Abstract
Replicative senescence is the state of irreversible proliferative arrest that occurs as a concomitant of progressive telomere shortening. By using cDNA microarrays and the gabriel system of computer programs to apply domain-specific and procedural knowledge for data analysis, we investigated global changes in gene transcription occurring during replicative senescence in human fibroblasts and mammary epithelial cells (HMECs). Here we report the identification of transcriptional “fingerprints” unique to senescence, the finding that gene expression perturbations during senescence differ greatly in fibroblasts and HMECs, and the discovery that despite the disparate nature of the chromosomal loci affected by senescence in fibroblasts and HMECs, the up-regulated loci in both types of cells show physical clustering. This clustering, which contrasts with the random distribution of genes down-regulated during senescence or up-regulated during reversible proliferative arrest (i.e., quiescence), supports the view that replicative senescence is associated with alteration of chromatin structure.
Keywords: telomere‖chromosomal clustering‖chromatin‖microarray‖replicative arrest
Normal somatic cells have a limited capacity for division in culture and eventually enter a state of irreversible proliferative arrest termed replicative senescence (1). The irreversibility of replicative senescence contrasts with the transient and reversible nature of physiologically induced arrest (i.e., quiescence). It has been suggested that the limited replicative potential of normal cells sets a barrier against the accumulation of the multiple mutations required for neoplastic transformation, and consequently, that senescence is a tumor suppression mechanism (2, 3). It also has been suggested that the loss of proliferative capacity during replicative senescence may represent aging at the cellular level (4, 5).
The occurrence of replicative senescence is determined by the number of times that a cell population divides, suggesting that a mitotic clock records cell divisions (6). In normal human somatic cells, which lack the activity of the telomere-lengthening enzyme telomerase (7, 8), telomere shortening may contribute to this clock (9). Progressive shortening of telomeres during each cell division has been proposed to eventually truncate telomeres to a critical length or to create an altered telomere state (9–11). It has been hypothesized that such shortened or altered telomeres lose protection by telomere-binding “capping” proteins and consequently signal activation of the events leading to senescence (10–12).
In human fibroblasts, replicative senescence of the population occurs after a finite mean number of cell divisions in culture, and expression of the telomerase catalytic subunit (hTERT) abrogates this arrest (13). Human mammary epithelial cells (HMECs) can experience two modes of proliferative arrest: M0 and M1 (14, 15). M0, which occurs after a few divisions of HMECs cultured under ordinary conditions, is not associated with telomere shortening (16) and not reversed by adventitious expression of hTERT (17). A small subset of M0 cells (10−4 to 10−5) inactivates p16INK4A in a process termed “selection” (18, 19) and continues to proliferate, experiencing progressive telomere shortening (16) and eventually entering a permanent state of proliferative arrest termed M1. M1, also termed “agonescence” (16), represents telomere-dependent senescence in HMECs and is abrogated by production of telomerase (17). When feeder layers are used to culture HMECs, M0 and p16INK4A inactivation do not occur, and proliferation of the population continues until telomere shortening leads to M1 (20, 21).
There is evidence that replicative senescence is a genetically dominant trait (22, 23), and studies using subtraction hybridization, differential display, or DNA microarrays have identified genes whose expression is altered in senescent cells (24–26). However, evidence that such alterations in gene expression are unique to senescence rather than being related nonspecifically to the cessation of cell proliferation, or even are a general feature of senescent cells, has been lacking. β-Galactosidase activity at pH 6 has been used as a marker for senescence (27), but it is induced also by a variety of other stress-promoting events (28) and its specificity has been questioned (20, 29).
We wished to determine whether telomere-dependent senescence is associated with specific transcriptional changes that globally can be used as a senescence “fingerprint,” to identify functions and locations of genes whose expression is altered during senescence, and to learn whether different cell types undergoing telomere-dependent arrest of proliferation show similar or disparate transcriptional changes. By using cDNA microarrays to concurrently compare the expression of 31,000 genes in senescent, quiescent, and early passage proliferating cells, we identified distinct transcriptional perturbations that characterize telomere-dependent irreversible proliferative arrest in human fibroblasts and HMECs. Remarkably, we found that while genes showing altered expression during replicative senescence differ dramatically in these two types of cells, up-regulated senescence-specific genes were chromosomally clustered in both types of cells.
