Abstract
Peroxisomes were visualized in living plant cells using a yellow fluorescent protein tagged with a peroxisomal targeting signal consisting of the SKL motif. Simultaneous visualization of peroxisomes and microfilaments/microtubules was accomplished in onion (Allium cepa) epidermal cells transiently expressing the yellow fluorescent protein-peroxi construct, a green fluorescent protein-mTalin construct that labels filamentous-actin filaments, and a green fluorescent protein-microtubule-binding domain construct that labels microtubules. The covisualization of peroxisomes and cytoskeletal elements revealed that, contrary to the reports from animal cells, peroxisomes in plants appear to associate with actin filaments and not microtubules. That peroxisome movement is actin based was shown by pharmacological studies. For this analysis we used onion epidermal cells and various cell types of Arabidopsis including trichomes, root hairs, and root cortex cells exhibiting different modes of growth. In transient onion epidermis assay and in transgenic Arabidopsis plants, an interference with the actin cytoskeleton resulted in progressive loss of saltatory movement followed by the aggregation and a complete cessation of peroxisome motility within 30 min of drug application. Microtubule depolymerization or stabilization had no effect.
The peroxisome is a ubiquitous and versatile subcellular organelle delimited by a single membrane. Peroxisomes are involved in various catabolic and anabolic pathways such as the photorespiration cycle, fatty acid β-oxidation, the glyoxalate cycle, ureide metabolism, and the generation of cellular messengers like superoxide radicals and nitric oxide (Tabak et al., 1999; Corpas et al., 2001). Unlike chloroplasts and mitochondria, peroxisomes do not possess their own DNA. As a consequence, all peroxisomal proteins are nuclear encoded and imported from the cytosol (Baker, 1996). Once synthesized on free polysomes, these proteins require correct targeting to the organelle. Peroxisomal targeting signals (PTS) have been identified at the carboxy terminus of some proteins (PTS-1) and at the amino terminus in others (PTS-2: Mullen et al., 1997; Flynn et al., 1998; Mullen and Trelease, 2000). The PTS-1 consists of the sequence Ser-Lys-Leu or a relatively conserved variant (Gould et al., 1989; Miura et al., 1992; Trelease et al., 1996). In plants, although a variety of signal sequences appear to lead proteins to peroxisomes, effective and unequivocal targeting using a PTS-1 was demonstrated in transgenic tobacco plants where the last six amino acids of glycolate oxidase were sufficient to target the β-glucuronidase protein to peroxisomes (Volokita, 1991).
In animal cells also, the addition of a PTS-1 consisting of the SKL motif to the carboxy terminus of the green fluorescent protein (GFP) from Aequorea victoria effectively targeted the protein to peroxisomes. Expression of the GFP-PTS1 construct was used to demonstrate that peroxisomal dynamics and distribution in living animal cells are dependent on the microtubular components of the cytoskeleton (Wiemer et al., 1997; Schrader et al., 2000). Furthermore, the inhibition of peroxisomal movement by overexpression of the dynactin subunit dynamitin (p50) suggested a role for dynein/dynactin motors in peroxisome motility and reaffirmed microtubules as exclusive tracks for peroxisomal movement in animal cells (Schrader et al., 2000). In plants, the random expression of GFP::cDNA fusions in Arabidopsis has led to the generation of at least 29 marker lines where GFP has been targeted to peroxisomes. A majority of these lines possess a PTS-1 at their carboxy terminus and carry the canonical tri-peptide sequence SKL or a close variant thereof (Cutler et al., 2000). However, despite their visualization in live cells (Mano et al., 1999; Cutler et al., 2000), details pertaining to the movement of peroxisomes, their choice of cytoskeletal tracks and motors have not been addressed so far in plant cells.
In the context of the intracellular organelle motility, two recent trends are noteworthy. First, contrary to the situation in animal cells, a number of subcellular organelles in plants have been shown to use actin and not microtubule tracks for their intracellular movement (Hawes and Satiat-Jeunemaitre, 2001). Second, reports in both animal (Rodionov et al., 1998; Rogers and Gelfand, 1998) and plant cells (Sato et al., 2000) have provided examples of conditional switching between actin and microtubule motors and tracks by some organelles, indicating that cytoskeletal components may adopt cooperative and not exclusive roles in organelle movement.
Keeping these trends in mind, we have assessed peroxisome motility in plants. Live onion (Allium cepa) epidermal cells and transgenic Arabidopsis plants have been transformed with a probe consisting of a PTS1 (SKL motif) fused to the carboxy terminus of a yellow fluorescent protein (EYFP) that is similar to the one used to study peroxisome movement in animal cells (Wiemer et al., 1997; Schrader et al., 2000). The EYFP-labeled peroxisomes were visualized simultaneously with EGFP-labeled actin microfilaments/microtubules. Peroxisome movement was analyzed in various cell types in transgenic Arabidopsis plants where drug-induced phenotypic alterations provided independent confirmation of the efficacy of drug treatment. The accumulated evidence strongly suggests that peroxisomes labeled with the EYFP-PTS1 fusion protein that had been shown to use microtubular tracks in animal cells move along actin microfilaments in plant cells.
RESULTS
The EYFP-Peroxi Fusion Protein Is Targeted to the Peroxisomal Compartment
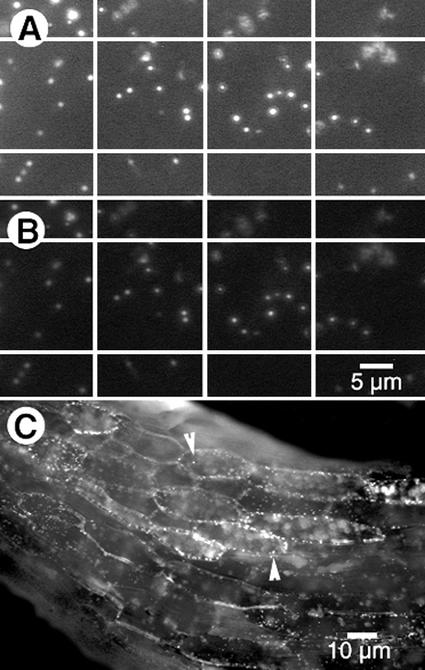
The EYFP-peroxi construct (catalog no. 6933-1, CLONTECH Laboratories, Palo Alto, CA) carries a PTS1 containing the SKL motif at its C terminus (Mullen et al., 1997). Although this is the first time, to our knowledge, that the EYFP-peroxi construct has been used in plants, an EGFP-PTS1 construct (CLONTECH) employing the same target sequence has already been shown to label peroxisomes in animal cells (Schrader et al., 2000). EYFP-peroxi cloned under a cauliflower mosaic virus 35S promoter in the pRT103 vector (Topfer et al., 1987) labeled numerous motile subcellular structures in both onion and Arabidopsis epidermal cells in a transient assay. Whole-mount immunocytochemistry of Arabidopsis seedlings transiently expressing EYFP-peroxi was carried out using antibodies directed against catalase, a peroxisomal matrix protein (Kunce et al., 1988), and a peroxisomal ascorbate peroxidase (Mullen and Trelease, 2000). The antibody localization for these peroxisome-specific proteins coincided with all GFP-labeled, yellow-green fluorescent, ovoid, 0.8- to 1.9-μm-long, motile bodies and identified them as peroxisomes (Fig. 1, A and B). After this confirmation of peroxisomal identity, stable transgenic Arabidopsis plants carrying the EYFP-peroxi construct were created via Agrobacterium tumefaciens-mediated transformation. Peroxisomes could be easily observed in all cells in stable EYFP-peroxi transgenic plants as bright, yellow-green motile dots and were clearly distinguished from the orange-red fluorescence of chloroplasts under blue light (approximately 490 nm) illumination (Fig. 1C).
Figure 1.
Structures labeled with the EYFP-peroxi protein react with an antibody directed against a peroxisomal protein. A, Transient expression of EYFP-peroxi construct in Arabidopsis epidermal cells labeled numerous motile microbodies that were visualized using a fluorescein isothiocyanate (FITC) filter (approximately 490 nm) after being processed for immunocytochemistry using anti-catalase antibodies. B, The same cell in A visualized upon illumination through a tetramethylrhodamine B isothiocyanate (TRITC) filter (approximately 550 nm) showed the secondary TRITC labeling of anti-cottonseed catalase antibody. A similar immunostaining pattern was obtained using the peroxisomal ascorbate peroxidase antiserum. Note that each EYFP-labeled structure in A was labeled red in B, demonstrating that each microbody expressing the EYFP-peroxi protein also expresses the specific peroxisomal protein. C, Upon illumination with blue light from an FITC filter, hypocotyl cells of Arabidopsis transgenic plants carrying the EYFP-peroxi construct displayed numerous bright, yellow-green motile peroxisomes. Chloroplasts appeared red under this illumination.
