Agrobactrium rhizogenes 1855. Plasmid 10:119-129. [DOI] [PubMed] [Google Scholar]
- 262.Porter, J. R. 1991. Host range and implications of plant infection by Agrobacterium rhizogenes. Crit. Rev. Plant Sci. 10:387-421. [Google Scholar]
- 263.Pu, X.-A., and R. N. Goodman. 1992. Induction of necrosis by Agrobacterium tumefaciens on grape explants. Physiol. Mol. Plant Pathol. 41:245-254. [Google Scholar]
- 264.Ramanathan, V., and K. Veluthambi. 1995. Transfer of non-T-DNA portions of the Agrobacterium tumefaciens Ti plasmid pTiA6 from the left terminus of TL-DNA. Plant Mol. Biol. 28:1149-1154. [DOI] [PubMed] [Google Scholar]
- 265.Rashid, H., S. Yokoi, K. Toriyama, and K. Hinata. 1996. Transgenic plant production mediated by Agrobacterium in Indica rice. Plant Cell Rep. 15:727-730. [DOI] [PubMed] [Google Scholar]
- 266.Ratcliff, F. G., B. Harrison, and D. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1550. [DOI] [PubMed] [Google Scholar]
- 267.Regensburg-Tuink, A. J. G., and P. J. J. Hooykaas. 1993. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature 363:69-71. [DOI] [PubMed] [Google Scholar]
- 268.Riker, A. J. 1926. Studies on the influence of some environmental factors on the development of crown gall. J. Agric. Res. 32:83-96. [Google Scholar]
- 269.Risseeuw, E., M. E. L. Franke-van Dijk, and P. J. J. Hooykaas. 1997. Gene targeting and instability of Agrobacterium T-DNA loci in the plant genome. Plant J. 11:717-728. [DOI] [PubMed] [Google Scholar]
- 270.Risseeuw, E., R. Offringa, M. E. I. Franke-van Dijk, and P. J. J. Hooykaas. 1995. Targeted recombination in plants using Agrobacterium coincides with additional rearrangements at the target locus. Plant J. 7:109-119. [DOI] [PubMed] [Google Scholar]
- 271.Ritchie, S. W., C.-N. Lui, J. C. Sellmer, Kononowicz, T. K. H. Hodges, and S. B. Gelvin. 1993. Agrobacterium tumefaciens-mediated expression of gusA in maize tissues. Transgenic Res. 2:252-265. [Google Scholar]
- 272.Robbs, S. L., M. C. Hawes, H.-J. Lin, S. G. Pueppke, and L. Y. Smith. 1991. Inheritance of resistance to crown gall in Pisum sativum. Plant Physiol. 95:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Rogowsky, P. M., T. J. Close, J. A. Chimera, J. J. Shaw, and C. I. Kado. 1987. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J. Bacteriol. 169:5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rossi, L., B. Hohn, and B. Tinland. 1993. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plants. Mol. Gen. Genet. 239:345-353. [DOI] [PubMed] [Google Scholar]
- 275.Rossi, L., B. Hohn, and B. Tinland. 1996. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 93:126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Rubin, R. A. 1986. Genetic studies on the role of octopine T-DNA border regions in crown gall tumor formation. Mol. Gen. Genet. 202:312-320. [Google Scholar]
- 277.Ruvkin, G. B., and F. M. Ausubel. 1979. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 278.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Salter, M. G., J. A. Paine, K. V. Riddell, I. Jepson, A. J. Greenland, M. X. Caddick, and A. B. Tomsett. 1998. Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J. 16:127-132. [Google Scholar]
- 280.Sanders, P. R., J. A. Winter, A. R. Barnason, S. G. Rogers, and R. T. Fraley. 1987. Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants. Nucleic Acids Res. 15:1543-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Scheiffele, P., W. Pansegrau, and E. Lanka. 1995. Initiation of Agrobacterium tumefaciens T-DNA processing: purified protein VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J. Biol. Chem. 270:1269-1276. [DOI] [PubMed] [Google Scholar]
- 282.Schena, M., A. M. Lloyd, and R. W. Davis. 1991. A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. USA 88:10421-10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Schoffl, F., and G. Baumann. 1985. Thermo-induced transcripts of a soybean heat shock gene after transfer into sunflower using a Ti plasmid vector. EMBO J. 4:1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schrammeijer, B., E. Risseeuw, W. Pansegrau, T. J. G. Regensburg-Tuink, W. L. Crosby, and P. J. J. Hooykaas. 2001. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11:258-262. [DOI] [PubMed] [Google Scholar]
