Abstract
The evolutionary origin of the conserved noncoding elements (CNEs) in the human genome remains poorly understood but may hold important clues to their biological functions. Here, we report the discovery of a CNE family with ≈124 instances in the human genome that demonstrates a clear signature of having been derived from an ancient transposon. The CNE family is also present in the chicken genome, although typically not at orthologous locations. The CNE family is closely related to the active transposon SINE3 in zebrafish and also to a previously uncharacterized transposon in the coelacanth, the so-called “living fossil” belonging to the lobe-finned fish lineage. The mammal, bird, zebrafish, and coelacanth families all share a highly similar core element of ≈180 bp but have important differences in their 5′ and 3′ ends. The core element has thus been preserved over 450 million years of evolution, implying an important biological function. In addition, we identify 95 additional CNE families that likely predate the mammalian radiation. The results highlight both the creative role of transposons and the importance of CNE families.
Keywords: coelacanth repeat, exapted repeat, SINE3, comparative genomics
Conserved noncoding elements (CNEs) represent ≈3.5% of the human genome and comprise the majority of the estimated 5% of the genome that has been subject to purifying selection throughout mammalian evolution (1–3). The functional significance of these elements remains largely unknown, although many of the most highly conserved elements have been implicated as regulators of key developmental genes based on their genomic location, and a handful have been shown experimentally to have such roles (1, 4–7).
Little is known about the evolutionary origin and history of mammalian CNEs. Only a tiny fraction (<0.1%) of mammalian CNEs are detectably conserved in the genome of the fish (5, 8), and none are recognizable within invertebrate such as insects and worms (5). The low rate of conservation outside mammals is striking because many of the key developmental genes that appear to be regulated by the most highly conserved CNEs have clear orthologs in all vertebrate and invertebrate genomes that are currently available.
One approach to investigating the evolution and function of CNEs is to focus on families of related CNEs. A pioneering study that attempted to cluster ≈700,000 human CNEs based on sequence similarity found that only ≈4% show significant similarity to any other CNE (9). The study identified ≈19,000 CNEs that can be grouped into ≈12,000 clusters; the vast majority of clusters are very small: typically, two or three members.
Several recent studies have sought to understand the evolutionary origin of CNE families and have suggested different mechanisms for their creation. One study reported that some families of vertebrate CNEs can be traced to ancient genomic duplication events (8). Of ≈2,300 mammalian CNEs conserved to pufferfish, the study identified 124 families of related CNEs, each with at most five members. In nearly every case, the family is associated with a set of paralogous genes that are typically involved in transcriptional and/or developmental regulation. These findings suggest that many of the most ancient vertebrate CNE families were created as a result of the same genomic duplication events that created paralogous copies of the genes that they currently regulate. However, these ancient CNEs represent only a small fraction of all mammalian CNE families. Most CNE families clearly must have been created by other means. Indeed, the large disparity between the number of CNEs and the number of gene families rules out genomic duplication as the sole or most common mechanism.
Attention has recently focused on the notion that mammalian CNE families may have been distributed in some cases by transposable elements, giving rise to dispersed repeats. We recently reported the existence of an unusual family of CNEs (MER121) consisting of nearly 1,000 dispersed repeats in the human genome (10). The vast majority of MER121 elements are highly conserved across both eutherian and marsupial mammals, but there are only a few copies in the chicken genome. Given the large number of instances in the human genome and fairly even distribution across all chromosomes, the MER121 family could not have been primarily distributed by genomic duplication. Instead, we proposed that MER121 was mobilized by an ancient transposable element around the time of the mammalian radiation. However, we were unable to find direct evidence to support this hypothesis: The MER121 family lacks any telltale signs of transposon-like sequence, possibly because the sequence has degenerated over 200 million years of evolution. [We also reported that other families of known mammalian ancient repetitive elements contain various instances that are significantly conserved across mammals (10).]
Recently, Bejerano et al. (11) reported a clear case of a CNE family derived from instances of an ancient transposable element. This CNE family (referred to as LF-SINE) has ≈245 copies in the human genome that are derived from an ancient SINE element. Interestingly, this SINE element (LF-SINE) appears to have been active recently in the genome of the coelacanth fish, implying that the transposon family has remained active for >400 million years. The conserved LF-SINE family members consist primarily of noncoding elements but also include 13 instances in which the sequence occurs within protein-coding sequence. By contrast, the MER121 family does not contain any coding sequences.