Peroxisomes Exhibit Different Kinds of Motility in Living Plant Cells
Observations on EYFP-labeled peroxisomes in onion and transgenic Arabidopsis epidermal cells allowed us to distinguish three distinct kinds of motile behavior (Figs. 2 and 3). These consisted of (a) a barely perceptible oscillation covering 1 to 2 μm where peroxisomes remained anchored to one position for long periods; (b) short-range, slow, to and fro movements covering 2 to 8 μm; and (c) saltations, i.e. bidirectional movements frequently covering large intracellular distances (Fig. 2). Our observations showed that individual peroxisomes within a cell were capable of independent movement where one peroxisome could remain static while its neighbor embarked on a long saltation of up to 250 μm (Fig. 2). Moreover, over time each peroxisome could change its state of motility, its velocity, and the direction of its movement (Fig. 2). At any given time nearly three-fourths (70% ± 2.67%) of the peroxisome population appeared to be resting (Fig. 2) and exhibited only Brownian oscillatory movements. Nearly 15% (12.3 ± 4.1) displayed movement over short distances of up to 8 to 10 μm at velocities ranging from 0.2 to 1.6 μm s−1 (mean velocity 0.7 ± 0.2 μm s−1). In peripheral regions and trans-vacuolar cytoplasmic strands, nearly 10% of the peroxisomes carried out saltations. These saltations could occur in a single burst of rapid movement (velocities up to 4 μm s−1) or with short resting/oscillation phases (velocity ranging from 0.7 to 2.8 μm s−1; Figs. 2 and 3). An attempt to correlate the variations in speed and distance covered with time by tracking more than 85 individual peroxisomes for up to 5 min failed to provide a clear pattern for movement. Thus, each peroxisome appeared to follow its own random program, making it difficult to predict its intracellular position or behavior at any given time (Fig. 3). In addition, it was observed that a varying subset (4.3% ± 2.6%) of the fast-moving peroxisomes could suddenly reverse their direction and continue movement at the same speed (data not shown). In every cell observed, peroxisomes interacted with each other and frequently engaged in transient contacts before continuing on their earlier or a changed path. At intervals, some peroxisomes appeared to fuse together or alternatively to divide and give rise to one or two smaller, independently moving bodies. In addition, in nearly all the cells observed and at apparently random time intervals, many peroxisomes moved toward and aggregated at the same intracellular location (Fig. 2) before re-embarking on their diverse paths.
Figure 2.
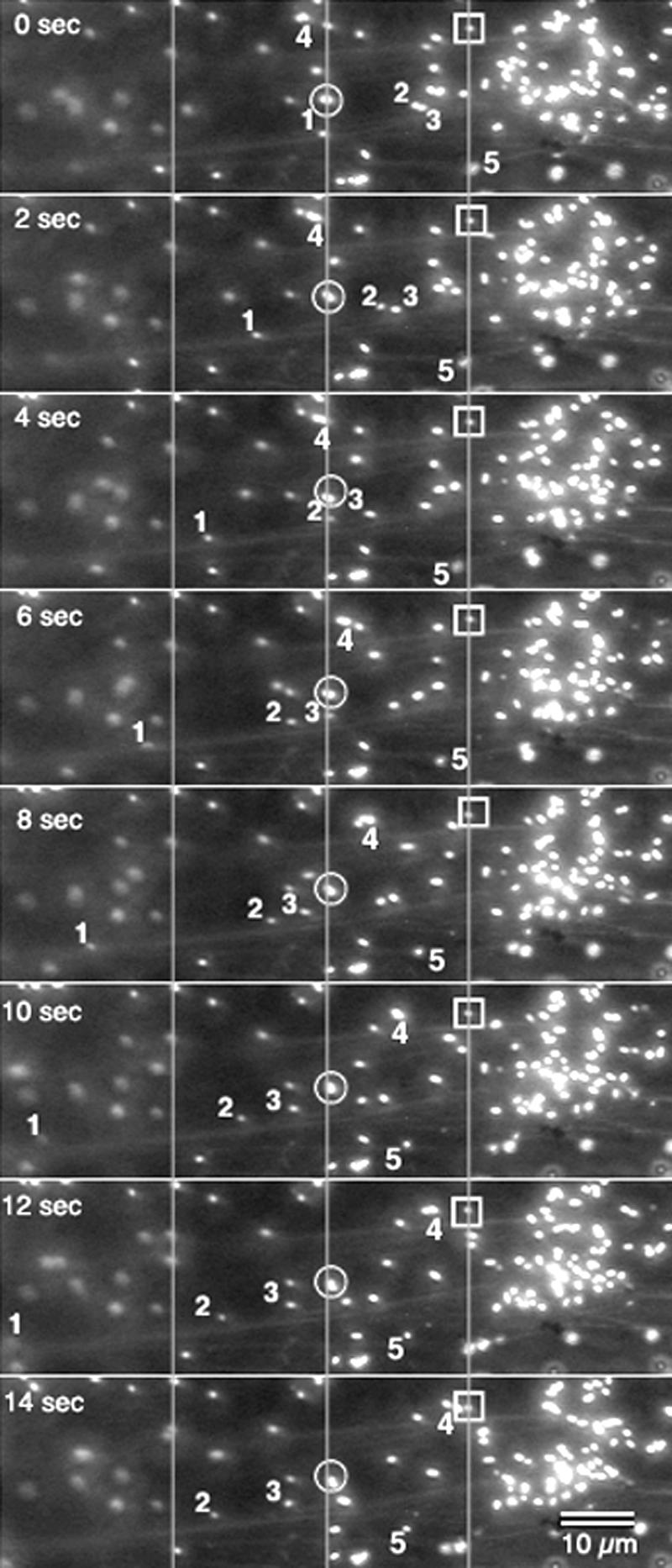
Sequential pictures taken at 2-s intervals allowed tracking of individual peroxisomes over 14 s and showed different kinds of peroxisomal motility in living cells. Two peroxisomes (enclosed □ and ○) displayed a barely perceptible Brownian movement and served as reference points for three other peroxisomes (designated 1, 2, and 3), which displayed more active movement. Peroxisome 1 moved rapidly and over a relatively larger distance (moving completely out of frame in the last panel) than 2 and 3, although all three peroxisomes exhibited saltatory movement in the same direction. Peroxisome 4 moved at varying speeds in a direction opposite to that of the other three. Peroxisome 5 displayed short-range movement during this time interval and during the last 4 s, showed very little movement. Other directions of movement may be discerned by observing unlabeled peroxisomes. The right side of the pictures shows an aggregation of peroxisomes from which individuals moved off in different directions at different time points. The pictures also show actin microfilaments in the background. Lines across the pictures act as reference points.
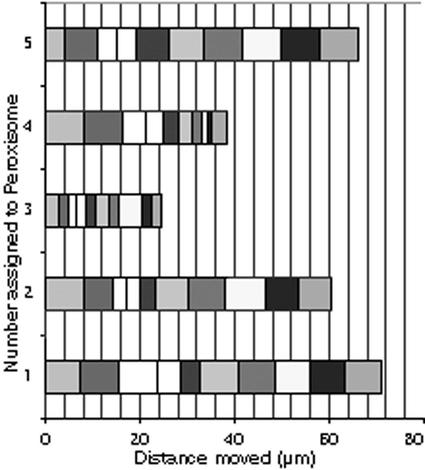
Figure 3.
Observation of five randomly chosen peroxisomes that had clearly embarked on saltations taken over 20 s revealed that the velocity and corresponding intracellular distance covered by each peroxisomes varied constantly over time. Each block represents the distance covered in 2 s. Thus, peroxisomes 1 and 5 moved the largest distances (nearly 70 μm), whereas peroxisome 3 covered a distance of only 24 μm in the same time.
These directed movements and sudden changes in velocity/directionality suggested that peroxisome movement did not result from a passive dragging-along in the cytoplasmic streaming motion but could involve an active, driving force. These observations prompted us to covisualize peroxisomes and cytoskeletal tracks before attempting to figure out the possible forces propelling peroxisomes along them.
Covisualization of Peroxisomes and Cytoskeletal Elements Reveals Peroxisome Alignment with Actin Microfilaments
Peroxisomes and actin microfilaments were visualized simultaneously by cobombarding EYFP-peroxi and GFP-mTalin. The GFP-mTalin construct has been shown to label filamentous (F)-actin strands specifically (Kost et al., 1998; Mathur et al., 1999). Peroxisomes and microtubules were visualized similarly by using EYFP-peroxi in combination with GFP-MBD, which has been shown to specifically label microtubules (Marc et al., 1998; Mathur and Chua, 2000).
