Abstract
Many regulatory elements in eukaryotic promoters do not correspond to optimal recognition sequences for the transcription factors that regulate promoter function by binding to the elements. The sequence of the binding site may influence the structural and functional properties of regulatory protein complexes. Fos-Jun heterodimers were found to bind nonconsensus AP-1 sites in a preferred orientation. Oriented Fos-Jun heterodimer binding was attributed to nonidentical recognition of the two half-sites by Fos and Jun. Jun bound preferentially to the consensus half-site, whereas Fos was able to bind nonconsensus half-sites. The orientation of heterodimer binding affected the transcriptional cooperativity of Fos-Jun-NFAT1 complexes at composite regulatory elements in mammalian cells. Jun dimerization with Fos versus ATF2 caused it to bind opposite half-sites at nonconsensus AP-1 elements. Similarly, ATF2 bound to opposite half-sites in Fos-ATF2-NFAT1 and ATF2-Jun-NFAT1 complexes. The orientations of nonconsensus AP-1 sites within composite regulatory elements affected the cooperativity of Fos-Jun as well as Jun-Jun binding with NFAT1. Since Jun homodimers cannot bind to AP-1 sites in a preferred orientation, the effects of the orientations of nonconsensus AP-1 sites on the stabilities of Jun-Jun-NFAT1 complexes are likely to be due to asymmetric conformational changes in the two subunits of the homodimer. Nonconsensus AP-1 site orientation also affected the synergy of transcription activation between Jun homodimers and NFAT1 at composite regulatory elements. The asymmetric recognition of nonconsensus AP-1 sites can therefore influence the transcriptional activities of Fos and Jun both through effects on the orientation of heterodimer binding and through differential conformational changes in the two subunits of the dimer.
The recognition sequences for individual transcription regulatory proteins vary among different promoter regions. The reasons for this diversity of recognition sequences are not well understood. The recognition sequences are frequently suboptimal binding sites for the regulatory proteins, which would appear to impede the already formidable task of transcription factors: to specify unique sites of transcription initiation in the genome. In some cases, replacement of nonconsensus recognition sequences by optimal binding sites eliminates their transcription regulatory functions (18). There are many possible explanations for this apparently imperfect nature of regulatory elements. One possibility is that they serve as binding sites for many different proteins and that the sequences of the elements serve to balance binding by alternative regulatory proteins. A second possibility is that each element contributes only a small fraction of the specificity required for promoter function and that the sequence variability prevents individual transcription factors from dominating the regulation of the promoter. A third possibility is that the sequence of the element influences the function of the protein(s) that binds to the element. Such functional effects might be mediated by changes in the conformation or the structural organization of nucleoprotein complexes (7, 14, 28, 32, 41, 44, 45).
In eukaryotic organisms, transcription initiation is regulated by the concerted action of many transcription factors that form multiprotein complexes at promoter and enhancer regions. Assembly of these nucleoprotein complexes can be affected by the orientations of the individual regulatory elements (26, 27, 39, 40). Some heterodimers that recognize palindromic DNA sequences also exhibit orientation-dependent transcriptional activities at regulatory elements that contain asymmetric base pairs either within the recognition site or in flanking sequences (6, 10, 35, 36). This orientation dependence can be mediated by cooperative DNA binding with transcription factors that recognize adjacent binding sites within composite regulatory elements (10, 35, 36).
Fos and Jun family bZIP proteins form homo- and heterodimers via a leucine zipper dimerization interface and bind to palindromic AP-1 recognition elements [TGA(C/G)TCA] via a basic DNA contact region. Fos and Jun make essentially identical contacts with the two half-sites in the X-ray crystal structure of the Fos-Jun-AP-1 complex (13). However, symmetry-related base substitutions in the two half-sites have distinct effects on Fos-Jun heterodimer binding (38). The differential effects of symmetry-related base substitutions indicate that they are recognized in the context of the asymmetric central base pair or flanking DNA sequences.
The consensus AP-1 element is not perfectly symmetrical, since the central CG base pair results in two different overlapping half-sites (TGAC and TGAG). Fos-Jun heterodimers and Jun homodimers have similar, though not identical, binding preferences for variants of the AP-1 recognition sequence (24). It remains unclear whether Fos and Jun have different DNA recognition specificities for the core AP-1 regulatory element and whether such differences influence the structural and functional characteristics of Fos-Jun heterodimers.
Fos-Jun heterodimers bind different AP-1 sites in opposite orientations that differ by an approximately 180° rotation about the dimer axis (29, 30, 33). Upon binding to the AP-1 site, Fos and Jun bend DNA in opposite directions through electrostatic interactions with the DNA flanking the AP-1 site (21, 25, 37). DNA bending by bZIP proteins has been questioned based on the lack of DNA bending in X-ray crystal structures (13) and the failure to detect bending in cyclization and minicircle binding assays (42, 43). Bending by bZIP proteins has been corroborated by conformationally sensitive gel electrophoresis (20), oligonucleotide cyclization (19), and fluorescence resonance energy transfer (15). The differences in DNA bending propensities between Fos and Jun on the one hand and sequences on opposite sides of the AP-1 site on the other exhibit a perfect correlation with the orientation of heterodimer binding (37). Both DNA bending by Fos and Jun and the orientation of heterodimer binding are affected by amino acid and nucleotide substitutions at a distance from the protein-DNA contact interface observed in the crystal structure (25, 29, 30). These data suggest that the orientation of heterodimer binding at sites with palindromic core recognition elements is determined by the difference in DNA bending propensities between sequences on opposite sides of the AP-1 site (29). These flanking DNA sequences are recognized through long-range electrostatic interactions between amino acid residues adjacent to the basic regions and the phosphate backbone of DNA (37).
The orientation of heterodimer binding can influence cooperative interactions with transcription factors that bind to adjacent regulatory elements (8, 10, 35, 36). Fos-Jun heterodimers can form cooperative complexes with NFAT1 at composite NFAT-AP-1 regulatory elements that activate cytokine gene expression in response to antigen presentation (17). The interaction between NFAT1 and Fos-Jun heterodimers requires Jun to occupy the half-site proximal to the NFAT binding site (3, 4, 8, 35, 36). The preferred orientation of Fos-Jun heterodimer binding influences the transcriptional cooperativity between Fos-Jun and NFAT1 at composite regulatory elements in vitro (35, 36). Fos-Jun heterodimers that favor the orientation of binding that allows Jun to bind the half-site proximal to the NFAT recognition element form more stable and transcriptionally active Fos-Jun-NFAT1 complexes than heterodimers that favor the opposite binding orientation (35, 36).
NFAT can also regulate transcription in concert with other bZIP family proteins (31). Jun-ATF2 heterodimers and NFAT1 participate in regulation of the tumor necrosis factor alpha promoter (11, 46). However, the proteins do not bind cooperatively to the nonadjacent sites in this promoter. Maf and NFAT bind and activate the interleukin 4 (IL-4) promoter in Th-2 cells, but it is not clear whether binding in this case is cooperative and whether Maf and NFAT act concurrently in the activation of IL-4 transcription (2, 16). The influence of the variation in the sequences of composite regulatory elements on the promoter selectivity of NFAT and bZIP family members remains to be determined.
Most composite NFAT-AP-1 regulatory elements contain nonconsensus AP-1 recognition sequences (5, 31). Asymmetric recognition of nonconsensus AP-1 sites by Fos and Jun may influence their cooperative interactions with other transcription factors at composite regulatory elements. We have studied the functional roles of asymmetric recognition of nonconsensus regulatory elements by investigating the cooperative interactions of Fos and Jun dimers with NFAT1.
MATERIALS AND METHODS
Preparation of fluorescent oligonucleotides and proteins.
Oligonucleotides with the sequence CTCATTGAGGAAATGATGACTCATCATATCGTCAATGAG and its complement (NC site) were labeled with fluorescein at their 5′ ends. This sequence contains an NFAT binding site (underlined) on the left side of a central AP-1 site (underlined, boldface). Base substitutions were made at the positions in the AP-1 site indicated in each figure. The eight base pairs on each end of each oligonucleotide were identical, in order to make the local environments of fluorophores linked to opposite ends of the duplexes identical. The oligonucleotide sequence CTCTAGATGCTGACTCAGCATCTAGAG (S site) was used to examine the effects of uracil substitutions. Duplexes were prepared between 5′ fluorescein-labeled and unlabeled complementary strands as described previously (34, 37). Proteins encompassing residues 139 to 200 of Fos, residues 257 to 318 of Jun, and residues 350 to 505 of ATF2 were expressed and affinity purified as described previously (1, 30). The FosRI and JunRI proteins contained the amino acid substitutions R155I and R273I, respectively. The proteins were labeled at unique cysteine residues introduced at positions 142 (Fos) and 260 (Jun), respectively, by incubation with Texas Red maleimide (Molecular Probes) and were purified as described previously (29, 34, 37). Fluorophore labeling had no detectable effect on the apparent binding affinities of the proteins, nor did it have any measurable effect on DNA bending by the heterodimers (37). The DNA binding domain of NFAT1 (residues 396 to 692) was expressed and purified as described previously (8).