Materials and Methods
Cell Cultures.
Human primary fibroblast cell lines, WS1 (from fetal skin), WI38 (from fetal lung), and BJ (from newborn foreskin), were purchased from American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Invitrogen). Two postselection HMEC lines, 48R and 184 (both from normal female breasts), were gifts from P. Yaswen and M. Stampfer (Lawrence Berkeley National Laboratory, Berkeley, CA) and were maintained in mammary epithelial cell growth medium (BioWhittaker) buffered with Hepes (Sigma) to pH 7.4. All cells were cultured at 37°C with 5% CO2. Contact inhibition was used to transiently arrest proliferation of early passage cells; briefly, cells were plated at a near-confluent density, cultured with daily medium change for 3 days, and harvested after 3 additional days.
cDNA Microarray Experiments.
cDNA microarrays on glass slides were made essentially as described (30). Detailed protocols are available at http://cmgm.stanford.edu/pbrown/array.html. We used microarrays containing 31,104 sequence-verified IMAGE clones corresponding to about 11,000 named genes and 20,000 unknown ESTs, ≈2,000 of which are highly homologous to known genes.
Poly(A)-RNA was isolated from cell cultures by using the FastTrack 2.0 kit (Invitrogen). Two-microgram aliquots of poly(A)-RNA were labeled by reverse transcription with Superscript II enzyme (Invitrogen) and oligo(dT)18 primer (New England Biolabs) in the presence of Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia). Slides were hybridized for about 16 h in a 65°C water bath and scanned at 10-μm resolution with a GENEPIX 4000B scanner (Axon Instruments, Union City, CA).
Data Analysis.
Microarray images were analyzed using GENEPIX PRO 3.0 software (Axon Instruments). The image files, GENEPIX settings, and GENEPIX data files were uploaded into and are available on the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD). Arrays were normalized by using a computer-based procedure that identified spots having >65% of pixels showing intensities at least 1.5 times greater than background for both the Cy3 (G) and Cy5 (R) channels. R/G ratios were calculated for these spots after subtraction of background. The mean log2(R/G) for the spots was set equal to zero, and this value was applied to the Cy5 channel to obtain its normalized intensity. Data filters excluded spots flagged by GENEPIX software as unsuitable for analysis and then selected spots showing net intensities in either channel that were at least twice the corresponding background intensity. Only spots that passed data filters in 75% or more of the arrays were used for analysis.
Data were analyzed by gabriel (http://gabriel.stanford.edu) (31). Pearson correlation coefficients were calculated between pairs of microarrays using all spots that satisfied filter parameters. A modified t-score (32) was used to determine the significance of transcriptional alterations. We permuted data from all spots randomly 5,000 times to determine the rate at which random data exceeded modified t-score thresholds and defined this value as the false positive rate (FPR). To estimate the false negative rate (FNR), we randomly selected 30 genes whose expression exceeded the modified t-score thresholds and added log2(R/G) ratios randomly selected from spots that passed data filters (i.e., “noise”); FNR was defined by how frequently the average expression level of these proband genes became negative after noise was added. Calculated FPR and FNR values assume that none of the changes in randomly selected data have biological significance. Additionally, as the selection of t-score thresholds that produced nonoverlapping gene groupings constrained the independence of FPRs and FNRs, the calculated FPR and FNR values for some gene groupings are overestimates.
Genes were classified according to their annotated role in biological processes or their cellular locations retrieved from SOURCE (http://source.stanford.edu). Enrichment of a particular class of genes was determined by comparison with randomly selected gene lists of the same size. For analysis of chromosomal clustering, we retrieved the chromosomal locations for all IMAGE clones from the human genome working draft (April 2002; http://genome.ucsc.edu) using their accession numbers. The number of chromosomal clusters and the number of genes in these clusters then were compared with the average number of clusters or clustered genes from randomly sampled lists of the same size.
Results
Gene Expression Fingerprints of Senescence in Human Fibroblasts.