- 286.Sen, P., G. J. Pazour, D. Anderson, and A. Das. 1989. Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J. Bacteriol. 171:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Shaw, C. H., M. D. Watson, G. H. Carter, and C. H. Shaw. 1984. The right hand copy of the nopaline Ti-plasmid 25 bp repeat is required for tumour formation. Nucleic Acids Res. 12:6031-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Shen, W.-H., J. Escudero, M. Schlappi, C. Ramos, B. Hohn, and Z. Koukolikova-Nicola. 1993. T-DNA transfer to maize cells: Histochemical investigation of β-glucuronidase activity in maize tissues. Proc. Natl. Acad. Sci. USA 90:1488-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Shimoda, N., A. Toyoda-Yamamoto, S. Aoke, and Y. Machida. 1993. Genetic evidence for an interaction between and VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J. Biol. Chem. 268:26552-26558. [PubMed] [Google Scholar]
- 290.Shurvinton, C. E., L. Hodges, and W. Ream. 1992. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. USA 89:11837-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Shurvinton, C. E., and W. Ream. 1991. Stimulation of Agrobacterium tumefaciens T-DNA transfer by overdrive depends on a flanking sequence but not on helical position with respect to the border repeat. J. Bacteriol. 173:5558-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Smarrelli, J., M. T. Watters, and L. H. Diba. 1986. Response of various cucurbits to infection by plasmid-harboring strains of Agrobacterium. Plant Physiol. 82:622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Song, Y.-N., M. Shibuya, Y. Ebzuka, and U. Sankawa. 1991. Identification of plant factors inducing virulence gene expression in Agrobacterium tumefaciens. Chem. Pharm. Bull. 39:2347-2350. [DOI] [PubMed] [Google Scholar]
- 294.Spector, D. L. 1990. Higher order nuclear organization: three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc. Natl. Acad. Sci. USA 87:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Spencer, P. A., and G. H. N. Towers. 1988. Specificity of signal compounds detected by Agrobacterium tumefaciens. Phytochemistry 27:2781-2785. [Google Scholar]
- 296.Springer, P. S., W. R. McCombie, V. Sundaresan, and R. A. Martienssen. 1995. Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268:877-880. [DOI] [PubMed] [Google Scholar]
- 297.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: applicaton to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Stachel, S. E., E. Messens, M. Van Montagu, and P. Zambryski. 1985. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624-629. [Google Scholar]
- 299.Stachel, S. E., and E. W. Nester. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Stachel, S. E., E. W. Nester, and P. C. Zambryski. 1986. A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc. Natl. Acad. Sci. USA 83:379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Stachel, S. E., B. Timmerman, and P. Zambryski. 1986. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature 322:706-712. [Google Scholar]
- 302.Stachel, S. E., B. Timmerman, and P. Zambryski. 1987. Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5′ virD gene products. EMBO J. 6:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Stachel, S. E., and P. C. Zambryski. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325-333. [DOI] [PubMed] [Google Scholar]
- 304.Stam, M., R. de Bruijn, S. Kenter, R. A. L. van der Hoorn, R. van Blokland, J. N. M. Mol, and J. M. Kooter. 1997. Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 12:63-82. [Google Scholar]
- 305.Steck, T. R., T. J. Close, and C. I. Kado. 1989. High levels of double-stranded transferred DNA (T-DNA) processing from an intact nopaline Ti plasmid. Proc. Natl. Acad. Sci. USA 86:2133-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Stief, A., D. M. Winter, W. H. Stratling, and A. E. Sippel. 1989. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341:343-345. [DOI] [PubMed] [Google Scholar]