Gel-based fluorescence resonance energy transfer (gelFRET) analysis of heterodimer orientation and oligonucleotide competition assays.
The orientation of heterodimer binding was determined by measuring the relative efficiencies of energy transfer from donor fluorophores (fluorescein) linked to opposite ends of an oligonucleotide to an acceptor fluorophore (Texas Red) linked to either subunit of the heterodimer (29, 34). Complexes were formed by incubation of 2 μM Fos-Jun, Fos-ATF2, or Jun-ATF2 heterodimers containing one labeled subunit with 500 nM oligonucleotides labeled with fluorescein at either end for 10 min at room temperature. Where indicated, NFAT1 was added to the binding reactions at 1 μM and unlabeled oligonucleotide competitors were added at 1, 5, or 25 μM. The binding buffer contained 20 mM HEPES (pH 7.6), 100 mM KCl, 5% glycerol, 1 mM EDTA, 5 mM dithiothreitol, 0.5 mg of bovine serum albumin/ml, 0.1% (wt/wt) NP-40, and 0.05 mg of poly(dI-dC)/ml. Heterodimer and homodimer complexes were separated from each other and from the free components by nondenaturing 8% polyacrylamide gel electrophoresis in 25 mM Tris-195 mM glycine gel buffer (pH 8.3) for 2 h at 300 V and 4°C. The gels were scanned using a FluorimagerSI (Molecular Dynamics) as described previously (34, 37).
The relative efficiencies of energy transfer from the two ends of the oligonucleotide (end preferences) are a function of the fraction of heterodimers bound in each orientation (34, 37). End preference values were calculated based on the ratio between Texas Red (TR) and fluorescein (FL) emissions when the donor fluorophore was placed on the left (L) or the right (R) end of the oligonucleotide as described previously (29):
 |
The free energy of heterodimer reorientation was calculated as described previously (36) by using the end preference values of Fos-Jun-NFAT1 complexes as calibration standards and making the assumption that these complexes were fully oriented.
Measurement of the rates of complex dissociation by using FRET.
Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes were prepared by incubation of 50 nM dimers formed between Fos and Texas Red-labeled Jun, 100 nM NFAT1, and 20 nM oligonucleotides labeled with fluorescein at one 5′ end and containing composite recognition elements for 10 min at 25°C. The binding buffer consisted of 20 mM Tris-Cl (pH 7.6), 50 mM NaCl, 5% (vol/vol) glycerol, 5 mM dithiothreitol, and 0.5 mg of bovine serum albumin/ml. Dissociated proteins were competed by manual injection of 1 mg of sonicated herring DNA ml−1 into a solution mixed with a stir bar. Previous experiments using the same method have indicated that the mixing time is approximately 1 s (35). The change in fluorescence resonance energy transfer (FRET) was monitored as a function of time by excitation of the fluorescein donor at 480 nm. The fluorescein emission was measured at 520 nm, and the Texas Red emission was measured at 610 nm. The time-dependent change in fluorescence emissions was fitted to a first-order exponential function: 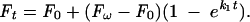
In this equation F0, Ft, and Fω are the fluorescences measured before addition of competitor, at time t after addition of competitor, and after equilibration, respectively. The half-lives of the complexes were calculated as ln 0.5/k1.
Dual-luciferase assay for comparison of the transcriptional activities of promoters with different structural organizations.
COS-1 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics at 37°C with 5% CO2. Reporter genes were constructed by inserting oligonucleotides containing variants of the NFAT-AP-1 composite regulatory element upstream of the minimal c-fos promoter linked to either the Renilla or the firefly luciferase reporter gene (35, 36). For each experiment, 0.5 μg (each) of the firefly and Renilla luciferase reporter plasmids was cotransfected with 0.9 μg of the SVβ-galactosidase internal control plasmid. The pcDNA-Fos and pcDNA-Jun expression vectors, as well as variants containing R155I and R273I mutations, respectively, were used at 0.5 μg for heterodimer experiments and 1.0 μg for homodimer experiments. The pEF-NFAT expression vector was used at 2.0 μg where indicated. Transfections were performed in six-well (30-mm-diameter) plates using FuGene 6 (Roche), and the cells were harvested 24 h posttransfection. Extracts were assayed for β-galactosidase and luciferase activities by using a dual-luciferase assay kit (Promega).
RESULTS
The effects of sequences flanking the AP-1 site on the orientation of heterodimer binding (29, 36, 37) suggested the possibility that Fos and Jun differentially recognize the two half-sites of the core AP-1 regulatory element despite their virtually identical DNA contacts in the X-ray crystal structure (3, 13). The orientation of heterodimer binding at sites containing asymmetric base substitutions is determined by the difference in DNA recognition specificities between the two subunits of the heterodimer. Analysis of the orientation of Fos-Jun heterodimer binding therefore provides information about the relative DNA recognition specificities of Fos and Jun.
Substitutions within the consensus AP-1 site influence the orientation of heterodimer binding.
To determine if Fos and Jun recognize the AP-1 site in an identical manner, we examined the effects of base substitutions on the orientation of heterodimer binding by using the gelFRET approach (Fig. 1) (29). We compared the relative efficiencies of energy transfer from donor fluorophores attached to opposite ends of a DNA oligonucleotide to an acceptor fluorophore attached to either subunit of the heterodimer (34). When the donor and acceptor fluorophores occupy the same side of the binding site, the efficiency of energy transfer is predicted to be higher than when they occupy opposite sides. The end preference value represents the relative efficiencies of energy transfer from opposite ends of the oligonucleotide and is a linear function of the fraction of heterodimers bound in each orientation. The end preference is not equivalent to the orientation fraction, since even fully oriented complexes exhibit some energy transfer from both ends of the oligonucleotide. For a perfectly symmetrical complex, the sum of the end preference values of heterodimers labeled on Fos and on Jun would be equal to 1. However, the fluorophores in the two subunits do not have to occupy symmetrical positions in order for this assay to be used for determination of relative orientation preferences at different binding sites (29). The absolute fraction of complexes bound in each orientation can be calculated if suitable standards for calibration of the relationship between end preference and orientation fraction are available (36).
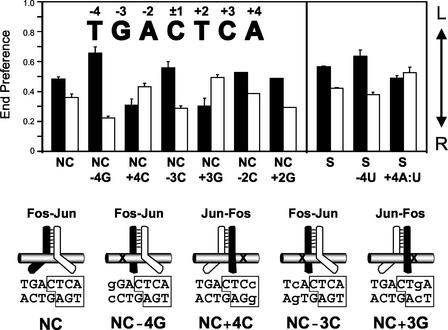
FIG. 1.
Effects of base substitutions within the consensus AP-1 site on the preferred orientation of Fos-Jun heterodimer binding. The orientation preferences of Fos-Jun heterodimers at AP-1 sites containing symmetry-related base substitutions were determined using the gelFRET approach. In this approach, heterodimers labeled with an acceptor fluorophore on either Fos or Jun were incubated with oligonucleotides labeled on opposite ends with a donor fluorophore, and the complexes were analyzed on a polyacrylamide gel. The relative efficiencies of energy transfer from opposite ends (end preferences, calculated as described in Materials and Methods) at AP-1 sites containing the base substitutions given below the graph are shown for heterodimers labeled on Fos (filled bars) and Jun (open bars). Standard deviations from three or more independent experiments, except at the NC−2C and NC+2G sites, are shown. A high end preference value indicates that the labeled subunit favors binding to the left half-site, whereas a low end preference value indicates a preference for the right half site (arrow on right). The numbering of the base pairs in the AP-1 site is indicated above the sequence. The diagrams below the graph depict the preferred orientations of Fos-Jun heterodimer binding at the different binding sites. Since the subunit that binds to the left half-site at the TGACTCA site can contact the central guanine on the lower strand, it is convenient to consider the central base on the lower strand to be part of the left half-site and the central base on the upper strand to be part of the right half-site. Base substitutions are shown in lowercase, and their positions are marked by an X in the diagrams.
The gelFRET approach for analysis of the orientation of heterodimer binding does not require determination of absolute distances between the fluorophores. Differences in protein and DNA conformations as well as in the mobilities of the fluorophores can also affect the end preference values. However, reciprocal changes in the relative efficiencies of energy transfer for complexes labeled on different subunits (FosTR-Jun versus Fos-JunTR) are most likely caused by a shift in the preferred orientation of heterodimer binding (34). Additional support for this interpretation is provided by the additive effects of asymmetric base substitutions and inversion of the central base pair (see below).
