Abstract
Under high-light conditions, photoprotective mechanisms minimize the damaging effects of excess light. A primary photoprotective mechanism is thermal dissipation of excess excitation energy within the light-harvesting complex of photosystem II (LHCII). Although roles for both carotenoids and specific polypeptides in thermal dissipation have been reported, neither the site nor the mechanism of this process has been defined precisely. Here, we describe the physiological and molecular characteristics of the Chlamydomonas reinhardtii npq5 mutant, a strain that exhibits little thermal dissipation. This strain is normal for state transition, high light–induced violaxanthin deepoxidation, and low light growth, but it is more sensitive to photoinhibition than the wild type. Furthermore, both pigment data and measurements of photosynthesis suggest that the photosystem II antenna in the npq5 mutant has one-third fewer light-harvesting trimers than do wild-type cells. The npq5 mutant is null for a gene designated Lhcbm1, which encodes a light-harvesting polypeptide present in the trimers of the photosystem II antennae. Based on sequence data, the Lhcbm1 gene is 1 of 10 genes that encode the major LHCII polypeptides in Chlamydomonas. Amino acid alignments demonstrate that these predicted polypeptides display a high degree of sequence identity but maintain specific differences in their N-terminal regions. Both physiological and molecular characterization of the npq5 mutant suggest that most thermal dissipation within LHCII of Chlamydomonas is dependent on the peripherally associated trimeric LHC polypeptides.
INTRODUCTION
In natural environments, photosynthetic organisms are exposed to a range of fluctuating light intensities. At low light intensities, an increase in photon flux density correlates with increased photosynthetic carbon fixation. However, above a certain threshold, carbon fixation becomes saturated and photosynthesis is incapable of using all of the energy absorbed by the light-harvesting complexes (LHCs). Under these conditions of excess light absorption, the chloroplast lumen becomes highly acidic, the electron transport chain becomes reduced, and excitation energy accumulates within the light-harvesting complexes of photosystem II (LHCII). Excess excitation of LHCII could result in an increase in the half-life of singlet chlorophyll a in the pigment bed and the consequent production of triplet chlorophyll a and singlet oxygen.
If not detoxified immediately, singlet oxygen can cause protein modification and lipid peroxidation. Furthermore, once initiated, lipid peroxidation becomes autocatalytic, resulting in massive membrane photodestruction (Niyogi, 1999). Excess light also depletes the NADP+ pool, causing an increase in the rate of electron flow from the donor side of photosystem I (PSI) to oxygen, generating superoxide and hydrogen peroxide (Asada, 1999).
Plants respond to the absorption of excess light with a suite of short-term and long-term photoprotective mechanisms that minimize damage. Over the short term, carotenoids have photoprotective functions in detoxifying and limiting the formation of singlet oxygen. The xanthophyll lutein occupies the L1 and L2 sites of LHC polypeptides and can quench ∼80% of the triplet chlorophyll generated within LHCII (Peterman et al., 1997). The remaining triplet chlorophyll can react with oxygen to generate singlet oxygen.
Singlet oxygen can be detoxified by lutein and neoxanthin within LHCII or by zeaxanthin and α-tocopherol in the thylakoid membranes (Croce et al., 1999b; Havaux et al., 2000). α-Tocopherol and zeaxanthin also disrupt autocatalytic membrane lipid peroxidation (Niyogi, 1999). Superoxides generated on the acceptor side of PSI are detoxified by a series of membrane-associated and stromal enzymes, including superoxide dismutase and ascorbate peroxide (Asada, 1999).
A first line of defense in photoprotection is the thermal dissipation of excess excitation energy in LHCII. This process decreases energy transfer to photosystem II (PSII) and reduces the formation of triplet chlorophyll in LHCII, diminishing the production of reactive oxygen species. Energy dissipation within LHCII or thermal dissipation results in nonphotochemical quenching of chlorophyll fluorescence (NPQ) that is reversed rapidly upon dissipation of the thylakoid membrane pH gradient (ΔpH) (Horton et al., 1996; Niyogi, 1999). Other processes that cause NPQ are state transitions and long-term inhibition of PSII photochemistry.
State transitions occur after a shift to high light and are caused by the occupancy of the quinone binding site of the cytochrome b6f complex by plastoquinol (Zito et al., 1999) and the subsequent movement of the peripheral LHCII antenna (LhcIIb) to PSI (Bassi et al., 1988; Vallon et al., 1991). The mechanism of inhibition of PSII photochemistry is not well understood, although there is evidence that a stable quenching center associated with the accumulation of zeaxanthin is involved (Färber et al., 1997; Verhoeven et al., 1998; Gilmore and Ball, 2000). The accumulation of photodamaged reaction centers also is responsible for part of the inhibition of PSII photochemistry (Horton et al., 1996).
Several investigators have demonstrated that zeaxanthin and lutein play critical roles in thermal dissipation. A strong correlation between thermal dissipation and zeaxanthin accumulation has been observed in a variety of plants (Demmig-Adams et al., 1996). Under light-limited conditions, the epoxidation of zeaxanthin results in the formation of violaxanthin, with antheraxanthin as an intermediate. Upon exposure of plants to excess excitation energy, the thylakoid lumen becomes acidic; this acidification activates violaxanthin deepoxidase and leads to the formation of zeaxanthin.
Mutants defective in violaxanthin deepoxidation in Chlamydomonas reinhardtii (npq1) and Arabidopsis (npq1) exhibit diminished levels of NPQ (Niyogi et al., 1997a, 1998). In wild-type Chlamydomonas, there is an initial rapid increase in NPQ (representing ∼70% of the total) and a second slower phase (representing ∼30%) after exposure of cells to high light; only the second phase is lacking in the Chlamydomonas npq1 mutant (Niyogi et al., 1997a). By contrast, the Arabidopsis npq1 mutant exhibits a decrease in reversible NPQ of ∼80% relative to wild-type plants (Niyogi et al., 1998). The Chlamydomonas lor1 and Arabidopsis lut2 mutants, both of which are unable to synthesize lutein, also exhibit defects in thermal dissipation (Niyogi et al., 1997b; Pogson et al., 1998). Specifically, zeaxanthin-independent thermal dissipation is absent.
The npq1 lor1 and npq1 lut2 double mutants of Chlamydomonas and Arabidopsis exhibit essentially no thermal dissipation (Niyogi et al., 1997b, 2001). Lutein might affect thermal dissipation directly and/or indirectly by causing a change in LHCII structure and composition (Niyogi et al., 1997b, 2001). Finally, in a mutant of Arabidopsis that accumulates zeaxanthin constitutively and is unable to make antheraxanthin and violaxanthin (npq2), the development of NPQ is more rapid than in wild-type cells, although the extent of NPQ remains unchanged (Niyogi et al., 1998).
Recent research has focused on whether the role of zeaxanthin in thermal dissipation is direct, with zeaxanthin quenching singlet chlorophyll (Gilmore et al., 1996a), or indirect, with zeaxanthin serving as an allosteric regulator of thermal dissipation (Horton et al., 1991, 2000). In the indirect model, the planar structure of zeaxanthin promotes a conformational change in LhcIIb (possibly aggregation) that favors quenching by altering chlorophyll–chlorophyll interactions (Phillip et al., 1996). In the direct model, the energy in singlet chlorophyll is transferred to and then dissipated thermally by zeaxanthin.
Recent measurements of the singlet excited energy states of zeaxanthin and violaxanthin suggest that both molecules are theoretically capable of accepting energy from singlet chlorophyll (Polívka et al., 1999; Frank et al., 2000). However, in vivo, the capacity for direct energy transfer is dependent on the proximity of the xanthophyll to the chlorophyll molecule and their respective orientations.
The actual site of thermal dissipation within LHCII and the polypeptides critical for this process are just beginning to be elucidated. The light-harvesting components associated with PSII are the core light-harvesting polypeptides (CP43 and CP47) and LHCII, which consists of both the monomeric or minor LHCII polypeptides (CP26, CP29, and CP24) and the peripheral trimers (LhcIIb), which constitute the bulk of the complex. Both the trimers (Horton et al., 1991) and the monomers (Bassi et al., 1993) have been hypothesized to play a role in thermal dissipation.
A number of investigators have suggested that lumen acidification facilitates the protonation of glutamates of the CP26 and CP29 polypeptides, thereby eliciting conformational changes that promote thermal dissipation (Walters et al., 1996; Bassi et al., 1997; Pesaresi et al., 1997). However, although CP26 and CP29 bind xanthophylls that undergo deepoxidation, the majority of the xanthophylls are bound to trimeric LHCII (LhcIIb) (Verhoeven et al., 1999). Furthermore, in experiments with vascular plants, the efficiency of xanthophyll deepoxidation in the trimeric LHCII (LhcIIb) was observed to be approximately equivalent to that in CP26 and greater than that in CP29 (Färber et al., 1997; Ruban et al., 1999).
Analyses of thermal dissipation in chlorophyll b–deficient mutants in barley, which accumulate fewer LHCII trimers, have been used to support the idea that thermal dissipation is associated with the minor LHCII polypeptides (Andrews et al., 1995; Gilmore et al., 1996b). However, some chlorophyll b–deficient strains exhibit reduced thermal dissipation (Härtel et al., 1996; Gilmore et al., 2000), accumulate less of several of the LHCII and LHCI polypeptides (Bossmann et al., 1997), and exhibit defects in thylakoid structure (Knoetzel and Simpson, 1991). The pleiotropic nature of these mutants complicate conclusions regarding the role of LHCII trimers in thermal dissipation.