- 307.Stomp, A.-M., C. Loopstra, W. S. Chilton, R. R. Sederoff, and L. W. Moore. 1990. Extended host range of Agrobacterium tumefaciens in the genus Pinus. Plant Physiol. 92:1226-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sundaresan, V., P. Springer, T. Volpe, S. Haward, J. D. G. Jones, C. Dean, H. Ma, and R. Martienssen. 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Gene Dev. 9:1797-1810. [DOI] [PubMed] [Google Scholar]
- 309.Sundberg, C., L. Meek, K. Carroll, A. Das, and W. Ream. 1996. VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 178:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Sundberg, C. D., and W. Ream. 1999. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol. 181:6850-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Suzuki, K., Y. Hattori, M. Uraji, N. Ohta, K. Iwata, K. Murata, A. Kato, and K. Yoshida. 2000. Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene 242:331-336. [DOI] [PubMed] [Google Scholar]
- 312.Szegedi, E., and P. Kozma. 1984. Studies on the inheritance of resistance to crown gall disease of grapevine. Vitis 23:121-126. [Google Scholar]
- 313.Thomashow, M. F., J. E. Karlinsey, J. R. Marks, and R. E. Hurlbert. 1987. Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J. Bacteriol. 169:3209-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Thomashow, M. F., R. Nutter, A. L. Montoya, M. P. Gordon, and E. W. Nester. 1980. Integration and organization of Ti plasmid sequences in crown gall tumors. Cell 19:729-739. [DOI] [PubMed] [Google Scholar]
- 315.Thomashow, M. F., C. G. Panagopoulos, M. P. Gordon, and E. W. Nester. 1980. Host range of Agrobacterium tumefaciens is determined by the Ti plasmid. 283:794-796.
- 316.Tighe, S. W., P. de Lajudie, K. Dipietro, K. Lindström, G. Nick, and B. D. W. Jarvis. 2000. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium species using the Sherlock Microbial Identification System. Int. J. Syst. E vol. Microbiol. 50:787-801. [DOI] [PubMed] [Google Scholar]
- 317.Tingay, S., D. McElroy, R. Kalla, S. Fieg, M. Wang, S. Thornton, and R. Brettell. 1997. Agrobacterium tumefaciens-mediated barley transformation. Plant J. 11:1369-1376. [Google Scholar]
- 318.Tinland, B., B. Hohn, and H. Puchta. 1994. Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 91:8000-8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tinland, B., Z. Koukolikova-Nicola, M. N. Hall, and B. Hohn. 1992. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl. Acad. Sci. USA 89:7442-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tinland, B., F. Schoumacher, V. Gloeckler, A. M. Bravo-Angel, and B. Hohn. 1995. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 14:3585-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Toki, S. 1997. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 15:16-21. [Google Scholar]
- 322.Toro, N., A. Datta, O. A. Carmi, C. Young, R. K. Prusti, and E. W. Nester. 1989. The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J. Bacteriol. 171:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Turk, S. C. H. J., L. S. Melchers, H. den Dulk-Ras, A. J. A. Regensburg-Tuink, and P. J. J. Hooykass. 1991. Environmental conditions differentially affect vir gene induction in different Agrobacterium strains. Role of the VirA sensor protein. Plant Mol. Biol. 16:1051-1059. [DOI] [PubMed] [Google Scholar]
- 324.Turk, S. C. H. J., R. P. van Lange, T. J. G. Regensburg-Tuink, and P. J. J. Hooykaas. 1994. Localization of the VirA domain involved in acetosyringone-mediated vir gene induction in Agrobacterium tumefaciens. Plant Mol. Biol. 25:899-907. [DOI] [PubMed] [Google Scholar]
- 325.Tzfira, T., and V. Citovsky. 2000. From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol. Plant Pathol. 1:201-212. [DOI] [PubMed] [Google Scholar]
- 326.Tzfira, T., and V. Citovsky. 2001. Comparison between nuclear localization of nopaline- and octopine-specific Agrobacterium VirE2 proteins in plant, yeast and mammalian cells. Mol. Plant Pathol. 2:171-176. [DOI] [PubMed] [Google Scholar]
- 327.Tzfira, T., and V. Citovsky. 2002. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12:121-128. [DOI] [PubMed] [Google Scholar]