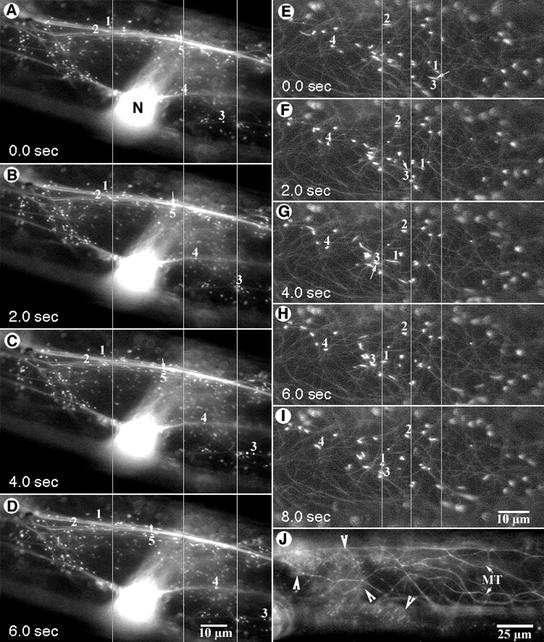
Although a high degree of spectral overlap exists between the GFP and EYFP chromophores, two different strategies distinguished peroxisomes from cytoskeletal elements. Thus, we relied on peroxisome movement and the relative nonmotility of cytoskeletal elements when using a fluorescence microscope. Under blue-light illumination through an FITC filter, peroxisomes appeared as bright motile spots, whereas cytoskeletal elements appeared as long fibrous structures (Fig. 4, A and G). Fine spectral differentiation achieved using a spectrophotometric confocal microscope (TCS-SP2, Leica) clearly separated the EGFP (used to label cytoskeletal elements) and EYFP (used to label peroxisomes) signals. The two fluorochromes were detected on separate channels and allocated false green and red colors, respectively (Fig. 4B). The combined observations showed that cortical F-actin strands in onion epidermal cells were relatively short and more compactly arranged (Fig. 4B) than the longitudinally stretching trans-vacuolar F-actin cables (Fig. 4A). Peroxisomes were closely aligned with actin strands, and their movement followed the contours defined by actin microfilaments (arrows in Fig. 4, A and B). In cortical regions, peroxisomal movement usually consisted of short-range movements and frequently involved crossing over to neighboring actin strands, which resulted in changes in movement directionality. Many actin filaments were observed within thick trans-vacuolar cytoplasmic strands (Fig. 4, C–F). However, in most cases, peroxisomes did not follow the general flow of cytoplasm and within a particular strand could even move in diametrically opposite directions (arrows in Fig. 4, C–F).
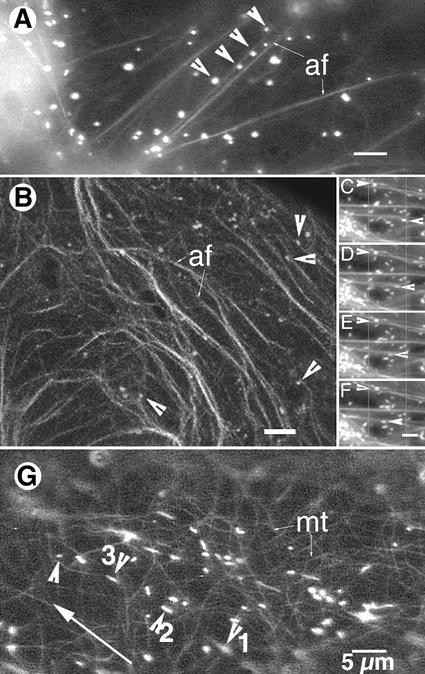
Figure 4.
Simultaneous visualization of peroxisomes and cytoskeletal elements. Lines across the pictures serve as points of reference. A, An onion epidermal cell transiently expressing both EYFP-peroxi and GFP-mTalin observed using a fluorescence microscope showed peroxisomes (arrows) aligned along longitudinally stretched, trans-vacuolar F-actin strands (af). B, Spectral differentiation between EYFP and EGFP using a spectroscopic confocal microscope (TCS-SP2, Leica Microsystems Vertrieb GmBH, Mannheim, Germany) allowed peroxisomes to be clearly distinguished (arrows) among the dense mesh of cortical F-actin strands (af). Peroxisomes were closely aligned with actin microfilaments (arrows). C through F, Time-lapse sequence (interval between successive images, 1 s) showed that within a thick transvacuolar cytoplasmic strand some peroxisomes (e.g. arrows) moved in diametrically opposite directions along separate F-actin strands. G, Simultaneous expression of EYFP-peroxi and GFP-MBD allowed the visualization of both peroxisomes and microtubules upon illumination through an FITC filter. Note that the directionality of peroxisome movement (long arrow) does not coincide with underlying cortical microtubules (mt) and that there are considerable gaps where microtubules are apparently not present (e.g. between peroxisomes 1 and 3).
Both cortical and subcortical microtubules labeled by the GFP-MBD assumed a net transverse orientation in the onion epidermal cells and the bright, yellow-green motile peroxisomes could be easily distinguished from them (Fig. 4G). In some instances, especially in the cortical regions, peroxisome motility appeared to follow microtubule contours, but tracking of individual peroxisomes showed gaps of up to 5 to 10 μm in each instance where the peroxisome clearly moved away from the microtubular structure and appeared to jump across clear cytoplasmic areas (Fig. 4G). Moreover, peroxisome movement, especially the long-range saltations, frequently occurred along the longitudinal axis of the cell and, in most cases, did not show any clear co-alignment with the transversely arranged microtubules (Fig. 4G). Because the level of gene expression between different cells differed in transient assays, we searched for cells expressing high levels of GFP-MBD, which is known to result in microtubule bundling (Olson et al., 1995; Marc et al., 1998). In these cells peroxisome movement was not affected despite the presence of pronounced microtubule bundles.
Taken together, the covisualization of peroxisomes and cytoskeletal elements indicated that peroxisome movement was linked to actin and not microtubule tracks. Further confirmation for this observation was sought by using specific microtubule- and micro-filament-interacting drugs.
Effect of Cytoskeletal Drugs on Peroxisomal Motility
Drug inhibitor studies were carried out initially on onion epidermal cells that transiently expressed EYFP-peroxi alone or in combination with GFP-mTalin or GFP-MBD. The drug treatments were extended subsequently to transgenic Arabidopsis plants carrying the EYFP-peroxi construct. Although all cells in these transgenic plants displayed fluorescent peroxisomes, for this study, we focused on three cell types exhibiting the two different modes of cell growth, namely tip and diffuse growth. These cell types, namely, trichomes and cortical and hair cells of the root, provide the additional advantage that they display clear morphological alterations upon treatment with microtubule- and actin-interacting drugs (Baskin et al.1994; Mathur et al., 1999; Baluska et al., 2001). Their use introduced an independent, visually scoreable morphological criterion for ensuring that each cell where peroxisomal motility was assessed had clearly sensed and responded to the respective drug treatment.
Specific and well-characterized microtubule-interacting drugs oryzalin, propyzamide (depolymerize microtubules), and paclitaxel (stabilizes microtubules) and actin-interacting drugs CD and Lat-B (both affecting actin polymerization) were used.
Drug Treatments of Onion Epidermal Cells Expressing Peroxisome and Cytoskeletal Marker Genes Transiently
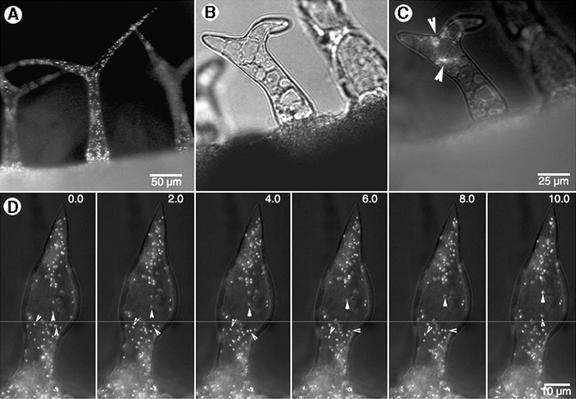
A treatment of onion epidermal cells expressing EYFP-peroxi either alone or in combination with GFP-mTalin with the drugs CD and Lat-B at 2 μm concentration resulted in a rapid, global arrest of peroxisomal movement (Fig. 5). Within 10 min of drug application, peroxisome movement became sluggish and long-range saltations decreased by 80% (Figs. 5 and 6). Peroxisomes started arranging themselves in large circular formations (Fig. 5A), and their movement gradually became restricted to short 1- to 4-μm oscillations (Fig. 5B). Movement became restricted earlier in Lat-B-treated cells (Fig. 6), whereas CD treatment allowed short-range movement to continue for an additional 10 to 15 min. At the end of 30 min, however, all traces of peroxisome movement were limited to Brownian oscillations, and the peroxisomes had assembled into loose globular aggregates (Fig. 5, C and D). CD-treated cells at this time displayed short F-actin filaments (Fig. 5C), whereas Lat-B-treated cells displayed a general, diffuse, green fluorescence. Removal of CD through five sequential washes over 30 min allowed a slow but complete recovery of peroxisomal movement. Nearly 30% of peroxisomes resumed saltatory motion by 5 h. Lat-B-induced inhibition of peroxisome motility was more lasting and could not be restored in more than 80% of the cells even after 12 h.