Senescent populations of the normal human fibroblast cell lines WS1, WI38, and BJ were obtained by serial passage until the cell population doubling time was greater than 2 weeks. Flow cytometry analysis indicated arrest mainly in G1/G0, with a small fraction of cells showing arrest in G2/M. Virtually all cells in senescent fibroblast populations stained positive for β-galactosidase activity at pH 6, as assessed microscopically. Transcript abundance in each senescent population was compared with abundance in isogenic early passage proliferating cells (24–32 population doublings) and also with abundance in early passage cells induced by contact inhibition to enter reversible arrest in G1/G0. Perturbations associated specifically with senescence were identified as described in Materials and Methods.
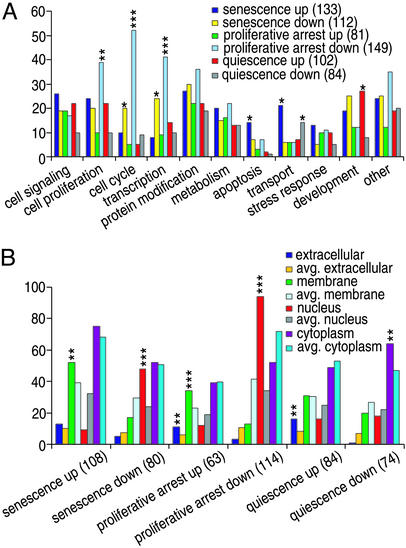
Initial examination of gene expression showed high Pearson correlation coefficients (0.6–0.8) for the three senescent human fibroblast cell lines we studied, and consequently, our analysis treated results from these lines as biological repeats. Changes in transcript abundance occurring during senescence in human fibroblasts, as compared with early passage proliferating cells or quiescent cells, were designated as senescence-specific (P < 0.001). The 376 up-regulated genes and 313 down-regulated genes (Table 1; annotated gene lists are shown in Tables 5 and 6, which are published as supporting information on the PNAS web site, www.pnas.org) define a fingerprint globally characteristic of replicative senescence in human fibroblasts. As seen in Fig. 1, genes encoding proteins having particular functions or cellular locations (function-related gene ontology annotations; http://www.geneontology.org) are represented differentially and disproportionately in the two groupings. For example, 48.1% of annotated senescence-specific up-regulated genes have been designated as membrane-associated proteins (P = 0.005), 10.5% relate to apoptosis (P = 0.017), and 15.8% to transport (P = 0.02). The proteins encoded by 48 of the 80 annotated genes down-regulated specifically during senescence (60%) are designated as being nuclear (P < 0.001), 17.9% (P = 0.02) are involved in cell cycle regulation, and 21.4% (P = 0.036) are involved in transcription.
Table 1.
Genes whose expression was altered during replicative senescence or quiescence in human fibroblasts and HMECs
| Source and phenotype | No. of genes | No. of IMAGE | FPR | FNR |
|---|---|---|---|---|
| Fibroblasts | ||||
| Up-regulation | ||||
| Senescence-specific | 376 | 445 | <0.001 | 0.358 |
| Proliferative-arrest | 246 | 272 | 0.004 | 0.481 |
| Quiescence-specific | 282 | 315 | 0.008 | 0.35 |
| Down-regulation | ||||
| Senescence-specific | 313 | 366 | <0.001 | 0.412 |
| Proliferative-arrest | 365 | 429 | 0.002 | 0.373 |
| Quiescence-specific | 250 | 285 | 0.014 | 0.495 |
| HMECs | ||||
| Up-regulation | ||||
| Senescence-specific | 16 | 17 | <0.001 | 0.624 |
| in 48R | 299 | 349 | 0.006 | 0.235 |
| in 184 | 159 | 183 | 0.008 | 0.425 |
| Proliferative-arrest | 26 | 27 | 0.011 | 0.36 |
| Quiescence-specific | 426 | 486 | 0.015 | 0.265 |
| Down-regulation | ||||
| Senescence-specific | 25 | 31 | <0.001 | 0.63 |
| in 48R | 349 | 406 | 0.014 | 0.384 |
| in 184 | 218 | 249 | 0.014 | 0.235 |
| Proliferative-arrest | 65 | 68 | 0.0096 | 0.501 |
| Quiescence-specific | 377 | 439 | 0.008 | 0.188 |
Gene numbers designate Unigenes whose altered expression was determined as described in Materials and Methods. FPR and FNR were calculated as similarly described.
