Abstract
The synthesis of ribosomes in Saccharomyces cerevisiae consumes a prodigious amount of the cell's resources and, consequently, is tightly regulated. The rate of ribosome synthesis responds not only to nutritional cues but also to signals dependent on other macromolecular pathways of the cell, e.g., a defect in the secretory pathway leads to severe repression of transcription of both rRNA and ribosomal protein genes. A search for mutants that interrupted this repression revealed, surprisingly, that inactivation of RPL1B, one of a pair of genes encoding the 60S ribosomal protein L1, almost completely blocked the repression of rRNA and ribosomal protein gene transcription that usually follows a defect in the secretory pathway. Further experiments showed that almost any mutation leading to a defect in 60S subunit synthesis had the same effect, whereas mutations affecting 40S subunit synthesis did not. Although one might suspect that this effect would be due to a decrease in the initiation of translation or to the presence of half-mers, i.e., polyribosomes awaiting a 60S subunit, our data show that this is not the case. Rather, a variety of experiments suggest that some aspect of the production of defective 60S particles or, more likely, their breakdown suppresses the signal generated by a defect in the secretory pathway that represses ribosome synthesis.
The biosynthesis of ribosomes consumes an enormous fraction of the resources of a rapidly growing cell of Saccharomyces cerevisiae (38). Consequently, a variety of mechanisms have evolved to control this process. The rate of ribosome synthesis is exquisitely sensitive to the carbon source and appears to be regulated through the ras/cyclic AMP/protein kinase A (PKA) pathway (22). The response to the nitrogen source is effected through the TOR pathway, and rapamycin causes a rapid repression of ribosome synthesis (25). But ribosome synthesis is responsive to more than just nutrients. Important elements of regulation strive to maintain balance during cell growth. As one example, we have found that any interruption in the secretory pathway leads to a rapid repression of ribosome synthesis (19). Since the secretory pathway is necessary for the growth of the plasma membrane and the cell wall, continued protein synthesis in a cell whose secretory pathway is compromised results in stretching of the plasma membrane, which in turn leads to a stress response detected by Wsc1p (7, 35) and mediated through protein kinase C (PKC) (13). One branch of the pathway downstream of PKC leads to the repression not only of ribosome synthesis but also of tRNA synthesis (16, 23).
These three pathways, and perhaps many more, in some way coordinately regulate the transcription both of rRNA genes by RNA polymerase I and of the 137 genes encoding ribosomal proteins (RPs) by RNA polymerase II (38). However, the downstream events by which the regulatory pathways interface with the ribosomal genes remain almost totally obscure. It is not known, for instance, whether they act through the same or different effectors of transcription, or even whether transcription of RP genes depends on concomitant transcription of rRNA genes or vice versa.
In an attempt to approach this question, we searched for mutants that interfered with the repression of ribosome biosynthesis in response to a defect in the secretory pathway. Surprisingly, one such mutant inactivated one of the two genes encoding RP L1 of the 60S ribosomal subunit. The primary effect of such a mutant is to cause an imbalance in the synthesis of the two ribosomal subunits, leading to the accumulation of excess 40S subunits. Further experiments showed that limiting production of 60S subunits by any of several means has the same effect. (During the preparation of this report, another report of this phenomenon appeared [18].) Limiting the production of 40S ribosomal subunits does not. Several lines of evidence suggest that it is not a simple deficiency of 60S subunits that is responsible for this phenotype, but rather it is the degradation of improperly assembled 60S subunits.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used are described in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 | MATα leu2-3,112 his3-11 trp1-1 ura3-1 ade2-1 can1-100 ssd1-1 | 33 |
| 312XX | W303 but sly1-1 | 19 |
| 312G418 | W303 but sly1-1 HIS3 URA3::L24-G418 | This work |
| sm9 | W303 but sly1ts HIS3 URA3::L24-G418 rpl1b-1 | This work |
| J1201 | MATα rps6aΔ::HIS3 leu2 | This work |
| J1203 | W303 but derived from tetrad with J1201 and J1204 | This work |
| J1204 | MATα rpl11bΔ::LEU2 his3 | This work |
| YZ86 | MATα sly1-1 rpl11bΔ::LEU2 | This work |
| YZ87 | MATα sly1-1 rps6aΔ::HIS3 leu2 | This work |
| TMY5-1 | W303 but tif5-1 | 17 |
| ts187 | MATagal1 ade1 ade2 ura1 his7 lys2 tyr1 prt1-1 | 9 |
| JWY4995 | ura3-52 lys2-801 trp1Δ101 leu2Δ1 his3Δ200 ade2-101 can1-100 nop4::TRP1 pRS315-nop4-6 | 32 |
| KSY603 | W303 but tif6Δ::HIS3 p[URA3 GAL10::Ub-TIF6] | 2 |
| AJY734 | MATα ade2 ade3 leu2 lys2 ura3 nmd3-4ts | 10 |
| PSY1082 | MATakap120Δ::HIS3 ura3-52 leu2Δ1 his3Δ200 | 31 |
Media.
Yeast cells were cultured in YPD medium (1% yeast extract, 2% Bacto Peptone, and 2% glucose) or synthetic complete minimal media (0.67% yeast nitrogen base without amino acids, 2% glucose supplemented with an amino acid mixture). In some experiments the glucose was replaced with 2% galactose (YPGal), 2% raffinose (YPR), or 2% glycerol plus 2% ethanol (YPGE). Selection of G418-resistant colonies was carried out on YPD plates containing 50 to 200 μg of G418 (Roche)/ml.
Plasmids and primers.