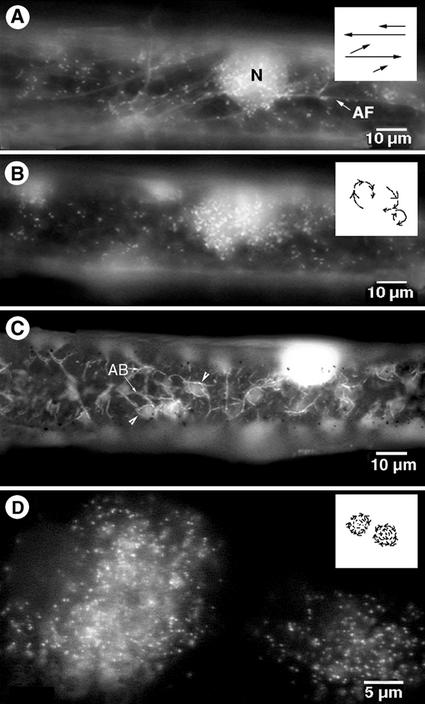
Figure 5.
Effects of actin-depolymerizing drugs cytochalasin D (CD) and latrunculin B (Lat-B) over time on peroxisome motility in onion epidermal cells. A, An onion epidermal cell transiently expressing EYFP-peroxi and GFP-mTalin 5 min after being treated with 2 μm Lat-B showed that peroxisome movement had slowed down. Although the distance covered by long-range saltations had become smaller, peroxisomes had not stopped moving totally at this stage (shown diagrammatically in box). Actin filaments (AF) and the nucleus (N) were clearly visible. B, The same onion cell as in (A) observed 15 min after Lat-B treatment showed that peroxisome movement no longer continued along straight paths and had assumed a circular track (shown diagrammatically in box). Peroxisomes now appeared to move in a slow funneling motion toward the center of the circle and resulted in increasingly compact accumulations of peroxisomes. Actin filaments were barely visible at this stage in LB-treated cells. C, An onion cell observed 30 min after treatment with 2 μm CD displayed thick, short bundles of actin (labeled AB) forming a polygonal mesh enclosing pockets of arrested peroxisomes (arrowheads). D, Enlarged view of globular aggregates of peroxisomes observed about 30 min after treatment with CD or Lat-B. The box provides a diagrammatic representation of this terminal stage in the series of events initiated in A and B.
Figure 6.
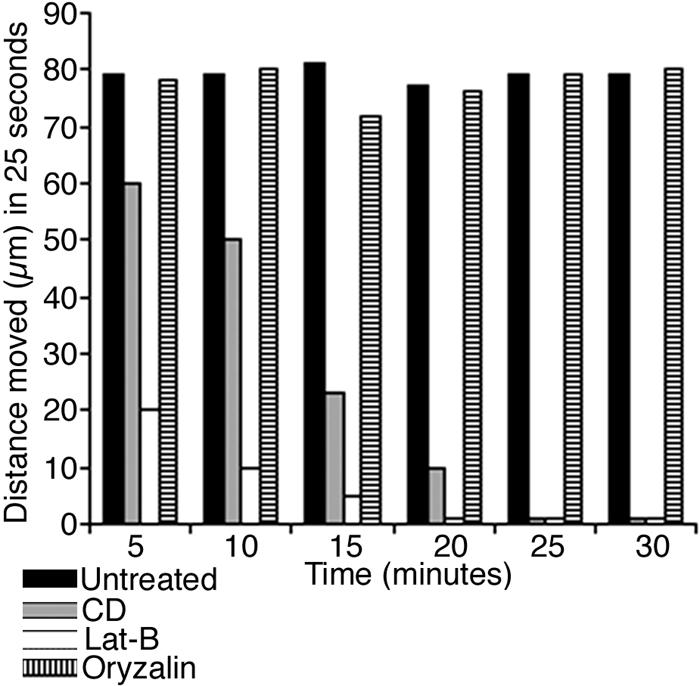
Effect of cytoskeletal drugs on peroxisome motility assessed over a total duration of 30 min. The time interval between each segment is 5 min, and the distance moved represents an average value from 10 peroxisomes obtained by tracking each peroxisome over 25 s. Peroxisome movement in both untreated and 30 μm oryzalin-treated cells remained unchanged, whereas it slowed down and finally stopped in cells treated with CD and Lat-B.
On the other hand, treatment of cells expressing EYFP-peroxi alone with the microtubule-depoly-merizing drugs oryzalin and propyzamide did not appear to affect peroxisome motility (Fig. 6). The same treatment was extended to cells co-expressing EYFP-peroxi and GFP-MBD, whereupon cortical microtubules followed by the thicker subcortical microtubule bundles broke down completely within 1 h of treatment with 30 μm oryzalin or propyzamide. Peroxisome motility remained unaffected over the next 12 h. No effect on peroxisome motility or actin filaments was observed in cells co-expressing EYFP-peroxi and GFP-mTalin treated with 30 μm oryzalin for 1 h (Fig. 7, A–D). Treatment with 5 μm of the microtubule-stabilizing drug paclitaxel similarly resulted in the formation of microtubule bundles within 180 min but did not affect peroxisomal motility (Fig. 7, E–I). However, control experiments consisting of Lat-B/CD treatment of cells co-expressing EYFP-peroxi and GFP-MBD displayed peroxisome aggregation with no apparent loss of microtubule integrity (Fig. 7J).
Figure 7.
Peroxisomal movement does not depend on the microtubular cytoskeleton in onion epidermal cells. Reference lines are drawn across the pictures. A through D, Time-lapse images of an onion cell expressing EYFP-peroxi and GFP-mTalin taken at 2-s intervals 1 h after treatment with 30 μm oryzalin. F-Actin strands (MF) with motile peroxisomes were seen. The movement of five peroxisomes was followed over 6 s and revealed that they were still moving actively. Note that peroxisomes labeled 1, 2, and 5 moved in a direction opposite to 3 and 4. N, Cell nucleus. E through I, An onion epidermal cell expressing EYFP-peroxi and GFP-MBD treated with 5 μm of the microtubule stabilizer paclitaxel for 2 h revealed thick microtubule bundles (MT) and motile peroxisomes. Tracking of individual peroxisomes (arrowheads) over 6 s showed their motility to be unaffected. Note that peroxisomes 1 and 3 (arrow) moved across nearly 10-μm areas that appeared devoid of microtubule bundles, whereas peroxisome 2 moved in an opposite direction to them. J, An onion epidermal cell expressing EYFP-peroxi and GFP-MBD treated with 2 μm of the actin drug Lat-B for 30 min showed peroxisomal aggregation (arrowheads), whereas microtubule bundles (MT) remained intact.
To check the global verity of our observations suggesting an actin-based peroxisome movement, further observations on drug effects were carried out in different cell types of transgenic Arabidopsis plants carrying the EYFP-peroxi construct.
Drug Treatments of Transgenic Arabidopsis-EYFP-Peroxi Plants
The three Arabidopsis cell types selected for distinct response to cytoskeletal drugs were trichomes, root hairs, and cortical cells of the root elongation zone. In Arabidopsis, these cells exhibit distinct morphological responses to microtubule- and micro-filament-interacting drugs (Baskin et al., 1994; Bibikova et al., 1999; Mathur et al., 1999; Baluska et al., 2001). The specific, drug-induced morphological responses of the three cell types were observed in more than 250 cells of each kind and always correlated with the respective observations on peroxisomal motility.