Furthermore, in recent experiments, the ability of intermittent light–grown plants transferred to continuous light to perform NPQ was correlated with increasing levels of chlorophyll b and trimeric LHCII (LhcIIb), suggesting a role for the trimers in thermal dissipation (Chow et al., 2000). Finally, Arabidopsis plants harboring antisense constructs for Lhcb4 (which encodes CP26) and Lhcb5 (which encodes CP29) exhibited only minor changes in chlorophyll fluorescence. The kinetics and capacity of NPQ development were essentially unaltered, and the authors concluded that the minor LHCII polypeptides (CP26 and CP29) are not sites of thermal dissipation (Andersson et al., 2001).
One polypeptide shown to be critical for thermal dissipation is PsbS, a PSII-associated polypeptide that is a member of the LHC superfamily of proteins. Arabidopsis plants null for psbS exhibited no thermal dissipation when exposed to high light, although the mutation had no effect on photosynthetic parameters in low light, LHCII polypeptide accumulation, or violaxanthin deepoxidation kinetics (Li et al., 2000). Recent site-directed mutagenesis experiments suggest that PsbS is protonated in high light and that this protonation is essential for its function (Li et al., 2002). Although the exact location of PsbS within the photosynthetic apparatus is not known, there is evidence that it is not tightly associated with the LHCII–PSII supercomplex, which contains the PSII core polypeptides (including D1, D2, CP47, and CP43), CP26, CP29, and one LHCII trimer (Boekema et al., 2000; Nield et al., 2000a, 2000b).
PsbS may interact with the more peripherally associated LHCII trimer pool (Harrer et al., 1998; Nield et al., 2000a). Protonation of PsbS in high light might stimulate conformational changes in LHCII that promote thermal dissipation. Alternatively, thermal dissipation might occur in a PsbS-containing complex that efficiently gathers and eliminates excitation energy harvested by LHCII. Interestingly, PsbS has not yet been identified in Chlamydomonas; no gene with high sequence identity or similarity to PsbS from vascular plants is represented in the Chlamydomonas cDNA database.
To identify the components important for thermal dissipation, we have characterized npq5, a Chlamydomonas npq mutant isolated originally in a screen based on video imaging of chlorophyll fluorescence (Niyogi et al., 1997a). Molecular and physiological analyses of this mutant suggest a role for LHCII trimers in thermal dissipation.
RESULTS
npq5 Is Defective Specifically in Thermal Dissipation
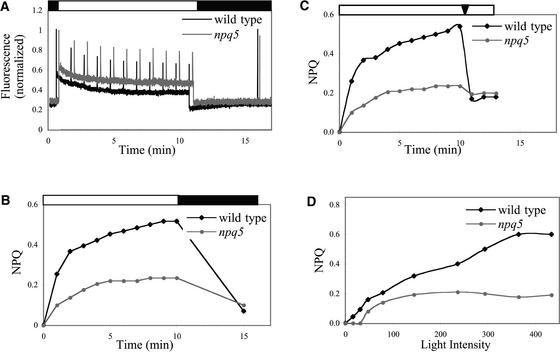
The induction kinetics and extent of NPQ in wild-type cells and the npq5 mutant were examined by measurements of modulated fluorescence. The data presented in Figure 1 demonstrate that npq5 is defective in thermal dissipation. After 10 min of high light (350 μmol·m−2·s−1; 3.5 times the light intensity used for growth), the level of NPQ in the npq5 mutant was 45% of that measured for wild-type cells. Furthermore, 85% of the NPQ that developed in wild-type cells in high light was reversed by a 5-min exposure to low-fluence far-red light, but only 50% was reversed in the npq5 strain (Figure 1B).
Figure 1.
Modulated Fluorescence and NPQ in Wild-Type and npq5 Mutant Cells.
(A) Chlorophyll fluorescence during induction and relaxation of NPQ.
(B) NPQ calculated from fluorescence as (Fm − Fm′)/Fm′.
(C) Relaxation of NPQ by the addition of nigericin.
(D) Light intensity dependence of NPQ.
The curves for wild-type cells and the npq5 mutant are shown in black and gray, respectively. The data shown in (B) to (D) are average values of experiments obtained with four separately grown wild-type and mutant cultures. The wild-type cultures were at cell concentrations between 1.6 × 106 and 1.8 × 106 cells/mL [2.8 to 3.2 μg chlorophyll (a + b)/mL], and the mutant cultures were at cell concentrations between 1.7 × 106 and 1.9 × 106 cells/mL [2.4 to 2.7 μg chlorophyll (a + b)/mL]. For (A) and (B), white bars above the graphs indicate illumination with 350 μmol·m−2·s−1, and black bars indicate illumination with ≈1 μmol·m−2·s−1 far-red light (690 to 710 nm). For (C), the arrowhead at top indicates the time at which nigericin was added. For (D), the level of NPQ attained after 10 min at each light intensity is shown. For (B) to (D), se values are <3%; representative traces are shown in (A).
The addition of the proton uncoupler nigericin to cells exposed to light for 10 min reversed 70% of the NPQ that developed in wild-type cells but only 10% of the NPQ that developed in the mutant strain (Figure 1C), supporting the conclusion that npq5 is defective specifically in the thermal dissipation component of NPQ. Finally, Figure 1D shows the levels of NPQ attained after exposing both mutant and wild-type cells to different light intensities. The npq5 mutant strain showed reduced NPQ at all intensities. It should be mentioned that the fluorescence maximum used for the determination of NPQ in both the wild-type and mutant strains was a “true” fluorescence maximum as determined by fast fluorescence induction (Kautsky curves) (data not shown).
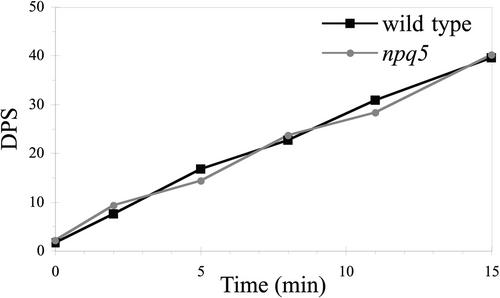
Because violaxanthin deepoxidation plays an important role in thermal dissipation, the levels of xanthophylls were quantified after exposure of cells to high light (1100 μmol·m−2·s−1) for 1 to 15 min. Wild-type cells and the npq5 strain showed no differences in the kinetics or extent of violaxanthin deepoxidation during exposure to high light (Figure 2). These results suggest that the kinetics of pH gradient generation in high light and the magnitude of this gradient are similar in wild-type cells and the npq5 strain. However, we cannot be certain that xanthophylls formed in the mutant strain are incorporated properly into the thylakoid membranes.
Figure 2.
Deepoxidation after Exposure of Cells to 1100 μmol· m−2·s−1.
Deepoxidation state (DPS) was calculated as (zeaxanthin + antheraxanthin)/(zeaxanthin + antheraxanthin + violaxanthin). The wild-type culture used was at a concentration of 1.65 × 106 cells/mL [2.82 μg chlorophyll (a + b)/mL], and the npq5 mutant culture was at 1.7 × 106 cells/mL [2.48 μg chlorophyll (a + b)/mL].
npq5 Is Capable of a State Transition
Because a state transition also can contribute to NPQ, the ability of the npq5 strain to perform state transitions was examined. Changes in maximum fluorescence were measured after the transition of cells from state 1 to state 2. Cells were maintained in state 1 by illumination with far-red light, which preferentially excites PSI and causes oxidation of the plastoquinone pool. A transition to state 2 was induced by placing cultures in the dark and adding Glc plus Glc oxidase to scavenge molecular oxygen (Bulté and Wollman, 1990). The transition to state 2 caused a 17% decrease in maximal fluorescence for both wild-type cells and the npq5 mutant (17.8% ± 0.4% for the wild type and 17.4% ± 0.7% for npq5 [n = 4]), suggesting that state transition is similar in the two strains.
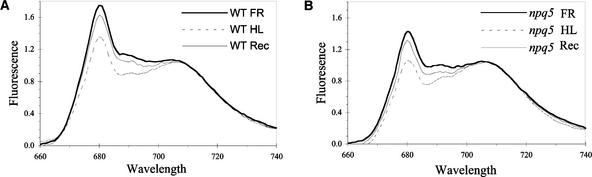
We also used 77K fluorescence emission analyses to evaluate state transitions. The ratio of fluorescence emitted at 680 nm to that emitted at 710 nm (F680/F710) reflects the ratio of antenna chlorophyll functionally associated with PSII relative to PSI. Measurements were performed on three samples: (1) treated with far-red light for 10 min; (2) treated with far-red light and then high light (350 μmol·m−2·s−1) for 10 min; and (3) treated with far-red light, high light, and then 10 min of far-red light.
As shown in Figure 3 and summarized in Table 1, the F680/F710 ratio for both wild-type and npq5 cells decreased by ∼20% after high-light treatment and recovered by >70% in far-red light. Recovery did not occur in the presence of 100 mM NaF, which inhibits a state 2-to-state 1 transition by inhibiting the LHCII phosphatase (data not shown). These results demonstrate that the npq5 mutant is capable of a high light–induced state transition. However, the F680/F710 ratios in both state 1 and state 2 were ∼20% lower in the npq5 mutant than in the wild-type strain, suggesting that the mutant has either a smaller PSII antenna or a lower PSII/PSI ratio.