The integrative plasmid pRS306-G418 carrying URA3 and an RPL24-G418r expression cassette was constructed as follows. An open reading frame encoding the bacterial G418r gene was amplified from pFA-kanMX2 (37) with primers P1 (5′-ATGGGTAAGGAAAAGACT-3′) and P2 (5′-TTAGAAAAACTCATCGAG-3′). The promoter and transcriptional terminator of RP RPL24A were also amplified from pRS-L24L30 (15) by using two primer sets, P3 (5′-GTGCACCACATATTTTTG-3′) and P4 (5′-AGTCTTTTCCTTACCCATTTTATCACTATATTA-3′) and P5 (5′-CTCGATGAGTTTTTCTAAGATTTATGCTCGAAC-3′) and P6 (5′-AAGCTTTAGATGCGGACA-3′). Secondary PCR was done with primers P3 and P6 from the mixture of the first PCR products with two consecutively overlapped regions. The PCR-amplified G418r expression cassette was inserted into pRS306 (30). pRS306-G418 was transformed into W303(SLY1) and 312XX(sly1-1) after linearization at the unique EcoRV site in URA3. Genomic PCR of RPL1A and RPL1B from the wild type and sm9 was performed with primers JW1394 (5′-AACAGTCAACCAGTCGTCCA-3′) and JW1395 (5′-CTATGAAAACACGTAATTATGC-3′) for RPL1A and primers JW1396 (5′-GGAAGACTAATTACATATCAT-3′) and JW1397 (5′-GTATTAATCGCTCGGAAGTGA-3′) for RPL1B.
Mutant selection.
A temperature-sensitive secretion defect mutant 312XX, carrying sly1-1 (19), was used as a parental strain to identify factors involved in the regulatory interconnection between the secretory pathway and ribosome synthesis. 312XX grows normally at a temperature of <30°C but grows at a reduced rate between 32 and 34°C, possibly due to a partial limitation of ribosome synthesis as a result of the secretion defect (Fig. 1B). We developed a window of selection for the cells with reduced repression of RP synthesis in the presence of a secretion defect by using semipermissive temperatures between 30 and 34°C and cells carrying a G418 selection marker under the control of the RPL24A promoter. Strain 312XX was first integratively transformed with EcoRV-digested pRS306-G418 containing the G418 resistance gene under the control of the RPL24A promoter and selected in synthetic complete minimal medium lacking uracil. A single colony was further tested for resistance to G418 on YPD plates with concentrations of G418 ranging from 0 to 200 μg/ml as a function of temperature (Fig. 1B). For the selection of spontaneous mutants that can grow under the conditions of nonpermissive temperature and G418 concentration, 5 × 108 cells were plated on a YPD plate with 100 μg of G418/ml and incubated at 32°C for 3 days.
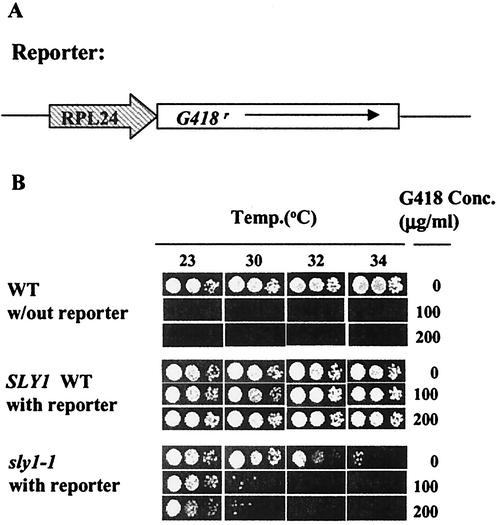
FIG. 1.
(A) The construct used in the screen. (B) Effects of G418 on signal mutants. Tenfold serial dilutions of the indicated strains were spotted on plates containing the indicated concentrations (Conc.) of G418 and incubated for 2 days at the indicated temperature. WT, wild type.
RNA isolation and Northern blot analysis.
Total yeast RNA was isolated from exponentially growing cells, and Northern blot analysis was carried out as described previously (15).
Polyribosome profiles.
Yeast strains were grown in 50 ml of YPD medium at 30°C to an optical density at 600 nm of 0.5 to 1.0. Cycloheximide was added to a 50-μg/ml final concentration, and the culture was chilled immediately on ice. The cells were harvested and washed twice with LHB solution (10 mM Tris [pH 7.4], 100 mM NaCl, 30 mM MgCl2, 50 μg of cycloheximide/ml, 200 μg of heparin/ml). The cell pellet was resuspended in 0.5 ml of LHB, glass beads were added, and the sample was vortexed twice for 30 s at 4°C. One milliliter of LHB was added, and the sample was centrifuged twice at 10,000 × g for 10 min. The supernatant was layered onto a 10 to 35% sucrose gradient in TMN buffer (50 mM acetyl Tris [pH 7.0], 50 mM NH4Cl, 12 mM MgCl2) and centrifuged at 38,000 rpm for 4 h in a Beckman SW41 rotor. All gradients were scanned at 254 nm from the top with an Isco gradient collector.
RESULTS
Isolation of a mutant.
Our aim was to select mutations that abolish the repression of ribosome synthesis in response to a defect in the secretory pathway. The inherent difficulty is that both the synthesis of ribosomes and a functional secretory pathway are essential for growth. The search for mutants therefore required a strategy that would skirt the edges of viability. It was based on the supposition that, under certain conditions where the secretory pathway was partially inhibited, the consequent repression of ribosome synthesis can be limiting for growth.
As a reporter for the synthesis of RPs, the G418 resistance gene was placed under the control of the promoter of the RP gene RPL24A (Fig. 1A) and integrated into strain 312XX, carrying a temperature-sensitive allele of SLY1, a component of the secretory pathway (see Materials and Methods). Titration of growth as a function both of temperature and of G418 concentration suggested that our supposition may be correct; the strain carrying sly1-1 was more sensitive to G418 even at a temperature at which its growth without drug was hardly compromised (Fig. 1B). To carry out the screen, we spread 5 × 108 cells on plates containing 100 μg of G418/ml and incubated them for 3 days at 32°C. Ten colonies grew up.