Peroxisomes in Trichomes before and after Drug Treatment
Wild-type Arabidopsis trichomes are large cells, having basal diameters of nearly 40 to 50 μm and extending out from the epidermis for up to 300 to 500 μm. In general, between 75 and 150 peroxisomes could be counted per trichome cell (Fig. 8A), and their movement is comprised largely of long-range saltations. Although a clear pattern of movement could not be deciphered from the movements of single peroxisomes within a trichome cell, nearly 40% of peroxisomes cycled around the cell within 3 min. The cycle could involve a straight path from the base of the trichome cell right to the tip of a branch followed by a reversal of direction and a foray into another branch. As an alternative, a peroxisome could cross over to an adjoining track in the middle of a saltation and continue in an altogether different direction. As observed in onion cells transiently expressing the EYFP-peroxi peroxisome, velocity varied greatly in different regions of the trichome cell and the saltation could be interrupted by numerous short stops and transient contacts with other peroxisomes.
Figure 8.
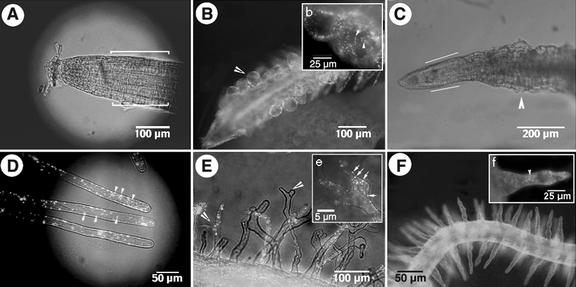
Peroxisomes in trichomes of EYFP-peroxi transgenic Arabidopsis plants before and after cytoskeletal drug treatment. A, A two branched trichome showed numerous brightly labeled peroxisomes. B, Treatment of seedlings with 2 μm of drugs interfering with actin polymerization resulted in stunted, distorted trichomes 24 to 48 h after drug treatment. C, The same trichome as in B under blue-light illumination showed that the previously motile peroxisomes (as in A) were totally arrested into distinct pockets (arrowheads). D, A 10-s time-lapse series of a trichome cell treated with 30 μm oryzalin for 48 h revealed that, although the trichome cell shape had changed (compared with A), peroxisome movement remained undisturbed. Arrowheads designate three peroxisomes that were tracked during the 10-s period. The diagonal across the pictures is a reference line.
Upon interference with the actin cytoskeleton, the unicellular, two to three branched, stellate trichomes of EYPF-peroxi transgenic Arabidopsis plants remained short but became distorted (Fig. 8B). Peroxisome distribution in the distorted trichomes was limited to small cytoplasmic pockets where peroxisomes performed minute Brownian oscillatory movements only (Fig. 8C). Treatment of EYFP-peroxi transgenic plants with microtubule drugs (30 μm oryzalin/propyzamide or 5 μm paclitaxel) for 48 h resulted in large, rounded trichome cells (Fig. 8D). In all microtubule-drug-affected trichomes, peroxisome motility remained unchanged, and peroxisomes continued longitudinal saltations and cyclic movements around the newly defined, rounded cell periphery (Fig. 8D). The observations clearly indicated that, in trichome cells, peroxisomal motility involves the actin cytoskeleton, whereas an interference with the microtubular cytoskeleton does not impede their movement.
Peroxisomes in Root Cortex and Hair Cells before and after Drug Treatments
Peroxisome movement in cortical cells of the root essentially occurred along the cell periphery. As described previously by Baskin et al. (1994), treatment with both microtubule-depolymerizing and -stabilizing drugs caused root cortical cells in the elongation zone to swell to nearly isodiametric dimensions (Fig. 9). However, peroxisome identity and movement remained unaffected in these cells (Fig. 9). Treatment with 2 μm Lat-B inhibited root elongation and resulted in small, unexpanded cells with arrested peroxisomes (Fig. 9).
Figure 9.
Peroxisomes in cortical and root hair cells of EYFP-peroxi transgenic Arabidopsis plants before and after cytoskeletal drug treatment. A, The elongation zone of the root consists of rectangular anisotropically expanding cells (between lines). B, A root treated with 30 μm oryzalin showed large, almost isodiametric bulging cells (arrowhead) in response to microtubule depolymerization. The box (b) shows an enlarged view of root cells with peroxisomes (arrowheads) clearly visible. Peroxisome identity and motility remained unchanged in such cells. C, A root treated with 2 μm Lat-B showed small unexpanded cells of the elongation zone (bordered by straight lines) with nonmotile aggregations of peroxisomes. D, A normal root hair showed numerous brightly labeled peroxisomes (arrowheads). E, Root hair treated with 30 μm oryzalin displayed branched tips (arrowheads) in response to microtubule depolymerization. Peroxisome movement remained unchanged in such cells. Box e shows a magnified view of the split hair tip with peroxisomes (arrows). F, Root hairs remained unextended and stumpy because of Lat-B-induced actin depolymerization. Box f shows a magnified view of a single root hair with arrested peroxisomes (arrowhead).
Peroxisome movement in wild-type root hairs consisted primarily of long saltations from the base of the hair to its tip with occasional short stops and transient contacts between peroxisomes (Fig. 9). The movement did not follow the reverse fountain type of cytoplasmic streaming described for Arabidopsis root hairs because peroxisomes frequently changed their tracks and direction of movement (data not shown). Treatment with both microtubule-depoly-merizing and -stabilizing drugs resulted in wavy, multibranched root hairs (Fig. 9). Peroxisome movement continued unaltered along the branched hair-cell periphery (Fig. 9). Again a treatment with the actin-depolymerizing drugs CD or Lat-B resulted in short, thick root hairs exhibiting distinct pockets of oscillating peroxisomes (Fig. 9).
Observations on different cell types in Arabidopsis and onion epidermal cells demonstrated that peroxisome movement clearly depended on the actin cytoskeleton but did not involve a passive streaming along in the general cytoplasmic flow. In an attempt to identify the driving force behind this actin cytoskeleton-dependent motion, the involvement of myosin motors was investigated.
Effect of the Myosin Inhibitors on Peroxisome Movement
Two common myosin inhibitors 2,3-butanedione monoxime (BDM) and N-ethylmaleimide (NEM) with varying levels of efficacy were used. BDM is a specific inhibitor of myosin ATPase (Herrmann et al., 1992; Samaj et al., 2000). NEM is an SH-blocking reagent that inhibits the actin-activated myosin ATPase by alkylating essential SH groups in the head region of the heavy chain of the myosin molecule but also displays nonspecific activity (Kohama et al., 1987).
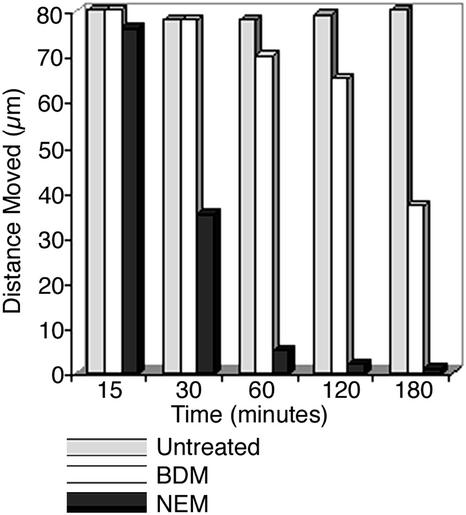
Following published literature (Nebenführ et al., 1999; Huber et al., 2000; Samaj et al., 2000), BDM was used at five different concentrations (2.5, 5, 10, 20, and 30 mm) but failed to arrest peroxisomal movement completely (Fig. 10). Peroxisomes continued moving at the same velocity as untreated controls at low concentrations (2.5–10 mm) of BDM. At the higher concentrations of 20 and 30 mm BDM, their velocity slowed down to nearly one-half over a period of 3 h (Fig. 10). Complete cessation of movement was not observed even 24 h after treatment with BDM in onion epidermal cells and seedlings of EYFP-peroxi transgenic plants.
Figure 10.
Effect of myosin inhibitors BDM (30 mm) and NEM (250 μm) on peroxisome movement over 3 h. The average distance moved by peroxisomes was obtained from 30 individual peroxisomes from five different cells, each tracked for 20 s. Whereas NEM arrested long distance movement completely within 60 min, only a slowing down of peroxisome movement was observed upon BDM treatment.
Treatment of onion epidermal cells and Arabidopsis transgenics expressing the EYFP-peroxi protein with three different concentrations (50, 100, and 250 μm) of NEM resulted in a global arrest of peroxisome movement within 180 min (Fig. 10). As with the actin-depolymerizing drugs, the peroxisomes grouped together into globular aggregates. The inhibitor was washed out over 50 min, and approximately 60% of the cells treated with 50 and 100 μm concentrations exhibited a complete recovery of peroxisome movement within 5 h. Peroxisomes in cells treated with 250 μm NEM did not regain motility even after 12 h.
