Abstract
The cytoplasmic domains (tails) of heterodimeric integrin adhesion receptors mediate integrins' biological functions by binding to cytoplasmic proteins. Most integrin β tails contain one or two NPXY/F motifs that can form β turns. These motifs are part of a canonical recognition sequence for phosphotyrosine-binding (PTB) domains, protein modules that are present in a wide variety of signaling and cytoskeletal proteins. Indeed, talin and ICAP1-α bind to integrin β tails by means of a PTB domain–NPXY ligand interaction. To assess the generality of this interaction we examined the binding of a series of recombinant PTB domains to a panel of short integrin β tails. In addition to the known integrin-binding proteins, we found that Numb (a negative regulator of Notch signaling) and Dok-1 (a signaling adaptor involved in cell migration) and their isolated PTB domain bound to integrin tails. Furthermore, Dok-1 physically associated with integrin αIIbβ3. Mutations of the integrin β tails confirmed that these interactions are canonical PTB domain–ligand interactions. First, the interactions were blocked by mutation of an NPXY motif in the integrin tail. Second, integrin class-specific interactions were observed with the PTB domains of Dab, EPS8, and tensin. We used this specificity, and a molecular model of an integrin β tail–PTB domain interaction to predict critical interacting residues. The importance of these residues was confirmed by generation of gain- and loss-of-function mutations in β7 and β3 tails. These data establish that short integrin β tails interact with a large number of PTB domain-containing proteins through a structurally conserved mechanism.
Integrin adhesion receptors are heterodimers of α and β subunits, which combine to form a large extracellular domain, two transmembrane domains (one for each subunit), and a cytoplasmic domain typically composed of the short α and β C-terminal cytoplasmic tails (1). Bidirectional signal transduction through integrin adhesion receptors is essential for a wide variety of functions, including cell adhesion and migration, and assembly and remodeling of the extracellular matrix. Binding of intracellular proteins to integrin cytoplasmic tails is an important step in the transduction of signals to and from integrin-adhesion receptors (2). Integrin β cytoplasmic tails, with the exception of those of β4 and β8, are short (<60 residues) and contain one or two NPXY or NPXY-like motifs (Fig. 1A), the first of which has the propensity to form a β turn (3). Such β turn-forming sequences frequently serve to bind to phosphotyrosine-binding (PTB) domains (4). NXXY motif-dependent binding of the Shc PTB domain to the large (>1,000 residues) β4 cytoplasmic tail has been observed (5) and molecular modeling studies suggested that the interaction of integrin cytoplasmic domain-associated protein (ICAP)1-α with β1A (6), and of talin with β3 (7), are mediated by PTB domain-like interactions. The solved crystal structure of a complex of a talin fragment with part of the β3 tail verified the predicted PTB domain-like interaction (8). Three recent publications (7, 9, 10) have hypothesized that other PTB domain-containing proteins may also interact with short integrin β tails. Thus, the PTB domain–ligand interaction is hypothesized to be a structural prototype used by a variety of integrin β tail-binding proteins.
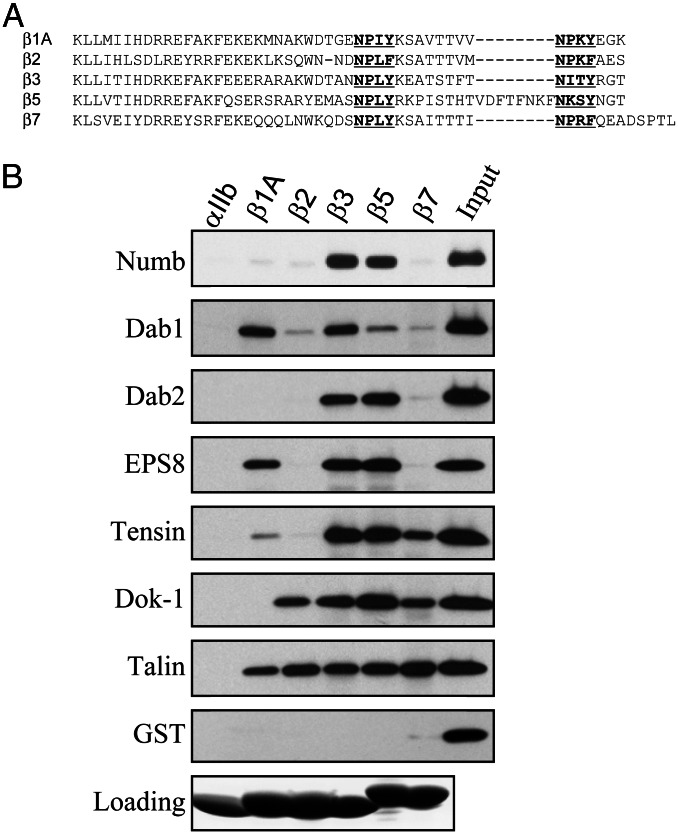
Figure 1.
PTB domains bind to β integrin cytoplasmic tails. (A) An alignment of the amino acid sequences of β integrin cytoplasmic tails. The NPXY or NPXY-like motifs are in bold and underlined. (B) PTB domains, which were expressed and purified as recombinant GST fusion proteins, were incubated with beads coated with recombinant αIIb, β1A, β2, β3, β5, and β7 cytoplasmic tails. Bound proteins were fractionated by SDS/PAGE and GST-PTB domains were detected by Western blotting with anti-GST antibodies. Loading of the recombinant integrin tails on the beads was assessed by Coomassie blue staining.