In this study, we report the discovery of a further family of CNEs that was clearly derived from an ancient transposable element. The family includes at least 120 instances in the human genome, most of which are present and highly conserved in orthologous locations in other mammals. There are also >200 instances in the chicken genome, although most are not in orthologous locations. The family members show high sequence similarity to a transposable element that is still active in zebrafish as the SINE3 element (12). Remarkably, we also discovered a related SINE repeat in ≈1.3 megabase (Mb) of genomic sequence of the coelacanth (Latimeria menadoensis). The mammalian CNE family, the zebrafish SINE3, and the previously uncharacterized coelacanth SINE all share a highly similar central core region but can have different 5′ and 3′ flanking ends, suggesting that the core region itself may have functional significance both to the transposable element and to the genomic host.
Based on these recent examples, we speculate that many other large CNE families may have been distributed by ancient transposable elements whose sequences are no longer present or recognizable after many tens of millions of years of neutral evolution. As a first step toward identifying such cases, we describe 95 additional CNE families that have the highest rates of sequence retention and sequence conservation within the mammalian lineage.
Results
Family of Mammalian CNEs Derived from a SINE Element That Is Still Active in Zebrafish.
To systematically identify CNEs potentially derived from ancient transposable elements, we aligned a list of 863,000 CNEs in the human genome not overlapping known human repeats against all known vertebrate transposable elements deposited in the RepBase database (13). The CNEs with the top-ranking alignment scores are shown in Table 1. The highest-scoring match aligns SINE3-1, an active SINE element in zebrafish (12), with a 340-bp CNE located on human chromosome 2 (nucleotides 164247319–164247658, hg17). The zebrafish SINE is notable in its own right because it is the only known SINE element that is derived from a 5S rRNA gene (most SINEs are derived from tRNAs or, less commonly, 7SL RNA genes). For clarity, we will refer to the SINE family in zebrafish (Danio rerio) as the DrSINE3 family and the matching human CNE sequence as HsSINE3-1.
Table 1.
Top 20 CNEs with greatest similarity to transposable elements
| CNE | Ancient repeat | Species of ancient repeat | Coordinates of aligned CNE | Aligned CNE size, bp | Alignment score |
|---|---|---|---|---|---|
| CNE-1 | SINE3-1 | D. rerio | chr 2, nt 164247345–164247579 | 235 | 7,072 |
| CNE-2 | PSLINE | Platemys spixii | chr 10, nt 10602610–10602815 | 206 | 6,497 |
| CNE-3 | DR000899 | D. rerio | chr X, nt 53055668–53055756 | 89 | 5,921 |
| CNE-4 | L1MM_F | Mus musculus | chr 4, nt 94875117–94875237 | 121 | 5,873 |
| CNE-5 | CR1-3_DR | D. rerio | chr 12, nt 58202066–58202231 | 166 | 5,503 |
| CNE-6 | PSLINE | P. spixii | chr 13, nt 72745011–72745257 | 247 | 5,150 |
| CNE-7 | MAUI | Takifugu rubripes | chr 2, nt 67450383–67450574 | 192 | 5,131 |
| CNE-8 | MER121 | Homo sapiens | chr 17, nt 14482101–14482285 | 185 | 5,105 |
| CNE-9 | SINE3-1 | D. rerio | chr 2, nt 103864038–103864244 | 207 | 5,000 |
| CNE-10 | PSLINE | P. spixii | chr 1, nt 97028090–97028385 | 296 | 4,826 |
| CNE-11 | PSLINE | P. spixii | chr 5, nt 97927609–97927921 | 313 | 4,823 |
| CNE-12 | L1ME4A | H. sapiens | chr 14, nt 68290000–68290207 | 208 | 4,816 |
| CNE-13 | CR1_F | G. gallus | chr X, nt 132219313–132219509 | 197 | 4,741 |
| CNE-14 | SINE3-1 | D. rerio | chr 10, nt 21399081–21399190 | 110 | 4,707 |
| CNE-15 | SINE3-1 | D. rerio | chr 2, nt 103864079–103864244 | 166 | 4,681 |
| CNE-16 | SWIMMER1 | Oryzias latipes | chr 4, nt 182348338–182348562 | 225 | 4,510 |
| CNE-17 | GYPSY-22-I | D. rerio | chr 5, nt 4719211–4719358 | 148 | 4,439 |
| CNE-18 | HER_LINE | Scyliorhinus torazame | chr 12, nt 16041054–16041286 | 233 | 4,408 |
| CNE-19 | DR000900 | D. rerio | chr 6, nt 101985517–101985606 | 90 | 4,397 |
| CNE-20 | MER121 | H. sapiens | chr X, nt 122030335–122030513 | 179 | 4,365 |
chr, chromosome on hg17.