Figure 3.
77K Fluorescence Emission Spectra of Wild-Type and npq5 Mutant Cells Treated with Far-Red Light, Far-Red Light followed by High-Light, or Far-Red Light, High Light, and Then Far-Red Light.
(A) Wild type (WT).
(B) npq5.
Samples were excited at 435 nm, and fluorescence emission was measured between 650 and 750 nm. The wild-type culture used was at a concentration of 1.75 × 106 cells/mL [2.95 μg chlorophyll (a + b)/mL], and the npq5 mutant culture was at 1.83 × 106 cells/mL [2.55 μg chlorophyll (a + b)/mL]. Spectra were measured two times and yielded essentially identical data, and data from one of the two replicates is shown. FR, far-red light; HL, far-red light followed by high light; Rec, far-red light, high light, and then far-red light.
Table 1.
State Transition in Wild-Type and npq5 Mutant Cells
| F680/F710
|
Change in F680/F710 (%) |
||||
|---|---|---|---|---|---|
| Sample | FR | HL | Recovery in FR | HL | Recovery in FR |
| Wild type | 1.66 | 1.29 | 1.55 | −22 | 70 |
| npq5 | 1.39 | 1.06 | 1.29 | −23 | 70 |
F680/F710 ratios correspond to those shown in Figure 3. Ratios are shown for wild-type and npq5 mutant cells treated with far-red light (FR), far-red light followed by high light (HL), and far-red light, high light, and then far-red light (Recovery in FR). The percentage change in the ratios after HL treatment, and the percentage of the initial decrease that recovers after the final far-red light exposure, also are shown.
Pigment Analysis Is Consistent with Fewer LHCII Trimers
The results of quantification of chlorophyll and carotenoid levels in the wild-type and npq5 mutant strains are consistent with reduced LHCII trimer accumulation in the mutant strain. As shown in Table 2, the npq5 mutant had a higher chlorophyll a/b ratio than wild-type cells. Because LHCII trimers have a lower chlorophyll a/b ratio than LHCI, CP26, or CP29 (Green and Durnford, 1996; Bassi et al., 1997), an increase in this ratio is consistent with fewer LHCII trimers relative to the rest of the photosynthetic apparatus. The 34% decrease in the absolute amount of neoxanthin and loroxanthin per cell in the mutant strain also is consistent with a significant reduction in LHCII trimer abundance, because >90% of thylakoid-bound neoxanthin is associated with these trimers (Croce et al., 1999a).
Table 2.
Pigment Levels in Wild-Type and npq5 Mutant Cells
| Parameter | Wild Type | npq5 | Ratio of npq5 to Wild Type |
|---|---|---|---|
| Chlorophyll a/b | 2.30 ± 0.08 | 2.55 ± 0.05 | 1.10 |
| Neo and Lor | 0.19 ± 0.01 | 0.11 ± 0.02 | 0.66 |
| Lutein | 0.20 ± 0.02 | 0.17 ± 0.02 | 0.82 |
| Chlorophyll b | 0.63 ± 0.05 | 0.46 ± 0.04 | 0.73 |
| Chlorophyll a | 1.40 ± 0.02 | 1.14 ± 0.06 | 0.82 |
| Xanthophyll pool | 0.12 ± 0.01 | 0.10 ± 0.01 | 0.85 |
Carotenoid and chlorophyll extracts from wild-type and npq5 cultures were analyzed. Chlorophyll was measured spectrophotometrically, and the carotenoids were quantified by HPLC. Levels of neoxanthin (Neo) and loroxanthin (Lor), lutein, chlorophyll b, chlorophyll a, and the xanthophyll pool (zeaxanthin + antheraxanthin + violaxanthin) are given in fmol/cell. The wild-type cultures used were at cell concentrations between 1.5 × 106 and 1.8 × 106 cells/mL [2.7 to 3.2 μg chlorophyll (a + b)/mL], and the npq5 mutant cultures were at cell concentrations between 1.6 × 106 and 1.9 × 106 cells/mL [2.3 and 2.7 μg chlorophyll (a + b)/mL]. Data shown are means ± se (n = 4). The ratio of the means also is shown.
Furthermore, the npq5 mutant contained 27% less chlorophyll b per cell than the wild-type strain; in wild-type cells, ∼80% of the chlorophyll b per cell was associated with the LHCII trimers. These data, along with the 77K fluorescence data and the photosynthetic parameters discussed below, suggest that the npq5 mutant has approximately one-third fewer LHCII trimers than wild-type cells. In wild-type cells, the LHCII trimers account for 50% of the chlorophyll associated with PSII, whereas the minor LHC polypeptides account for 15%, and CP43 and CP47 of the PSII core account for 35%. Thus, the reduction in LHCII trimers would result in a reduction in PSII antenna chlorophyll by ∼20%, in agreement with the decreased F680/F710 ratio observed in the mutant strain.
Photosynthetic Parameters and Growth
Photosynthesis was monitored as the level of oxygen evolution and by modulated fluorescence. As shown in Table 3, photosynthetic efficiency, as estimated by variable PSII fluorescence/maximum PSII fluorescence ratio (Fv/Fm) was similar in wild-type and npq5 mutant cells. Oxygen evolution measurements demonstrated that on a per cell basis, neither the maximum photosynthetic rate (measured at >200 μmol·m−2·s−1) nor photosynthetic efficiency (measured between 0 and 50 μmol·m−2·s−1) was significantly different in npq5 and wild-type cells. However, the se values generated for these measurements were between 5 and 20%; therefore, the mutant and wild-type strains could differ in photosynthetic efficiency by up to 25%.
Table 3.
Photosynthetic Parameters in Wild-Type and npq5 Mutant Cells
| Sample | Pmaxa | Φb | Fv/Fm | Doubling Time (h) |
|---|---|---|---|---|
| Wild type | 248 ± 42 | 1.72 ± 0.1 | 0.785 ± 0.01 | 13.2 ± 1.1 |
| npq5 | 289 ± 20 | 1.57 ± 0.1 | 0.783 ± 0.02 | 13.7 ± 0.8 |
Pmax (maximal photosynthetic rate), Φ (photosynthetic efficiency), Fv/Fm, and doubling time at 100 μmol·m−2·s−1 were determined for the wild-type and npq5 mutant strains. For measurements of Pmax, Φ, and Fv/Fm, the wild-type cultures used were at cell concentrations ranging from 1.6 × 106 to 1.8 × 106 cells/mL [2.8 to 3.2 μg chlorophyll (a + b)/mL], and the npq5 mutant cultures were at cell concentrations ranging from 1.6 × 106 to 1.9 × 106 cells/mL [2.3 to 2.8 μg chlorophyll (a + b)/mL]. Data shown are means ± se (n = 6 for Pmax, Φ, and Fv/Fm, n = 3 for doubling time).
Measured in μmol O2·h−1·10−12 cells.
Measured in μmol O2·h−1·(μmol photon)−1·10−12 cells.
If reported on a per chlorophyll basis, maximum photosynthetic rate was significantly greater for the npq5 mutant than for wild-type cells, suggesting that the mutant contains more PSII reaction centers per chlorophyll (i.e., a smaller antenna). Similarly, a higher maximum photosynthetic rate per chlorophyll was noted for other mutant strains having smaller LHCII antennae (Polle et al., 2001). Data presented in Table 3 show that photoautotrophic growth rates were not significantly different between mutant and wild-type cells under moderate light. The doubling time of photoautotrophically grown cultures in moderate light was ∼13.5 h for both wild-type and npq5 mutant cells.
Photoinhibition Is Greater in npq5
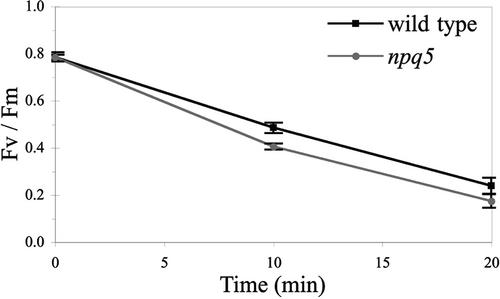
Because thermal dissipation reduces the half-life of singlet excited chlorophyll and decreases the amount of excitation energy transferred to the reaction center under high-light conditions, the mutant strain was expected to be more sensitive to photoinhibition than wild-type cells. We monitored photoinhibition (defined as a decrease in Fv/Fm) in both the wild-type and mutant strains after 10 and 20 min of exposure to 1100 μmol·m−2·s−1 in the presence of lincomycin (which prevents protein synthesis in the chloroplast, which is essential for the repair of PSII reaction centers).
To eliminate the effects of thermal dissipation and state transition on Fv/Fm, cells were exposed to low-fluence far-red light for 4 min before fluorescence measurements. As shown in Figure 4, upon exposure of the cells to 1100 μmol·m−2·s−1, the loss of PSII activity was significantly faster in the npq5 strain than in the wild-type strain. Photosynthetic efficiency decreased, with half-times of 14 and 12 min for wild-type and npq5 mutant cells, respectively.
Figure 4.
Photoinhibition in the Wild-Type and npq5 Strains.