To test this hypothesis, we first queried sequence databases for predicted PTB domains and identified a number of PTB domain proteins implicated in integrin-related functions such as talin and ICAP-1α. We then examined the binding of a series of recombinant PTB domains to a panel of integrin β tails. We found that Numb (a negative regulator of Notch signaling) and Dok-1 (a signaling adaptor involved in cell migration) and their isolated PTB domain bound to integrin tails. Additional integrin-class specific interactions were observed with the PTB domains of Dab (a downstream target of c-Abl), EPS8 (a regulator of Rac signaling), and tensin, a focal adhesion protein. Finally, by modeling the interaction of an integrin β tail with a PTB domain based on the NMR solution structure of the Numb/Numb-associated kinase (Nak) complex (11) we predicted that critical determinants of the specificity of the integrin interaction were an uncharged residue at position −5 and a polar residue at position +2 relative to the Tyr of the NPXY motif. This prediction was confirmed by gain- and loss-of-function mutations. Hence, in vitro binding assays indicate that a majority of integrin β tails interact with many PTB domain-containing proteins through a structurally conserved mechanism.
Materials and Methods
Antibodies and DNAs.
Monoclonal anti-GST antibody B14 and polyclonal anti-Dok antibody M-276 were obtained from Santa Cruz Biotechnology. pGEX constructs encoding GST fusion protein of PTB domains from mouse Shc, Dab-1, Dab-2/DOC-2, Numb, and the talin PTB-like domain (309–405) [Residue numbers refer to SWISS-PROT entry TALI_MOUSE (p26039)], rat X11α and X11β, Caenorhabditis elegans Lin10, and human JIP have been described (7, 12, 13). EST clones encoding human EPS8, EB-1/E2A-PBX1-associated protein, and CED6 [I.M.A.G.E. Consortium ID nos: 2459720 (human), 1684718 (human), and 2207248 (human)], were obtained from American Type Culture Collection. cDNA encoding human tensin 2 (KIAA1075) was obtained from Kazusa DNA Research Institute (Kisarazu, Japan). cDNA encoding human ICAP-1α, human RGS12, human GAPCenA, and rat insulin receptor substrate (IRS)-1 were provided by D. Siderovski (University of Michigan), B. Goud (Institut Curie, Paris), and M. White (Howard Hughes Medical Institute, Harvard Medical School, Boston), respectively. To generate expression constructs for GST fusion proteins of PTB domains, the PTB portions of human EPS8-(59–199) [residue numbers refer to SWISS-PROT entry EPS8_HUMAN (Q12929)], mouse Dok-1-(149–256) [SWISS-PROT: DOK1_MOUSE (P97465)], rat IRS-1-(153–257) [SWISS-PROT: IRS1_RAT (P35570)], human CED6-(20–160) [SWISS-PROT: Q9UBP9 (Q9UBP9)], human ICAP-1α-(61–138) [SWISS-PROT: ITP1_HUMAN (O14713)], human RGS-(224–376) [SWISS-PROT: RGSC_HUMAN (O14924)], human GAPCenA-(69–206) [SWISS-PROT: Q9Y3P9 (Q9Y3P9)], human EB–(762–904) [SWISS-PROT: Q9Y5K9 (Q9Y5K9)], and human tensin 2-(1365–1505) [SWISS-PROT: BAA83027 (BAA83027)] were amplified by PCR and cloned, in frame, into the bacterial expression vector pGEX-4T. Mammalian expression constructs encoding full-length mouse Numb cDNA in pcDNA3.1 (13) and hemagglutinin (HA)-tagged full-length mouse Dok-1 cDNA and Dok-1 lacking the pleckstrin homology (PH) domain [Dok-1(ΔPH)] in pRC/CMV (14) (generously provided by T. Noguchi, Kobe University School of Medicine, Kobe, Japan) have been described. A cDNA encoding mouse Dok-1 lacking the PTB domain (amino acids 152–254) was generated by splice-overlap PCR and cloned into pCMV-Tag3B (Stratagene) to allow expression of N-terminally c-myc-tagged Dok-1(ΔPTB).
Bacterial expression constructs encoding recombinant His-tagged integrin cytoplasmic tail model proteins have been described (15, 16). Specific β tail mutations were introduced by QuikChange site-directed mutagenesis (Stratagene) and mutations were confirmed by DNA sequencing.
Homology Search for PTB Domains.
To search for proteins containing putative PTB domains or related amino acid sequences, psi-blast searches (available at www.ncbi.nlm.nih.gov/blast/) were performed by using the peptide sequences from the PTB domain of human p72 Numb-(34–175) [residue numbers refer to SWISS-PROT entry NUMB_HUMAN (P49757)] or the PTB domain of human IRS-1-(160–262) [SWISS-PROT: IRS1_HUMAN (P35568)] as queries. These searches were performed with Expect = 50, Inclusion Threshold = 0.02, and all other parameters at basic search default values. To evaluate the similarity of individual proteins identified in the psi-blast searches to the consensus sequences of Shc- or IRS-1-related PTB domains, Conserved Domain searches (available at www.ncbi.nlm.nih.gov/blast/) were performed. For Conserved Domain searches, the partial sequences that exhibited homology to PTB domains in the psi-blast searches were used as queries.
Binding Assays.
Binding assays using recombinant integrin tail model proteins were performed as described (7, 15, 16). For binding of intact proteins, Chinese hamster ovary (CHO) cells were transiently transfected with 4 μg of expression vector encoding full-length mouse Dok-1, or Dok-1 mutant by using Lipofectamine (Life Technologies, Grand Island, NY). Cells were harvested 48 h after transfection and lysed as described (16), and binding assays were preformed. Bound proteins were eluted in SDS-sample buffer, fractionated by SDS/PAGE, and detected by Western blotting with anti-Dok antibodies.
Immunoprecipitation.