Figure 1.
Genes showing altered transcription in human fibroblasts during senescence or quiescence. Biological processes (A) and cellular locations (B) associated with products of genes bearing gene ontology annotations are shown. The number of genes in each group is given in parentheses. The likelihood of enrichment of genes in a group due to random events (P value) is indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Certain transcripts differed in abundance in quiescent fibroblasts relative to senescent and proliferating cells (Table 1; annotated gene lists are shown in Tables 7 and 8, which are published as supporting information on the PNAS web site), consistent with evidence that senescence and quiescence are initiated by different biological events (33). However, certain transcripts were affected in common by these two types of proliferative arrest and were termed proliferative-arrest-associated (Table 1; annotated gene lists are shown in Tables 9 and 10, which are published as supporting information on the PNAS web site). As had been observed for senescence-specific transcripts, certain functions and cellular locations were represented disproportionately in proliferative-arrest-associated up- and down-regulated gene groupings (Fig. 1).
Clustering of Chromosomal Loci Specifically Up-Regulated During Senescence.
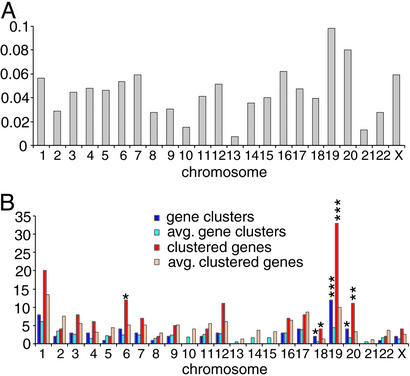
During our analysis of senescence-specific gene expression in human fibroblasts, we observed that the incidence of genes up-regulated during senescence varied substantially on different chromosomes and was independent of chromosome length (Fig. 2A). To investigate a possible relationship between senescence-specific gene expression and gene location, chromosomal positions for IMAGE clones were retrieved from the human genome working draft (April 2002; http://genome.ucsc.edu); IMAGE clones corresponding to the same Unigene cluster ID were considered as the same gene. A chromosomal gene cluster was defined as two or more genes (minimal number indicated by n) located within a specified linear distance (d). The incidence of clusters and clustered genes was compared with values generated from randomly sampled lists of the same size using a range of values for n and d and the procedures described in Materials and Methods.
Figure 2.
Chromosomal clustering of fibroblast genes up-regulated specifically during senescence. (A) Normalized numbers of genes having expression that was up-regulated specifically during senescence in human fibroblasts on each chromosome were obtained by dividing up-regulated genes by total genes on that chromosome having measurements that satisfied filter parameters (Materials and Methods). (B) Numbers for chromosomal gene clusters and clustered genes on each chromosome were compared with the average values estimated from randomly selected lists containing the same number of genes. P value limits are indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
By using 1 Mb as the distance defining cluster boundaries (i.e., d; Table 2), clustering of 150 of the 376 genes up-regulated specifically during senescence in human fibroblasts was observed; the incidence of clustering in this gene group was ≈30% higher than the average incidence of clustering among 376 randomly selected genes (P = 0.002) analyzed by using the same parameters (Table 2). A comparable number of genes down-regulated during senescence showed no evidence of clustering (Tables 2 and 3). The 150 clustered up-regulated genes were present in a total of 60 groupings of 2 to 6 genes. Five clusters containing a total of 14 genes included members of annotated gene families. Clustering of up-regulated senescence-specific genes also was observed using d values down to 1 Kb or n values of 2 to 4 to define a cluster (Tables 2 and 3). As chromosomal locations of IMAGE clones represent segments of full-length genes, calculated intergeneic distances are overestimates of actual distances; consequently, clustering frequencies of up-regulated senescence-specific genes are likely to be underestimates.
Table 2.