Photosynthetic efficiency (Fv/Fm) was measured after 10 and 20 min of high-light exposure (1100 μmol·m−2·s−1) in the presence of lincomycin. Before measuring Fv/Fm, the cultures were exposed to low-fluence far-red light for 4 min. To calculate the half-time of photosynthetic efficiency in high light, linear trend lines were added (R2 > 0.96 for both). The half-time of efficiency for the wild-type and npq5 strains were 14.1 ± 1.1 min and 12.2 ± 0.4 min, respectively. Points are means ± se (n = 4). The wild-type cultures used were at cell concentrations between 1.5 × 106 and 1.7 × 106 cells/mL [2.7 to 3.1 μg chlorophyll (a + b)/mL], and the npq5 mutant cultures were at cell concentrations between 1.6 × 106 and 1.8 × 106 cells/mL [2.3 and 2.6 μg chlorophyll (a + b)/mL].
npq5 Has an Insertion in a Major LHCII Gene
The npq5 mutant was generated using insertional mutagenesis (Niyogi et al., 1997a). The parental strain, CC-425, a mutant at the ARG7 locus (which encodes argininosuccinate lyase, an enzyme essential for Arg biosynthesis), was transformed with the linearized plasmid pJD67, which contains a wild-type ARG7 gene and pBluescript KS+ vector (Davies et al., 1994; Purton and Rochaix, 1994). Transformants were selected on medium devoid of Arg, grown photoautotrophically to ensure photosynthetic competence, and screened by video imaging of chlorophyll fluorescence (Niyogi et al., 1997a). Because exogenously introduced DNA integrates into the nuclear genome of Chlamydomonas by nonhomologous recombination, mutations generated often were the result of the integration of pJD67 DNA into the genome (the lesions are tagged with insert DNA).
To determine whether the mutant phenotype cosegregated with the introduced wild-type ARG7 gene in the npq5 mutant strain, a mating-type-plus-npq5 strain was crossed to a mating-type-minus-arg7 strain. All of the 280 progeny tested exhibited cosegregation of the npq5 phenotype (assayed by video imaging) and Arg-independent growth, suggesting that the NPQ defect in npq5 was generated by the insertion of pJD67. DNA gel blot hybridizations using genomic DNA from the npq5 mutant strain demonstrated that the mutant has a single insertion of pJD67 and that the bacterial origin of replication from pBluescript was lost during insertion (data not shown).
Chlamydomonas genomic DNA flanking the pJD67 insertion was isolated by screening a genomic library generated from npq5 DNA with sequences from pBluescript and ARG7 (see Methods). Flanking DNA was sequenced and used to isolate both wild-type genomic and cDNA clones. Insertion of pJD67 in npq5 was in a novel gene with strong sequence identity to genes encoding the major LHCII polypeptides of vascular plants that constitute the LHCII trimers. The gene was designated Lhcbm1 (Lhc for light-harvesting complex, b as by convention for PSII-associated LHC polypeptides, m for major, and 1 because it is the Lhcb gene that was most highly represented in the nonnormalized cDNA sampling of the Kazusa EST database).
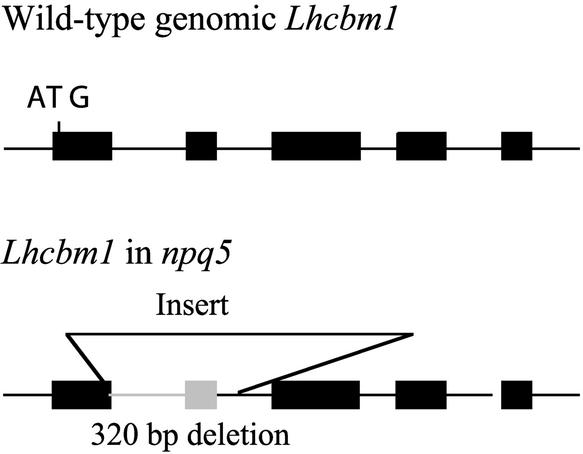
Recent biochemical evidence in which Chlamydomonas LHCII trimers and monomers were resolved by PAGE confirmed that all of Lhcbm1 is present in the trimers (D. Elrad and A.R. Grossman, unpublished data). Figure 5 depicts the Lhcbm1 locus in the wild-type and npq5 mutant strains. The wild-type Lhcbm1 gene contains five exons and four introns. In npq5, pJD67 was inserted into the first exon, generating a 320-nucleotide deletion. Full-length cDNA clones are 999 nucleotides long with a coding region of 768 nucleotides. The predicted 256–amino acid precursor polypeptide contains an N-terminal domain characteristic of thylakoid transit peptides. Cleavage would generate a mature protein of 226 amino acids, with a predicted molecular mass of 25.1 kD and a pI of 5.02. The cleavage site was deduced based on homology with Lhcb2 polypeptides from vascular plants (Figure 6; see Discussion).
Figure 5.
Scheme of the Lhcbm1 Locus in the Wild-Type and npq5 Mutant Strains.
Lhcbm1 has five exons and four introns. In the npq5 mutant, an incomplete pJD67 plasmid was inserted in the first exon of the gene, generating a 320-nucleotide deletion at the site of insertion. Black boxes represent exons, and the gray box represents genomic DNA within the Lhcbm1 gene of npq5 that was deleted during the insertion of pJD67.
Figure 6.
Alignment of the Amino Acid Sequences of the Chlamydomonas Lhcbm Polypeptides and Lhcb2.1 from Pea (Ps).
Residues identical to the consensus sequence are shown in black, and stars mark potential Thr phosphorylation sites.
(A) Predicted transit peptides.
(B) Predicted mature proteins.
The Defect in NPQ Is Complemented by a Lhcbm1 Genomic Clone
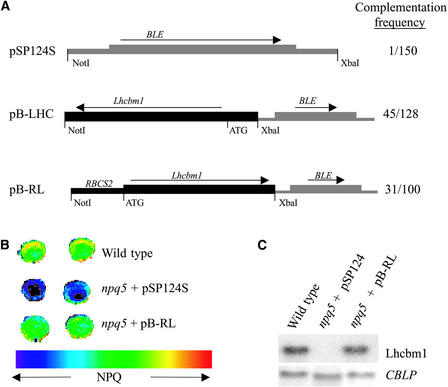
To determine if the NPQ defect in the mutant was caused by the insertion in Lhcbm1, we introduced the wild-type Lhcbm1 gene into the mutant and analyzed NPQ in the resulting transformants. Three constructs were transformed into npq5: (1) the negative control pSP124S, a vector that contains the Ble gene, a selectable marker conferring resistance to Zeocin (Cayla, Toulouse, France; Lumbreras et al., 1998); (2) pB-LHC, a 4.4-kb genomic fragment containing Lhcbm1 ligated into pSP124S (the fragment includes 2 kb 5′ of the transcriptional start site and 400 bp 3′ of the polyadenylation signal); and (3) pB-RL, a 3.7-kb Lhcbm1 genomic fragment joined at the ATG site to a 173-bp RbcS2 promoter fragment inserted in pSP124S.
The mutant strain was transformed with these constructs by the glass bead transformation method, and transformants were selected on medium containing Zeocin (Lumbreras et al., 1998). After 10 days of growth, single colonies were transferred to solid minimal medium, allowed to grow at 100 μmol·m−2·s−1, and screened for the npq phenotype by video imaging. Figure 7A shows a scheme of the three constructs and the frequency with which transformants with a wild-type NPQ phenotype were recovered. Although <1% of the colonies transformed with the pSP124S negative control appeared complemented using the video-imaging assay, >30% of the colonies transformed with either pB-LHC or pB-RL appeared complemented.
Figure 7.
Transformation with the Genomic Lhcbm1 Sequence Complements the Mutant Phenotype in npq5.
(A) Scheme of the constructs used for complementation, and complementation frequencies.
(B) False-color image of NPQ in the wild-type strain, the npq5 mutant strain, and a complemented strain.
(C) Levels of Lhcbm1 RNA expression in strains used in (B). The CBLP transcript level was used as a loading control.
Figure 7B shows a false-color image of NPQ in colonies of wild-type cells, the npq5 mutant containing the control vector (pSP124S), and a transformant harboring the Lhcbm1 gene (from pB-RL) in which the mutant Npq phenotype is complemented. As shown by the RNA gel blot experiment presented in Figure 7C, Lhcbm1 mRNA accumulated in wild-type cells and the complemented strain but not in the mutant strain. Complementation of the mutant phenotype was retested after growth in liquid medium by measurements of modulated fluorescence. Many apparently complemented strains exhibited completely wild-type NPQ development, whereas others exhibited partial complementation. The degree of complementation generally correlated with the level of mRNA accumulation (data not shown).
The npq5 mutant transformed with pB-RL generally exhibited higher levels of Lhcbm1 mRNA accumulation than npq5 transformed with pB-LHC. Finally, complementation of the Npq phenotype was accompanied by complementation of the other phenotypes associated with the mutant strain (e.g., changes in 77K fluorescence and pigment levels) (data not shown).
Ten Genes Encode Major LHCII Polypeptides in Chlamydomonas
The finding that the mutation in npq5 resided in the Lhcbm1 gene suggested that LHCII trimers play a functional role in thermal dissipation. In vascular plants, the trimers are composed of Lhcb1 and Lhcb2, two highly related polypeptides that are encoded by four or more genes each (Jansson, 1999). To further elucidate the role of Lhcbm1 in NPQ, we identified other Chlamydomonas genes that potentially encode polypeptides associated with the LHCII trimers. Members of the Lhc gene family of Chlamydomonas were identified initially by performing a TBLASTX search (Altschul et al., 1997) on the Kazusa EST database and the cDNA database of the National Science Foundation–funded Chlamydomonas genome project with the conserved transmembrane domain that is present in all LHC polypeptides and the entire Lhcbm1 polypeptide sequence.