CHO cells stably expressing integrin αIIbβ3 (17), chimeric αIIbβ3 where the β3 tail was replaced with that of β7 (αIIbβ3β7) (18), or αIIbβ3 lacking the β3 cytoplasmic tail (αIIbβ3Δ728) (17) were transfected with 4 μg of HA-tagged Dok-1. Cells were lysed in ice-cold lysis buffer [50 mM Tris, pH 7.4/50 mM NaCl/10 mM NaF/1 mM sodium orthovanadate/0.5% Igepal CA-630 (a Nonidet P-40 substitute)] with Complete protease inhibitor mixture (Roche Diagnostics). The lysates were centrifuged (10,000 × g for 30 min at 4°C) to remove insoluble material. Supernatant containing 1 mg of protein was incubated overnight at 4°C with anti-αIIbβ3 antibody, D57, in a total volume of 500 μl. Fifteen microliters of protein G Sepharose beads (Amersham Pharmacia) was then added and mixed at 4°C for 1 h longer. Beads were washed three times with lysis buffer, and bound proteins were eluted by boiling in SDS-sample buffer and were fractionated by SDS/PAGE. Dok-1 and αIIb were detected by immunoblotting.
Structural Modeling of Integrin β Tail–Numb PTB Domain Interactions.
The NMR structure of a peptide derived from Nak in complex with the Numb PTB domain [Protein Data Bank (PDB) ID code 1DDM (11)] was used as a template to model the bound β-integrin tails. Initial models were built by replacement of the side chains in the Nak peptide with those of integrin residues in positions −6 to +4 with respect to the Tyr or Phe of the NPXY/F motif. The ligands where then subject to conjugate energy minimization in the presence of Numb PTB domain by using the program cns (19).
Results and Discussion
Identification of PTB Domains in Integrin-Associated Proteins.
Two specific PTB domain–short integrin β tail interactions have been predicted: β1A–ICAP-1α (6) and β3–talin F3 (7). We and others (7, 9, 10) have hypothesized that PTB domain–NPXY interactions may mediate interactions of many signaling molecules with integrin β cytoplasmic domains; however, the generality of this interaction had not been demonstrated. To identify PTB domains for study, we queried protein databases. There are two structural classes of PTB domains: Shc- and IRS-related PTB domains. These classes lack significant primary sequence similarity, but adopt similar structures and have similar, but not identical, binding specificities (20–22). We performed psi-blast searches (23) by using the peptide sequences from the PTB domain of human p72 Numb-(34–175), which is a Shc-related PTB domain, or the PTB domain of human IRS-1 (160–262), an IRS-related PTB domain. We also performed Conserved Domain (24) searches to evaluate the similarity of individual identified proteins to the consensus sequences of PTB domains. A number of proteins implicated in integrin function, as integrin-binding proteins, cytoskeletal proteins, or proteins involved in migration or adhesion-mediated signaling, were identified (Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org).
One group of PTB domain-containing proteins includes Shc (5, 25), ICAP-1α (26), and talin (27), which are known to bind integrin β tails. The interactions of ICAP-1α and talin are mediated by the binding of a PTB domain to an NPXY-containing peptide sequence (6, 7). Another group of proteins physically interact or colocalize with integrins, but their binding to the β tails has not been established. These include tensin and Dab1 (Disabled-1) (28, 29). Others are downstream targets of integrin signaling, such as Dok-1, which is phosphorylated following integrin-mediated adhesion and mediates cell migration (14, 30). Thus, a number of PTB domain-containing proteins have functional links with, and in some cases bind directly to, integrins.
Integrin Class-Specific Binding of PTB Domains to Integrin β Tails.
The presence of NPXY motifs in integrin β tails, and of PTB domains in proteins that either bind to integrin β tails or are likely to be involved in integrin function, led us to survey the interaction of a number of other recombinant PTB domains with a matrix of recombinant integrin-cytoplasmic tail-model proteins. We found that Numb and Dok-1 PTB domains bound to integrin tails. Additional interactions were observed with the PTB domains of Dab1 (a downstream target of c-Abl), EPS8, and tensin. There was considerable specificity to the interactions. None of the PTB domains bound to the αIIb tail, and none of the β tails bound to purified GST (Fig. 1B). Most of the PTB domains bound selectively to particular integrin β tails (Fig. 1B; see Table 3, which is published as supporting information on the PNAS web site). β1A bound PTB domains from Dab1, EPS8, and talin and more weakly to the tensin PTB domain. In contrast, β7 bound specifically to tensin, Dok-1, and talin PTB domains. β2 bound only talin and Dok-1, and bound weakly to Dab1 PTB domains. In contrast, the β3 and β5 tails bound to all of the PTB domains tested. Thus, there was considerable specificity to the interaction of PTB domains with integrin β tails.
The foregoing studies established that PTB domains bound specifically to integrin β tails. If these interactions were canonical PTB domain–peptide interactions, then they would depend on the formation of a stable β turn at the NPXY motif (20, 21). To test this hypothesis, we examined the effect of Tyr to Ala substitutions within these NPXY motifs because this mutation is known to disrupt β-turn structure in the β3 cytoplasmic domain (3). Mutation of the first NPXY/F motif in β1A [β1A(Y788A)], β7 [β7(Y778A)], or β3 [β3(Y747A)] disrupted binding of the Numb, EPS8, Dab1, Dok-1, and talin PTB domains (Fig. 2 A–C), indicating that this region is important for integrin–PTB domain interactions. Most integrin β tails have two NPXY motifs and the more C-terminal motif is implicated in binding to the ICAP-1α PTB domain (6). Consistent with a PTB domain-binding function for the second NPXY-like motif, the binding of Dab1 and Dab2 PTB domains to β3 tails was largely unaffected by a Tyr-747 to Ala mutation in the first NPXY motif of β3, but was inhibited by Tyr-759 to Ala mutations in the second NPXY-like motif (Fig. 2D). This inhibition was not complete, suggesting that the N-terminal motif may have some residual binding activity when the second motif is lost. Thus, the interaction of PTB domains with integrin β tails requires the integrity of NPXY motifs in the integrin tails and is therefore likely to resemble the interactions of PTB domains with other peptide ligands.