At least three different types of mutants could be predicted using such a screen: mutants that suppress the sly1-1 mutation, spontaneous mutants providing G418 resistance, and our goal, mutants that compromise the signaling pathway. To distinguish among these alternatives we first tested the cells for growth at 37°C to rule out reversion or suppression of the sly1-1 mutation. Secondary screening was then carried out with tunicamycin, which inhibits protein glycosylation within the endoplasmic reticulum and induces a secretory defect, as shown by induction of the unfolded protein response (3). Treatment with tunicamycin also represses the transcription of RP genes, with the same signaling as the temperature-sensitive signal mutant (23). Therefore, a true signal mutant should render RP transcription insensitive to tunicamycin. One mutant strain, sm9, also passed this test.
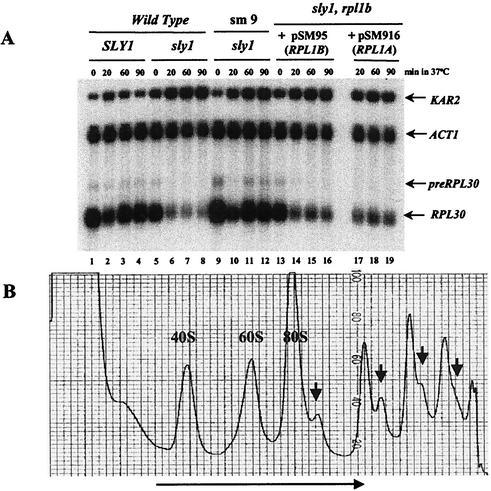
The critical phenotype of strain sm9 is shown in Fig. 2A. As we have found previously, upon a shift to 37°C in a wild-type cell, there is a temporary inhibition of transcription of RPL30, representing the RP genes (Fig. 2A, lane 2) (15). There is no such effect on the ACT1 mRNA. In cells carrying the temperature-sensitive sly1-1 allele, transcription of the RP gene declines precipitously and does not recover (Fig. 2A, lanes 5 to 8). Actin remains unaffected. The induced transcription of KAR2, encoding a chaperone that is induced by the unfolded protein response, serves as a positive control for the distress of the secretory pathway. In strain sm9, it is clear that the repression of RP synthesis that usually accompanies a shift to the nonpermissive temperature in a sly1-1 strain no longer occurs (Fig. 2A, lanes 9 to 12). Note, however, that the temporary heat shock-induced inhibition of transcription of RPL30, apparent at the 30-min time point, is not affected by the mutation. Furthermore, the decline in the RP mRNA in Fig. 2A, lane 10, shows that the mutation in strain sm9 does not alter the half-life (t1/2) of the RP mRNA.
FIG. 2.
Identification of a mutant that suppresses the repression of ribosome synthesis in response to a defect in the secretory pathway. (A) Cells with the indicated genotypes (note that sm9 is rpl1b-1/sly1-1) were shifted from 30 to 37°C, and samples were taken for RNA at the indicated times. RNA was prepared and analyzed on Northern blots as described previously (15). The two panels on the right show the results for strain sm9 transformed with plasmids carrying either RPL1B or RPL1A (see the text). (B) Sucrose gradient analysis of ribosomes and polyribosomes of strain sm9. Sedimentation was from left to right as indicated by the long arrow. Half-mers are indicated by small arrows. Note that the sedimentation was extended in order to better resolve the half-mers.
Identity of the mutant gene.
Strain sm9 has a slow-growth phenotype on YPD at 30°C. The mutant gene was cloned by searching for a suppressor of this phenotype by using a yeast library in plasmid pRS315 (26). Two fast-growing colonies were selected, and their total DNA was isolated and back transformed into Escherichia coli, yielding plasmids pSM95 and pSM916. Retransformation of either plasmid into sm9 complemented the slow-growth phenotype. Sequencing of pSM95 and pSM916 revealed that the inserts consisted of genomic fragments from bp 253,643 to 256,312 of chromosome VII and from bp 135,452 to 140,160 of chromosome XVI. The common feature is that pSM916 carries RPL1A and pSM95 carries RPL1B, two genes that encode the RP L1. This suggested that sm9 may grow slowly due to a deficiency of L1. RPL1A and RPL1B were recovered from the sm9 genome by PCR, and the products were sequenced. RPL1B, with 218 codons, was found to have a single base deletion that resulted in a frame shift at the 162nd codon and premature termination at the 183rd codon. We term this allele rpl1b-1. The suppression of rpl1b-1 by either of the wild-type genes suggests that the mutant protein is not functional. Subsequently, experiments with a strain carrying a deletion of RPL1B (24) gave results similar to those for strain sm9 (data not shown).
Strain sm9 with a plasmid carrying an intact RPL1B gene or an RPL1A gene grows at a normal rate and shows the usual repression of RP gene expression upon transfer to the nonpermissive temperature (Fig. 2, lanes 14 to 19), confirming that the original phenotype is due to a deficiency of RP L1.
Effects on rRNA transcription.
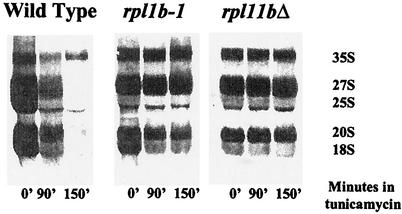
The major cost of ribosome synthesis is the transcription of rRNA. A defect in the secretory pathway leads to repression not only of RP genes but also of rRNA genes (19). We asked whether the limitation of 60S biosynthesis due to L1 deficiency also prevented the repression of rRNA transcription. Figure 3 shows the precursor and mature rRNAs labeled with a 3-min pulse of [C3H3]methionine. Disturbance of the secretory pathway, in this case with tunicamycin, brings about a severe repression of rRNA transcription in a wild-type cell, evident by 90 min and almost complete by 150 min. However, in strain sm9, carrying rpl1b-1, tunicamycin has relatively little effect on rRNA transcription, although rRNA processing is slowed somewhat. Thus, limitation of 60S subunits interferes with the repression of ribosome synthesis at the levels of both RNA polymerase I and RNA polymerase II.