Effect of boundary distance (d) on the determination of chromosomal clustering for genes up-regulated (376 genes) or down-regulated (313 genes) in human fibroblasts specifically during senescence
| d | No. of clusters
|
P value | No. of clustered genes
|
P value | ||
|---|---|---|---|---|---|---|
| OBS | RE | OBS | RE | |||
| Up-regulation | ||||||
| 1 kb | 1 | 0.17 | 0.016 | 2 | 0.34 | 0.016 |
| 2 kb | 2 | 0.43 | 0.006 | 4 | 0.85 | 0.006 |
| 5 kb | 6 | 1.11 | <0.001 | 12 | 2.23 | <0.001 |
| 10 kb | 8 | 1.80 | <0.001 | 16 | 3.60 | <0.001 |
| 20 kb | 12 | 2.93 | <0.001 | 24 | 5.88 | <0.001 |
| 50 kb | 21 | 6.05 | <0.001 | 43 | 12.2 | <0.001 |
| 100 kb | 28 | 10.5 | <0.001 | 59 | 21.3 | <0.001 |
| 200 kb | 36 | 17.8 | <0.001 | 78 | 36.3 | <0.001 |
| 500 kb | 48 | 34.5 | 0.001 | 110 | 72.3 | <0.001 |
| 1 Mb | 60 | 52.9 | 0.082 | 150 | 114.7 | 0.002 |
| 2 Mb | 76 | 74.1 | 0.33 | 196 | 169.6 | <0.001 |
| 5 Mb | 88 | 96.2 | 0.944 | 258 | 249.8 | 0.179 |
| Down-regulation | ||||||
| 1 kb | 0 | 0.10 | 0.097 | 0 | 0.20 | 0.097 |
| 2 kb | 2 | 0.26 | 0.003 | 4 | 0.52 | 0.003 |
| 5 kb | 4 | 0.78 | 0.001 | 8 | 1.57 | 0.001 |
| 10 kb | 5 | 1.28 | 0.002 | 10 | 2.56 | 0.002 |
| 20 kb | 6 | 2.08 | 0.003 | 12 | 4.16 | 0.003 |
| 50 kb | 6 | 4.34 | 0.146 | 12 | 8.73 | 0.155 |
| 100 kb | 7 | 7.25 | 0.447 | 14 | 14.6 | 0.468 |
| 200 kb | 11 | 12.6 | 0.624 | 22 | 25.6 | 0.651 |
| 500 kb | 22 | 24.8 | 0.698 | 46 | 51.5 | 0.714 |
| 1 Mb | 37 | 38.9 | 0.626 | 77 | 83.1 | 0.715 |
| 2 Mb | 56 | 56.0 | 0.471 | 120 | 125.6 | 0.7 |
| 5 Mb | 75 | 77.0 | 0.629 | 184 | 192.3 | 0.813 |
A chromosomal cluster was defined as two or more genes located within the specified distance. Observed (OBS) values indicate the number of chromosomal clusters and clustered genes identified using the parameters shown. Randomly expected (RE) values indicate the average number of false-positive clusters or clustered genes estimated by the bootstrapping algorithm described in Materials and Methods. P values indicate the probability that the difference between the observed and calculated incidence of clusters or clustered genes is due to random chance.
Table 3.
Effect of variation in minimal number of genes (n) used to define a chromosomal cluster of genes whose expression was altered during senescence in human fibroblasts
| d | No. of clusters (n ≥ 3)
|
P value | No. of clusters (n ≥ 4)
|
P value | ||
|---|---|---|---|---|---|---|
| OBS | RE | OBS | RE | |||
| Up-regulation | ||||||
| 50 kb | 1 | 0.08 | 0.001 | |||
| 100 kb | 3 | 0.23 | <0.001 | |||
| 200 kb | 5 | 0.73 | <0.001 | 1 | 0.03 | <0.001 |
| 500 kb | 9 | 3.00 | <0.001 | 3 | 0.23 | <0.001 |
| 1 Mb | 19 | 7.66 | <0.001 | 7 | 1.00 | <0.001 |
| 2 Mb | 24 | 17.1 | 0.01 | 9 | 3.56 | 0.001 |
| 5 Mb | 41 | 37.3 | 0.136 | 15 | 13.5 | 0.234 |
| Down-regulation | ||||||
| 500 kb | 1 | 1.78 | 0.538 | 1 | 0.13 | 0.01 |
| 1 Mb | 2 | 4.65 | 0.87 | 1 | 0.49 | 0.078 |
| 2 Mb | 6 | 11.2 | 0.959 | 2 | 2.03 | 0.312 |
| 5 Mb | 27 | 26.5 | 0.386 | 7 | 8.40 | 0.622 |
The number of genes required to constitute a chromosomal cluster is indicated. Parameters are as described for Table 2.