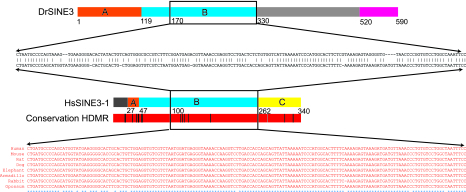
The similarity of the DrSINE3 consensus to HsSINE3-1 is confined largely to the central region (Fig. 1). SINE elements can typically be parsed into three segments: a 5′ end containing the promoter (in this case, related to 5S rRNA), a 3′ end related to a LINE element (which is presumed to be responsible for reverse transcription of the SINE element), and a central region (whose function, if any, is unclear). The region of similarity with the DrSINE3 consensus covers ≈235 bp (bases 27–261) of the human element and aligns primarily to the central region of the DrSINE3 consensus and only a small part (20 bp) of the 5′ end. Within the central region, a region of 162 bp (bases 100–261) shows sequence identity of 71%, which is remarkably high given the vast evolutionary separation between the species (see Fig. 1).
Fig. 1.
Comparison of the zebrafish DrSINE3 transposable element and HsSINE3-1 as well as the orthologous conservation of HsSINE3-1 across various mammals. Related portions are shown in the same color. Most of the similarity between the human sequence and DrSINE3 lies within the central core region (shown in blue and labeled “B”); the best aligning portion of 162 bp in humans is indicated by the rectangle and shown as a pairwise alignment. For conservation across HDMR, colors are as follows: red, identical 4-way; black, nonidentical 4-way; gray, <4 bases align. The corresponding orthologous sequence in seven other mammals is shown as a multiple alignment in red.
The HsSINE3-1 element itself is present at orthologous loci in many mammalian species and is extremely well conserved at the nucleotide level. The orthologous sequence is of identical size in human, dog, mouse, and rat (HDMR), apart from a 1-bp deletion in the dog. Strikingly, 96% of the bases present in all four species show perfect four-way identity (Fig. 1). The conservation rate is similar in the portion matching DrSINE3 as in the remainder of the element (97% vs. 95%). The rate of perfect conservation remains at 96% when elephant, armadillo, and rabbit are included in the comparison and falls only slightly to 93% when sequence from the marsupial Monodelphis domestica is included (Fig. 1).
The HsSINE3-1 element is located 41 kb past the 3′ end of the fidgetin gene (FIGN) and lies within a larger region of 1.5 Mb that contains no other annotated genes. FIGN belongs to the AAA superfamily of ATPases, which is associated with a wide variety of cellular activities, including membrane fusion, proteolysis, and DNA replication, and plays an important role in mammalian development (14, 15).
We noticed that several other human CNEs also showed similarity to the DrSINE3 family (see Table 1), suggesting the existence of an entire family of related CNEs in humans. We therefore defined the HsSINE3 family as the collection of all sequences in the human genome with significant similarity to HsSINE3-1. When we searched the human genome, we identified a family of 124 related; and see Fig. 4, which is published as supporting information on the PNAS web site elements (including HsSINE3-1 itself) by using a similarity threshold chosen such that a randomized control yields only a single hit (described in Methods; and see Fig. 4, which is published as supporting information on the PNAS web site). The full list of 124 instances is presented as Table 2, which is published as supporting information on the PNAS web site. None of these instances overlaps known protein-coding sequence [in contrast to the situation for LF-SINE (11)].
When we took the alternative approach of searching the human genome for sequences that are similar to the DrSINE3 consensus, we identified essentially the same instances as when searching for similarity to HsSINE3-1. Using the same methodology and thresholds, we found only 13 additional instances, most of which showed matches to HsSINE3-1 that fell just below threshold in the initial search.
With only a single exception, the elements found by alignment to HsSINE3-1 could not be further extended by alignment to the DrSINE3 consensus, indicating that they do not have sequences similar to the 5′ and 3′ regions of the SINE3 elements. However, the sole exception proved to be informative: This instance on chromosome 12 contains an additional 125 bases at the 5′ end. The alignment extends nearly to the 5′ start of the DrSINE3 consensus and includes the portion that is derived from the 5S rRNA gene. This instance provides unambiguous proof that the HsSINE3 family is derived from the same ancestral transposon that gave rise to the DrSINE3 elements but that the 5′ end has been lost in the vast majority of human instances. (A multiple alignment of this human instance, the 5S rRNA sequence, and the DrSINE3 consensus is supplied in Fig. 5, which is published as supporting information on the PNAS web site).