FIG. 3.
rRNA synthesis is also protected by a shortage of 60S subunits. Strains carrying the indicated mutations were grown for several generations in minimal medium without methionine. To each culture was added tunicamycin to a final concentration of 5 μg/ml. Each lane represents RNA from a culture that had been labeled with [C3H3]methionine for 3 min (see Materials and Methods) at the indicated time after the addition of tunicamycin.
The effect is not limited to L1 mutants.
The defective RPL1B gene causes strain sm9 to grow slowly; complementation with another copy of RPL1B permits normal growth. Therefore, growth appears to be limited by the supply of 60S ribosomal subunits. This is apparent in Fig. 2B, which shows a sucrose gradient analysis of the ribosomes. The presence of half-mers, polysomes with an additional 43S initiation complex awaiting a limiting 60S subunit, is evidence of insufficient 60S subunits (27). Although some free 60S subunits are present, these may be defective due to the mutant L1 protein.
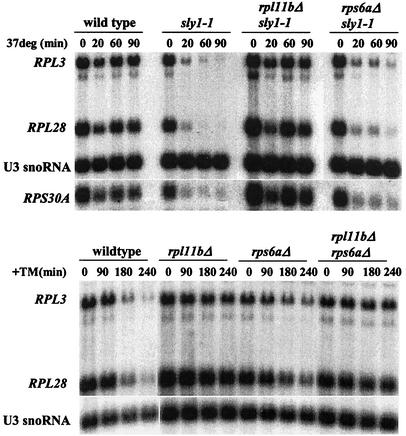
We then asked whether limitation of ribosomal subunits in general could counteract the repression of ribosome synthesis due to a defect in the secretory pathway, by using cells with a deletion either of a gene encoding a small subunit protein, RPS6A, or of a gene encoding a different large subunit protein, RPL11B. In each case, cell growth is limited by the loss of one of a pair of RP genes, with doubling times of 160 and 180 min, respectively, compared to 90 min for the wild type. In cells lacking RPS6A, large amounts of free 60S subunits accumulate; in cells lacking RPL11B, 40S subunits and half-mers accumulate to a substantially greater extent than in sm9 cells (data not shown). After cells are shifted to the nonpermissive temperature, the repression of RP gene transcription in cells bearing sly1-1 is independent of the presence of RPS6A, and it is almost abolished in cells lacking RPL11B (Fig. 4A). Note that the transcriptional response of a gene encoding a 40S protein, RPS30A, is the same as that of genes encoding 60S proteins. The deletion of RPL11B also protects transcription of rRNA from repression by tunicamycin (Fig. 3), showing the generality of the phenomenon.
FIG. 4.
Insufficient 60S protein, but not insufficient 40S protein, protects ribosome synthesis from repression due to a defect in the secretory pathway. (A) The indicated strains were grown to log phase in YPD at 30°C. An aliquot was harvested, the rest of the culture was shifted to 37°C, and aliquots were harvested as indicated. Total RNA was isolated, and 5 μg of RNA was analyzed by Northern blotting. Individual RNAs were detected by using oligonucleotide probes as described in Materials and Methods. (B) The same experiment as described for panel A, except that the indicated strains were grown at 30°C and treated with tunicamycin (TM) at a concentration of 2.5 μg/ml.
Parallel experiments with strains with a deletion of RPL20B, RPL34B, or RPS4B give the same results: deficiency of a 40S RP leading to accumulation of free 60S subunits has little if any effect on the repression of RP mRNA synthesis, and deficiency of a 60S RP that leads to accumulation of 40S subunits and half-mers renders RP mRNA synthesis resistant to repression upon treatment with tunicamycin.
To test the possibility that the effect of deficient 60S subunits is mediated through the imbalance of the 40S and 60S subunits, we generated two sets of double-mutant strains missing genes encoding both RPL11B and RPS6A or RPS23A. The growth rate of these strains differs little from that of the single mutants, and the level of half-mers is reduced, though not eliminated. Nevertheless, the RP genes are still protected from repression by tunicamycin (Fig. 4B, four right lanes, and Table 2, compare the last two lines). Thus, we conclude that an imbalance in subunit concentration is not responsible for this effect.
TABLE 2.
Effect of deletion of genes encoding both a large and a small subunit proteina
| Strain |
RPL0/U3 ratio in expt no.:
|
|
|---|---|---|
| 1 | 2 | |
| Wild type | 24 | 29 |
| RPS6AΔ or RPS23BΔ | 11 | 28 |
| RPL11BΔ | 80 | 84 |
| Both deletion mutants | 94 | 76 |
Cells of the indicated genotype (in experiment 1, RPS6A was the 40S gene deletion mutant, and in experiment 2, RPS23B was the 40S gene deletion mutant) were treated with tunicamycin for 3 h. RNA was prepared from samples before and after treatment with tunicamycin, subjected to Northern analysis, probed with sequences from RPL30 and U3 snoRNA, and quantitated by PhosphorImager analysis. The values in the table represent the RPL30/U3 ratio at 180 min as a percentage of the ratio at zero time.
Mutants that block 60S formation have the same effect.
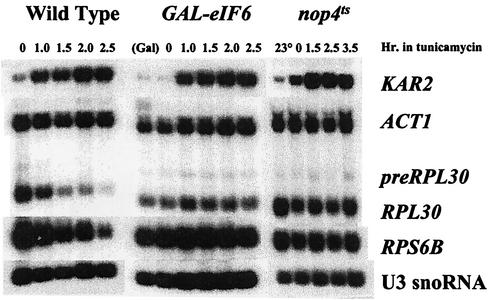
Insufficiency of an RP could effect its phenotype either by causing a deficiency of 60S subunits or in some more-direct manner. To distinguish these alternatives, we took advantage of some of the large number of non-RPs needed to produce a 60S subunit (14). A strain carrying a temperature-sensitive allele of NOP4 cannot make 60S subunits at a nonpermissive temperature (32). As in the case of a deficiency of a 60S RP, an imbalance in subunits develops; 40S subunits and half-mers accumulate (32). When such a strain is shifted to the nonpermissive temperature 1 h before the addition of tunicamycin, the repression of RP synthesis is greatly reduced (Fig. 5). The protein eukaryotic initiation factor 6 (eIF-6p), a product of the TIF6 gene, is essential for a late step in 60S synthesis (2, 29). In its absence, 27S pre-rRNA is formed but is rapidly degraded and no 25S mature rRNA appears. Taking advantage of a strain in which ubiquitin-tagged eIF-6p, under the control of the GAL10 promoter, decays rapidly after the cells are shifted to glucose, we find that cells given tunicamycin 2 h after the addition of glucose no longer repress RP synthesis (Fig. 5). From these two experiments, we conclude that any defect in the production of 60S subunits reduces or blocks the repression of RP synthesis due to an interruption in the secretory pathway.