Figure 2.
Tyr to Ala mutations in β integrin cytoplasmic tails inhibit binding of PTB domains. The binding of recombinant GST-PTB domain fusion proteins to recombinant β1A (A), β3 (B), and β7 (C) integrin cytoplasmic tails with Tyr to Ala mutations in their N-terminal NPXY motifs [β1A(Y787A), β3(Y747A), and β7(Y778A)] was assessed as described in the legend to Fig. 1. (D) The binding of GST-PTB domains to recombinant β3 cytoplasmic tails containing mutations in either the N-terminal [β3(Y747A)] or C-terminal [β3(Y759A)] NPXY-like motif was assessed as described in B.
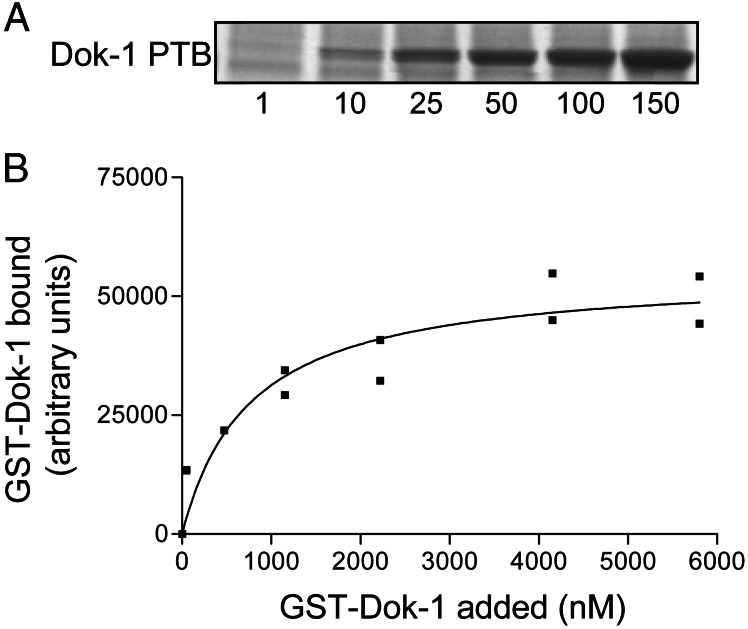
We used affinity chromatography to quantify the interaction of one of the PTB domain–integrin β tail interactions identified above (Fig. 3A). Dok-1 binding to the β3 tail was quantified by Coomassie blue staining, followed by scanning densitometry and a dose–response curve was plotted (Fig. 3B). The Dok-1 PTB domain bound with an EC50 of 760 nM. This finding indicates a lower affinity than that determined for the talin head domain in a similar assay format (EC50 = 130 nM; ref. 3), or the isolated PTB-like domain of talin, measured by surface plasmon resonance (Kd = 130 nM; ref. 7). However, it is comparable in affinity to the interaction of full-length talin with β3 integrin tails (31, 32), and appears to be of higher affinity than the β3 integrin–filamin interactions (32). Furthermore the EC50 of the Dok-1 PTB domain–β3 tail interaction is within the range of affinities reported for other PTB domain–ligand interactions, e.g., X11–βAPP (33), Dab1–APP (34), Shc–middle T antigen (4), and Numb–Nak (35, 36). Thus, the binding of the Dok-1 PTB domain to the β3 integrin tail is comparable with known PTB domain–ligand interactions and to other known integrin-binding proteins.
Figure 3.
The Dok-1 PTB domain binds integrin β3 tails. (A) Various amounts (1–150 μg as indicated) of recombinant GST-Dok-1 PTB domains were mixed with beads coated with recombinant β3 tails. Bound proteins were fractionated by SDS/PAGE and detected by Coomassie blue staining. (B) Bound GST-Dok-1 fusion protein was quantified by scanning densitometry and the amount bound was plotted against the input concentration.
Interactions of Native Proteins with Integrin β Tails and Intact Integrins Are Mediated by PTB Domains.