The substantial majority of members of the HsSINE3 family (85% or 106 of 124) could be aligned to orthologous sequence in the HDMR genomes, indicating that the sequences have largely been retained in all four species (see Table 2). Of the instances present in all four species, nearly three-quarters (74% or 80 of 106) also showed significant conservation at the nucleotide level. The average rate of perfect four-way identity for aligned copies is 72%, which is significantly higher than the background genomic rate (49%; P < 10−494) and only slightly lower than the rate for coding exons (78%). Notably, some of the copies show extremely high levels of sequence similarity: There are 13 instances with a rate of perfect four-way identity >90%; these instances range in length from 62 to 340 bp in humans. We did not observe a strong correlation between the rate of orthologous conservation of each copy and the similarity of the copy to HsSINE3-1 (see Fig. 6, which is published as supporting information on the PNAS web site).
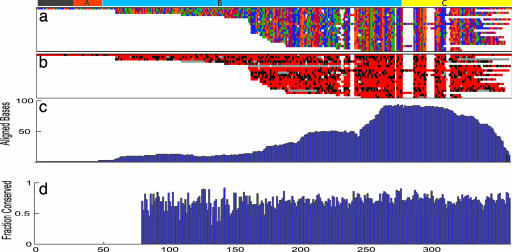
Interestingly, the members of the HsSINE3 family show a higher level of retention in the terminal region (the 79 bp at the 3′ end, which show no similarity to the zebrafish SINE) than in the central region (the 215 bp that show strong similarity to DrSINE3) (Fig. 1). The terminal region is retained in 88% of cases (at least 30 bases remaining), whereas the central region is retained in 63% of cases; this finding is all of the more significant given the smaller size of the region. The preferential retention of terminal region can be seen directly from multiple-sequence alignment of the top 20 instances most similar to HsSINE3-1 (Fig. 2b). When all 124 family members are aligned, the highest level of retention is seen at the juncture of terminal and central regions (Fig. 2c). However, the differences in the retention rate do not translate into differences in the nucleotide conservation rate. When retained, the central and terminal regions show a similarly high conservation rate (Fig. 2 b and d). (Figs. 7–9, which are published as supporting information on the PNAS web site, show the association between the rate of perfect four-way identity and the extent of central and terminal overlap for all aligned instances).
Fig. 2.
A multiple alignment of the top 20 human instances most similar to HsSINE3-1 was carried out. (a) The multiple alignment by color-coding bases (A, blue; G, green; T, red; C, orange). (b) The orthologous conservation profile across HDMR (perfect four-way identity, red; four bases aligned imperfectly, black; fewer than four bases aligning, gray). All 124 members of HsSINE3 were aligned to the reference instance HsSINE3-1. (c) How often a base was mapped to a given position along the reference. (d) The rate of perfect HDMR conservation along the reference for mapped human bases.
The analysis above focuses on 124 members of the HsSINE3 family. However, it is likely that the human genome contains many additional family members that are more distantly diverged. For example, we detect 785 human matches to HsSINE3-1 by using a more permissive threshold for which randomized controls yield ≈89 hits (see Fig. 4). However, most of these lower-scoring matches are short (median ≈ 58 bp) and show only modest cross-species conservation (the retention rate is 33% across the four species, HDMR, and the rate of perfect four-way identity is 64% within retained copies); these values are not substantially higher than those observed for the random control sequence. Accordingly, these instances are likely to consist primarily of relics of SINE3 insertions that are nonfunctional and thus not under purifying selection (that is, that are not CNEs).
A Family Related to HsSINE3 Is also Present in the Chicken Genome.
The conservation of the HsSINE3 family across mammalian genomes indicates that the ancient SINE elements transposed before the mammalian radiation and that a subset of the copies was exapted (that is, acquired a functional role). To better understand the evolutionary history of these events, we studied the chicken genome (16) for evidence of SINE3-related activity. We first searched for instances in the chicken genome similar to HsSINE3-1 by using the same strategy used to define the HsSINE3 family. We found 233 instances in chicken (Table 3, which is published as supporting information on the PNAS web site); this collection defines the GalSINE3 (Gallus gallus SINE3) family. Most instances have retained at least 30 bp of the terminal region (78% or 182 of 233) and central region (71% or 166 of 233) of HsSINE3-1. We also aligned the DrSINE3 consensus to the chicken genome and discovered only 24 additional instances, of which all but 6 overlap matches to HsSINE3-1 that score just below the threshold used to define GalSINE3.