We examined the effects of symmetry-related single-base-pair substitutions within the core AP-1 site on the orientations of heterodimers formed by the bZIP domains of Fos and Jun (Fig. 1). At a consensus AP-1 site with a central CG base pair, Fos exhibited a slight preference for the left half-site and Jun exhibited a slight preference for the right half-site (Fig. 1, NC) (30, 37). Replacement of the TA base pair at the −4 position in the AP-1 site by a GC base pair caused reciprocal changes in the end preference values of Fos-Jun heterodimers labeled on Fos versus Jun. Jun bound preferentially to the consensus half-site on the right, whereas Fos bound to the nonconsensus half-site on the left (Fig. 1, NC−4G). When the symmetry-related base substitution was made in the opposite half-site, the preferred orientation of heterodimer binding was reversed (Fig. 1, NC+4C). Similar results were obtained at binding sites where the base pairs at the −4 and +4 positions were replaced by AT and TA base pairs, respectively (see #1 Fig. 2, NC−4A and NC+4T). Substitution of the GC base pair at the −3 position by a CG base pair also resulted in a change in orientation preference (Fig. 1, NC−3C). Again, Jun bound preferentially to the consensus half-site, whereas Fos bound to the nonconsensus half-site. The symmetry-related base substitution at the +3 position shifted the orientation preference in the opposite direction (Fig. 1, NC+3G). Similar results were obtained at binding sites containing AT and TA or TA and AT substitutions at the −3 and +3 positions, respectively (data not shown). In contrast, neither the replacement of the AT base pair at the −2 position by a CG base pair nor the symmetry-related replacement of the TA base pair at the +2 position by a GC base pair had a significant effect on the orientation of Fos-Jun heterodimer binding (Fig. 1, NC−2C and NC+2G). These base substitutions at the ±2 positions therefore did not have differential effects on DNA binding by Fos and Jun. The orientations of Fos-Jun binding at these nonconsensus AP-1 sites indicate that base substitutions at the ±4 and ±3 positions cause a greater loss of binding energy for Jun than for Fos. The free energies of reorientation (ΔΔGORI) for these complexes ranged from less than 1 kJ/mol to about 5 kJ/mol (36). In the context of the Fos-Jun heterodimer, Jun therefore exhibits a greater preference for binding to the consensus AP-1 half-site than Fos.
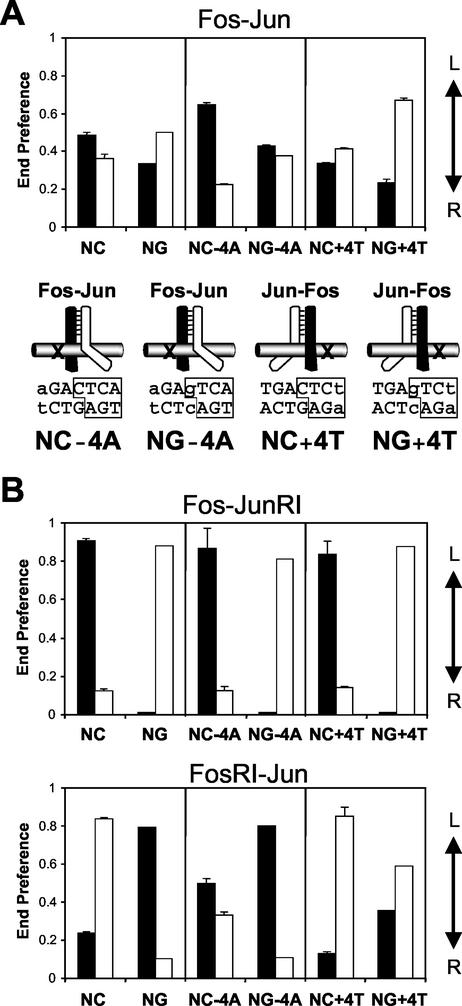
FIG. 2.
Combined effects of contacts to the central base pair and asymmetric base substitutions on the orientation of heterodimer binding. (A) Comparison of the orientation preferences of Fos-Jun heterodimers at the binding sites indicated below the bars. The end preferences of Fos-Jun heterodimers at AP-1 sites with a central CG base pair (NC sites) and at AP-1 sites with a central GC base pair (NG sites) were compared. Diagrams below the graph depict the orientation preferences of Fos-Jun heterodimers at AP-1 sites containing the indicated base substitutions. (B) Comparison of the orientation preferences of Fos-JunRI heterodimers and FosRI-Jun heterodimers at the binding sites indicated below the bars. Filled bars show end preferences for heterodimers labeled on Fos or FosRI, and open bars show end preferences for heterodimers labeled on Jun or JunRI. Standard deviations from three or more independent experiments are shown for complexes at the NC, NC−4A, and NC+4T sites.
Both Fos and Jun contact the methyl group on the thymine at the ±4 positions of the AP-1 site (3, 13). To determine if this methyl group was recognized identically by Fos and Jun, we examined the orientation preferences at binding sites in which the thymine was replaced by uracil (Fig. 1, S−4U and S+ 4A:U). Removal of the methyl group in opposite half-sites had reciprocal effects on the orientation preferences of Fos-Jun heterodimers. Removal of the methyl group had approximately 50% of the effect of other base pair substitutions at the ±4 positions. Fos and Jun therefore differentially recognized the methyl groups on the thymines at the ±4 positions, and the differential recognition of these methyl groups was a major determinant of the asymmetric recognition of the ±4 positions of the AP-1 site by Fos and Jun.
To determine if the central base pair and base substitutions in the two half-sites affected the orientation of Fos-Jun heterodimer binding independently, we examined the effect of inversion of the central base pair in combination with base substitutions at the ±4 positions of the AP-1 site (Fig. 2A ). At consensus AP-1 sites, inversion of the central base pair reversed the preferred orientation of Fos-Jun heterodimer binding (Fig. 2A, NC versus NG). At AP-1 sites containing an AT base pair at position −4, inversion of the central CG base pair reduced the orientation preference, whereas at sites containing the symmetry-related TA base substitution in the opposite half-site, inversion of the central CG base pair increased the orientation preference (Fig. 2A). At the NC−4A and NG+4T sites, the effects of the central base pair and the substituted base pair cooperated to increase the orientation preference, whereas at the NG−4A and NC+4T sites, these effects counteracted each other and reduced the orientation preference. The −4A and +4T base substitutions caused changes in orientation preference that were comparable in magnitude regardless of the identity of the central base pair. The additive effects of the base substitutions at these sites suggest that the central base pair and the base substitutions at the ±4 positions independently affected the orientation of Fos-Jun heterodimer binding.
The central asymmetric base pair has a modest effect on the orientation preference of wild-type Fos-Jun heterodimers, since both Fos and Jun contain homologous arginine residues that can contact the central guanine (13, 30). However, mutation of the arginine residue in one subunit causes the central base pair to have a dominant effect on the orientation preferences of heterodimers at binding sites containing palindromic core recognition elements (29, 30). To determine the relative effects of these arginine residues that can contact the central base pair and the asymmetric recognition of the two half-sites, we examined the effects of the base substitutions on the orientation preferences of the mutated heterodimers (Fig. 2B). Heterodimers in which the arginine in Jun was mutated exhibited comparable orientation preferences at both consensus AP-1 sites and AP-1 sites containing base substitutions at the ±4 positions (Fig. 2B, Fos-JunRI). Contacts between the arginine in Fos and the central base pair therefore had a greater effect on the orientation preference of Fos-JunRI than the unequal recognition of the two half-sites. In contrast, heterodimers in which the arginine in Fos was mutated exhibited different orientation preferences at AP-1 sites containing base substitutions at the ±4 positions (Fig. 2B, FosRI-Jun). At the NC−4A and NG+4T sites, where the effects of the asymmetric base substitutions counteract the effect of the arginine in Jun, the end preferences of FosRI-Jun heterodimers were more similar to those of wild-type Fos-Jun heterodimers at these asymmetric sites than to the end preferences of FosRI-Jun at the symmetrical NC and NG sites. The unequal recognition of the two half-sites therefore had a greater effect on the orientation preference of FosRI-Jun than contacts between the arginine in Jun and the central base pair. The greater influence of the arginine in Fos compared with the arginine in Jun on heterodimer orientation preference is consistent with the orientation preference of wild-type Fos-Jun at the consensus AP-1 site. The effects of the asymmetric base substitutions on heterodimer orientation preference were greater for FosRI-Jun than for wild-type Fos-Jun. The differential recognition of the ±4 positions was therefore enhanced by mutation of the arginine in Fos. Since this arginine is located at a distance from the ±4 positions in the X-ray crystal structure (13), it is likely that this mutation affected recognition of the ±4 positions through a change in basic region structure.