A total of 1100 sequences were identified and placed into 10 contigs that encode polypeptides with high sequence identity to Lhcb1 and Lhcb2 from vascular plants. Four of the contigs were identical to the previously described Chlamydomonas Lhc genes Cab II-1, Lhcb2, Lhcb3, and CabII-2 (Imbault et al., 1988; Somanchi et al., 1998; J.H. Mussgnug and O. Kruse, unpublished data). Three other contigs were identical to genes identified in a recently published EST search (Teramoto et al., 2001).
Unlike in vascular plants, the distinction between Lhcb1 and Lhcb2 in Chlamydomonas is not clear (Green and Durnford, 1996). Therefore, we have designated all of the genes that encode potential constituents of the LHCII trimers (based on sequence similarity to Lhcb1 and Lhcb2 of vascular plants) Lhcbm. The genes were given numerical suffixes (Lhcbm1 to Lhcbm10) based on EST frequency in the Kazusa database (as of December 2000) (the lower the number, the more frequently the sequence appeared in the EST database). This naming scheme precisely defines each of these Lhcb sequences and is meant to provide a simple and consistent set of gene designations.
The Lhcbm gene mutated in npq5 exhibited the highest EST frequency; therefore, it was designated Lhcbm1 (NPQ5). Lhcbm9 and Lhcbm10 were far less represented in the EST library than the other Lhcb sequences. Interestingly, Lhcbm9 was represented only by reads from the cDNA library constructed with mRNA isolated after growth in sulfur-deficient medium, and Lhcbm10 was represented by only two reads.
The contigs available in the public databases did not represent full-length cDNA sequences. To obtain full-length sequences, missing fragments of the specific Lhcbm genes were amplified from a cDNA library by PCR and sequenced (see Methods). Figure 6 shows an alignment of the 10 predicted Lhcbm polypeptides. To determine if the Lhcbm1 deletion caused an alteration in the expression of other Lhcbm genes, gene-specific probes for each were generated and RNA gel blot hybridizations were performed. Expression levels for all of these genes (except Lhcbm1) were essentially identical in the wild-type and npq5 mutant strains (data not shown).
DISCUSSION
To gain a better understanding of thermal dissipation in photosynthetic organisms, a screen for Chlamydomonas mutants defective in this process was performed (Niyogi et al., 1997a). Mutants generated were placed into three categories: (1) those defective for photosynthetic electron transport that were unable to generate the ΔpH necessary for thermal dissipation; (2) those abnormal for the xanthophyll cycle (npq1 and npq2); and (3) those that were not obviously impaired in photosynthesis or the xanthophyll cycle. In this article, we present the physiological and molecular characterization of npq5, a mutant from the third category.
Physiological and Molecular Characterization of npq5
The npq5 mutant developed less than one-third as much reversible NPQ as wild-type cells, and nearly all of the NPQ that did develop was not reversed by the addition of nigericin, suggesting that the diminished NPQ is a result of a defect in thermal dissipation. Furthermore, although the mutant strain was capable of high light–induced violaxanthin deepoxidation and of undergoing state transitions, it appeared to have one-third fewer LHCII trimers (e.g., a 20% smaller PSII antenna) than wild-type cells. Assuming equivalent numbers of PSII reaction centers in mutant and wild-type strains, when grown in moderate light, npq5 cells would have four trimers per PSII reaction center compared with six trimers per reaction center in wild-type cells.
In accord with the mutant's reduced ability for thermal dissipation, photoinhibition was significantly more rapid in npq5 than in wild-type cells upon exposure to high light (1100 μmol·m−2·s−1), suggesting that thermal dissipation is important for protecting PSII from damage at very high light intensities. It also is possible that the increased rate of photoinhibition is not a direct result of the absence of thermal dissipation but is an indirect result of the mutation. Photoinhibition was not significantly faster at a light intensity of 350 μmol·m−2·s−1 (data not shown). These results suggest that there are other photoprotective processes that compensate for the lower capacity for thermal dissipation in the mutant strain.
The npq5 strain showed no significant defect in photosynthetic efficiency, even though the PSII antenna size appeared to be reduced. Although consistent results were obtained when photosynthetic efficiency was measured repeatedly for the same culture, there was considerable variation among both wild-type and npq5 cultures. This variation would obscure differences in photosynthetic efficiency of up to 25%. Growth at 100 μmol·m−2·s−1 was not significantly different in the npq5 and wild-type strains. An increase in the doubling time might be expected for the mutant strain because it has a smaller PSII antenna. However, unlike the npq5 mutant, wild-type cells develop NPQ at 100 μmol· m−2·s−1, which would result in a somewhat decreased photosynthetic efficiency and slower growth. At 100 μmol·m−2· s−1, the consequences of reduced antennae cross-section and less NPQ might cancel each other with respect to the growth rates.
Because integration of exogenous DNA into the nuclear genome of Chlamydomonas occurs by nonhomologous recombination, transformation with a selectable marker can result in lesions that are tagged with the introduced DNA, facilitating molecular cloning of the altered gene. After demonstrating that the npq5 mutant phenotype was linked to the ARG7 insertion, the genomic DNA flanking the insertion site was characterized. A single incomplete transformation vector was inserted into a gene with high sequence identity to Lhcb1 genes of vascular plants; we have designated this gene Lhcbm1.
To prove that the npq5 phenotype was caused by the insertion in Lhcbm1, we transformed the mutant with a wild-type copy of Lhcbm1 and analyzed the fluorescence phenotype of the transformants. The introduction of either the entire Lhcbm1 gene with 2 kb 5′ of the transcriptional start site or the Lhcbm1 gene fused to the RbcS2 promoter was able to complement the mutant Npq phenotype in ∼30% of transformants, as analyzed by video imaging on solid medium. However, when complementation was retested by measurements of modulated fluorescence in liquid medium, only some of the strains originally scored as complemented exhibited complete restoration of the wild-type phenotype.
Many of the putatively complemented strains attained NPQ levels that were between those exhibited by wild-type cells and the npq5 mutant. RNA gel blot hybridizations with total RNA isolated from these cultures demonstrated that the degree of complementation was correlated with the level of Lhcbm1 mRNA accumulation (D. Elrad and A.R. Grossman, unpublished data). Although measurements of protein levels are required to test this conclusion, it is likely that the degree of complementation reflects polypeptide accumulation that is linked to the level of Lhcbm1 transcript.
This is different from the conclusions drawn from studies of antisense tobacco plants. Although there was a 95% reduction in Lhcb1 mRNA in tobacco plants containing antisense Lhcb1 constructs, the level of the Lhcb1 protein was not altered, suggesting that in tobacco, Lhcb1 protein levels are regulated primarily by post-transcriptional processes (Flachmann and Kühlbrandt, 1995). The variation in Lhcbm1 mRNA levels in complemented strains probably reflects position effects (differences in the site of integration of the exogenous DNA) on the rate of transcription. On average, higher expression levels were observed in transformants harboring the RbcS2 promoter fusion, suggesting that the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit promoter is stronger or less dependent on position effects than the Lhcbm1 promoter.
The Lhcbm Gene Family: Predicted Cleavage and Thr Phosphorylation Sites
We identified other Chlamydomonas genes that potentially encode polypeptides associated with the LHCII trimers by searching the Chlamydomonas databases. Alignment of the predicted Lhcbm polypeptides and Lhcb2.1 from pea is shown in Figure 6. Although the cleavage sites between the transit peptides and mature proteins cannot be predicted with certainty, probable cleavage sites are given. The cleavage sites in Lhcbm2 and Lhcbm8 were deduced from the N-terminal sequence of a mature LHCII polypeptide (Hippler et al., 2000). The N-terminal sequence of this Chlamydomonas LHC (5′-IEXYGPDRPKFLGPFR-3′) is identical (with the exception of the last R, which probably is a mistake generated during N-terminal sequencing) to sequences near the beginning of the deduced Lhcbm2 and Lhcbm8 polypeptides.
The cleavage sites of Lhcbm3, Lhcbm4, Lhcbm6, Lhcbm7, and Lhcbm9 were predicted based on homologies with Lhcbm2 (and Lhcbm8) near the predicted cleavage site. The sequences of Lhcbm1 and Lhcbm10 do not show identity to Lhcbm2 (from Chlamydomonas) in the region at which the Lhcbm2 precursor is predicted to be cleaved. The cleavage sites of Lhcbm1 and Lhcbm10 were predicted from homologies to Lhcb2 polypeptides of vascular plants. The cleavage site of the Lhcbm5 precursor protein is more difficult to predict: the two most probable sites are between Q30 and K31 and between G45 and N46. These predictions can be tested by N-terminal sequencing of the mature polypeptides.
The alignment presented in Figure 6 demonstrates that the Lhcbm polypeptides are highly conserved in Chlamydomonas and that differences are greatest in the N-terminal region, which extends into the stroma and may function in Lhcb trimerization and state transition. Lhcbm1 (the gene disrupted in npq5), Lhcbm10, and possibly Lhcbm5 encode polypeptides with extended N-terminal regions, similar to Lhcb1 and Lhcb2 of vascular plants. Furthermore, like Lhcb2 of vascular plants, the predicted mature proteins of Lhcbm1, Lhcbm10, and possibly Lhcbm5 contain Thr residues that have the potential to be phosphorylated, based on the NetPhos (Blom et al., 1999) predictive program and alignments with Lhcb2 polypeptides from vascular plants with known phosphorylation sites (Michel et al., 1991).