- 328.Tzfira, T., Y. Rhee, M.-H. Chen, T. Kunik, and V. Citovsky. 2000. Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu. Rev. Microbiol. 54:187-219. [DOI] [PubMed] [Google Scholar]
- 329.Tzfira, T., M. Vaidya, and V. Citovsky. 2001. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20:3596-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Tzfira, T., M. Vaidya, and V. Citovsky. 2002. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc. Natl. Acad. Sci. USA 99:10435-10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ulker, P., G. C. Allen, W. F. Thompson, S. Siker, and A. K. Weissinger. 1999. A tobacco matrix attachment region reduces the loss of transgene expression in the progeny of transgenic tobacco plants. Plant J. 18:253-263. [Google Scholar]
- 332.Unger, L., S. F. Ziegler, G. A. Huffman, V. C. Knauf, R. Peet, L. W. Moore, M. P. Gordon, and E. W. Nester. 1985. New class of limited-host-range-Agrobacterium mega-tumor-inducing plasmid lacking homology to the transferred DNA of a wide-host-range, tumor-inducing plasmid. J. Bacteriol. 164:723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Vain, P., B. Worland, A. Kohli, J. W. Snape, P. Christou, G. C. Allen, and W. F. Thompson. 1999. Matrix attachment regions increase transgene expression levels and stability in transgenic rice plants and their progeny. Plant J. 18:233-242. [Google Scholar]
- 334.Van der Geest, A. H. M., G. E. Hall, S. Spiker, and T. C. Hall. 1994. The beta-phaseolin gene is flanked by matrix attachment regions. Plant J. 6:413-423. [Google Scholar]
- 335.van Haaren, M. J. J., N. J. A. Sedee, H. A. de Boer, R. A. Schilperoort, and P. J. J. Hooykaas. 1988. Bidirectional transfer from a 24 bp border repeat of Agrobacterium tumefaciens. Nucleic Acids Res. 16:10225-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.van Haaren, M. J. J., N. J. A. Sedee, H. A. de Boer, R. A. Schilperoort, and P. J. J. Hooykaas. 1989. Mutational analysis of the conserved domains of a T-region border repeat of Agrobacterium tumefaciens. Plant Mol. Biol. 13:523-531. [DOI] [PubMed] [Google Scholar]
- 337.van Haaren, M. J. J., N. J. A. Sedee, R. A. Schilperoort, and P. J. J. Hooykaas. 1987. Overdrive is a T-region enhancer which stimulates T-strand production in Agrobacterium tumefaciens. Nucleic Acids Res. 15:8983-8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Van Haute, E., H. Joos, M. Maes, G. Warren, M. Van Montagu, and J. Schell. 1983. Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for the reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J. 2:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Van Larebeke N., G. Engler, M. Holsters, S. Van den Elsacker, I. Zaenen, R. A. Schilperoort, and J. Schell. 1974. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252:169-170. [DOI] [PubMed] [Google Scholar]
- 340.Van Slogteren, G. M. S., P. J. J. Hooykaas, and R. A. Schilperoort. 1984. Silent T-DNA genes in plant lines transformed by Agrobacterium tumefaciens are activated by grafting and by 5-azacytidine treatment. Plant Mol. Biol. 3:333-336. [DOI] [PubMed] [Google Scholar]
- 341.van Wordragen, M. F., and H. J. M. Dons. 1992. Agrobacterium tumefaciens-mediated transformation of recalcitrant crops. Plant Mol. Biol. Rep. 10:12-36. [Google Scholar]
- 342.Vaudequin-Dransart, V., A. Petit, C. Poncet, C. Ponsonnet, X. Nesme, J. B. Jones, H. Bouzar, W. S. Chilton, and Y. Dessaux. 1995. Novel Ti plasmids in Agrobacterium strains isolated from fig tree and chrysanthemum tumors and their opine-like molecules. Mol. Plant-Microbe Interact. 8:311-321. [DOI] [PubMed] [Google Scholar]
- 343.Velten, J., L. Velten, R. Hain, and J. Schell. 1984. Isolation of a dual plant promoter fragment from the Ti plasmid of Agrobacterium tumefaciens. EMBO J. 3:2723-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Veluthambi, K., R. K. Jayaswal, and S. B. Gelvin. 1987. Virulence genes A, G, and D mediate the double-stranded border cleavage of T-DNA from the Agrobacterium Ti plasmid. Proc. Natl. Acad. Sci. USA 84:1881-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Veluthambi, K., W. Ream, and S. B. Gelvin. 1988. Virulence genes, borders, and overdrive generate single-stranded T-DNA molecules from the A6 Ti plasmid of Agrobacterium tumefaciens. J. Bacteriol. 170:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Venkateswarlu, K., and R. N. Nazar. 1991. Evidence for T-DNA mediated gene targeting to tobacco chloroplasts. Bio/Technology 9:1103-1105. [DOI] [PubMed] [Google Scholar]