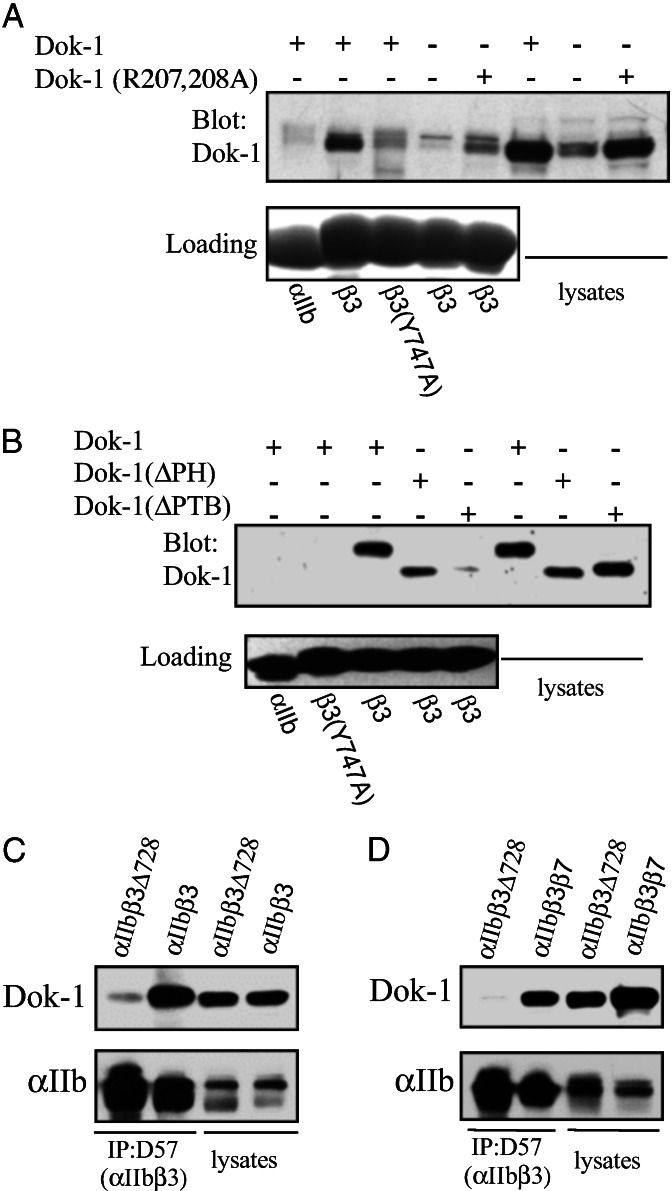
The preceding experiments demonstrate that isolated PTB domains can interact with integrin β tails. Full-length talin and ICAP-1α interact with integrin β cytoplasmic domains (16, 26). To determine whether the binding of isolated PTB domains predicted binding of the intact protein, we examined one of the integrin–PTB domain interactions identified above: Dok-1. Recombinant full-length mouse Dok-1 was expressed in CHO cells, cell lysates were prepared, and the binding of Dok-1 to β3 tails was assessed by affinity chromatography. Dok-1 bound to β3 but not to αIIb tails (Fig. 4A). The binding was likely to be mediated by a canonical PTB domain–peptide ligand interaction because it was inhibited by mutations in the PTB domain [Dok-1(R207,208A); ref. 37], or in the β3 tail [β3(Y747A)], predicted to disrupt such interactions. Furthermore, Dok-1 mutants lacking the PH domain, but not those lacking the PTB domain, could bind β3 tails (Fig. 4B), demonstrating the requirement for the Dok-1 PTB domain, but not its PH domain for binding to integrin β tails. The NPXY-dependent binding of full-length Numb to integrin β tails was also observed (data not shown). Thus, full-length PTB domain-containing proteins bind to integrin β tails by means of their PTB domains.
Figure 4.
Intact Dok-1 binds to integrin β tails. (A) Lysates from CHO cells transiently transfected with Dok-1, Dok-1 containing a mutation that reduces PTB domain ligand binding [Dok-1 (R207,208A; ref. 37], or from mock-transfected cells were incubated with beads coated with recombinant αIIb, β3, or β3(Y747A) cytoplasmic domains. Bound proteins were fractionated by SDS/PAGE and Dok-1 was detected by Western blotting with anti-Dok-1 antibodies. Note the specific binding of a band in the untransfected cells, presumably that of endogenous hamster Dok-1. (B) Lysates from CHO cells transiently transfected with epitope-tagged Dok-1, Dok-1 lacking its PH domain [Dok-1(ΔPH)], or Dok-1 lacking its PTB domain [Dok-1(ΔPTB)] were incubated with beads coated with recombinant αIIb, β3, or β3(Y747A) cytoplasmic domains. Bound proteins were fractionated by SDS/PAGE and Dok-1 was detected by Western blotting with antibodies against the epitope tags. (C) CHO cells stably expressing αIIbβ3 or αIIbβ3Δ728 were transfected with Dok-1, and 24 h later cells were lysed and the integrins were precipitated with the mAb D57. Immunoprecipitated αIIb and coimmunoprecipitated Dok-1 were detected by Western blotting. (D) CHO cells stably expressing αIIbβ3β7 or αIIbβ3Δ728 were transfected with Dok-1, and 24 h later cells were lysed and the integrins were precipitated with the mAb D57. Immunoprecipitated αIIb and coimmunoprecipitated Dok-1 were detected by Western blotting.
To demonstrate that interactions identified by using recombinant models of integrin β tails also occur with intact integrins, we examined the association of Dok-1 with integrins in coimmunoprecipitation experiments. Dok-1 coimmunoprecipitated with αIIbβ3, but not with αIIbβ3 lacking the β3 cytoplasmic tail (αIIbβ3Δ728; Fig. 4C). Furthermore, if the β3 tail was replaced with that of β7, Dok-1 could also be coimmunoprecipitated with the chimeric αIIbβ3β7 integrin (Fig. 4D). Thus, full-length Dok-1 bound to intact integrins in a β tail-dependent manner.
Molecular Modeling and Mutagenesis Establish That Integrin β Tail Binding Is Mediated by Canonical PTB Domain–Ligand Interactions.
The interaction of PTB domains with integrin β tails requires the integrity of NPXY motifs in the integrin tails. However, while the NPXY motifs are highly conserved among integrin β tails (Fig. 1A), integrin–PTB domain interactions exhibit specificity (Fig. 1B). This result suggests that additional residues contribute to the selectivity of integrin–PTB domain interactions. As the first step toward identifying these residues, we produced molecular models of integrin β tails bound to a PTB domain.