Although no Thr residues are predicted to be phosphorylated in the other mature Lhcbm proteins, the mature proteins contain conserved Thr residues, and there are putative Thr phosphorylation sites in the predicted transit sequences. Thr phosphorylation mediates state transitions. When the plastoquinone pool is reduced, LHCII trimer polypeptides are phosphorylated, stimulating trimer detachment from PSII and attachment to PSI. The npq5 mutant is competent to perform state transitions, suggesting that the phosphorylation of an Lhcbm other than Lhcbm1 is involved in this process. In vascular plants, Lhcb2 is phosphorylated more rapidly and to a greater extent than Lhcb1. This has led to the speculation that the inner trimers contain only Lhcb1 and do not detach from PSII, whereas the peripheral trimers contain Lhcb2 and can be mobilized during state transition (Walters and Horton, 1999).
Does Lhcbm1 Have a Specific Role in Thermal Dissipation or Is the Reduced Thermal Dissipation Caused by Decreased Trimer Accumulation?
Four hypotheses can account for the effect of the Lhcbm1 lesion on thermal dissipation. (1) The reduced antennae size in the npq5 strain might result in an inability of the mutant to generate a ΔpH large enough to elicit thermal dissipation. This is unlikely because violaxanthin deepoxidation occurred with the same kinetics in the wild-type and npq5 mutant strains. Furthermore, the light used to elicit NPQ development was at least twofold the intensity of that needed to saturate photosynthesis. (2) The capacity for thermal dissipation is reduced because the npq5 mutant contains fewer trimers and dissipation occurs in the trimers. This seems unlikely because the decrease in the number of trimers was ∼30%, whereas the decrease in the capacity for thermal dissipation was at least 65%. (3) The function of Lhcbm1 in thermal dissipation is indirect, and a loss of this polypeptide causes reduced thermal dissipation in monomeric Lhc polypeptides. Although possible, this is not supported by the finding that mutants devoid of specific monomeric LHCII polypeptides perform thermal dissipation (Andersson et al., 2001). (4) Lhcbm1 serves a specific function in the development of thermal dissipation. From the biochemical/physiological data presented, this possibility seems most likely. If Lhcbm1 has a specific role in thermal dissipation, features of its N terminus might be required. Site-directed mutagenesis and the introduction of altered Lhcbm1 genes into the npq5 mutant could help reveal the features of Lhcbm1 that might be important for thermal dissipation.
Although xanthophylls have a critical role in thermal dissipation in both Chlamydomonas and vascular plants, a role for the specific polypeptide PsbS has been demonstrated only in vascular plants. It is still unclear whether Chlamydomonas contains PsbS; although >8500 unique genes have been identified by the Chlamydomonas genome project, none encodes a polypeptide with high sequence similarity to PsbS. Also, although the inability to deepoxidize violaxanthin in high light causes a defect in thermal dissipation in both organisms, the defect is significantly greater in Arabidopsis. By contrast, the inability to synthesize lutein causes a greater defect in thermal dissipation in Chlamydomonas. Thus, there are likely to be differences in the manner in which Chlamydomonas and Arabidopsis promote thermal dissipation of excess absorbed light energy (Horton et al., 2000; Li et al., 2000).
If Chlamydomonas contains PsbS (or a functional analog of this polypeptide), it could function in energy dissipation by a mechanism similar to that used by vascular plants. Excessive lumen acidification may induce protonation of PsbS as well as the formation of zeaxanthin. Protonated PsbS plus zeaxanthin could modulate the structure of the LHCII trimers to promote thermal dissipation of singlet excited chlorophyll. This process may depend on a specific association between PsbS and Lhcbm1. Alternatively, PsbS plus zeaxanthin might be capable of gathering and efficiently dissipating energy harvested by LHCII in a Lhcbm1-dependent manner.
METHODS
Strains and Media
The Arg auxotroph CC-425 (arg7-8 cw15 mt + sr-u-2-60) was the parental strain of Chlamydomonas reinhardtii used for insertional mutagenesis to generate npq5 (Niyogi et al., 1997a). A random transformant, KN53.22, was used as the control in all physiological experiments. Cells were grown photoautotrophically in minimal high salt (HS) medium or photoheterotrophically in acetate-containing Tris-acetate-phosphate medium (Harris, 1989). Liquid cultures were grown with constant shaking at 26°C at 100 μmol·m−2·s−1 continuous white light.
Modulated fluorescence, 77K fluorescence, oxygen evolution, and pigment analyses were performed with midlogarithmic-phase cells (1.3 to 2 × 106 cells/mL) grown photoautotrophically. For video imaging, cells were grown on HS-agar plates at 100 μmol·m−2·s−1. When necessary, the medium was supplemented with 50 μg/mL Arg or 1 μg/mL Zeocin. Genetic crosses were performed as described by Harris (1989). All strains used are available from the corresponding author.
Pulse-Amplitude Modulated Fluorometry
Fluorescence characteristics were determined with a pulse-amplitude modulated fluorometer (model OS-100; PP Systems, Haverville, MA). Samples were stirred continuously and maintained at 26°C in a water-jacketed chamber. The intensity of the measuring beam was <0.1 μmol·m−2·s−1. The actinic light source was incandescent (halogen lamp, 12 V, 20 W; EZX), and saturating pulses of 0.4 s exceeded 1500 μmol·m−2·s−1. A far-red light filter (LE-700-S-676N; ThermoCorion, Franklin, MA) was used to generate ≈1 μmol·m−2·s−1 at 690 < λ < 710. Conventional fluorescence nomenclature is used (VanKooten and Snel, 1990), and NPQ was calculated as (Fm − Fm′)/Fm′. To induce anaerobic conditions, 20 mM Glc and 2 mg/mL Glc oxidase (final concentrations) were added to cultures in the dark (Bulté and Wollman, 1990). Nigericin was added at a final concentration of 10 μM (Niyogi et al., 1997a).
77K. Fluorescence Emission Spectra
Fluorescence emission spectra were determined with a single-beam fluorometer (Photon Technology International, New Brunswick, NJ). Samples were submerged in liquid nitrogen and excited with a quartz cylinder at 435 nm (2.8 μmol·m−2·s−1; bandwidth = 5 nm); fluorescence emission was measured for 1 s at every 1 nm (bandwidth = 1 nm) between 650 and 750 nm. The emission spectra generated for HS medium was subtracted from all sample spectra. Before measurements, cells were dark adapted for 3 min to allow for the relaxation of thermal dissipation.
Oxygen Evolution
Respiration and photosynthesis were quantified with the Oxygraph System (Hansatech, Norfolk, UK) connected to a personal computer. Cultures were maintained at 26°C by a circulating water bath. To determine photosynthetic efficiency, oxygen evolution was measured at 0, 15.7, 25.6, 31.2, 39.7, and 47.5 μmol·m−2·s−1 white light. Photosynthetic efficiency was determined as the slope of the curve generated by plotting oxygen evolution relative to light intensity. Maximal photosynthesis was determined as oxygen evolution at 350 μmol·m−2·s−1 plus dark respiration.
Growth
Growth rates were determined for cells in liquid HS medium. Midlogarithmic cultures were diluted to 1 × 105 cells/mL and maintained under standard growth conditions for 16 h before the first cell count. After the 16-h incubation, cell concentrations were determined at 0, 8, 24, and 32 h using a hemocytometer after dilution with an equal volume of Lugol (Sigma, St. Louis, MO).
Pigment Determination
Chlorophyll concentration was determined spectrophotometrically after extraction with 90% acetone (Jeffrey and Humphrey, 1975). HPLC analysis of carotenoids and chlorophyll was performed as described previously (Niyogi et al., 1997a).
Video Imaging
Video imaging of Chlamydomonas colonies was performed according to Niyogi et al. (1998). Saturating pulses of light were given before and 6 min after illumination with 700 μmol·m−2·s−1. Fluorescence images of Fm and Fm′ were captured during saturating pulses, and false-color images of NPQ were generated.
Isolation of DNA Flanking the Insertion
To isolate DNA flanking the insertion, a library of npq5 genomic DNA was constructed in λ-dash (Stratagene, La Jolla, CA). Approximately 20 μg of genomic DNA from the mutant strain was partially digested with 1.5 units of Sau3A for 20 min followed by 20 min at 65°C. The digested DNA was treated with shrimp alkaline phosphatase, extracted with phenol followed by chloroform, and then precipitated with ethanol. The DNA was ligated to λ-dash arms digested with BamHI according to protocols provided by Stratagene and packaged using Gigapack II Plus Packaging Extract (Stratagene).
A total of 500,000 recombinant phage were recovered and screened for hybridization with pBluescript and a 700-nucleotide PstI-SalI fragment of ARG7. DNA from phage isolated using the pBluescript probe was sequenced directly with a primer from pBluescript (BSL2232, 5′-TGGCGAACTTACTCTA-3′). DNA from phage isolated using the ARG7 probe was sequenced directly with a primer generated to the 3′ end of ARG7 (Arg8494u, 5′-GGCGGGAGGGACAGCACTGA-3′).