The ambiguity of these results led us to perform identical experiments with transgenic plants carrying GFP targeted to mitochondria (Logan and Leaver, 2000). Mitochondria have been shown to associate with the actin cytoskeleton (Olyslaegers and Verbelen, 1998). In our drug experiments, mitochondria behaved in a manner almost identical to peroxisomes (data not shown). BDM treatment of GFP-mitochondria transgenics with 30 mm BDM slowed down but did not stop their movement, whereas 250 μm NEM arrested it completely (data not shown).
DISCUSSION
Peroxisome Labeling and Covisualization with Cytoskeletal Elements
The GFP has revolutionized live-cell imaging (Hanson and Köhler, 2001; Hawes and Satiat-Jeunemaitre, 2001). In plants, specific targeting of GFP has allowed the visualization of individual subcellular structures such as endoplasmic reticulum (Haseloff et al., 1997), Golgi stacks (Boevink et al., 1998; Nebenführ et al., 1999), mitochondria (Köhler et al., 1997b), nuclei (Grebenok et al., 1997), plastids (Köhler et al., 1997a; Sidorov et al., 1999), plasma membrane (Bischoff et al., 2000), vacuoles (Di Sansebastiano et al., 1998; Mitsuhashi et al., 2000), cell wall (Scott et al., 1999), actin microfilaments (Kost et al., 1998), and microtubules (Marc et al., 1998; Ueda et al., 1999). GFP-labeled peroxisomes have been visualized by Cutler et al. (2000) and Mano et al. (1999).
Although direct visualization of an organelle moving on actin or microtubule tracks in live plant cells has not been reported so far, to our knowledge, many of the studies mentioned above have used a combination of immunolocalization techniques and pharmacological treatments to link the respective organelle behavior with cytoskeletal components.
This study used a commercially available EYFP-peroxi construct (CLONTECH) carrying a well-characterized PTS1 at the carboxy terminus. Although this is the first time, to our knowledge, that this construct has been used in plants, the same PTS1 consisting of the SKL motif fused to GFP has been shown to label peroxisomes in animal cells (Schrader et al., 2000). Following the method of Cutler et al. (2000), we reconfirmed the targeting of the EYFP-peroxi construct to peroxisomes through immunocytochemistry. Antibodies against catalase, a peroxisomal matrix protein, and a peroxisomal membrane-bound ascorbate peroxidase faithfully localized to all GFP-labeled structures confirming them as peroxisomes. Thus, this study adds to the number of live-cell probes available for visualizing subcellular components. In addition, the covisualization of peroxisomes and cytoskeletal elements was achieved for the first time, to our knowledge, in living plant cells. Although the spectrally well-separated GFP variants CFP and YFP are the favored fluorescent proteins for two-color imaging (Tsien, 1998), this study uses spectral differentiation between two GFP variants with a high degree of spectral overlap and very close emission peaks (510 and 524 for EGFP and EYFP, respectively). The clear separation between GFP and YFP was achieved using a spectrophotometric confocal microscope (TCS-SP2, Leica) and clearly visualized the close association between peroxisomes and actin microfilaments.
Actin Microfilaments and Not Microtubules Are Involved in the Spatial Organization and Motility of Peroxisomes
Contrary to the expectations developed from reports in animal cells, the movement of organelles such as nuclei and Golgi elements in plants is actin and not microtubule based (for review, see Hawes and Satiat-Jeunemaitre, 2001). Although earlier reports in both animal and plant systems supported exclusive roles for microtubules or microfilaments as cytoskeletal tracks for organelle motility (Williamson, 1993; Cole and Lippincott-Schwartz, 1995) more recent studies indicate a cooperative scenario, wherein subcellular structures such as melanophores (Rodionov et al., 1998), mitochondria (Morris and Hollenbeck, 1995), and chloroplasts (Sato et al., 2000) may conditionally choose and switch motors and tracks between microfilaments and microtubules. Thus, a certain degree of flexibility and functional redundancy appears to be associated with subcellular organelle movement.
Studies on peroxisomal motility in animal cells have demonstrated microtubules as the preferred tracks for peroxisome movement (Rapp et al., 1996; Schrader et al., 1996; Wiemer et al., 1997). Observations leading to this conclusion in chicken hamster ovary cells (Rapp et al., 1996; Huber et al., 2000) included evidence from transmission electron microscopy, which revealed a close association between peroxisomes and microtubules. Further evidence involved a complete cessation of saltations in response to the antimicrotubule drugs nocodazole and colcemid. However, Rapp et al. (1996) conceded that the intracellular movement of peroxisomes was influenced not only by nocodazole treatment but also, to a lesser extent, by the actin polymerization inhibitor CD. Thus, saltations were still observed in CD-treated cultures, but their velocity was reduced by nearly 25%. It was unclear whether this CD-induced effect pointed to indirect or direct interaction with the actin cytoskeleton. As a consequence, Rapp et al. (1996) suggested that the actin-dependent organelle motility that seemed to be coordinated with the microtubule system possibly fulfilled a subservient role in organelle transport. In view of the more recent reports of cooperative interaction between subcellular organelles and actin and microtubule motors, one is inclined to believe that peroxisome movement in animal cells may also favor a cooperative rather than an exclusive track system.
On the contrary, our observations in plant cells indicate that actin filaments and not microtubules act as exclusive tracks for peroxisome motility. We prove this by direct observation of peroxisome movement along F-actin microfilaments and clearly not along microtubular tracks. In cortical areas, peroxisome movement sometimes appeared to follow microtubule tracks, but tracking of individual peroxisomes identified areas in their path where they departed clearly from the underlying microtubule strands. In onion cells, a considerable degree of microtubule and actin filaments run parallel to each other (J. Mathur and M. Hülskamp, unpublished data), and such closely laid tracks could easily create the illusion of microtubules being the preferred/alternative tracks for peroxisomal movement. To provide a certain degree of specificity to the kind of cytoskeletal element involved, drug treatments have frequently formed an integral part of studies relating to organelle motility (Rapp et al., 1996; Nebenführ et al., 1999; Huber et al., 2000). We applied similar parameters to seek further confirmation for our observations. Treatment of onion epidermal cells transiently co-expressing the different markers, with a variety of cytoskeletal drugs, known for their specific but differing modes of action confirmed that peroxisome movement is actin based. To verify the global veracity of results obtained with onion epidermal cells, we extended our observations to three cell types exhibiting different modes of growth in Arabidopsis plants. Thus, trichomes and root cortical cells generally elongate by a diffuse growth mechanism involving the microtubule cytoskeleton. Root hairs, on the other hand, develop by an actin cytoskeleton-based tip-growing mechanism. Because these cell types respond in characteristic ways to drugs that interact with the actin and microtubule cytoskeleton (Baskin et al., 1994; Bibikova et al., 1999; Mathur et al., 1999; Baluska et al., 2001), the success of our drug treatments was verified by an independent observation of altered cell morphology that were made before assessing the state of peroxisome motility in these cells. In each case, an interference with actin polymerization led to the slowing down and eventual arrest of peroxisome motility. On the contrary, even long-term treatments spanning up to 2 weeks with microtubule-depolymerizing/stabilizing drugs created numerous alterations in cell morphology but did not affect peroxisome movement. These observations lead us to conclude that in plant cells peroxisomes move along actin and not microtubule tracks.
Peroxisome Movement May Not Involve Myosin Motors and Be an Actin Polymerization-Based Process
In plants, inhibitors and immunological techniques have provided convincing evidence for an actin-based system of movement (Hawes and Satiat-Jeunemaitre, 2001). The involvement of myosin motors has, in most cases, been presented as a major possibility rather than a proven fact (Olyslaegers and Verbelen, 1998; Kandasamy and Meagher, 1999). The well-analyzed movement of Golgi units in plants has relied on the use of two drugs, NEM and BDM, for associating myosin motors with Golgi motility (Boevink et al., 1998; Nebenführ et al., 1999). Thus, Boevink et al. (1998) used 0.5 mm NEM and observed complete inhibition of Golgi movement within 5 min of application. Nebenführ et al. (1999) used a high concentration (30 mm) of BDM and were able to inhibit Golgi movement. Considering peroxisome movement specifically, Huber et al. (2000) demonstrated that treatment of Chinese hamster ovary cells with up to 10 mm BDM had no effect on peroxisomal movement. This particular observation was used as additional support for a role for microtubules in peroxisomal movement (Huber et al., 2000). We, therefore, treated both onion epidermal peels (transiently expressing EYFP-peroxi) and transgenic EYFP-peroxi Arabidopsis seedlings with the reported concentrations of myosin inhibitors (Boevink et al., 1998; Nebenführ et al., 1999; Huber et al., 2000).