We previously found that asymmetric base pairs flanking the AP-1 site affect the orientation of heterodimer binding through electrostatic interactions with charged amino acid residues adjacent to the basic DNA contact region (30, 37). However, mutation of these amino acid residues had no detectable effect on heterodimer orientation at binding sites with asymmetric base pairs within the core AP-1 recognition element (data not shown). Combined base substitutions within the core AP-1 site and in flanking DNA sequences had approximately additive effects on orientation preference. The additive effects of multiple base substitutions indicate that the individual substitutions did not eliminate specific recognition of other base pairs in the half-site. Hence, asymmetric base pairs within the core and in flanking sequences can simultaneously affect heterodimer orientation and are recognized by distinct mechanisms. Nonconsensus base pairs within the AP-1 site also affected the orientation preferences of heterodimers formed by full-length Jun with the bZIP domain of Fos (37) (data not shown). The effects characterized here by use of modified bZIP domains therefore are likely to apply to heterodimers formed by the native proteins.
The preferred orientation of heterodimer binding at composite regulatory elements influences transcriptional synergy with NFAT1.
The orientation preference of Fos-Jun heterodimer binding can influence the stability and transcriptional activity of Fos-Jun-NFAT1 complexes in vitro (35, 36). To investigate the influence of the preferred orientation of heterodimer binding on transcription activation in cultured cells, we compared the transcriptional activities of heterodimers with opposite orientation preferences alone and in the presence of NFAT1 at composite NFAT-AP-1 regulatory elements (Fig. 3). Two promoters containing composite NFAT-AP-1 regulatory elements that differ only by inversion of the central base pair of the AP-1 site were linked to different reporter genes to enable comparison of promoter activities in the same cells (Fig. 3A, NG-renilla and NC-firefly). The reporter genes were cotransfected into cells with expression vectors encoding heterodimers with a fixed orientation preference and NFAT1. The efficiencies of transcription activation by Fos-Jun, FosRI-Jun, and Fos-JunRI heterodimers in the presence and absence of NFAT1 were measured concurrently at the two promoters (Fig. 3B).
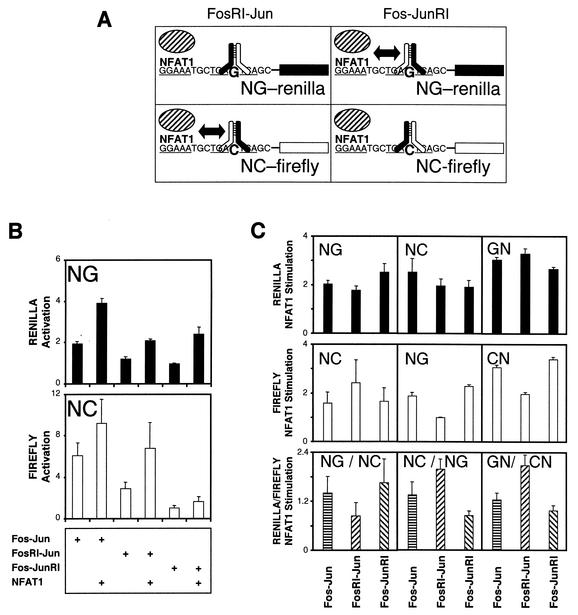
FIG. 3.
Effects of the preferred orientation of heterodimer binding on transcription activation by Fos-Jun-NFAT1 complexes. (A) Diagrams illustrate the preferred orientations of FosRI-Jun and Fos-JunRI heterodimer binding at the NG-Renilla and NC-firefly reporter genes. Double-headed arrows indicate the heterodimer orientation that favors cooperative DNA binding with NFAT1 (8, 35, 36). The two promoters were linked to different reporter genes that can be assayed in parallel in the same cell extract (Renilla and firefly luciferase genes). (B) Activation of NG-Renilla (filled bars) and NC-firefly (open bars) reporter gene transcription by Fos-Jun, FosRI-Jun, or Fos-JunRI in the presence or absence of NFAT1. Data shown are averages and standard deviations from four parallel transfection experiments for each complex. Each transfection mixture contained equal amounts of NG-Renilla and NC-firefly reporter plasmids, together with expression vectors encoding the proteins indicated below the bars. (C) Stimulation of transcription by NFAT1 in the presence of different heterodimers. Enhancement by NFAT1 (NFAT1 stimulation) of the transcriptional activity of each promoter was calculated based on the ratio between its transcriptional activities in the presence of a given heterodimer, indicated at the bottom of the figure, with and without NFAT1. Stimulation of promoters linked to Renilla luciferase is shown in the upper panels (solid bars), and stimulation of promoters linked to firefly luciferase is shown in the center panels (open bars). The ratio between the effects of NFAT1 on transcription of the Renilla and firefly reporter genes in the presence of a given heterodimer is shown in the lower panels (striped bars). Data are representative of four independent experiments in which the absolute levels of NFAT1 stimulation were variable but the relative effects of NFAT1 on the activities of heterodimers with opposite orientation preferences were reproducible.
All of the proteins exhibited greater activation of the firefly reporter gene than of the Renilla reporter gene. This difference may be due to a higher basal level of Renilla luciferase activity in cells transfected with the reporter plasmids alone or to a different rate-limiting step in the synthesis of Renilla and firefly luciferases. To determine the effect of heterodimer orientation preference on the synergy with NFAT1, we compared the stimulation of the transcriptional activities of the two promoters by NFAT1 in the presence of different heterodimers (Fig. 3C). In the presence of Fos-Jun, NFAT1 stimulated the NG-Renilla reporter slightly more than the NC-firefly reporter. In the presence of FosRI-Jun, NFAT1 preferentially stimulated the NC-firefly reporter, whereas in the presence of Fos-JunRI, NFAT1 preferentially stimulated the NG-Renilla reporter. NFAT1 alone had no detectable effect on the activities of the reporter genes. These differences in transcriptional synergy with NFAT1 between heterodimers with opposite orientation preferences are consistent with a role of the orientation of heterodimer binding in transcription activation.
To confirm that the differences in transcriptional synergy were caused by the opposite orientation preferences of the heterodimers at the two promoters, we exchanged the NG and NC promoters between the Renilla and firefly reporters and measured the stimulation of NC-Renilla and NG-firefly transcription by NFAT1 in the presence of the different heterodimers (Fig. 3C, center panels [top to bottom]). In the presence of wild-type Fos-Jun, the effects of NFAT1 on transcriptional activity were unaffected by exchange of the promoters. These promoters differ by a single base pair substitution that has little effect on the orientation preference of wild-type Fos-Jun (Fig. 2). In contrast, the relative effects of NFAT1 on the transcriptional activities of the two promoters were reversed in the presence of FosRI-Jun or Fos-JunRI heterodimers. These heterodimers favor opposite binding orientations at the two promoters, consistent with an effect of the preferred orientation of heterodimer binding on transcription activation.
The influence of heterodimer orientation preference on transcriptional activity at these promoters was likely caused by differences in cooperative DNA binding with NFAT1 (35, 36). To confirm that the influence of the preferred orientation of heterodimer binding on transcriptional activity depends on cooperative interactions with NFAT1, we compared the transcriptional activities of promoters where the NFAT site was moved to the opposite side of the AP-1 site (Fig. 3C, right panels, GN-Renilla and CN-firefly). NFAT1 stimulated the GN-Renilla reporter more than the CN-firefly reporter in the presence of FosRI-Jun, whereas the opposite was the case in the presence of Fos-JunRI. Significantly, transfer of the NFAT site to the opposite side of the AP-1 site reversed the relative effects of NFAT1 on the transcriptional activities of the two promoters in the presence of oriented heterodimers. These results constitute the first evidence that the orientation preference of Fos-Jun heterodimer binding can influence transcriptional synergy with NFAT1 in mammalian cells.
Dimerization with different partners can result in binding to opposite half-sites at nonconsensus AP-1 sites and in association with cooperative binding partners.
bZIP family proteins can form heterodimers in many different combinations. Since the orientation of heterodimer binding is determined by the difference in DNA binding specificities between the two subunits of the heterodimer, dimerization with different partners can result in preferential binding to different half-sites. Fos and Jun can form heterodimers with ATF2, and Jun-ATF2 heterodimers can bind to different cyclic AMP response element (CRE) sites in opposite orientations (10, 22). We examined the effects of asymmetric base substitutions on the orientation preferences of Fos-ATF2 and Jun-ATF2 heterodimers at AP-1 sites (Fig. 4). Fos-ATF2 and Jun-ATF2 bound with lower orientation preferences to the palindromic NC site, indicating that the central base pair had less effect on the orientation preferences of these heterodimers (Fig. 4B). The −4G, +4C, and +3G base substitutions had opposite effects on the end preferences of JunTR-Fos and JunTR-ATF2 heterodimers. Whereas Jun favored binding to the consensus half-site in JunTR-Fos heterodimers, it bound to the nonconsensus half-site in JunTR-ATF2 heterodimers. Thus, Jun bound to opposite half-sites in heterodimers with Fos and with ATF2. In contrast, the same base substitutions had comparable effects on the end preferences of FosTR-Jun and FosTR-ATF2 heterodimers. Thus, ATF2 bound to the consensus half-site in both Fos-ATF2 and Jun-ATF2 heterodimers. ATF2 therefore had a greater preference for the consensus AP-1 half-site than either Fos or Jun in these heterodimers.