Isolation of Wild-Type Genomic and cDNA Lhcbm1
A fragment of Chlamydomonas DNA flanking the site of insertion that encoded the first exon and the 5′ untranslated region (UTR) of the Lhcbm1 gene was used to screen a cDNA library provided by M. Goldschmidt-Clermont (University of Geneva, Switzerland) and a wild-type genomic library (Davies et al., 1994). A 4.4-kb NotI-XbaI fragment of the genomic clone was subcloned into pBluescript (pLHC4.4), sequenced, and used to construct vectors for complementation experiments. Three identical cDNA clones also were sequenced completely.
Construction of pB-LHC and pB-RL
To construct pB-LHC, pLHC4.4 was digested with NotI and EcoRI and the 4.4-kb insert was cloned into pBkBle digested with NotI and EcoRI. pBkBle is a plasmid containing the Ble gene in the reverse orientation of pSP124S. To construct pB-RL, the RbcS2 promoter region was amplified by PCR from pSP124S with the primers Rnco25 (5′-CGTCGACTCACCTGGCCATGGTAAG-3′) and T3 (5′-AATTAA-CCCTCACTAAAGGG-3′). Primer Rnco25 was designed to alter the RbcS2 sequence to generate a NcoI site at the start codon; a perfectly matching primer would have been 5′-CGTCGACTCACCTGG-TTTTGGTAAG-3′. The RbcS2 PCR product then was digested with NcoI and NotI.
The DNA fragment containing Lhcbm1 was amplified by PCR with the primers RlNcoI (5′-TACCCACCAGTCACCATGGCCT-3′) and Lin2lw (5′-GGAAAGCAAGTAAGGGTGTG-3′). Primer RlNcoI was designed to alter the Lhcbm1 sequence to generate a NcoI site at the start codon; a perfectly matching primer would have been 5′-TACCCACCAGTCAAAATGGCCT-3′. However, unlike the change in the RbcS2 promoter sequence, these changes were not in the final vector. The Lhcbm1 PCR product then was digested with NcoI and SphI. pLHC4.4 was digested with SphI and NotI. The digested vector, RbcS2 promoter fragment, and amplified Lhcbm1 fragment were ligated (three-way ligation) to generate pRlhc. pRlhc then was digested with EcoRI and EcoRV, and the 3.7-kb insert was ligated into pBkBle digested with EcoRI and EcoRV to generate pB-RL.
DNA and RNA Hybridization
Hybridizations to DNA and RNA gel blots were performed as described previously except that UltraHybe (Ambion, Austin, TX) was used as the hybridization buffer for hybridizations to RNA (Davies et al., 1999).
Identification of Lhcbm Genes
To identify the Lhcb genes of Chlamydomonas, the Kazusa EST database and sequences deposited in GenBank by the National Science Foundation–funded Chlamydomonas genome project were TBLASTX searched (Altschul et al., 1997) with the conserved transmembrane domain (5′-ELIHARWAMLGALGCITPE-3′) and the complete Lhcbm1 amino acid sequence. Sequences of the 1100 top scoring reads were entered into SeqMan (DNASTAR, Madison, WI). SeqMan was directed to generate contigs from ESTs with >98% identity. Ten contigs with similarity to Lhcb1 and Lhcb2 of vascular plants were generated. Because the contigs did not represent full-length gene sequences, the missing gene fragments were amplified from a cDNA library (provided by M. Goldschmidt-Clermont) and sequenced.
For each Lhcbm gene, gene-specific probes were generated from the 3′ UTR region or from the first exon, which encodes the thylakoid transit peptide region (the least conserved region of the protein). Probes were tested for specificity by hybridization to Chlamydomonas genomic DNA (data not shown). The primers used to generate the specific probes for each Lhcbm gene were as follows: Lhcbm1 (NPQ5) (5′ UTR and first exon), Lcdup1 (5′-TGGACGCCTTAA-ATACTCAG-3′) and Lcdlw1 (5′-GGCGAGCTACACACCTGTCC-3′); Lhcbm2 (3′ UTR), C32up3 (5′-TGGAGTAGGTGTGCTGCTTGA-3′) and C32lw3 (5′-TCGAGACCCATGTCCCTGTAT-3′); Lhcbm3 (3′ UTR), C45up1 (5′-AATCAGTCAGTAACGGGCATT-3′) and C45lw1 (5′-TGCCCGTTACTGACTGATTGA-3′); Lhcbm4 (3′ UTR), L2up1 (5′-CGGTTTGTTGCTGGGGCTCTA-3′) and L2lw1 (5′-ATGGGGGCA-CTCTTGTGTC-3′); Lhcbm5 (5′ UTR and first exon), Lhcb3up (5′-ACTCACCGAGTACCGTGTATA-3′) and Lhcb3lw (5′-GGGAAC-TCGCCAGTCAGGTAG-3′); Lhcbm6 (3′ UTR), Cabup1 (5′-CGGCGT-ATATTGGCACTTTGA-3′) and Cablw1 (5′-AGACTTTGGAATGGG-CTCTTC-3′); Lhcbm7 (3′ UTR), C39up1 (5′-GCGCTGCCGACCTGG-ACAAGT-3′) and C39lw1 (5′-CCCCAAGGCACGGCGAAGTAG-3′); Lhcbm8 (3′ UTR), L8up1 (5′-TGAATGTACTGGCGTGATTGA-3′) and L8lw1 (5′-ACCAGTGGCCGTCAAGCCATTT-3′); Lhcbm9 (3′ UTR), L9up1 (5′-CCACGCCTGTGCCTGAATGTTT-3′) and L9lw1 (5′-CAC-GCATGGCACACTGTCTTCT-3′); Lhcbm10 (3′ UTR), L10up1 (5′-CGCCTTCGCCACCAAGTTCAC-3′) and L10lw1 (5′-ATTTGCGCGCACCACGGGACC-3′).
Accession Numbers
Accession numbers and previous names for previously reported LhcII genes are as follows: Lhcbm1 is LhcII-4 (AB051210 and AB051206); Lhcbm2 is LhcII-3 (AB051209 and AB051205); Lhcbm3 is LhcII-1.3 (AB051208 and AB051204); Lhcbm4 is Lhcb2 (AF104630); Lhcbm5 is Lhcb3 (AF104631); Lhcbm6 is CabII-1 (M24072.1); and Lhcbm8 is CabII-2 (AF330793). Lhcbm7, Lhcbm9, and Lhcbm10 represent novel sequences and have been entered into GenBank with accession numbers AF479779, AF479778, and AF479777, respectively. The genomic sequence of Lhcbm1 and the cDNA sequence of CabII-1 were entered into GenBank with accession numbers AF495473 and AF495472, respectively. Information concerning all of the Lhcbm genes is available in the supplementary data online. The accession number for Lhcb2.1 from Pisum sativum is P27520.
Supplementary Material
Acknowledgments
We thank Saul Purton for the pSP124S construct and LHCI, and M. Goldschmidt-Clermont for the cDNA library. We also thank John Christie, Qingfang He, Chung Soon Im, Govindjee, and Olle Björkman for helpful discussions. We thank the National Science Foundation for supporting this work (Grant INT 0084189 awarded to A.R.G.). The Chlamydomonas genome project, represented by a consortium of researchers including A.R.G. (Grant MCB 9975765), enabled us to identify and compare all of the Lhcb genes in Chlamydomonas.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002154.