- 347.Vergunst, A. C., and P. J. J. Hooykaas. 1998. Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol. Biol. 38:393-406. [DOI] [PubMed] [Google Scholar]
- 348.Vergunst, A. C., L. E. T. Jansen, and P. J. J. Hooykaas. 1998. Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res. 26:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. T. de Vlaam, T. J. G. Regensburg-Tuink, and P. J. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 350.Verheijen, R., W. van Venrooij, and F. Rameekers. 1988. The nuclear matrix: structure and composition. J. Cell Sci. 90:11-36. [DOI] [PubMed] [Google Scholar]
- 351.Vogel, A. M., and A. Das. 1992. Mutational analysis of Agrobacterium tumefaciens virD2: tyrosine 29 is essential for endonuclease activity. J. Bacteriol. 174:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wang, K., L. Herrera-Estrella, M. Van Montagu, and P. Zambryski. 1984. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell 38:455-462. [DOI] [PubMed] [Google Scholar]
- 353.Wang, K., S. E. Stachel, B. Timmerman, M. Van Montagu, and P. C. Zambryski. 1987. Site-specific nick in the T-DNA border sequence as a result of Agrobacterium vir gene expression. Science 235:587-591. [DOI] [PubMed] [Google Scholar]
- 354.Wang, K., C. Genetello, M. Van Montagu, and P. Zambryski. 1987. Sequence context of the T-DNA border repeat element determines its relative activity during T-DNA transfer to plant cells. Mol. Gen. Genet. 210:338-346.
- 355.Ward, E. R., and W. M. Barnes. 1988. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242:927-930. [Google Scholar]
- 356.Wenck, A., M. Czako, I. Kanevski, and L. Marton. 1997. Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol. Biol. 34:913-922. [DOI] [PubMed] [Google Scholar]
- 357.Wenck, A. R., M. Quinn, R. W. Whetten, G. Pullman, and R. Sederoff. 1999. High-efficiency Agrobacterium-mediated transformation of Norway spruce (Picea abies) and loblolly pine (Pinus taeda). Plant Mol. Biol. 39:407-416. [DOI] [PubMed] [Google Scholar]
- 358.White, F. F., and E. W. Nester. 1980. Hairy root: plasmid encodes virulence traits in Agrobacterium rhizogenes. J. Bacteriol. 141:1134-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Winans, S. C. 1991. An Agrobacterium two-component regulatory system for the detection of chemicals released from plant wounds. Mol. Microbiol. 5:2345-2350. [DOI] [PubMed] [Google Scholar]
- 360.Wirawan, I. G. P., H. W. Kang, and M. Kojima. 1993. Isolation and characterization of a new chromosomal virulence gene of Agrobacterium tumefaciens. J. Bacteriol. 175:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wirawan, I. G. P., and M. Kojima. 1996. A chromosomal virulence gene (acvB) product of Agrobacterium tumefaciens that binds to a T-strand to mediate its transfer to host plant cells. Biosci. Biotechnol. Biochem. 60:44-49. [Google Scholar]
- 362.Wirawan, I. G. P., and M. Kojima. 1996. Distribution of a chromosomal virulence gene, acvB, of Agrobacterium tumefaciens among various bacteria. Biosci. Biotechnol. Biochem. 60:50-53. [DOI] [PubMed] [Google Scholar]
- 363.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M.-J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z.-Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J.-F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 364.Xiang, C., P. Han, I. Lutziger, K. Wang, and D. J. Oliver. 1999. A mini binary vector series for plant transformation. Plant Mol. Biol. 40:711-717. [DOI] [PubMed] [Google Scholar]
- 365.Xing, A., Z. Zhang, S. Sato, P. Staswick, and T. E. Clemente. 2000. The use of the two T-DNA binary system to derive marker-free transgenic soybeans. In Vitro Cell. Dev. Biol—Plant 36:456-463. [Google Scholar]