Interestingly, the chicken and human elements are hardly ever found in orthologous locations. Of the 63% of chicken elements for which it is possible unambiguously to define the orthologous region in the human genome, one case has a HsSINE3 element in the orthologous location. This one case is a chicken element on chromosome 24 (nucleotides 2222133–2222205, galGal2) that is orthologous to a HsSINE3 family member on chromosome 11 (nucleotides 132510948–132511036, hg17). To search for cases of additional orthologous elements so diverged that they were not included within the HsSINE3 family, we used a more permissive threshold that detects many additional human sequences similar to HsSINE3-1. (The expanded list contains 3,913 copies, with ≈15% expected to be false positives based on the fact that a randomized control sequence yields 631 comparable alignments in the human genome). Even with this permissive threshold, we were able to detect only 11 further possible instances of orthologous sequence.
Taken together, the results above suggest that HsSINE3 and GalSINE3 originated from a common amniote ancestor >300 million years ago but that different copies have been preferentially retained in the two genomes. We conjecture that the chicken copies will also show a high degree of conservation within birds, although we currently lack appropriate genome sequence with which to perform such an analysis.
One particular chicken instance is extremely informative (chromosome 2, nucleotides 86373901–86374233, galGal2) because it has retained sequence similar to the terminal region of HsSINE3-1 and most of the sequence from the 5′ end of the DrSINE3 element, spanning both the central region and the 5S rRNA-related region of DrSINE3 (Fig. 10, which is published as supporting information on the PNAS web site). This instance represents a clear link between the amniote and zebrafish forms of SINE3.
Central Region of DrSINE3 Defines a Previously Uncharacterized SINE Element in Coelacanth.
Multiple copies of the DrSINE3 retrotransposon have been reported previously only in the zebrafish genome. We searched for its presence in several other ray-fin fishes for which draft genomes are available, including Fugu, tetraodon, or stickleback, but could not find any cases of multiple significant matches. (We did observe singleton instances in the small amount of sequence available from Atlantic salmon (Fig. 11, which is published as supporting information on the PNAS web site) and freshwater eel (see Fig. 12 and Supporting Text, which are published as supporting information on the PNAS web site). These fish species are more closely related to one another than they are to the zebrafish; the failure to identify DrSINE3 in these species could be attributed to inactivation of the ancestral repeat after the split from the zebrafish lineage (a phylogenetic tree is shown in Fig. 13, which is published as supporting information on the PNAS web site) followed by deletion and divergence of the repeat remnants.
We then searched for related elements in other vertebrate species. Strikingly, we found many matches in the coelacanth (L. menadoensis) despite the fact that only a tiny fraction of the genome sequence (≈1 Mb) is available. The fish lineage bifurcated ≈500 million years ago into two lineages: ray-finned fish (actinopterygii), which gave rise to most modern fish, and lobe-finned fish (sarcopterygii), which gave rise to tetrapods ≈410 million years ago (17, 18). The coelacanth and lungfish are the only two extant groups of lobe-finned fishes. [Coelacanths were thought to have become extinct ≈70 million years ago until one was discovered in 1938 off South Africa (19); they are colloquially referred to as “living fossils.”]
A repeat closely related to DrSINE3 is clearly present in high copy number in the genome of the coelacanth. The ≈1 Mb of available genomic sequence contains a total of 29 distinct instances. If this density were representative, it would imply that ≈90,000 copies may be present in the coelacanth (assuming a genome size of ≈3 gigabases; see ref. 18). The coelacanth sequences show a wide range of sequence variation when compared with one another: The rate of nucleotide identity ranges from 63% to 92%, with a median value of 76%, which suggests that some copies of the repeat have been active in relatively recent evolutionary time.
We created multiple alignments of all instances, together with varying amounts of flanking sequence, to derive a repeat consensus consisting of 301 bases (see Methods). When the derived consensus is aligned to the DrSINE3 consensus, there is remarkable similarity over the internal region: 83% identity over 162 bp. By contrast, the 5′ ends show little similarity. A multiple alignment of the internal regions of DrSINE3, HsSINE3-1, and the coelacanth consensus is shown in Fig. 14b, which is published as supporting information on the PNAS web site.
The coelacanth element itself is clearly a unique SINE element, which we have named LmSINE3 (L. menadoensis SINE3). We compared the LmSINE3 consensus to all repeats in RepBase. Surprisingly, the 5′ end is a clear match to SINE elements derived from tRNAs. The characteristic B box element found in the polymerase III promoters of tRNA genes is clearly recognizable and conserved (20). The 5′ end of the LmSINE3 consensus is most similar to the 5′ end of the SINE_TE element, which is also tRNA derived; this element is found in many fish species and is a member of the V-SINE superfamily (13, 21).