Peroxisome movement ceased in onion epidermal cells within 30 min of 50 μm NEM application but could be restarted upon removal of the inhibitor. However, NEM is a nonspecific sulfahydryl-poisoning agent that also inhibits myosin motors along with other effects (Kohama et al., 1987). On the contrary, even 30 mm BDM, acknowledged as a more specific inhibitor of myosins (Samaj et al., 2000), failed to arrest peroxisome movement completely. Movement velocity did slow down considerably, but it is unclear whether this was because of a specific effect of BDM on myosins or an artifact resulting from its high concentration. These experiments suggested that, although peroxisome motility is actin based, it does not seem to involve myosin motors. To test whether this observation is peroxisome specific or is generally true for other organelles that display an actin-based motility, we assessed the motility of mitochondria (Olyslaegers and Verbelen, 1998). Transgenic plants carrying GFP-labeled mitochondria (Logan and Leaver, 2000) were used. Mitochondria behaved in a manner similar to peroxisomes. Their movement also was not totally arrested by BDM. One possible explanation for this unexpected finding is that the movement of these two organelles is driven by actin polymerization. Mitochondrial motility in yeast (Saccharomyces cerevisiae) has been shown to be driven by actin polymerization and involves the ARP2/3 complex (Boldogh et al., 2001). This mechanism is well studied for the motility of neutrophils (Weiner et al., 1999), endocytic vesicles (Merrifield et al., 1999), and microbial pathogens like Listeria monocytogenes and Shigella spp. (May et al., 1999; Frischknecht and Way, 2001).
Although, at this stage, it is unclear whether subcellular organelles like mitochondria and peroxisomes in plants also use a similar ARP2/3 complex-based mechanism for their movement, the following observations from our actin drug experiments are highly suggestive. The two actin drugs CD and Lat-B used by us differ in their mode of action. Lat-B associates and competes with actin monomers that are required for efficient polymerization, whereas CD affects the kinetics of actin polymerization by binding at the ends of an actin filament (Brown and Spudich, 1979; Morton et al., 2000). Because of its direct action on monomeric actin, Lat-B behaves as a more potent inhibitor compared with CD. An actin polymerization-driven process would therefore be expected to stop much earlier upon Lat-B treatment than CD treatment. Our data showing a rapid arrest of peroxisome movement upon Lat B treatment compared with CD treatment are consistent with actin polymerization-driven movement.
Thus, in addition to the direct revelations about peroxisome motility, this work also opens up the exciting possibility of investigating an actin polymerization-based mechanism for organelle motility in plants.
MATERIALS AND METHODS
Transient Transfections
Gold particles (1 μm, Bio-Rad, Hercules, CA) were coated with EYFP-peroxi DNA or its combinations (1:1 [w/v]mixture) with GFP-mTalin and GFP-MBD DNA following the manufacturer's directions. Particles were delivered into onion (Allium cepa)/Arabidopsis epidermal cells using the Biolistic PDS-1000/He system (Bio-Rad) with 1,100-pounds inch−2 rupture discs under a vacuum of 25 inches of Hg. After bombardment, tissue was maintained on moist filter paper in parafilm-sealed plastic petri dishes. Fluorescence microscopy was carried out on tissue mounted in tap water 24 to 72 h after bombardment.
Transgenic EYFP-Peroxi Plants and Culture Conditions
Flowering plants of Arabidopsis (ecotype Landsberg) were transformed with EYFP-peroxi by dipping them for approximately 20 s in a suspension of Agrobacterium tumefaciens strain GV3101-carrying p35S-EYFP-peroxi construct in the pCAMBIA 1300 vector (kindly provided by Richard Jefferson, Commonwealth Scientific and Industrial Research Organization, Canberra, Australia), in 5% (w/v) Suc solution containing 0.05% (v/v) Silwet. Seeds were harvested after 6 weeks and selected for the transgene by germination on Murashige and Skoog (1962) medium plates containing 20 mg L−1 Hygromycin-B (Boehringer Mannheim, Basel).
Immunocytochemistry
Whole-mount immunocytochemistry on onion epidermal peels and 4-d-old Arabidopsis seedlings bombarded with EYFP-peroxi construct was done following the methods described by Boudonck et al. (1998) and Cutler et al. (2000). Primary antisera raised against a peroxisomal matrix protein, catalase (Kunce et al., 1988), and cucumber (Cucumis sativus) peroxisomal ascorbate-peroxidase (Mullen and Trelease, 2000) were kindly provided by Dick Trelease (Arizona State University, Tucson). Triton X-100-permeabilized and bovine serum albumin-blocked tissue was incubated overnight in 1:50 (v/v) dilutions of antisera in phosphate-buffered saline. The primary antibody was detected using TRITC-conjugated secondary goat-anti rabbit antibodies (Sigma, St. Louis). Images were acquired sequentially using TRITC and FITC filters on a DMRE fluorescent microscope (Leica) and the DISKUS frame grabbing software (version 4.20 Technisches Buro, Konigswinter, Germany).
Treatment with Drugs Affecting the Cytoskeleton
Microtubule-depolymerizing drugs oryzalin and propyzamide (Crescent Chemical Co., Singapore), the micro-tubule-stabilizing drug paclitaxel (Molecular Probes, Eugene, OR), and actin depolymerizing drugs CD (Sigma) and Lat-B (Calbiochem, San Diego) were dissolved in ethanol/dimethyl sulfoxide according to the manufacturer's instructions and diluted directly to the required concentration in tap water (for transient assays) or Murashige and Skoog liquid medium (for treatment of transgenic Arabidopsis plants). The drug solution was directly applied to onion epidermal peels on the glass slide. Observations on epidermal peels started soon after drug application and continued over the next 72 h. For every drug treatment, 10 epidermal peels with approximately 250 cells expressing the DNA probes were analyzed. Transgenic seedlings were treated with drugs after the first leaf primordia were visible and were maintained in 5-mL plastic petri dishes. They were first immersed in the drug solution for 2 min and then placed in an upright position on a filter paper that dipped in 2 mL of drug solution. Observations on the treated plants were initiated 48 h after the initiation of the drug treatment and continued over 7 d. For each experiment, 25 plants were analyzed, and every experiment was carried out at least three times.
Live Cell Microscopy
Living plant cells were observed using an epifluorescence microscope (LEICA-DMRE) equipped with a high-resolution KY-F70 3-CCD camera (JVC, Freidberg, Germany) and the filters GFP-LP, dual band filter FITC/Texas red, and a TRITC filter. Confocal microscopy was carried out on a TCS SP2 system (Leica). A krypton-argon laser (488-, 568-, and 647-nm lines) was used to discriminate between the EGFP and EYFP fluorochromes. The bandwidth mirror settings for discriminating between the two signals were 493/518 (EGFP) and 585/612 (EYFP). The two channels were allocated false green (EGFP) and red (EYFP) colors. Image stacks were processed using Photoshop 6.0 software (Adobe Systems, Mountain View, CA).
Computerized Image Analysis
Digital images from the fluorescence microscope were captured using the DISKUS frame grabbing software. Using the sequential image grabbing function, an image was acquired every second, allowing the tracking of individual peroxisomes for different time periods. The measurement function in the DISKUS program was then used for calculating the velocity and distance covered by each peroxisome. Thus, average values were acquired for 50 peroxisomes executing different kinds of movements. Data were processed using the Excel 2001 program (Microsoft, Redmond, WA). All images were processed using Adobe Photoshop 5.5 software. Quick-time movies of peroxisome movement were generated from sequential images acquired at 1-s intervals and threaded together into a movie sequence using QuickTime 5.0 program.
Supplementary Material
Time-lapse movies of peroxisome movement in living plant cells can be viewed at http://www.plantphysiol.org.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bhylahalli Purushottam Srinivas (University of Köln, Germany) for his critical comments on the manuscript, Anshudeep Mathur (University of Köln) for help with the computer work, Prof. Wolfgang Werr (Institute for Developmental Biology, Cologne, Germany) for permission to use the Biolistic apparatus, Dorothee Schroth (Leica Microsystems Vertrieb GmBH) for help with confocal microscopy, Prof. Dick Trelease (Arizona State University, Tucson) for the antibodies, Prof. Richard Cyr (Pennsylvania State University, University Park) for the GFP-MBD construct, and Prof. Nam Hai Chua (Rockefeller University, New York) for the GFP-mTalin construct.
Footnotes
This work was supported by a Volkswagen Stiftung grant (to M.H.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011018.