- 366.Yadav, N. S., J. Van der Leyden, D. R. Bennett, W. M. Barnes, and M.-D. Chilton. 1982. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc. Natl. Acad. Sci. USA 79:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Yanofsky, M., B. Lowe, A. Montoya, R. Rubin, W. Krul, M. Gordon, and E. Nester. 1985. Molecular and genetic analysis of factors controlling host range in Agrobacterium tumefaciens. Mol. Gen. Genet. 201:237-348. [Google Scholar]
- 368.Yanofsky, M. F., and E. W. Nester. 1986. Molecular characterization of a host-range-determining locus from Agrobacterium tumefaciens. J. Bacteriol. 168:244-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Yanofsky, M. F., S. G. Porter, C. Young, L. M. Albright, M. P. Gordon, and E. W. Nester. 1986. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47:471-477. [DOI] [PubMed] [Google Scholar]
- 370.Yi, H. C., K. S. Mysore, and S. B. Gelvin. 2002. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 32:285-298. [DOI] [PubMed]
- 371.Yibrah, H. S., R. Gronroos, A. Lindroth, H. Franzen, D. Clapham, and S. von Arnold. 1996. Agrobacterium rhizogenes-mediated induction of adventitious rooting from Pinus contorta hypocotyls and the effect of 5-azacytidine on transgene activity. Transgenic Res. 5:75-85. [Google Scholar]
- 372.Yoder, J. I., and A. P. Goldsbrough. 1994. Transformation systems for generating marker free transgenic plants. Bio/Technology 12:263-267. [Google Scholar]
- 373.Young, C., and E. W. Nester. 1988. Association of the VirD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 170:3367-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Young, J. M., L. D. Kuykendall, E. Martinez-Romero, A. Kerr, and H. Sawada. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Sys. Evol. Microbiol. 51:89-103. [DOI] [PubMed] [Google Scholar]
- 375.Yusibov, V. M., T. R. Steck, V. Gupta, and S. B. Gelvin. 1994. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 91:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Zaenen I., N. Van Larebeke, M. Van Montagu, and J. Schell. 1974. Supercoiled circular DNA in crown-gall inducing Agrobacterium strains. J. Mol. Biol. 86:109-127. [DOI] [PubMed] [Google Scholar]
- 377.Zahm, P., C. Hohmeyer, and K. Geider. 1984. Site-specific mutagenesis of the Ti plasmid by transformation of Agrobacterium tumefaciens with mutagenized T-DNA fragments cloned in E. coli plasmids. Mol. Gen. Genet. 194:188-194. [Google Scholar]
- 378.Zambryski, P., M. Holsters, K. Kruger, A. Depicker, J. Schell, M. Van Montagu, and H. M. Goodman. 1980. Tumor DNA structure in plant cells transformed by A. tumefaciens. Science 209:1385-1391. [DOI] [PubMed] [Google Scholar]
- 379.Zambryski, P., P. H. Joos, C. Genetello, J. Leemans, M. Van Montagu, and J. Schell. 1983. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 2:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Zhou, X.-R., and P. J. Christie. 1999. Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 181:4342-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Ziemienowicz, A., D. Gorlich, E. Lanka, B. Hohn, and L. Rossi. 1999. Import of DNA into mammalian nuclei by proteins originating from a plant pathogenic bacterium. Proc. Natl. Acad. Sci. USA 96:3729-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ziemienowicz, A., T. Merkle, F. Schoumacher, B. Hohn, and L. Rossi. 2001. Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13:369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Zupan, J. R., V. Citovsky, and P. Zambryski. 1996. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci. USA 93:2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zupan, J., T. R. Muth, O. Draper, and P. Zambryski. 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23:11-28. [DOI] [PubMed] [Google Scholar]
- 385.Zupan, J., and P. Zambryski. 1997. The Agrobacterium DNA transfer complex. Crit. Rev. Plant Sci. 16:279-295. [Google Scholar]
- 386.Zyprian, E., and C. I. Kado. 1990. Agrobacterium-mediated plant transformation by novel mini-T vectors in conjunction with a high-copy vir region helper plasmid. Plant Mol. Biol. 15:245-256. [DOI] [PubMed] [Google Scholar]