As shown in Fig. 2B, a disproportionately high number of clustered up-regulated senescence-specific genes were located on chromosomes 19 (P < 0.001), 20 (P < 0.01), 18, and 6 (P < 0.05). This disproportionality was independent of the density of expressed and/or ascertained genes on these chromosomes. For example, chromosomes 19 and 17 contained a similar number of genes (407 and 420, respectively) giving valid expression measurements as defined in Materials and Methods, but twice the number of genes on chromosome 19 showed up-regulation during senescence (40 vs. 20); 82.5% of these genes were located within 1 Mb of another up-regulated gene and were distributed in 12 chromosomal clusters. We also observed clustering of genes whose transcripts were up-regulated in both senescence and quiescence, but not for genes whose up-regulation was restricted to quiescence (shown in Table 11, which is published as supporting information on the PNAS web site). Increasing the number of genes required to define a cluster did not change the clustering status of these groups (shown in Table 12, which is published as supporting information on the PNAS web site). Interestingly, whereas no clustering of genes down-regulated during senescence in fibroblasts had been observed, down-regulated quiescence-specific genes showed nonrandom chromosomal distribution (shown in Table 11).
Disparate Gene Expression Profiles in Senescent HMECs.
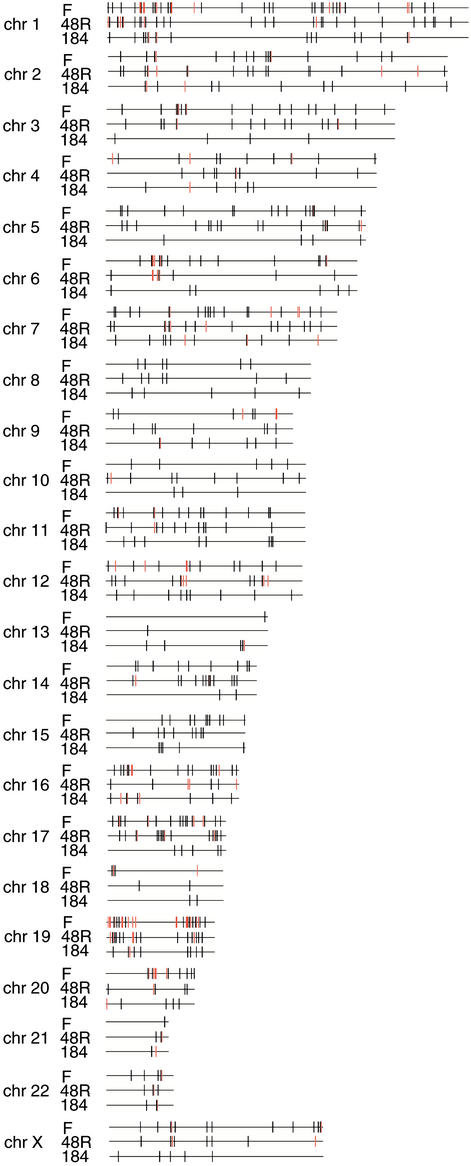
Senescence in postselection HMECs (i.e., M1), like senescence in human fibroblasts, is accompanied by telomere shortening (16) and is reversed by ectopic expression of telomerase (17). We isolated senescent populations of HMEC lines 48R and 184 as described for fibroblasts; these stained positive for β-galactosidase activity at pH 6 and were found by flow cytometry to be arrested in either G1/G0 or G2/M. To learn whether HMECs show senescence-specific transcriptional fingerprints similar to those of fibroblasts and/or gene clustering, we compared transcript abundance in senescent HMECs with abundance in isogenic proliferating early passage HMECs (passage 7–10) and HMECs subjected to contact inhibition, which like M1 leads to HMEC growth arrest in G1/G0 or G2/M. In contrast to the expression perturbations and clustering patterns observed in common among the three senescent human fibroblast cell lines we studied, relatively few genes showed expression changes common to both HMEC lines (overall Pearson correlation coefficient ≈0.3) (Table 1 and Tables 13 and 14, which are published as supporting information on the PNAS web site). Genes affected in individual HMEC lines during senescence are listed