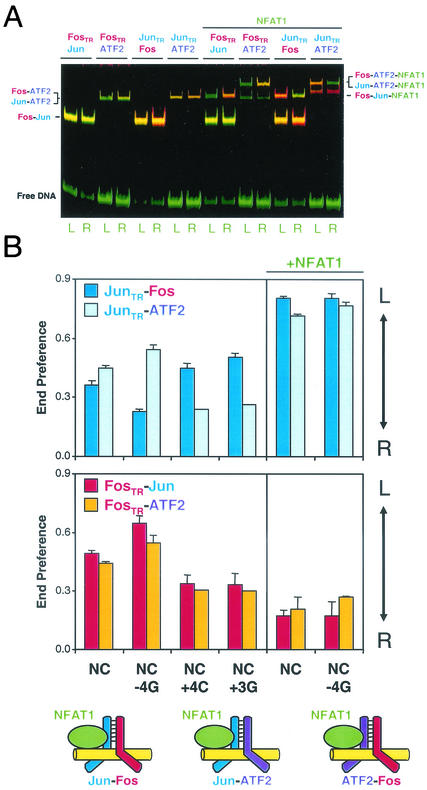
FIG. 4.
Effects of base substitutions and interactions with NFAT1 on the binding orientations of ATF2 heterodimers with Fos and Jun. (A) gelFRET analysis of the binding orientations of Fos-Jun, Fos-ATF2, and Jun-ATF2 heterodimers in the absence and the presence of NFAT1. The proteins indicated above the lanes were incubated with NC site oligonucleotides labeled with fluorescein at either the left (L) or the right (R) end, and the complexes were separated by polyacrylamide gel electrophoresis. The gel was scanned using 488-nm laser excitation, and the emissions from donor (green) and acceptor (red) fluorophores were measured at each position in the gel and were superimposed to produce the image (29, 34). (B) Comparison of the effects of asymmetric base substitutions on the preferred orientations of Fos-Jun, Fos-ATF2, and Jun-ATF2 binding. The end preferences of heterodimers, alone or in complex with NFAT1, were measured at the binding sites given below the bars. The subunit labeled with Texas Red is indicated by the subscript TR. Standard deviations from three or more independent experiments, except for dimers formed by ATF2 at the NC+4C and NC+3G sites, are shown. Diagrams below the graphs depict the orientations of Fos-Jun, Fos-ATF2, and Jun-ATF2 heterodimers in quaternary complexes with NFAT1.
Fos-Jun heterodimers bind cooperatively to composite regulatory elements with NFAT1, but ATF2 has not been reported to form cooperative complexes with NFAT family proteins. We examined interactions between the Fos-ATF2 or Jun-ATF2 heterodimer and NFAT1 by using the gelFRET approach (Fig. 4). Both Fos-ATF2 and Jun-ATF2 heterodimers exhibited cooperative binding with NFAT1 at several different composite regulatory elements. The quaternary Fos-ATF2-NFAT1 and Jun-ATF2-NFAT1 complexes were formed at concentrations at which neither of the individual complexes was observed (data not shown). Moreover, both the interaction of NFAT1 with Fos-ATF2 and that with Jun-ATF2 affected the orientation of heterodimer binding (Fig. 4B). Fos-ATF2 heterodimers were reoriented to bind with ATF2 occupying the proximal AP-1 half-site in complexes with NFAT1. In contrast, Jun-ATF2 heterodimers were reoriented to bind with ATF2 occupying the distal AP-1 half site in complexes with NFAT1. Thus, cooperative DNA binding by NFAT1 with Fos-ATF2 and Jun-ATF2 resulted in Fos and Jun binding to the same half-sites as in the Fos-Jun-NFAT1 complex. In contrast, ATF2 bound to opposite half-sites in the Fos-ATF2-NFAT1 and Jun-ATF2-NFAT1 complexes. These results demonstrate that many bZIP protein heterodimers are reoriented by NFAT1 and that the half-site occupied by a protein can be affected by its dimerization partner.
The orientation of nonconsensus AP-1 sites within composite regulatory elements influences cooperative DNA binding by Fos-Jun-NFAT1 as well as Jun-Jun-NFAT1 complexes.
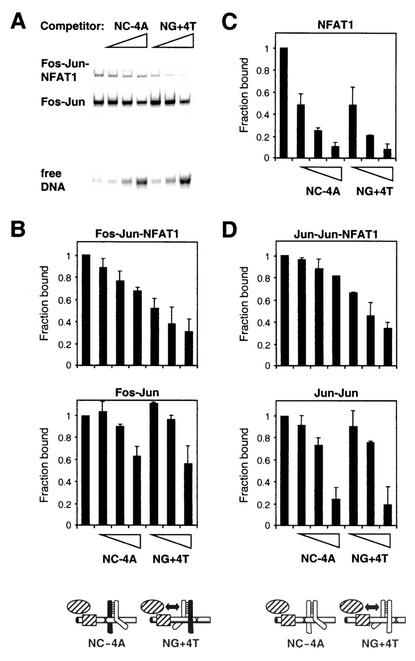
The asymmetric recognition of AP-1 sites may influence interactions with other transcription regulatory proteins through mechanisms in addition to the orientation of heterodimer binding. To examine the effects of nonconsensus AP-1 site orientation on cooperative DNA binding by Fos, Jun, and NFAT1, we compared the efficiencies of competition by NC− 4A and NG+4T oligonucleotides for binding of Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes (Fig. 5). These oligonucleotides contained the same AP-1 site placed in opposite orientations relative to the NFAT site. To eliminate the possibility that sequences flanking the AP-1 site affected the results, the flanking sequences within 8 bp from the center of the AP-1 site were symmetrical on these oligonucleotides. We compared competition for the quaternary Fos-Jun-NFAT1 complex with competition for the ternary Fos-Jun complex in the same reaction by using limiting NFAT1 concentrations and separating the complexes by gel electrophoresis (Fig. 5A). The Fos-Jun-NFAT1 complex was preferentially competed by the NG+4T oligonucleotide, whereas the Fos-Jun complex was competed equally by the NC−4A and NG+4T oligonucleotides (Fig. 5B). Identical results were obtained whether levels of competition for Fos-Jun-NFAT1 binding and Fos-Jun binding were compared in the same reaction or in separate reactions. The higher efficiency of competition for Fos-Jun-NFAT1 complexes relative to Fos-Jun complexes by the NG+4T oligonucleotide indicates that the interaction with NFAT1 enhances Fos-Jun binding at the NG+4T site more than at the NC site. NFAT1 alone was competed with equal efficiencies by the two oligonucleotides, demonstrating that the base substitutions within the AP-1 site did not influence NFAT1 binding (Fig. 5C). The difference between the efficiencies of competition for Fos-Jun-NFAT1 complexes by the NC−4A and NG+4T oligonucleotides was therefore caused by differences in cooperative interactions between NFAT1 and Fos-Jun at composite regulatory elements containing nonconsensus AP-1 sites in opposite orientations.
FIG. 5.
Effects of nonconsensus AP-1 site orientation within composite regulatory elements on the efficiency of competition for Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes. (A) Comparison of the efficiencies of competition by oligonucleotide competitors for Fos-Jun-NFAT1 and Fos-Jun complexes. Different concentrations (1, 5, or 25 μM) of the competitor oligonucleotides indicated above the lanes containing the same AP-1 site in opposite orientations relative to the NFAT site were incubated with Fos, Jun, a limiting concentration of NFAT1, and the NC site oligonucleotide labeled with fluorescein. The complexes were separated by gel electrophoresis, and the fluorescence emission at each position in the gel was measured using a fluorescence imager. (B) Comparison of the relative efficiencies of competition for Fos-Jun-NFAT1 complexes (upper graph) and Fos-Jun complexes (lower graph) by NC−4A and NG+4T competitor oligonucleotides. The relative amounts of complexes formed in the presence of different competitors are plotted as fractions of the amounts of these complexes formed in the absence of competitors. Diagrams below the graphs depict the asymmetric recognition of the NC−4A and NG+4T sites by Fos-Jun heterodimers. The double-headed arrow indicates the preferential configuration for interactions with NFAT1. (C) Relative efficiencies of competition for NFAT1 by NC−4A and NG+4T competitor oligonucleotides. (D) Relative efficiencies of competition for Jun-Jun-NFAT1 complexes (upper graph) and Jun homodimers (lower graph) by NC−4A and NG+4T competitor oligonucleotides. Standard deviations from three or more independent experiments are shown. Diagrams below the graphs depict the asymmetric recognition of the NC−4A and NG+4T sites by Jun homodimers and their differential interactions with NFAT1.