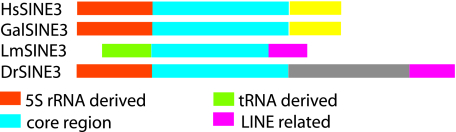
The presence in coelacanth of a 5′ end derived from a tRNA contrasts with the presence in zebrafish, chicken, and mammals of a 5′ end derived from a 5S RNA-derived 5′ end. Given the structure of the evolutionary tree, it seems that the SINE element acquired a new 5′ end in the lineage leading to coelacanth. A comparison of the various SINE3 families is shown in Fig. 3.
Fig. 3.
A summary of the SINE3-related families found in different species. Related portions are shown in the same color. HsSINE3 is found in humans, GalSINE3 in chickens, LmSINE3 in the coelacanth, and DrSINE3 in the zebrafish. Only the core region (blue) is shared across families; different combinations of 5′ and 3′ ends characterize each family.
The 3′ end of LmSINE3 is clearly related to the 3′ ends of other SINE elements. The closest relative is the 3′ end of a SINE element (SINE_AFC) in cichlid fish, which is also the closest relative to the 3′ end of the zebrafish DrSINE3 family, consistent with earlier reports (12). A multiple alignment of the 3′ ends is shown in Fig. 14a. We interpret the shared similarity among all three SINEs as a reflection of their dependence on unrelated LINE elements.
We note that when DrSINE3 was first characterized (12), the researchers noted that the internal region was similar to two sequences in coelacanth. Because the flanking sequence of these two instances revealed no similarity to any known repeats, the researchers speculated that the central region was itself a distinct transposable element that most likely had spread by horizontal rather than vertical transfer (12). It is now clear, however, that the central region has been an integral part of the transposon since at least the divergence of fish and mammals some 500 million years ago.
Finally, we note that there is a single strong match in one distant organism: the human blood fluke (Schistosoma japonicum). This instance shares 88% sequence identity with the core region SINE3 over 72 bp (see Fig. 15, which is published as supporting information on the PNAS web site). Given the large evolutionary distance between fluke and vertebrates (>600 million years), this single instance should not be assumed to imply vertical descent from a common ancestor; the possibility of horizontal transfer must be considered. Resolving this question will require extensive genomic sequence from platyhelminthes. If the element in blood flukes does represent vertical descent from a common ancestor, then the core element must be very old indeed.
Identification of Additional Ancient Mammalian CNE (AMCNE) Families.
With the discovery of the MER121 and LF-SINE families and now the HsSINE3 family, it is clear that ancient CNE families are likely to be of biological interest and importance. We therefore analyzed the human CNE families generated by Bejerano et al. (9) (by clustering CNEs based on sequence similarity) to identify those most likely to represent ancient mammalian families. Specifically, we extended each CNE family by identifying paralogous sequences in the human genome that were not retained and conserved in mouse, rat, or dog [that is, lineage-specific instances (see Methods)]. We reasoned that CNE families whose genome-wide distribution substantially preceded the mammalian radiation should have a low proportion of lineage-specific copies, because most would have degenerated during mammalian evolution.
We found 96 families with at least 10 instances and <25% lineage-specific instances (see Methods). We refer to these families as AMCNE families. The list includes the three families discussed above (MER121, LF-SINE, and HsSINE3), with MER121 being the largest family on the list. The second largest family (designated AMCNE2) contains 186 elements, with a consensus sequence of ≈130 bp and with >90% of cases being conserved across mammals (Fig. 16, which is published as supporting information on the PNAS web site). The 186 instances do not include known protein-coding sequences, have no significant overlap with ESTs or mRNAs, and show no evidence of conserved secondary structure; thus, we suspect that they play a regulatory role. The 96 AMCNE families are described, along with certain genomic properties, in Table 4, which is published as supporting information on the PNAS web site.
Discussion
With this report, three examples of large families of ancient CNEs have now been discovered within the past year.
We reported the existence of the MER121 family, with ≈1,000 copies in the human genome. The copies show considerable sequence variation among themselves, but the vast majority are retained at orthologous locations in mammals, and the orthologs at each genomic location tend to show high sequence similarity. We speculated that the MER121 family arose as a functional sequence distributed by a transposon, which underwent positive selection to optimize its local function and was then retained while the transposon sequence degenerated.
Bejerano et al. (11) recently reported an unambiguous case of a large CNE family that arose through a transposon, termed LF-SINE. There are ≈245 instances in the human genome with ≈5% overlapping coding exons and the remainder being noncoding. They showed experimentally that at least some instances act as distal enhancers. Notably, the family is related to a SINE element that has been active in coelacanth.