FIG. 5.
Mutant non-RPs involved in 60S subunit assembly also protect ribosome synthesis from repression due to a defect in the secretory pathway. Strain W303 (wild type) was treated with tunicamycin at a concentration of 5 μg/ml, and samples were taken at the indicated times for the preparation of RNA. Strain KSY603, carrying a ubiquitin-tagged eIF-6 protein under the control of the GAL10 promoter (2) and growing in YPGal medium, was transferred to YPD medium to repress transcription and to permit the decay of the eIF-6 protein. A sample was taken for RNA. After 2 h in YPD, another sample was taken. Tunicamycin was added to the rest of the culture, and samples were taken at the indicated times. Strain JWY4995, carrying a temperature-sensitive allele of NOP4, was grown at 23°C and shifted to 37°C. One hour later, tunicamycin was added, and the culture was maintained at 37°C. Samples were taken at 23°C, after 1 h at 37°C, and at the indicated times after tunicamycin treatment.
Deletion of RPL11B affects the response to tunicamycin even when L11 is not limiting for growth.
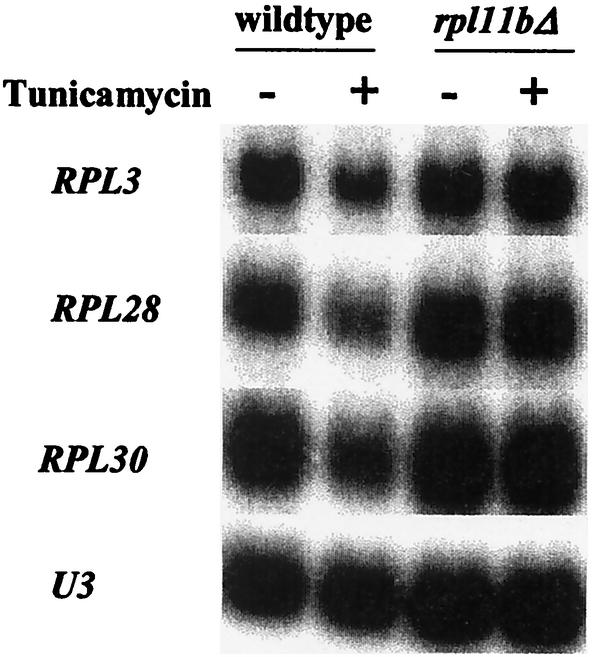
In YPGE medium, where cells utilize the nonfermentable carbon sources glycerol and ethanol, the wild-type and rpl11bΔ strains grow at nearly the same rate, with a doubling time of about 4 h. Thus, L11 is no longer limiting for growth, although it is limiting for translation, as half-mers occur to the same extent as in YPD medium (data not shown). Nevertheless, treatment of the two strains with tunicamycin leads to repression of RP mRNA in wild-type cells but not in rpl11bΔ cells (Fig. 6). This result demonstrates that a partial block in the synthesis of 60S ribosomal subunits can influence the control of ribosome synthesis even when they are not limiting for growth.
FIG. 6.
A defect in 60S synthesis affects regulation of ribosome synthesis even when it does not affect growth rate. Cultures of strains J1203 (wild type) and J1204 (rpl11bΔ) were grown in YPGE at 30°C, in which they have the same doubling time, for about 4 h. Samples were taken before and 4 h after treatment with 2.5 μg of tunicamycin/ml. RNA was prepared and subjected to Northern analysis. The presence of tunicamycin reduced the RP mRNA by about 50% in wild-type cells but not at all in cells carrying rpl11bΔ.
How a deficiency of 60S subunits could influence the regulation of ribosome synthesis.
Defective ribosome synthesis could generate a signal at one of three levels: (i) insufficiency of ribosomes could have a feedback effect, stimulating the synthesis of ribosomes; (ii) interference with the initiation of translation, at least in certain ways, could counteract the repression of ribosome synthesis; or (iii) improper assembly of ribosomal subunits could itself provide a feedback to counteract the repression of ribosome synthesis. Each of these has been tested.
Deletion of RPL11B does not lead to an increase in the mRNA level of other RPs.
Cells might compensate for the deficiency of 60S subunits by increasing the overall synthesis of ribosomes. However, comparison of the level of RP mRNAs in mutant and wild-type strains revealed no such stimulation (Table 3). The level of RP mRNA, compared to that of the stable U3 snoRNA, generally reflects the growth rate of the cells. In cells growing more slowly due to a less favorable carbon source, the overall level of RP mRNA is reduced. In cells growing more slowly due to deletion of RPL11B, the level of RP mRNA is also reduced. In YPGE medium, where the growth rates of the two strains are similar, the levels of the RP mRNAs are also similar. Thus, the phenotype of the strain carrying rpl11bΔ, in reducing the effect of a defect in the secretory pathway on ribosome synthesis, is not due to a general increase in ribosome synthesis. Furthermore, since much of the response to the carbon source is mediated through the ras-PKA pathway (22), these data suggest that this pathway is unaffected by a deficiency of 60S subunits.
TABLE 3.