Surprisingly, Jun-Jun-NFAT1 complexes were also differentially competed by the NC−4A and NG+4T oligonucleotides (Fig. 5D). The efficiency of competition by the NG+4T oligonucleotide was higher than that by the NC−4A oligonucleotide. As predicted, no difference in competition was observed for Jun homodimers alone. Since Jun homodimers cannot bind to AP-1 sites in a preferred orientation, there must exist additional mechanisms that mediate the effect of nonconsensus AP-1 site orientation on Jun cooperativity with NFAT1. These mechanisms may also influence cooperative interactions between Fos-Jun and NFAT1 at nonconsensus AP-1 sites. We propose that the base substitutions in the AP-1 site cause asymmetric conformational changes in the two subunits of the bZIP dimer that influence their interactions with NFAT1.
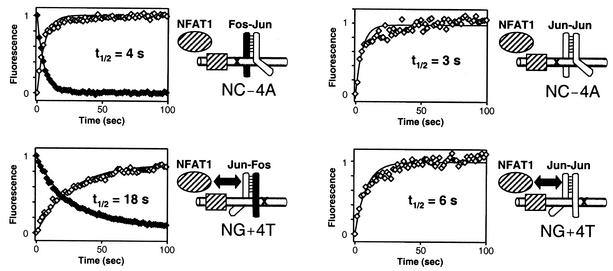
To measure directly the stabilities of complexes at binding sites containing symmetry-related base substitutions within the AP-1 site, we compared the dissociation rates of Fos-Jun-NFAT1 as well as Jun-Jun-NFAT1 complexes from oligonucleotides containing identical AP-1 sites in opposite orientations relative to the NFAT site (Fig. 6). We monitored complex dissociation by measuring the time-dependent change in FRET (increase in fluorescein donor and decrease in Texas red acceptor fluorescence) following addition of unlabeled DNA competitor (35). We detected the dissociation of only the labeled Jun protein directly, but this represents the dissociation rate of the entire complex as shown previously using fluorescence anisotropy (35). The dissociation rates of Jun-Jun-NFAT1 complexes were determined based on the increase in fluorescein donor emission, since the acceptor emission was affected by the close proximity of the fluorophores linked to the two Jun subunits.
FIG. 6.
Effects of nonconsensus AP-1 site orientation within composite regulatory elements on the stabilities of Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes. Shown is a comparison of the dissociation rates of Fos-Jun-NFAT1 (left panels) and Jun-Jun-NFAT1 (right panels) at the NC−4A and NG+4T sites containing the same AP-1 site in opposite orientations relative to the NFAT site. Jun was labeled with Texas Red, and the oligonucleotides were labeled with fluorescein. Changes in fluorescence emissions from fluorescein (open symbols) and Texas Red (filled symbols) were monitored after addition of an excess of competitor DNA to Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes at the binding sites shown to the right of each graph. Changes in fluorescence were normalized to the same range to allow comparison of the rates. Data for each complex were fitted to a first-order exponential function (R < 0.98 for all complexes), and the half-life (t1/2) was calculated from the best fit. Diagrams indicate the influence of the asymmetric recognition of AP-1 sites on interactions with NFAT1 (double-headed arrows).
The dissociation rate of Fos-Jun-NFAT1 at the NC−4A site was fivefold higher than that at the NG+4T site (Fig. 6). Both the increase in donor emission and the decrease in acceptor emission displayed the same difference in dissociation rates. The dissociation rate of Jun-Jun-NFAT1 at the NC−4A site was twofold higher than that at the NG+4T site. The dissociation rates of both Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes at these composite elements containing nonconsensus AP-1 sites were considerably higher than those observed at elements containing consensus AP-1 sites (35). This reflects the effects of the base substitutions within the core AP-1 element on the stabilities of these complexes. These results corroborate the differences between the efficiencies of competition by these oligonucleotides for Fos-Jun-NFAT1 and Jun-Jun-NFAT1 complexes (Fig. 5). Similar results were obtained at other composite elements containing the nonconsensus AP-1 sites with base substitutions at the ±4 or ±3 positions listed in Fig. 1 (data not shown). The asymmetric recognition of the AP-1 site therefore influences cooperative interactions with NFAT1 at many different composite regulatory elements.
The orientation of nonconsensus AP-1 sites within composite regulatory elements influences the transcriptional synergy of Jun homodimers with NFAT1.
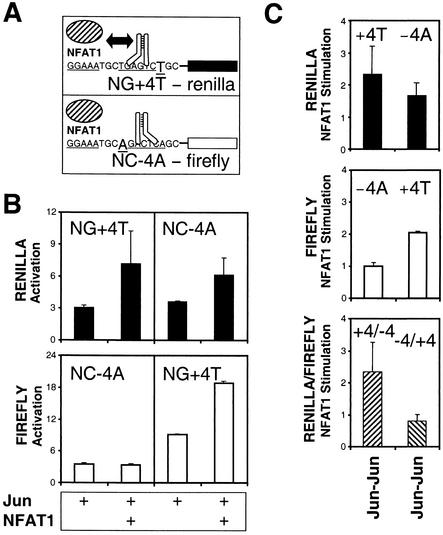
To investigate the influence of the asymmetric recognition of nonconsensus AP-1 sites on transcription activation by Jun homodimers and NFAT1, we compared the transcriptional activities of two promoters containing composite regulatory elements with the same nonconsensus AP-1 sites in opposite orientations relative to the NFAT site (Fig. 7). The two promoters were linked to different reporter genes to enable direct comparison of their transcriptional activities in the same cells (Fig. 7A, NG+4T-Renilla and NC−4A-firefly). These reporter plasmids were cotransfected into cells with expression vectors encoding Jun with or without NFAT1, and the efficiencies of transcription activation were measured for each reporter (Fig. 7B). Jun alone activated the two reporter genes to similar extents, but Jun and NFAT1 preferentially activated the NG+4T-Renilla reporter. NFAT1 therefore exhibited greater stimulation of the NG+4T-Renilla reporter than of the NC−4A-firefly reporter in the same cells. To confirm that the preferential activation of the NC+4T promoter was caused by asymmetric recognition of the nonconsensus AP-1 site, we exchanged the promoters between the two reporter genes (Fig. 7B, right panels, and 7C). Exchange of the promoters resulted in greater stimulation of the NG+4T-firefly reporter than of the NC−4A-Renilla reporter by NFAT1. The preferential activation of reporters linked to the NC+4T promoter was therefore determined by the orientation of the nonconsensus AP-1 site within the composite regulatory element. Similar results were obtained for Fos-Jun heterodimers by using the same reporter genes as well as for both Fos-Jun heterodimers and Jun homodimers when reporter genes containing NC−4A and NC+4T regulatory elements were used (data not shown). Jun homodimers activated the NC and NG promoters to the same extent, as did Fos-Jun heterodimers (Fig. 3). The influence of nonconsensus AP-1 site orientation on transcription activation was therefore caused by the asymmetrical half-sites. Hence, the transcriptional synergy of Fos and Jun with NFAT1 is modulated by the asymmetric recognition of nonconsensus AP-1 sites within composite regulatory elements.
FIG. 7.
Effects of nonconsensus AP-1 site orientation within composite regulatory elements on the transcriptional synergy between Jun homodimers and NFAT1. (A) Diagrams illustrate asymmetric recognition of the nonconsensus AP-1 sites within the composite regulatory elements in the NG+4T-Renilla and NC−4A-firefly reporter genes. The promoters contain the same AP-1 site in opposite orientations relative to the NFAT site. A double-headed arrow indicates the AP-1 site orientation that favors cooperative interactions between Jun homodimers and NFAT1 based on the results from oligonucleotide competition and dissociation analyses (Fig. 5 and 6). (B) Comparison of transcription activation by Jun alone and by Jun with NFAT1 at composite regulatory elements containing nonconsensus AP-1 sites in opposite orientations (NG+4T and NC−4A) linked to Renilla (filled bars) and firefly (open bars) reporters. The reporter constructs indicated in each vertical set of graphs were cotransfected into cells with expression vectors encoding the proteins indicated below the graphs, and reporter gene activities were assayed in the same cell extract. Data shown are averages and standard deviations from four parallel transfection experiments for each complex. (C) Effects of NFAT1 on the transcriptional activities of the NG+4T and NC−4A promoters in the presence of Jun homodimers. Stimulation by NFAT1 was calculated based on the ratio between the transcriptional activities of the reporter genes in the presence of Jun and NFAT1 and activities in the presence of Jun alone. Data are representative of five independent experiments, two of which were performed using the NG+4T and NC−4A reporters, and three of which were performed using the NC+4T and NC−4A reporters.