Footnotes
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J., Walters, R.G., Horton, P., and Jansson, S. (2001). Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 13, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, J.R., Fryer, M.J., and Baker, N.R. (1995). Consequences of LHCII deficiency for photosynthetic regulation in chlorina mutants of barley. Photosynth. Res. 44, 81–91. [DOI] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygen and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Giacometti, G.M., and Simpson, D.J. (1988). Changes in the organization of stroma membranes induced by in vivo state- 1-state-2 transition. Biochim. Biophys. Acta 935, 152–165. [Google Scholar]
- Bassi, R., Pineau, B., Dainese, P., and Marquardt, J. (1993). Carotenoid-binding proteins of photosystem II. Eur. J. Biochem. 212, 297–303. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Sandonà, D., and Croce, R. (1997). Novel aspects of chlorophyll a/b-binding proteins. Physiol. Plant. 100, 769–779. [Google Scholar]
- Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., van Breemen, J.F.L., van Roon, H., and Dekker, J.P. (2000). Conformational changes in photosystem II supercomplexes upon removal of extrinsic subunits. Biochemistry 39, 12907–12915. [DOI] [PubMed] [Google Scholar]
- Bossmann, B., Knoetzel, J., and Jansson, S. (1997). Screening of chlorina mutants of barley (Hordeum vulgare L.) with antibodies against light-harvesting proteins of PS I and PS II: Absence of specific antenna proteins. Photosynth. Res. 52, 127–136. [Google Scholar]
- Bulté, L., and Wollman, F.-A. (1990). Stabilization of state-I and state-II by p-benzoquinone treatment of intact cells of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1016, 253–258. [Google Scholar]
- Chow, W.S., Funk, C., Hope, A.B., and Govindjee (2000). Greening of intermittent-light-grown bean plants in continuous light: Thylakoid components in relation to photosynthetic performance and capacity for photoprotection. Indian J. Biochem. Biophys. 37, 395–404. [PubMed] [Google Scholar]
- Croce, R., Remelli, R., Varotto, C., Breton, J., and Bassi, R. (1999. a). The neoxanthin binding site of the major light harvesting complex LHCII from higher plants. FEBS Lett. 456, 1–6. [DOI] [PubMed] [Google Scholar]
- Croce, R., Weiss, S., and Bassi, R. (1999. b). Carotenoid binding sites of the major light-harvesting complex II of higher plants. J. Biol. Chem. 274, 29613–29623. [DOI] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A.R. (1994). Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A.R. (1999). Sac3, an Snf1-like serine threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams, B., Gilmore, A.M., and Adams, W.W. (1996). Carotenoids. 3. In vivo functions of carotenoids in higher plants. FASEB J. 10, 403–412. [DOI] [PubMed] [Google Scholar]
- Färber, A., Young, A.J., Ruban, A.V., Horton, P., and Jahns, P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants: The relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching. Plant Physiol. 115, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachmann, R., and Kühlbrandt, W. (1995). Accumulation of plant antenna complexes is regulated by posttranscriptional mechanisms in tobacco. Plant Cell 7, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, H.A., Bautista, J.A., Josue, J.S., and Young, A.J. (2000). Mechanism of non-photochemical quenching in green plants: Energies of the lowest excited singlet states of violaxanthin and zeaxanthin. Biochemistry 39, 2831–2837. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M., and Ball, M.C. (2000). Protection and storage of chlorophyll in overwintering evergreens. Proc. Natl. Acad. Sci. USA 97, 11098–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.M., Hazlett, T.L., Debrunner, P.G., and Govindjee (1996. a). Comparative time-resolved photosystem II chlorophyll a fluorescence analyses reveal distinctive differences between photoinhibitory reaction center damage and xanthophyll cycle-dependent energy dissipation. Photochem. Photobiol. 64, 552–563. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M., Hazlett, T.L., Debrunner, P.G., and Govindjee (1996. b). Photosystem II chlorophyll a fluorescence lifetimes and intensity are independent of the antenna size differences between barley wild-type and chlorina mutants: Photochemical quenching and xanthophyll cycle-dependent nonphotochemical quenching of fluorescence. Photosynth. Res. 48, 171–187. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M., Itoh, S., and Govindjee (2000). Global spectral-kinetic analysis of room temperature chlorophyll a fluorescence from light-harvesting antenna mutants of barley. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 355, 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B.R., and Durnford, D.G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714. [DOI] [PubMed] [Google Scholar]
- Harrer, R., Bassi, R., Testi, M.G., and Schafer, C. (1998). Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L liverwort which contains reaction center and antenna proteins. Eur. J. Biochem. 255, 196–205. [DOI] [PubMed] [Google Scholar]
- Harris, E. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press).
- Härtel, H., Lokstein, H., Grimm, B., and Rank, B. (1996). Kinetic studies on the xanthophyll cycle in barley leaves: Influence of antenna size and relations to nonphotochemical chlorophyll fluorescence quenching. Plant Physiol. 110, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M., Bonfils, J.P., Lutz, C., and Niyogi, K.K. (2000). Photodamage of the photosynthetic apparatus and its dependence on the leaf developmental stage in the npq1 Arabidopsis mutant deficient in the xanthophyll cycle enzyme violaxanthin de-epoxidase. Plant Physiol. 124, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler, M., Biehler, K., Kriegerliszkay, A., Vandillewjin, J., and Rochaix, J.-D. (2000). Limitation in electron transfer in photosystem I donor side mutants of Chlamydomonas reinhardtii: Lethal photo-oxidative damage in high light is overcome in a suppressor strain deficient in the assembly of the light harvesting complex. J. Biol. Chem. 275, 5852–5859. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., Rees, D., Pascal, A.A., Noctor, G., and Young, A.J. (1991). Control of the light-harvesting function of chloroplast membranes by aggregation of the LhcII chlorophyll protein complex. FEBS Lett. 292, 1–4. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Walters, R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Wentworth, M. (2000). Allosteric regulation of the light-harvesting system of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbault, P., Wittemer, C., Johanningmeier, U., Jacobs, J.D., and Howell, S.H. (1988). Structure of the Chlamydomonas reinhardtii CabII-1 gene encoding a chlorophyll a/b-binding protein gene. Gene 73, 397–407. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jeffrey, S.W., and Humphrey, G.F. (1975). New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher-plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194. [Google Scholar]
- Knoetzel, J., and Simpson, D. (1991). Expression and organization of antenna proteins in the light-sensitive and temperature-sensitive barley mutant Chlorina-104. Planta 185, 111–123. [DOI] [PubMed] [Google Scholar]
- Li, X.I., Björkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Phippard, A., Pasari, J., and Niyogi, K.K. (2002). Structural-functional analysis of Photosystem II subunit S (PsbS) in vivo. Funct. Plant Biol., in press. [DOI] [PubMed]
- Lumbreras, V., Stevens, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441–447. [Google Scholar]
- Michel, H., Griffin, P.R., Shabanowitz, J., Hunt, D.F., and Bennett, J. (1991). Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II: Proteolytic cleavage, acetylation, and phosphorylation. J. Biol. Chem. 266, 17584–17591. [PubMed] [Google Scholar]
- Nield, J., Funk, C., and Barber, J. (2000. a). Supermolecular structure of photosystem II and location of the PsbS protein. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1337–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nield, J., Orlova, E.V., Morris, E.P., Gowen, B., van Heel, M., and Barber, J. (2000. b). 3D map of the plant photosystem II supercomplex obtained by cryoelectron microscopy and single particle analysis. Nat. Struct. Biol. 7, 44–47. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Björkman, O., and Grossman, A.R. (1997. a). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Björkman, O., and Grossman, A.R. (1997. b). The roles of specific xanthophylls in photoprotection. Proc. Natl. Acad. Sci. USA 94, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Björkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Shih, C., Chow, W.S., Pogson, B.J., DellaPenna, D., and Björkman, O. (2001). Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 67, 139–145. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Sandonà, D., Giuffra, E., and Bassi, R. (1997). A single point mutation (E166Q) prevents dicyclohexylcarbodiimide binding to the photosystem II subunit CP29. FEBS Lett. 402, 151–156. [DOI] [PubMed] [Google Scholar]
- Peterman, E.J.G., Gradinaru, C.C., Calkoen, F., Borst, J.C., van Grondelle, R., and van Amerongen, H. (1997). Xanthophylls in light-harvesting complex II of higher plants: Light harvesting and triplet quenching. Biochemistry 36, 12208–12215. [DOI] [PubMed] [Google Scholar]
- Phillip, D., Ruban, A.V., Horton, P., Asato, A., and Young, A.J. (1996). Quenching of chlorophyll fluorescence in the major light-harvesting complex of photosystem II: A systematic study of the effect of carotenoid structure. Proc. Natl. Acad. Sci. USA 93, 1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson, B.J., Niyogi, K.K., Björkman, O., and DellaPenna, D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 95, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polívka, T., Herek, J.L., Zigmantas, D., Åkerlund, H.E., and Sundstrom, V. (1999). Direct observation of the forbidden S-1 state in carotenoids. Proc. Natl. Acad. Sci. USA 96, 4914–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle, J.E.W., Niyogi, K.K., and Melis, A. (2001). Absence of lutein, violaxanthin and neoxanthin affects the functional chlorophyll antenna size of photosystem-II but not that of photosystem-I in the green alga Chlamydomonas reinhardtii. Plant Cell Physiol. 42, 482–491. [DOI] [PubMed] [Google Scholar]
- Purton, S., and Rochaix, J.-D. (1994). Characterization of the ARG7 gene of Chlamydomonas reinhardtii and its application to nuclear transformation. Eur. J. Phycol. 30, 141–148. [Google Scholar]
- Ruban, A.V., Lee, P.J., Wentworth, M., Young, A.J., and Horton, P. (1999). Determination of the stoichiometry and strength of binding of xanthophylls to the photosystem II light harvesting complexes. J. Biol. Chem. 274, 10458–10465. [DOI] [PubMed] [Google Scholar]
- Somanchi, A., Handley, E.R., and Moroney, J.V. (1998). Chlamydomonas reinhardtii cDNAs upregulated in low-CO2 conditions: Expression and analyses. Can. J. Bot. 76, 1003–1009. [Google Scholar]
- Teramoto, H., Ono, T., and Minagawa, J. (2001). Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42, 849–856. [DOI] [PubMed] [Google Scholar]
- Vallon, O., Bulté, L., Dainese, P., Olive, J., Bassi, R., and Wollman, F.-A. (1991). Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88, 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKooten, O., and Snel, J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25, 147–150. [DOI] [PubMed] [Google Scholar]
- Verhoeven, A.S., Adams, W.W., and Demmig-Adams, B. (1998). Two forms of sustained xanthophyll cycle-dependent energy dissipation in overwintering Euonymus kiautschovicus. Plant Cell Environ. 21, 893–903. [Google Scholar]
- Verhoeven, A.S., Adams, W.W., Demmig-Adams, B., Croce, R., and Bassi, R. (1999). Xanthophyll cycle pigment localization and dynamics during exposure to low temperatures and light stress in Vinca major. Plant Physiol. 120, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, R.G., and Horton, P. (1999). Structural and functional heterogeneity in the major light-harvesting complexes of higher plants. Photosynth. Res. 61, 77–89. [Google Scholar]
- Walters, R.G., Ruban, A.V., and Horton, P. (1996). Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. USA 93, 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito, F., Finazzi, G., Delosme, R., Nitschke, W., Picot, D., and Wollman, F.-A. (1999). The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 18, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.