Relative levels of RP mRNAsa
| mRNA | Relative level of RP mRNA in strain grown in medium (doubling time [min])
|
|||||
|---|---|---|---|---|---|---|
| Wild type
|
rpl11bΔ
|
|||||
| YPD (100 ± 15) | YPR (215 ± 25) | YPGE (245 ± 15) | YPD (180 ± 15) | YPR (220 ± 10) | YPGE (260 ± 10) | |
| RPL3 | 1.00 | 0.34 | 0.45 | 0.48 | 0.35 | 0.28 |
| RPL28 | 1.00 | 0.38 | 0.72 | 0.75 | 0.84 | 0.65 |
| RPL30 | 1.00 | 0.52 | 0.64 | 0.59 | 0.62 | 0.50 |
| RPS26 | 1.00 | 0.40 | 0.56 | 0.85 | 0.83 | 0.61 |
The quantities of RPL3, RPL28, RPL30, and RPS26 mRNA measured with specific oligonucleotide probes are expressed as relative quantities, with U3 snoRNA as the internal loading control. The quantities of RP mRNA of the wild-type strain grown in YPD were arbitrarily set to 1.00. The values of RP genes are the means of two independent experiments, with variations of usually less than 10%.
Translation initiation factors and repression of RP mRNAs in response to secretory defect.
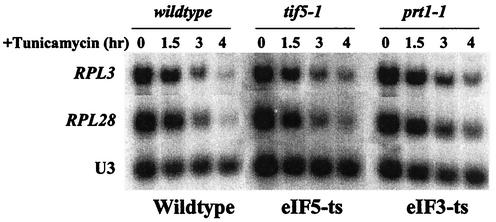
An imbalance in the 60S/40S ratio leads to a slowing of the translational initiation process. Perhaps the cell senses the reduced rate of initiation, either by the presence of sparsely translated mRNAs or by the accumulation of half-mers, which could sequester initiation factors that are generally at a far lower concentration than ribosomes. This could lead to a signal to override the repression due to a defect in the secretory pathway. Therefore, we examined the effects of two mutants that inhibit different steps in the initiation of translation: prt1-1, a temperature-sensitive allele of a subunit of eIF-3 that promotes binding of Met-tRNAiMet and mRNA to 40S subunits (4), and tif5-1, a temperature-sensitive allele of eIF-5 that promotes eIF-2 GTPase activity at the AUG initiation site (17). The prt1-1 and tif5-1 cells were grown at a semipermissive temperature (30°C), at which their doubling time is about 200 min. In both strains, tunicamycin caused the RP mRNA levels to decrease substantially (Fig. 7), as in wild-type cells. Thus, inhibition of either an early or a late step of translation initiation does not behave like a limitation of 60S subunits.
FIG. 7.
Regulation of ribosome biosynthesis in response to a defect in the secretory pathway is not affected by reduced initiation of translation. Strains TMY5-1 (tif5-1ts) and ts187 (prt1-1ts) were grown at the semipermissive temperature of 30°C, where they have a doubling time of about 200 min. A sample was taken, tunicamycin was added to a concentration of 2.5 μg/ml, and additional samples were taken at the indicated times. RNA was prepared and subjected to Northern analysis.
Effect of 60S deficiency may be a nuclear rather than a cytoplasmic phenomenon.
The production of defective ribosomal particles due to insufficient supply of an essential protein, or to a mutant processing factor, has three consequences. The first is an insufficiency of functional ribosomes. The second is the presence of defective particles. The third is the degradation of the defective particles, since they rarely, if ever, accumulate to a significant degree. We have argued above that the effects on translation of insufficient ribosomal particles are not responsible for the phenotype of strain sm9.
Perhaps some aspect of the generation of defective 60S subunits, or their degradation, e.g., the accumulation of some intermediate breakdown product, influences the regulation of ribosome synthesis. To try to distinguish whether it is the generation of defective 60S particles or their attendant degradation that contributes to the phenotype that we observe, we have examined the effects of tunicamycin on strains carrying mutations in three genes encoding proteins implicated in late steps of 60S production. Indeed, each has been linked circumstantially with the export pathway. Tif6p (28, 29) and Nmd3p (11) appear to be shuttling proteins that accompany the newly formed 60S particle as it is exported to the cytoplasm. Kap120p is one of the karyopherins involved in 60S export (31). Mutations in the genes encoding these three proteins all bring about a diminution of 60S synthesis, an imbalance in ribosomal subunits, and the appearance of half-mers. However, the immediate phenotypes differ. In cells lacking eIF-6p, newly formed 25S rRNA is not observed, either because the 27S pre-rRNA is degraded or because the 25S rRNA is degraded with a vanishingly short t1/2 (2). In cells with temperature-sensitive Nmd3p, 25S rRNA is formed but decays with a t1/2 of 3 to 4 min (10). In the absence of the karyopherin Kap120p, cells generate apparently mature 25S rRNA that is stable for the period of a 10- to 15-min pulse-chase, although eventually it must be degraded since the steady-state molar ratio of 25S/18S is about 0.5 (31).
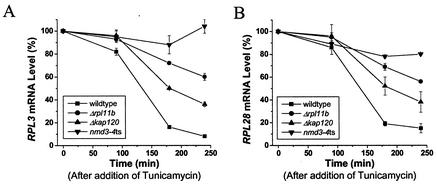
As seen in Fig. 5, depletion of eIF-6p protects RP genes from the repression caused by tunicamycin. Temperature-sensitive nmd3-4 cells grown at a semipermissive temperature limit the cells for 60S subunits and cause the accumulation of half-mers. These cells appear even less responsive to tunicamycin than the nop4ts cells or the rpl11bΔ cells (Fig. 8). However, kap120Δ cells, with the same growth rate and accumulation of half-mers, are substantially more responsive to tunicamycin, though less so than wild-type cells (Fig. 8). We suggest that the difference between the kap120Δ strain and the nmd3-4 strain is due to a difference in the rate, and perhaps the mechanism, of the degradation of defective (or untransportable) 60S subunits.
FIG. 8.