DISCUSSION
Fos-Jun heterodimers and Jun homodimers recognize the same optimal DNA sequence element (23). Fos and Jun also make virtually identical contacts to the AP-1 site in the X-ray crystal structure (3, 13). The preferred orientation of heterodimer binding at AP-1 sites containing asymmetric base substitutions revealed a difference in the DNA binding specificities between Fos and Jun. Jun had a greater preference for binding to the consensus AP-1 half-site than Fos. In heterodimers formed among Fos, Jun, and ATF2, ATF2 had the greatest preference for the consensus recognition sequence, whereas Fos had the lowest preference. The relative binding preferences of these heterodimers were consistent at six different binding sites, suggesting that they reflect differences in the overall DNA binding specificities of the proteins.
Fos and Jun differentially recognized the methyl group on the thymine at the ±4 positions of the AP-1 site. This methyl group is contacted by two alanines and a serine (substituted for the native cysteine) that are conserved in Fos and Jun (3, 13). The differential recognition of the methyl group must therefore be due to differences in the geometries of these residues or to additional contacts that are not observed in the X-ray crystal structure. Remarkably, these amino acid residues as well as a majority of the other residues that make base-specific contacts in the Fos-Jun-AP-1 complex are also conserved among other bZIP proteins, including proteins with distinct DNA recognition specificities. The differential DNA recognition specificities of Fos and Jun are therefore consistent with the variation in DNA recognition specificities among bZIP proteins that share the same DNA contact residues. This variation in DNA recognition specificities is likely to be, at least in part, due to conformational differences among the basic regions of different bZIP proteins (9, 12).
Fos-Jun heterodimers bind cooperatively with NFAT1 to composite NFAT-AP-1 regulatory elements. Fos-ATF2 and Jun-ATF2 heterodimers also exhibited cooperative DNA binding with NFAT1. The interactions with NFAT1 caused ATF2 to bind to opposite half-sites in Fos-ATF2-NFAT1 and ATF2-Jun-NFAT1 complexes. Fos and Jun occupied the same half-sites in these complexes as they occupy in the Fos-Jun-NFAT1 complex (3). In contrast, ATF2 occupied opposite half-sites in the two complexes. Since only one of the amino acid residues in Fos and in Jun that contact NFAT1 in the crystal structure is conserved in ATF2, either the interactions with NFAT1 are mediated primarily by Fos and by Jun, respectively, in these complexes, or ATF2 contains structurally distinct interaction interfaces that interact with NFAT1 in these complexes.
Jun-ATF2 heterodimers regulate expression of the beta interferon promoter cooperatively with IRF-3 at the PRDIV and PRDIII elements (10). Jun and ATF2 cross-link preferentially to photoreactive groups on opposite sides of the PRDIV element only in the presence of IRF-3 or IRF-1 (10). In these quaternary complexes ATF2 binds to the nonconsensus half-site whereas Jun binds to the consensus half-site (10). The differences in orientation preferences between Jun-ATF2 heterodimers alone and Jun-ATF2 heterodimers in association with IRF-1 or IRF-3 are likely to be due to protein interactions in these complexes as well as the sequence of the PRDIV element. The Fos-ATF2-NFAT1 and ATF2-Jun-NFAT1 complexes differ from the Jun-ATF2-IRF-3 and Jun-ATF2-IRF-1 complexes both in the ability of NFAT1 to alter the orientation preferences of Fos-ATF2 and Jun-ATF2 heterodimers and in the flexible positioning of ATF2 in different heterodimers in association with NFAT1. The complexes analyzed here containing NFAT1 were formed at nonconsensus AP-1 recognition elements, whereas the complexes containing IRF-3 and IRF-1 analyzed previously were formed at variants of the CRE site (10).
Cooperative DNA binding by Fos-Jun heterodimers and NFAT1 requires Jun to bind the AP-1 half-site proximal to the NFAT recognition element and Fos to bind the distal half-site (3, 4, 8, 35, 36). Our studies demonstrate that the preferred orientation of heterodimer binding influences the cooperativity of complex formation as well as the transcriptional activity of the complex in vitro and in transfected cells (35, 36). Since Fos-Jun heterodimers bind to nonconsensus AP-1 sites in a preferred orientation, it follows that the orientation of nonconsensus AP-1 sites within composite regulatory elements is likely to influence the stability and the transcriptional activity of Fos-Jun-NFAT1 complexes. In agreement with this prediction, the stabilities and transcriptional activities of Fos-Jun-NFAT1 complexes were affected by the orientations of nonconsensus AP-1 sites within composite regulatory elements. Surprisingly, the orientations of nonconsensus AP-1 sites also affected the stabilities and transcriptional activities of Jun-Jun-NFAT1 complexes. The asymmetric recognition of AP-1 sites therefore influences bZIP protein interactions with NFAT1 through mechanisms in addition to the orientation of heterodimer binding.
The functional asymmetry of Jun homodimers at nonconsensus AP-1 sites implies a functionally important structural asymmetry of the homodimer complex. This asymmetry may reflect a difference in DNA and/or protein conformations between the two half-sites. It was previously found that base substitutions at the ±4 positions can prevent the α-helical conformational change that accompanies DNA binding by Fos and Jun (9). We infer that the conformations of the two subunits are likely to be different at AP-1 sites containing asymmetric base substitutions. Formation of the Fos-Jun-NFAT1 complex requires changes in both DNA and protein conformations and involves distinct interactions between the two subunits and NFAT1 (3). We propose that the conformational asymmetry of the bZIP dimer at nonconsensus AP-1 sites influences the cooperativity of complex formation with NFAT1 either by directly altering interactions with NFAT1 or by affecting the conformational changes required for cooperative complex formation.
The effects of asymmetric recognition of nonconsensus AP-1 sites on Jun homodimer interactions with NFAT1 do not negate the influence of the preferred orientation of heterodimer binding on Fos-Jun cooperativity with NFAT1. The reciprocal effects of inversion of the central base pair on the transcriptional activities of Fos-JunRI-NFAT1 and FosRI-Jun-NFAT1 complexes cannot be easily explained based on conformational differences between heterodimers bound to the consensus AP-1 site. Even in the unlikely event that inversion of the central base pair might have reciprocal effects on the conformations of these heterodimers, it would not explain why inversion of this base pair has no effect on the characteristics of wild-type Fos-Jun-NFAT1 complexes. The perfect correspondence between heterodimer orientation, cooperative DNA binding, and synergistic transcription activation constitutes strong evidence in favor of a role of the orientation of heterodimer binding in transcription activation in mammalian cells.
Many composite NFAT-AP-1 sites contain nonconsensus AP-1 recognition sequences that contain base substitutions in the half-site proximal to the NFAT binding site. Examples include the GM550 composite site of the granulocyte-macrophage colony-stimulating factor enhancer, the −595 site in the IL-3 promoter, the −115 site in the IL-5 promoter, and the −195 site in the CTLA4 promoter (5). The nonconsensus AP-1 recognition sequences at these elements may be essential to prevent activation of these promoters by Fos-Jun in the absence of NFAT1. Alternatively, they may be necessary to modulate the stabilities of cooperative complexes at these elements, possibly to allow disruption of the complexes during downregulation of promoter activity. Finally, the differences in structural organization of bZIP protein complexes formed at these regulatory elements may contribute to selective interactions between particular members of both the Fos-Jun and NFAT families at different regulatory elements. The influence of nonconsensus AP-1 site orientation on cooperative DNA binding and transcription activation by Fos and Jun with NFAT1 demonstrates that differences in conformation and binding orientation can contribute to the combinatorial regulation of gene expression in mammalian cells.
Acknowledgments
We thank the members of the Kerppola laboratory and David Engelke for helpful discussions. We gratefully acknowledge Timothy Liao for performing the gelFRET experiments using uracil-substituted oligonucleotides and Shaohua Xiao for performing gelFRET experiments using additional nonconsensus AP-1 binding sites.
V.R.-C. was supported by a Rackham predoctoral fellowship.