Here, we describe another human CNE family that arose through transposition of a SINE element. The family contains at least 124 copies, which are mostly retained at orthologous locations and significantly conserved across mammals. There are also at least 233 copies in the chicken, although most lack a human ortholog. The sequences are unmistakably related to the SINE3 repeat that is active in zebrafish and to a SINE element in coelacanth that we term LmSINE3. The mammalian, chicken, zebrafish, and coelacanth elements all share the core element but have clear differences in the 5′ and 3′ ends. The 5′ end is derived from a tRNA gene in coelacanth and a 5S RNA gene in the other lineages, whereas the 3′ end is shared between mammal and bird but differs from that seen in fish. The preservation of the core element across 450 million years of evolution argues strongly for its functional importance. [We note that Nishihara et al. (22) have independently discovered the SINE3 family and characterized its properties; we thank these researchers for sharing findings before publication.]
These examples suggest that transposable elements may provide a common mechanism by which large regulatory elements that evolve in one location may be disseminated, with some insertions conferring evolutionary benefit and being retained. The functional sequences may simply be passengers accidentally acquired and passively transported by the transposon. Or, the regulatory sequences may be advantageous to certain transposons, perhaps by increasing transcription. Such an advantage may explain the long life of the LF-SINE and SINE3 families relative to other similar transposons (23).
We suspect that many other CNE families have also arisen through distribution by an ancient transposon. The challenge in establishing their origin is that the transposon-related sequences would be expected to degenerate to the point of being absent or unrecognizable. Only in rare cases may there be a “smoking gun,” such as the presence of a recently active version of the transposon in other species (as for LF-SINE and SINE3).
We also note the curious fact that both LmSINE3 and LF-SINE recently have been active in the coelacanth. This observation suggests that it will be valuable to sequence the coelacanth to study lobe-finned fish, which are more closely related to amniotes than the better-studied ray-finned fish.
Whatever their origin, ancient CNE families are likely to be important. In addition to the MER121, LF-SINE, and HsSINE3 families, we have identified 93 AMCNE families that have experienced little evolutionary turnover since their creation before the mammalian radiation. All of these families have at least 10 instances in the human genome; the second largest (after MER121) has at least 186 instances. It is thus clear that the human genome harbors extensive families of conserved noncoding elements, with at least some having been distributed by transposable elements. This realization should give us renewed respect for the creative role of transposons, which are often dismissed as “junk” DNA.
The obvious challenge now is to determine the function of these families of elements. It is unclear whether one should expect a unified biological function for each family or whether each copy within a family may have its own distinctive role. At present, functional evidence is available for only a few instances of LF-SINE (11). In these cases, the CNE sequence has been exapted into several alternatively spliced exons in one case and into a distal regulatory element in another; these uses appear unrelated, but the exonic sequences likely have extensive regulatory sequences controlling their splicing. New methods will likely be required to study the functions of AMCNEs in a manner that is efficient enough to see the larger patterns that lead to a general understanding of these important but mysterious features.
Methods
Identification of CNEs Similar to Transposable Elements.
We started with a collection of ≈1.6 million conserved elements identified by the phastCons program (24) and downloaded from the University of California, San Diego, CA (UCSC) Genome Bioinformatics web site (hg17). We removed those elements overlapping protein-coding exons, known RNA genes, or sequences annotated as mammalian transposable elements. We further removed those that are not present in the orthologous regions of the mouse genome. This process of elimination left 863,000 elements (CNEs) covering of 85 Mb of the human genome. We aligned all CNEs to the RepBase 10.05 database of transposable elements (13) by using the Blastz aligner (25) (default parameters) and ranked all alignments by alignment score.
Instances in Human and Chicken Genomes.
We identified sequences similar to HsSINE3-1 by standard Smith–Waterman alignment against the human (hg17) and chicken (galGal2) genomes (match, 10; mismatch, −10; gap open, −40; gap extend, −5). As a control, we aligned a reshuffled consensus. Alignments were ranked by alignment score. The same protocol was followed when aligning the SINE3-1 repeat consensus (from the RepeatMasker database) (26).
Conservation Across HDMR.
We analyzed four-way alignment of genome sequence from human (hg17), mouse (mm5), dog (dog1), and rat (rn3) as provided by the UCSC Genome Bioinformatics web site and based on Blastz/Multiz alignments. We identified retained instances across HDMR by requiring that at least 50 bp be present in each of the four species to allow for partial deletion. For retained instances, we then identified human bases that align to bases in all four species. The rate of perfect four-way identity is defined as the proportion of four-way aligned bases that are identical across all four species. To estimate the background (i.e., neutral) rate, we applied the same procedure to the ancient repeat elements that were studied in the initial analysis of the mouse genome (2) and arrived at rate of 49%. Instances showing significant conservation were defined as those with a significant conservation above background (z > 2.0, normal distribution; N = number of aligned bases).
Gene Set.