LITERATURE CITED
- Baker A. Biogenesis of plant peroxisomes. In: Smallwood M, Knox JP, Bowles DJ, editors. Membranes: Specialized Functions in Plants. Oxford, UK: BioScientific; 1996. pp. 421–440. [Google Scholar]
- Baluska F, Jasik J, Edelmann HG, Salajova T, Volkmann D. Latrunculin B induced plant dwarfism: Plant cell elongation is F-actin dependent. Dev Biol. 2001;231:113–124. doi: 10.1006/dbio.2000.0115. [DOI] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 1994;35:935–942. [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulates tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Bischoff F, Vahlkamp L, Molendijk A, Palme K. Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGD11 from Arabidopsis. Plant Mol Biol. 2000;42:515–530. doi: 10.1023/a:1006341210147. [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa-Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Yang HC, Nowakowski WD, Karmon SL, Hays LG, Yates JR, III, Pon LA. ARP2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc Natl Acad Sci USA. 2001;98:3162–3167. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters. J Cell Sci. 1998;111:3687–3694. doi: 10.1242/jcs.111.24.3687. [DOI] [PubMed] [Google Scholar]
- Brown S, Spudich JA. Cytochalasin inhibits the rate of elongation of actin filament fragments. J Cell Biol. 1979;83:657–662. doi: 10.1083/jcb.83.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Rio LA. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 2001;6:145–150. doi: 10.1016/s1360-1385(01)01898-2. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Erhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of sub-cellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano GP, Paris N, Marc-Martin S, Neuhaus JM. Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 1998;15:449–457. doi: 10.1046/j.1365-313x.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- Flynn CR, Mullen RT, Trelease RN. Mutational analysis of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import in tobacco BY2 glyoxysomes. Plant J. 1998;16:709–720. doi: 10.1046/j.1365-313x.1998.00344.x. [DOI] [PubMed] [Google Scholar]
- Frischknecht F, Way M. Surfing pathogens and the lessons learned for actin polymerization. Trends Cell Biol. 2001;11:30–38. doi: 10.1016/s0962-8924(00)01871-7. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok RJ, Lambert GM, Galbraith DW. Characterization of the targeted nuclear accumulation of GFP within the cells of transgenic plants. Plant J. 1997;12:685–696. [Google Scholar]
- Hanson MR, Köhler RH. GFP imaging: methodology and application to investigate cellular compartmentation in plants. J Exp Bot. 2001;52:529–539. [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes CR, Satiat-Jeunemaitre B. Trekking along the cyto-skeleton. Plant Physiol. 2001;125:119–122. doi: 10.1104/pp.125.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, Wray J, Travers F, Barman T. Effect of 2,3-butanedione monoxime on myosin and myofibrillar ATPases: an example of an uncompetitive inhibitor. Biochemistry. 1992;31:12227–12232. doi: 10.1021/bi00163a036. [DOI] [PubMed] [Google Scholar]
- Huber CM, Saffrich R, Gorgas K, Just WW. Organelle motility regulated by the cell's environment: dissection of signalling pathways regulating movements of peroxisomes. Protoplasma. 2000;213:18–27. [Google Scholar]
- Kandasamy MK, Meagher RB. Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton. 1999;44:110–118. doi: 10.1002/(SICI)1097-0169(199910)44:2<110::AID-CM3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kohama K, Kohama T, Kendrick-Jones J. Effect of N-ethylmaleimide on Ca-inhibition of Physarum myosin. J Biochem. 1987;102:17–25. doi: 10.1093/oxfordjournals.jbchem.a122029. [DOI] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. Exchange of protein molecules through connections between higher plant plastids. Science. 1997a;276:2039–2042. doi: 10.1126/science.276.5321.2039. [DOI] [PubMed] [Google Scholar]
- Köhler RH, Zipfel WR, Webb WW, Hanson MR. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 1997b;11:613–621. doi: 10.1046/j.1365-313x.1997.11030613.x. [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kunce CM, Trelease RN, Turley RB. Purification and biosynthesis of cottonseed (Gossypium hirusutum L.) catalase. Biochem J. 1988;251:147–155. doi: 10.1042/bj2510147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ. Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot. 2000;51:865–871. [PubMed] [Google Scholar]
- Mano S, Hayashi M, Nishimura M. Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 1999;17:309–320. doi: 10.1046/j.1365-313x.1999.00378.x. [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell. 1998;10:1927–1939. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua NH. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell. 2000;12:465–477. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua NH. The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development. 1999;126:5559–5568. doi: 10.1242/dev.126.24.5559. [DOI] [PubMed] [Google Scholar]
- May RC, Hall ME, Higgs HN, Polard TD, Chakraborty T, Wehland J, Machesky LM, Sechi AS. The ARP2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Moss SE, Ballestrem C, Imhof BA, Giese G, Wunderlich I, Almer W. Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat Cell Biol. 1999;1:72–74. doi: 10.1038/9048. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N, Shimada T, Mano S, Nishimura M, Hara-Nishimura I. Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 2000;41:993–1001. doi: 10.1093/pcp/pcd040. [DOI] [PubMed] [Google Scholar]
- Miura S, Kasuya-Arai I, Mori H, Miyazawa S, Osumi T, Hashimoto T, Fujiki Y. Carboxy terminal consensus ser-lys-leu related tripeptide of peroxisomal proteins functions in vitro as a minimal peroxisome-targeting signal. J Biol Chem. 1992;267:14405–14411. [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerisation. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Flynn CR, Trelease RN. Diverse amino acid residues function within the type I peroxisomal targeting signal. Plant Physiol. 1997;115:881–889. doi: 10.1104/pp.115.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN. The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J Biol Chem. 2000;275:16337–16344. doi: 10.1074/jbc.M001266200. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–797. [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehlin LA. Stop and go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 1999;121:1127–1141. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR, McIntosh RJ, Olmsted JB. Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olyslaegers G, Verbelen JP. Improved staining of F-actin and co-localization of mitochondria in plant cells. J Microsc. 1998;192:73–77. [Google Scholar]
- Rapp S, Saffrich R, Anton M, Jäkle U, Ansorge W, Gorgas K, Just WW. Microtubule-based peroxisomal movement. J Cell Sci. 1996;109:837–849. doi: 10.1242/jcs.109.4.837. [DOI] [PubMed] [Google Scholar]
- Rodionov VI, Hope AJ, Svitkina TM, Borisy GG. Functional coordination of microtubule-based and actin-based motility in melanophores. Curr Biol. 1998;8:165–168. doi: 10.1016/s0960-9822(98)70064-8. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Gelfand VI. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol. 1998;8:161–164. doi: 10.1016/s0960-9822(98)70063-6. [DOI] [PubMed] [Google Scholar]
- Samaj J, Peters M, Volkmann D, Baluska F. Effects of myosin ATPase inhibitor 2,3-butanedione 2-monoxime on distribution of myosins, F-actin, microtubules, and cortical endoplasmic reticulum in maize root apices. Plant Cell Physiol. 2000;41:571–582. doi: 10.1093/pcp/41.5.571. [DOI] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A. Choice of tracks, microtubules and/or actin filaments for chloroplast photo movement is differentially controlled by phytochrome and a blue light receptor. J Cell Sci. 2000;114:269–279. doi: 10.1242/jcs.114.2.269. [DOI] [PubMed] [Google Scholar]
- Schrader M, Burkhardt JK, Baumgart E, Lüers G, Spring H, Völkl A, Fahimi HD. Interaction of microtubules with peroxisomes: tubular and spherical peroxisomes in HepG2 cells and their alterations induced by microtubule-active drugs. Eur J Cell Biol. 1996;69:24–35. [PubMed] [Google Scholar]
- Schrader M, King SJ, Stroh TA, Schroer TA. Real time imaging reveals a peroxisomal reticulum in living cells. J Cell Sci. 2000;113:3663–3671. doi: 10.1242/jcs.113.20.3663. [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques. 1999;26:1125–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- Sidorov V, Kasten D, Pang SZ, Hajdukiewicz TJ, Staub JM, Nehra NS. Stable chloroplast transformation in potato, use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Tabak HF, Braakman I, Distel B. Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol. 1999;9:447–453. doi: 10.1016/s0962-8924(99)01650-5. [DOI] [PubMed] [Google Scholar]
- Topfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH. A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 1987;15:5890–5893. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease RN, Lee MS, Banjoko A, Bunkelmann J. C-terminal polypeptides are necessary and sufficient for in-vivo targeting of transiently-expressed proteins to peroxisomes in suspension-cultured cells. Protoplasma. 1996;195:156–167. [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma. 1999;206:201–206. [Google Scholar]
- Volokita M. The carboxy-terminal end of glycolate oxidase directs a foreign protein into tobacco leaf peroxisomes. Plant J. 1991;1:361–366. doi: 10.1046/j.1365-313x.1991.t01-4-00999.x. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR. Spatial control of actin polymerisation during neutrophil chemotaxis. Nat Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Wenzel T, Deerinck TJ, Ellisman MH, Subramani S. Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J Cell Biol. 1997;136:71–80. doi: 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RE. Organelle movements. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:181–202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.