Degradation of malformed 60S may attenuate repression of ribosome synthesis. The wild-type strain and strains J1204 (rpl11bΔ) and PSY1082 (kap120Δ) were grown to log phase at 30°C; strain AJY734 (nmd3-4ts) was grown to log phase at the semipermissive temperature of 34°C, at which its doubling time is about 200 min. In each case, samples were taken before and at the indicated times after the addition of tunicamycin to a concentration of 2.5 μg/ml. RNA was prepared and subjected to Northern analysis, where the blots were probed with several RP genes and with U3 snoRNA. After PhosphorImager analysis, the values for the RP genes were normalized with the U3 RNA and compared to those at zero time.
Taken together, the results presented above suggest (i) that the effect of a mutant such as rpl1b-1 is more likely due to the production of defective 60S subunits in the nucleus than to the deficiency of 60S subunits in the cytoplasm and (ii) that perhaps some product of the degradation of defective subunits can counteract the signal for repression of ribosome synthesis that arises from a defect in the secretory pathway.
DISCUSSION
While the mutant search that initiated this work has not revealed direct participants in the regulatory scheme downstream of Pkc1p, as we had hoped, it has uncovered an intriguing case of autoregulation within the process of ribosome synthesis, itself. What is clear from the initial isolation of rpl1b-1, followed by the use of strains with deletions of other RPL genes and of mutants defective in 60S formation, is that some aspect of a deficiency of 60S ribosomal subunits largely suppresses the repression of ribosome synthesis due to a defect in the secretory pathway. This effect must be at the heart of the regulatory process because it suppresses transcription not only of RP genes but of rRNA genes as well (Fig. 3). Surprisingly, a deficiency of 40S subunits does not have this effect. Perhaps this is related to the recent, rather surprising observation that the maturations of the 40S and 60S subunits occur almost entirely separately (6). Could the specific effect of malassembly of 60S subunits reflect the fact that it is the 60S subunit that interacts with the endoplasmic reticulum membrane?
We have endeavored to determine which of the many manifestations of a deficiency of 60S subunits is responsible for the observed effect.
A deficiency of 60S subunits reduces the rate of protein synthesis, as evident from the slower growth rate of the initial sm9 mutant. Supplementation of the cell with another copy of either RPL1A or RPL1B overcomes the slow-growth phenotype, demonstrating that insufficient L1 protein is limiting for cell growth. As an alternative way of reducing protein synthesis and growth rate, we used temperature-sensitive mutants affecting two proteins of the translational initiation machinery, Prt1p, which is part of eIF-3 and acts at a relatively early stage of initiation (21), and Tif5p, a GTPase-activating protein for the GTPase of eIF-2p that induces GTP hydrolysis when the 43S initiation complex reaches the initiator AUG of the mRNA at a relatively late stage (17). In each case, the strains were grown at a semipermissive temperature at which translation initiation is limiting and the growth rate was approximately the same as that of sm9 cells. For both the translation initiation mutants, tunicamycin brought about the normal repression of ribosome synthesis (Fig. 7). These results, together with the observation that deficient 40S subunits have no effect on regulation (Fig. 4), suggest that neither reduced protein synthesis nor reduced growth rate is the cause of the suppression of repression that we observe.
A deficiency of 60S subunits brings about the formation of half-mers, where 43S initiation complexes are stalled at the initiator AUG, waiting for limiting 60S subunits (27). While this reduces the rate of initiation, as dealt with in the previous paragraph, it also disrupts the dynamic stoichiometry of initiation factors and 40S subunits. The initiation factors are present at only 5 to 10% of the level of ribosomes (based on mRNA levels) (12), a situation that is efficient if most of the ribosomes are elongating, as is usually the case. However, if a substantial fraction of the 40S subunits are tied up as half-mers, then their associated initiation factors may become limiting, a situation that could send a signal to restore ribosome synthesis. Therefore, we considered whether the accumulation of half-mers could overcome the repressive signal. However, this appears not to be the case. When cells in which deletion of RPL11B is combined with deletion of RPS6A or RPS23B, the balance of subunits is largely, though not completely, restored, and the level of half-mers is reduced. Nevertheless, the lack of a 60S protein appears dominant, as ribosome synthesis in these cells is resistant to repression by a defective secretory pathway (Fig. 4B and Table 2). Furthermore, although the level of half-mers is consistently higher in cells carrying rpl11bΔ than in sm9 cells or in cells carrying rpl1bΔ, the effect of tunicamycin is suppressed to almost the same degree in all of these strains. Therefore, we conclude that the formation of half-mers, per se, is not responsible for the effects of a deficiency of 60S subunits on ribosome synthesis.
The manufacture of a 60S ribosomal subunit is extraordinarily complex, involving nearly 200 auxiliary protein and RNA molecules and many serial steps of processing the rRNA together with assembly and disassembly of RPs and non-RPs (1, 8, 14). The data presented above show that improper assembly of 60S subunits, whether due to insufficiency of L1, L11, L20, or L34, to a lack of eIF-6p, to a temperature-sensitive Nop4p, or to a temperature-sensitive Nmd3p, substantially prevents the repression of ribosome synthesis in response to a defect in the secretory pathway. These results suggest that it is not a specific step of assembly that impacts the regulation of ribosome synthesis but rather the existence of malformed 60S subunits and/or their degradation.
Since the RNA components of 60S and 40S subunits, except for 5S RNA, derive from a common 35S pre-rRNA precursor, they start their lives as equimolar. Furthermore, many measurements suggest that the synthesis of RPs is roughly equimolar (reviewed in reference 39). Therefore, cells in which half the supply of one essential RP is eliminated have a serious disposal problem. Cells carrying rpl1b-1 destroy more than half of their 27S pre-rRNA and more than half of their 60S RPs, representing some 15% of the total transcription and 5% of the total translation of the cell. Little is known of this process, but it must be efficient because little if any accumulation of partially assembled ribosomal subunits or of RPs has been detected. We wondered if some aspect of the degradation process could be responsible for the effects on the transcription of ribosomal components.