REFERENCES
- 1.Abdel-Hafiz, H. A., L. E. Heasley, J. M. Kyriakis, J. Avruch, D. J. Kroll, G. L. Johnson, and J. P. Hoeffler. 1992. Activating transcription factor-2 DNA-binding activity is stimulated by phosphorylation catalyzed by p42 and p54 microtubule-associated protein kinases. Mol. Endocrinol. 6:2079-2089. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., O. Avni, and A. Rao. 2000. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12:643-652. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L., J. N. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392:42-48. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., M. G. Oakley, J. N. Glover, J. Jain, P. B. Dervan, P. G. Hogan, A. Rao, and G. L. Verdine. 1995. Only one of the two DNA-bound orientations of AP-1 found in solution cooperates with NFATp. Curr. Biol. 5:882-889. [DOI] [PubMed] [Google Scholar]
- 5.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 6.Chytil, M., B. R. Peterson, D. A. Erlanson, and G. L. Verdine. 1998. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc. Natl. Acad. Sci. USA 95:14076-14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary, M. A., P. S. Pendergrast, and W. Herr. 1997. Structural flexibility in transcription complex formation revealed by protein-DNA photocrosslinking. Proc. Natl. Acad. Sci. USA 94:8450-8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diebold, R. J., N. Rajaram, D. A. Leonard, and T. K. Kerppola. 1998. Molecular basis of cooperative DNA bending and oriented heterodimer binding in the NFAT1-Fos-Jun-ARRE2 complex. Proc. Natl. Acad. Sci. USA 95:7915-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dlakic, M., A. V. Grinberg, D. A. Leonard, and T. K. Kerppola. 2001. DNA sequence-dependent folding determines the divergence in binding specificities between Maf and other bZIP proteins. EMBO J. 20:828-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falvo, J. V., B. S. Parekh, C. H. Lin, E. Fraenkel, and T. Maniatis. 2000. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-Jun heterodimer orientation. Mol. Cell. Biol. 20:4814-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falvo, J. V., A. M. Uglialoro, B. M. Brinkman, M. Merika, B. S. Parekh, E. Y. Tsai, H. C. King, A. D. Morielli, E. G. Peralta, T. Maniatis, D. Thanos, and A. E. Goldfeld. 2000. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 20:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, Y., T. Shimizu, T. Toda, M. Yanagida, and T. Hakoshima. 2000. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 7:889-893. [DOI] [PubMed] [Google Scholar]
- 13.Glover, J. N., and S. C. Harrison. 1995. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373:257-261. [DOI] [PubMed] [Google Scholar]
- 14.Hall, J. M., D. P. McDonnell, and K. S. Korach. 2002. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol. Endocrinol. 16:469-486. [DOI] [PubMed] [Google Scholar]
- 15.Hardwidge, P. R., J. Wu, S. L. Williams, K. M. Parkhurst, L. J. Parkhurst, and L. J. Maher. 2002. DNA bending by bZIP charge variants: a unified study using electrophoretic phasing and fluorescence resonance energy transfer. Biochemistry 41:7732-7742. [DOI] [PubMed] [Google Scholar]
- 16.Ho, I. C., M. R. Hodge, J. W. Rooney, and L. H. Glimcher. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 85:973-983. [DOI] [PubMed] [Google Scholar]
- 17.Jain, J., P. G. McCaffrey, Z. Miner, T. K. Kerppola, J. N. Lambert, G. L. Verdine, T. Curran, and A. Rao. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365:352-355. [DOI] [PubMed] [Google Scholar]
- 18.Kamachi, Y., M. Uchikawa, A. Tanouchi, R. Sekido, and H. Kondoh. 2001. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 15:1272-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerppola, T. K. 1996. Fos and Jun bend the AP-1 site: effects of probe geometry on the detection of protein-induced DNA bending. Proc. Natl. Acad. Sci. USA 93:10117-10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerppola, T. K. 1997. Comparison of DNA bending by Fos-Jun and phased A tracts by multifactorial phasing analysis. Biochemistry 36:10872-10884. [DOI] [PubMed] [Google Scholar]
- 21.Kerppola, T. K., and T. Curran. 1991. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell 66:317-326. [DOI] [PubMed] [Google Scholar]
- 22.Kerppola, T. K., and T. Curran. 1993. Selective DNA bending by a variety of bZIP proteins. Mol. Cell. Biol. 13:5479-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerppola, T. K., and T. Curran. 1994. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9:675-684. [PubMed] [Google Scholar]
- 24.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 25.Kerppola, T. K., and T. Curran. 1997. The transcription activation domains of Fos and Jun induce DNA bending through electrostatic interactions. EMBO J. 16:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurokawa, R., J. DiRenzo, M. Boehm, J. Sugarman, B. Gloss, M. G. Rosenfeld, R. A. Heyman, and C. K. Glass. 1994. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371:528-531. [DOI] [PubMed] [Google Scholar]
- 27.Kurokawa, R., M. Soderstrom, A. Horlein, S. Halachmi, M. Brown, M. G. Rosenfeld, and C. K. Glass. 1995. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377:451-454. [DOI] [PubMed] [Google Scholar]
- 28.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885-888. [DOI] [PubMed] [Google Scholar]
- 29.Leonard, D. A., and T. K. Kerppola. 1998. DNA bending determines Fos-Jun heterodimer orientation. Nat. Struct. Biol. 5:877-881. [DOI] [PubMed] [Google Scholar]
- 30.Leonard, D. A., N. Rajaram, and T. K. Kerppola. 1997. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc. Natl. Acad. Sci. USA 94:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macian, F., C. Lopez-Rodriguez, and A. Rao. 2001. Partners in transcription: NFAT and AP-1. Oncogene 20:2476-2489. [DOI] [PubMed] [Google Scholar]
- 32.Misra, V., S. Walter, P. Yang, S. Hayes, and P. O'Hare. 1996. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol. Cell. Biol. 16:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajaram, N., and T. K. Kerppola. 1997. DNA bending by Fos-Jun and the orientation of heterodimer binding depend on the sequence of the AP-1 site. EMBO J. 16:2917-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Carrozzi, V. R., and T. K. Kerppola. 2001. Gel-based fluorescence resonance energy transfer (gelFRET) analysis of nucleoprotein complex architecture. Methods 25:31-43. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez-Carrozzi, V. R., and T. K. Kerppola. 2001. Dynamics of Fos-Jun-NFAT1 complexes. Proc. Natl. Acad. Sci. USA 98:4893-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez-Carrozzi, V. R., and T. K. Kerppola. 2001. Control of the orientation of Fos-Jun binding and the transcriptional cooperativity of Fos-Jun-NFAT1 complexes. J. Biol. Chem. 276:21797-21808. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez-Carrozzi, V. R., and T. K. Kerppola. 2001. Long-range electrostatic interactions influence the orientation of Fos-Jun binding at AP-1 sites. J. Mol. Biol. 305:411-427. [DOI] [PubMed] [Google Scholar]
- 38.Risse, G., K. Jooss, M. Neuberg, H. J. Bruller, and R. Muller. 1989. Asymmetrical recognition of the palindromic AP1 binding site (TRE) by Fos protein complexes. EMBO J. 8:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrader, M., K. M. Muller, S. Nayeri, J. P. Kahlen, and C. Carlberg. 1994. Vitamin D3-thyroid hormone receptor heterodimer polarity directs ligand sensitivity of transactivation. Nature 370:382-386. [DOI] [PubMed] [Google Scholar]
- 40.Schrader, M., S. Nayeri, J. P. Kahlen, K. M. Muller, and C. Carlberg. 1995. Natural vitamin D3 response elements formed by inverted palindromes: polarity-directed ligand sensitivity of vitamin D3 receptor-retinoid X receptor heterodimer-mediated transactivation. Mol. Cell. Biol. 15:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scully, K. M., E. M. Jacobson, K. Jepsen, V. Lunyak, H. Viadiu, C. Carriere, D. W. Rose, F. Hooshmand, A. K. Aggarwal, and M. G. Rosenfeld. 2000. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 290:1127-1131. [DOI] [PubMed] [Google Scholar]
- 42.Sitlani, A., and D. M. Crothers. 1996. Fos and Jun do not bend the AP-1 recognition site. Proc. Natl. Acad. Sci. USA 93:3248-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sitlani, A., and D. M. Crothers. 1998. DNA-binding domains of Fos and Jun do not induce DNA curvature: an investigation with solution and gel methods. Proc. Natl. Acad. Sci. USA 95:1404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staal, A., A. J. van Wijnen, J. C. Birkenhager, H. A. Pols, J. Prahl, H. DeLuca, M. P. Gaub, J. B. Lian, G. S. Stein, J. P. van Leeuwen, and J. L. Stein. 1996. Distinct conformations of vitamin D receptor/retinoid X receptor-α heterodimers are specified by dinucleotide differences in the vitamin D-responsive elements of the osteocalcin and osteopontin genes. Mol. Endocrinol. 10:1444-1456. [DOI] [PubMed] [Google Scholar]
- 45.Tomilin, A., A. Remenyi, K. Lins, H. Bak, S. Leidel, G. Vriend, M. Wilmanns, and H. R. Scholer. 2000. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103:853-864. [DOI] [PubMed] [Google Scholar]
- 46.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfeld. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]