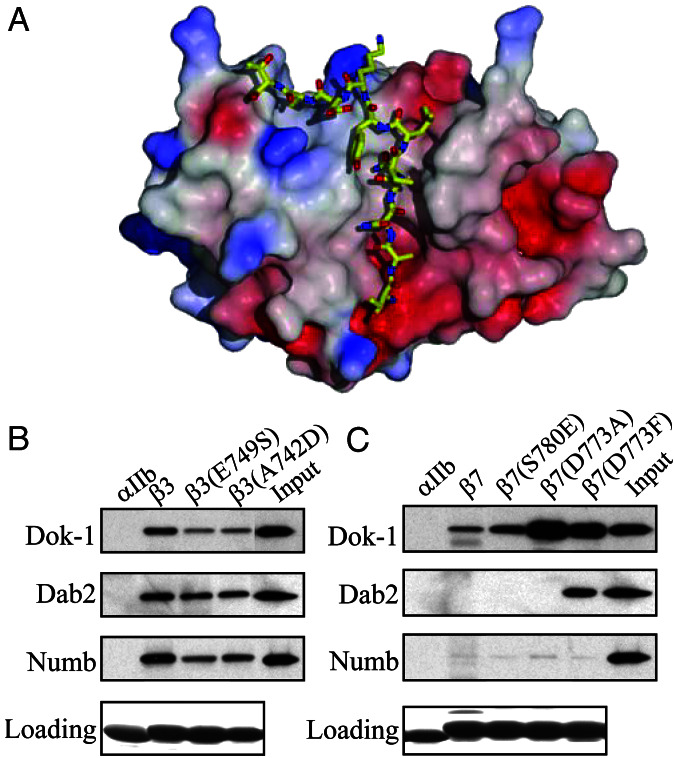
The Numb PTB domain binds β3 and β5 tails, but binds very weakly to β1A, β2, and β7 tails (Fig. 1B). The high-resolution structures of the Numb PTB domain in complex with two cognate ligands are known (11). The Nak peptide contains a NMSF sequence and binds to Numb PTB in a similar fashion to the canonical NPXY-PTB recognition mode (21). We used the structure of the bound Nak as a template to predict the structure of bound integrin β tails. Binding of Numb to β3 relies on the N-terminal NPXY motif of the β3 tail; therefore, we modeled the structure of the region 741–751 of β3 (Fig. 5A).
Figure 5.
Mutations in β integrin cytoplasmic tails alter PTB domain-binding specificity. (A) Structural modeling of the interaction between the Numb PTB domain and the β3 integrin cytoplasmic tail. A model of β3 residues Thr-741 to Thr-751 docked in the Numb-PTB domain binding site was generated based on the structure of the Numb–Nak complex. (B) Introduction of a charged residue at the −5 position and a noncharged residue at the +2 position relative to the Tyr or the β3 NPXY motif inhibit PTB domain binding. GST-PTB domain fusion proteins were incubated with beads coated with recombinant β3, β3(E749S), or β3(A742D) tails. Bound proteins were fractionated by SDS/PAGE and GST-PTB domains were detected by Western blotting with anti-GST antibodies. Loading of the recombinant integrin tails on the beads was assessed by Coomassie blue staining. (C) Introduction of an uncharged residue at the −5 position and a charged residue at the +2 position relative to the Tyr or the β7 NPXY motif enhances PTB domain binding. The binding of recombinant GST-PTB domain fusion proteins to β7, β7(S780E), β7(D773A), or β7(D773F) integrin cytoplasmic tails was assessed as described in B.
The most critical residues for the binding of the Nak peptide to the Numb PTB domain are Asn at −3 and Phe at +0 within the NMSF motif of the Nak peptide (numbers are relative to the conserved Tyr or Phe of the NPXY/F motif; ref. 11). In our model, Tyr-747 in the β3 tail corresponds to Phe at +0 of the Nak peptide, and mutation of this residue disrupts Numb–β3 integrin interactions. However, the β1A, β2, and β7 tails all contain Asn at the −3 position and Tyr at the +0 position, yet bind weakly to Numb, indicating that these residues do not contribute to the selectivity of binding. We therefore examined residues upstream and downstream of the NPXY/F motif.
Upstream of the NPXY/F motif, the aromatic ring of the Phe residue at −5 of the Nak peptide inserts in a hydrophobic cavity, and mutation of this residue to Ala produces a moderate reduction in the affinity of the interaction (11), suggesting that this contact is important. None of the integrin β subunits contain a large hydrophobic residue in this position. However, whereas the Numb-binding integrins, β3 and β5, have an Ala residue in position −5, the nonbinding integrins β2 and β7 contain a polar or charged side chain, and β1A contains a Gly, suggesting that a nonpolar residue in this position is necessary for Numb binding. The side chains of residues −6 and −4 are predicted to be oriented toward the solvent, and are polar in all integrin tails, with the exception of Met at −6 in β5. Therefore, positions −6 and −4 are not expected to be determinants of integrin selectivity in Numb binding.
Nak residues C-terminal to the NMSF motif adopt a β-turn conformation and mutation of Asp at +2 into Ala severely reduces binding of Nak to Numb (11). In the Numb-binding integrins, β3 and β5, position +2 is occupied by residues with charged groups, whereas in the nonbinding integrins it is a Ser.
Therefore, modeling suggests that residues in positions −5 and +2 with respect to the Tyr of the NPXY/F motif contribute to PTB-domain interactions and may play a role in the selectivity of integrin PTB domain recognition. To test this prediction we introduced mutations at the −5 and +2 positions of β3 and β7 tails and examined their PTB domain-binding activities.
Loss-of-function mutations confirmed the importance of the +2 and −5 positions for integrin–PTB domain interaction. β3 tails containing Glu-749 to Ser substitutions at the +2 position, or Ala-742 to Asp substitutions at the −5 position, exhibited reduced binding to Dok-1 and Numb PTB domains (Fig. 5B), indicating that the +2 and −5 residues of β3 tails contribute to PTB domain binding. Little effect on Dab1 and Dab2 binding was observed (Fig. 5B and data not shown), consistent with their binding to β3 primarily by means of its C-terminal NITY motif (Fig. 2D).