Results with the strain carrying kap120Δ support this view. In that case, assembly of the 60S subunits appears to be essentially complete, but the subunits are exported only inefficiently and accumulate within the nucleus (31). As a result, cytoplasmic polysomes have abundant half-mers, and protein synthesis is slowed. However, these cells are substantially more responsive to tunicamycin than are cells carrying other mutants affecting the synthesis of 60S subunits (Fig. 8). By contrast, cells limited for a different export factor, Nmd3p, produce apparently mature 25S rRNA but degrade it rapidly (10). Ribosome synthesis in these cells is resistant to the effects of tunicamycin. Thus, we conclude that it is more likely the degradation rather than the malassembly of 60S subunits that blocks the signal from PKC that a defective secretory pathway is endangering the cell and that ribosome synthesis should be shut down. Why a similar effect is not seen for malassembled 40S subunits is not clear. One possibility is that since the final steps of 40S maturation take place in the cytoplasm (20, 34), perhaps the quality control of 40S subunits is a cytoplasmic phenomenon while that of 60S subunits is a nuclear phenomenon that can more readily be utilized for regulation. However, since there remains some ambiguity regarding the cellular compartment in which Nmd3p and Kap120p function, one should be cautious with such an interpretation.
As pointed out above, little is known about the turnover of malassembled ribosomal subunits. However, we can draw two conclusions. First, Fig. 2A and 5 provide striking evidence that the components of 60S subunits lacking an essential protein, or improperly assembled, are not recycled. Any excess of the RP L30 can bind to the unspliced transcript of its own gene and prevent its splicing, leading to accumulation of unspliced transcript (5, 36). Recycling of L30 derived from improperly assembled subunits would lead to an excess of L30 and consequently to the accumulation of unspliced RPL30 transcript. Since little, if any, such accumulation is evident in Fig. 2A or 5 (pre-RPL30), we conclude that malassembled subunits are degraded in their entirety. Second, the growth of cells missing an RP gene seems to be roughly proportional to the availability of that protein. This suggests that the turnover of malassembled subunits does not include the associated proteins involved in ribosome maturation (1, 8). Since these are present in much smaller amounts (as estimated from transcriptome data) (12), their degradation would bring ribosome synthesis to a halt.
In summary, we suggest that the influence of mutations that reduce the biosynthesis of 60S subunits on the transcription of ribosomal genes is not due to the reduction of protein synthesis or growth rate caused by insufficient 60S subunits, nor to some interference with translational initiation mediated through the formation of half-mers, nor to the malassembly of 60S subunits per se, but is brought about by the process or the products of the degradation of malformed 60S subunits.
We know embarrassingly little about the interface between the signal transduction pathways regulating ribosome synthesis, the heat shock, the PKA, the PKC, or the TOR pathway, with the apparatus modulating the transcription of rRNA genes by polymerase I and of RP genes by polymerase II. The interesting aspect of our observations is that problems with the synthesis of 60S subunits affect only the repression of ribosome biosynthesis that is regulated through the PKC pathway. Cells with a defect in 60S production are as sensitive as wild-type cells to heat shock (Fig. 2) and to rapamycin (data not shown) while their RP mRNA level has a normal ras-PKA-mediated response to the carbon source (Table 3). Moreover, deficient and/or defective 60S subunits do not lead to a generalized stimulation of ribosome synthesis. These considerations suggest that the regulation of ribosome synthesis is even more subtle and more attuned to the needs of the cell than we had originally imagined.
Acknowledgments
We are grateful to U. Maitra, J. Woolford, A. Johnson, and P. Silver for strains and to I. Willis and C. Query for careful readings of the manuscript.
This work was supported in part by grant GM25532 to J.R.W. and grant CA13330 to the Albert Einstein Cancer Center and by a Human Frontiers grant to J.R.W.
REFERENCES
- 1.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 2.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox, J. S., C. E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197-1206. [DOI] [PubMed] [Google Scholar]
- 4.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 5.Eng, F. J., and J. R. Warner. 1991. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell 65:797-804. [DOI] [PubMed] [Google Scholar]
- 6.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 7.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell, L. H., and C. S. McLaughlin. 1969. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl. Acad. Sci. USA 62:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, J. H., and A. W. Johnson. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 13.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 14.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of transcription of ribosomal protein synthesis. Mol. Cell. Biol. 19:5393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., R. D. Moir, I. K. Sethi-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiti, T., S. Das, and U. Maitra. 2000. Isolation and functional characterization of a temperature-sensitive mutant of the yeast Saccharomyces cerevisiae in translation initiation factor eIF5: an eIF5-dependent cell-free translation system. Gene 244:109-118. [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi, K., R. Tsujii, H. Yoshida, Y. Maki, A. Wada, Y. Matsui, E. Toh, and K. Mizuta. 2002. Normal assembly of 60S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 277:18334-18339. [DOI] [PubMed] [Google Scholar]
- 19.Mizuta, K., and J. R. Warner. 1994. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 14:2493-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moy, T. I., and P. A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13:2118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naranda, T., S. E. MacMillan, and J. W. Hershey. 1994. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J. Biol. Chem. 269:32286-32292. [PubMed] [Google Scholar]
- 22.Neuman-Silberberg, F. S., S. Bhattacharya, and J. R. Broach. 1995. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol. 15:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in S. cerevisiae. J. Biol. Chem. 274:13235-13241. [DOI] [PubMed] [Google Scholar]
- 24.Petitjean, A., N. Bonneaud, and F. Lacroute. 1995. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal proteins. Mol. Cell. Biol. 15:5071-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signalling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rameau, G., K. Puglia, A. Crowe, I. Sethy, and I. Willis. 1994. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol. 14:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotenberg, M. O., M. Moritz, and J. L. Woolford, Jr. 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2:160-170. [DOI] [PubMed] [Google Scholar]
- 28.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 29.Si, K., and U. Maitra. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 34.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 35.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for the maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilardell, J., and J. R. Warner. 1994. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 8:211-220. [DOI] [PubMed] [Google Scholar]
- 37.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 38.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 39.Woolford, J. L., Jr., and J. R. Warner. 1991. The ribosome and its synthesis, p. 587-626. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.