We used the Ensembl set of gene predictions (hg17, downloaded from the UCSC Genome Bioinformatics web site) for both the conservation analysis and to establish overlap with instances of the CNE families. To estimate the rate of perfect four-way identity of coding regions, we analyzed the coding exons as described above.
Identification of LmSINE3 Repeats in Coelacanth.
We downloaded all currently available genomic sequence (≈1.3 Mb) for coelacanth (see Supporting Text). We aligned the DrSINE3 consensus and identified 34 high-scoring alignments, of which 5 appeared to be in duplicated genomic regions, leaving 29 unique instances. We created a multiple alignment of all instances, together with varying amounts of flanking sequence (10, 50, and 100 bp). In each case, we derived a consensus sequence by identifying bases with a clear majority (≥40%) within multiple alignment columns with at least 14 aligned bases. The ends of the consensus were then manually edited, and the three consensus sequences were checked for consistency. Variations in the cutoffs did not significantly affect the consensus. The LmSINE3 consensus we derived has 301 bp and appears in Supporting Text.
Identifying AMCNE Families.
We parsed the human vs. human whole-genome alignments available from the UCSC Genome Bioinformatics web site genome browser (the hg17 “self-chain” alignments) and identified all sequences that can be aligned to each of the 863,000 CNEs. For each of the CNEs, we counted the number of aligned instances (N) in the entire human genome and the number of aligned instances (K) overlapping any one of the 863,000 CNEs. We accepted those CNEs with K ≥ 10 and with <25% human-specific instances [that is, (N − K)/N < 0.25]. The list of families is shown in Table 4.
Supplementary Material
Abbreviations
- CNE
conserved noncoding element
- Mb
megabase
- DrSINE3
Danio rerio SINE3
- HsSINE3-1
human sequence SINE3-1
- HDMR
human, dog, mouse, and rat
- GalSINE3
Gallus gallus SINE3
- LmSINE3
Latimeria menadoensis SINE3
- AMCNE
ancient mammalian CNE.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lindblad-Toh K., Wade C. M., Mikkelsen T. S., Karlsson E. K., Jaffe D. B., Kamal M., Clamp M., Chang J. L., Kulbokas E. J., Zody M. C., et al. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Dermitzakis E. T., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S. E. Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- 4.Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., Mattick J. S., Haussler D. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 5.Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K., et al. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dermitzakis E. T., Reymond A., Antonarakis S. E. Nat. Rev. Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 7.Boffelli D., Nobrega M. A., Rubin E. M. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 8.McEwen G. K., Woolfe A., Goode D., Vavouri T., Callaway H., Elgar G. Genome Res. 2006;16:451–465. doi: 10.1101/gr.4143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejerano G., Haussler D., Blanchette M. Bioinformatics. 2004;20(Suppl. 1):I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- 10.Kamal M., Xie X., Lander E. S. Proc. Natl. Acad. Sci. USA. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bejerano G., Lowe C. B., Ahituv N., King B., Siepel A., Salama S. R., Rubin E. M., James Kent W., Haussler D. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 12.Kapitonov V. V., Jurka J. Mol. Biol. Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- 13.Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 14.Beyer A. Protein Sci. 1997;6:2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox G. A., Mahaffey C. L., Nystuen A., Letts V. A., Frankel W. N. Nat. Genet. 2000;26:198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
- 16.Hillier L. W., Miller W., Birney E., Warren W., Hardison R. C., Ponting C. P., Bork P., Burt D. W., Groenen M. A., Delany, et al. Nature. 2004;432:695–716. [Google Scholar]
- 17.Noonan J. P., Grimwood J., Danke J., Schmutz J., Dickson M., Amemiya C. T., Myers R. M. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danke J., Miyake T., Powers T., Schein J., Shin H., Bosdet I., Erdmann M., Caldwell R., Amemiya C. T. J. Exp. Zool. A Comp. Exp. Biol. 2004;301:228–234. doi: 10.1002/jez.a.20024. [DOI] [PubMed] [Google Scholar]
- 19.Thompson K. S. Living Fossil: The Story of the Coelacanth. New York: W. W. Norton; 1992. [Google Scholar]
- 20.Paule M. R., White R. J. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogiwara I., Miya M., Ohshima K., Okada N. Genome Res. 2002;12:316–324. doi: 10.1101/gr.212302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishihara H., Smit A. F., Okada N. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shedlock A. M., Okada N. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S., et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz S., Kent W. J., Smit A., Zhang Z., Baertsch R., Hardison R. C., Haussler D., Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit A. F. A., Hubley R., Green P. RepeatMasker Open. Seattle: Univ. of Washington; 2005. Version 3.0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.