Conversely, we mutated the same positions in the β7 tail to assess whether we could generate gain of PTB domain-binding function. We first replaced Asp-773 at the −5 position of β7 tails with more hydrophobic residues. Substitution with the uncharged Ala [β7(D773A)] dramatically increased Dok PTB domain binding to β7 tails, but had little effect on Numb or Dab2 binding (Fig. 5C). However, substitution with the more hydrophobic Phe [β7(D773F)] also increased binding of the Dab2 (Fig. 5C) and Dab1 (data not shown) PTB domains. Substitution of Glu for Ser-780 at the +2 position of β7 [β7(S780E)] tails increased Dok-1 PTB domain binding (Fig. 5C), but had little effect on binding to Numb or Dab2. Therefore, both gain- and loss-of-function experiments confirmed that the residues at −5 and +2 positions of the integrin β tails contribute to the PTB domain-binding interaction. Notably, the ability of substitutions at the −5 position of PTB domain ligands has previously been shown to switch binding specificity for PTB domains, both in vitro and in vivo (4, 38–40), reinforcing our conclusion that the binding of integrin β tails to PTB domains resembles classical PTB domain–ligand interactions.
Tyrosine phosphorylation of the NPXY motifs in integrin β tails by Src family kinases is important in cell migration (41), hemostasis (42), and transformation (43). PTB domain recognition of peptide ligands can be promoted or inhibited by NPXY phosphorylation (21). For integrin–PTB domain interactions, the binding of talin F3 subdomain (7) and ICAP-1α (6) to integrin β tails is phosphorylation-independent, whereas the binding of Shc to the β3 tail is promoted by tyrosine phosphorylation (25). Thus, integrin β tail tyrosine phosphorylation may serve as switch that controls which of the alternating groups of PTB-containing proteins can bind to the integrin. A structural determinant important for PTB domain phosphate recognition is the presence of a strongly basic pocket (11). This pocket typically contains two arginine residues that coordinate the phosphate moiety of the NPXpY ligand. Integrin β tail phosphorylation blocks interactions with talin (44), and the pocket in the talin PTB domain lacks these arginines. Instead, β3 Tyr-747 points into an acidic and hydrophobic pocket formed by the ends of two β strands of the PTB domain on one side, and a reverse turn of the integrin β tail on the other (8). Thus, the nature of this binding pocket in the PTB domain protein is predicted to specify the effect of integrin-tyrosine phosphorylation on the particular integrin-binding partner.
Here we have demonstrated that PTB domains from 17 different proteins (Table 3) can bind integrin β tails in vitro, and furthermore, full-length PTB domain-containing proteins can bind integrin β tails in pull-down and coimmunoprecipitation assays. There are a large number of PTB domain-containing proteins [at least 58 in humans (45)], plus many FERM (band 4.1 and ezrin/radixin/moesin) domain, PTB-like domain-containing proteins [85 in humans (45)], in addition to those studied here. Thus, the remarkable conservation of the NPXY/F motif in integrin β subunits and its role in interaction with a number of PTB domains suggests that this interaction is paradigmatic for a wide variety of integrin-signaling events. Further work will be needed to test the roles of PTB domain–integrin interactions in vivo, using endogenous proteins. However, based on the results reported here, we propose general principles of integrin–cytoplasmic protein interaction: (i) Integrin β cytoplasmic domains use their conserved NPXY/F motifs to bind to cytoplasmic proteins that contain PTB (or PTB-like) modules. (ii) The nature of the integrin β tail residues at the −5 and +2 positions (relative to the Tyr/Phe of NPXY/F motif) specifies preferential interaction with PTB domains. (iii) Integrin phosphorylation-regulated changes in PTB domain-binding specificity are molecular toggle switches that designate biological responses to integrin-dependent adhesion. These general principles of integrin interaction with cytoplasmic proteins may provide useful algorithms for deciphering the integrin-signaling code.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Cell Migration Consortium, the Susan G. Komen Breast Cancer Foundation, and the American Heart Association. This is publication no. 15268-VB from The Scripps Research Institute.
Abbreviations
- CHO
Chinese hamster ovary
- PTB
phosphotyrosine binding
- IRS
insulin receptor substrate
- ICAP
integrin cytoplasmic domain-associated protein
- PH domain
pleckstrin homology domain
- Nak
Numb-associated kinase
References
- 1.Hynes R O. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 2.Liu S, Calderwood D A, Ginsberg M H. J Cell Sci. 2000;113:3563–3571. doi: 10.1242/jcs.113.20.3563. [DOI] [PubMed] [Google Scholar]
- 3.Ulmer T S, Yaspan B, Ginsberg M H, Campbell I D. Biochemistry. 2001;40:7498–7508. doi: 10.1021/bi010338l. [DOI] [PubMed] [Google Scholar]
- 4.Trub T, Choi W E, Wolf G, Ottinger E, Chen Y, Weiss M, Shoelson S E. J Biol Chem. 1995;270:18205–18208. doi: 10.1074/jbc.270.31.18205. [DOI] [PubMed] [Google Scholar]
- 5.Dans M, Gagnoux-Palacios L, Blaikie P, Klein S, Mariotti A, Giancotti F G. J Biol Chem. 2001;276:1494–1502. doi: 10.1074/jbc.M008663200. [DOI] [PubMed] [Google Scholar]
- 6.Chang D D, Hoang B Q, Liu J, Springer T A. J Biol Chem. 2002;277:8140–8145. doi: 10.1074/jbc.M109031200. [DOI] [PubMed] [Google Scholar]
- 7.Calderwood D A, Yan B, de Pereda J M, Alvarez B G, Fujioka Y, Liddington R C, Ginsberg M H. J Biol Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Alvarez B, de Pereda J M, Calderwood D A, Ulmer T S, Critchley D R, Campbell I D, Ginsberg M H, Liddington R C. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 9.Liddington R C, Ginsberg M H. J Cell Biol. 2002;158:833–839. doi: 10.1083/jcb.200206011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes R. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 11.Zwahlen C, Li S C, Kay L E, Pawson T, Forman-Kay J D. EMBO J. 2000;19:1505–1515. doi: 10.1093/emboj/19.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borg J P, Straight S W, Kaech S M, Taddeo-Borg M, Kroon D E, Karnak D, Turner R S, Kim S K, Margolis B. J Biol Chem. 1998;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- 13.Dho S E, French M B, Woods S A, McGlade C J. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi T, Matozaki T, Inagaki K, Tsuda M, Fukunaga K, Kitamura Y, Kitamura T, Shii K, Yamanashi Y, Kasuga M. EMBO J. 1999;18:1748–1760. doi: 10.1093/emboj/18.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaff M, Liu S, Erle D J, Ginsberg M H. J Biol Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- 16.Calderwood D A, Zent R, Grant R, Rees D J, Hynes R O, Ginsberg M H. J Biol Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 17.O'Toole T E, Mandelman D, Forsyth J, Shattil S J, Plow E F, Ginsberg M H. Science. 1991;254:845–847. doi: 10.1126/science.1948065. [DOI] [PubMed] [Google Scholar]
- 18.Calderwood D A, Huttenlocher A, Kiosses W B, Rose D M, Woodside D G, Schwartz M A, Ginsberg M H. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 19.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Margolis B, Borg J P, Straight S, Meyer D. Kidney Int. 1999;56:1230–1237. doi: 10.1046/j.1523-1755.1999.00700.x. [DOI] [PubMed] [Google Scholar]
- 21.Forman-Kay J D, Pawson T. Curr Opin Struct Biol. 1999;9:690–695. doi: 10.1016/s0959-440x(99)00031-7. [DOI] [PubMed] [Google Scholar]
- 22.Wolf G, Trub T, Ottinger E, Groninga L, Lynch A, White M F, Miyazaki M, Lee J, Shoelson S E. J Biol Chem. 1995;270:27407–27410. doi: 10.1074/jbc.270.46.27407. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Panchenko A R, Shoemaker B A, Thiessen P A, Geer L Y, Bryant S H. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowan K J, Law D A, Phillips D R. J Biol Chem. 2000;275:36423–36429. doi: 10.1074/jbc.M004068200. [DOI] [PubMed] [Google Scholar]
- 26.Chang D D, Wong C, Smith H, Liu J. J Cell Biol. 1997;138:1149–1157. doi: 10.1083/jcb.138.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz A, Duggan K, Buck C A, Beckerle M C, Burridge K. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- 28.Dulabon L, Olson E C, Taglienti M G, Eisenhuth S, McGrath B, Walsh C A, Kreidberg J A, Anton E S. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 29.Lo S H, An Q, Bao S, Wong W K, Liu Y, Janmey P A, Hartwig J H, Chen L B. J Biol Chem. 1994;269:22310–22319. [PubMed] [Google Scholar]
- 30.Hosooka T, Noguchi T, Nagai H, Horikawa T, Matozaki T, Ichihashi M, Kasuga M. Mol Cell Biol. 2001;21:5437–5446. doi: 10.1128/MCB.21.16.5437-5446.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan B, Calderwood D A, Yaspan B, Ginsberg M H. J Biol Chem. 2001;276:28164–28170. doi: 10.1074/jbc.M104161200. [DOI] [PubMed] [Google Scholar]
- 32.Goldmann W H. Biochem Biophys Res Commun. 2000;271:553–557. doi: 10.1006/bbrc.2000.2653. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Lee C H, Mandiyan V, Borg J P, Margolis B, Schlessinger J, Kuriyan J. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell B W, Lanier L M, Frank R, Gertle F B, Cooper J A. Mol Cell Biol. 1999;19:5179–5188. doi: 10.1128/mcb.19.7.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S C, Songyang Z, Vincent S J, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman-Kay J, Pawson T. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S C, Zwahlen C, Vincent S J, McGlade C J, Kay L E, Pawson T, Forman-Kay J D. Nat Struct Biol. 1998;5:1075–1083. doi: 10.1038/4185. [DOI] [PubMed] [Google Scholar]
- 37.Songyang Z, Yamanashi Y, Liu D, Baltimore D. J Biol Chem. 2001;276:2459–2465. doi: 10.1074/jbc.M005504200. [DOI] [PubMed] [Google Scholar]
- 38.Isakoff S J, Yu Y P, Su Y C, Blaikie P, Yajnik V, Rose E, Weidner K M, Sachs M, Margolis B, Skolnik E Y. J Biol Chem. 1996;271:3959–3962. doi: 10.1074/jbc.271.8.3959. [DOI] [PubMed] [Google Scholar]
- 39.van der Geer P, Wiley S, Lai V K, Olivier J P, Gish G D, Stephens R, Kaplan D, Shoelson S, Pawson T. Curr Biol. 1995;5:404–412. doi: 10.1016/s0960-9822(95)00081-9. [DOI] [PubMed] [Google Scholar]
- 40.Borg J P, Ooi J, Levy E, Margolis B. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai T, Jove R, Fassler R, Mosher D F. Proc Natl Acad Sci USA. 2001;98:3808–3813. doi: 10.1073/pnas.240456398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Law D A, DeGuzman F R, Heiser P, Ministri-Madrid K, Killeen N, Phillips D R. Nature. 1999;401:808–811. doi: 10.1038/44599. [DOI] [PubMed] [Google Scholar]
- 43.Datta A, Shi Q, Boettiger D E. Mol Cell Biol. 2001;21:7295–7306. doi: 10.1128/MCB.21.21.7295-7306.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapley P, Horwitz A, Buck C A, Duggan K, Rohrschneider L. Oncogene. 1989;4:325–333. [PubMed] [Google Scholar]
- 45.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.