in Tables 15–18, which are published as supporting information on the PNAS web site. Moreover, gene expression changes in senescent HMECs were dramatically different from those in senescent fibroblasts (Fig. 3; also shown in Tables 5, 6, and 13–18), despite the common role of telomere shortening in precipitating replicative arrest in these cell types. Only five genes up-regulated and seven genes down-regulated in both HMEC cell lines during senescence showed similar regulation in senescent fibroblasts. However, notwithstanding the distinctly different identities of genes up-regulated in the two senescent HMEC lines, the up-regulated genes showed chromosomal clustering in both (Fig. 3 and Table 4). As had been observed for genes down-regulated during senescence in fibroblasts, genes down-regulated in each HMEC lines during senescence showed no physical association beyond random chance (shown in Table 19, which is published as supporting information on the PNAS web site).
Figure 3.
Chromosomal distribution of genes up-regulated in common during replicative senescence in fibroblasts (F) or two HMEC lines. Black lines indicate the chromosomal positions of up-regulated genes, and red lines designate gene clusters.
Table 4.
Chromosomal clustering analysis of genes whose expression was altered specifically during senescence (M1) in two individual HMEC lines (48R and 184)
| d | No. of clusters
|
P value | No. of clustered genes
|
P value | ||
|---|---|---|---|---|---|---|
| OBS | RE | OBS | RE | |||
| Up-regulation in 48R | ||||||
| 1 kb | 2 | 0.11 | <0.001 | 4 | 0.21 | <0.001 |
| 2 kb | 5 | 0.27 | <0.001 | 10 | 0.55 | <0.001 |
| 5 kb | 8 | 0.68 | <0.001 | 16 | 1.36 | <0.001 |
| 10 kb | 12 | 1.15 | <0.001 | 24 | 2.31 | <0.001 |
| 20 kb | 14 | 1.90 | <0.001 | 29 | 3.81 | <0.001 |
| 50 kb | 16 | 3.75 | <0.001 | 34 | 7.53 | <0.001 |
| 100 kb | 21 | 6.64 | <0.001 | 44 | 13.4 | <0.001 |
| 200 kb | 24 | 11.2 | <0.001 | 53 | 22.8 | <0.001 |
| 500 kb | 33 | 22.6 | 0.004 | 72 | 46.9 | <0.001 |
| 1 Mb | 45 | 35.6 | 0.016 | 100 | 75.6 | 0.002 |
| 2 Mb | 59 | 52.1 | 0.054 | 144 | 115.7 | 0.001 |
| 5 Mb | 76 | 73.0 | 0.192 | 209 | 179.6 | <0.001 |
| Up-regulation in 184 | ||||||
| 1 kb | 2 | 0.05 | <0.001 | 4 | 0.09 | <0.001 |
| 2 kb | 2 | 0.08 | <0.001 | 4 | 0.15 | <0.001 |
| 5 kb | 4 | 0.21 | <0.001 | 8 | 0.42 | <0.001 |
| 10 kb | 5 | 0.33 | <0.001 | 10 | 0.66 | <0.001 |
| 20 kb | 6 | 0.60 | <0.001 | 12 | 1.21 | <0.001 |
| 50 kb | 7 | 1.11 | <0.001 | 14 | 2.23 | <0.001 |
| 100 kb | 7 | 1.83 | <0.001 | 14 | 3.69 | <0.001 |
| 200 kb | 8 | 3.23 | 0.008 | 16 | 6.56 | 0.008 |
| 500 kb | 12 | 6.92 | 0.009 | 24 | 14.1 | 0.016 |
| 1 Mb | 18 | 11.5 | 0.006 | 37 | 23.7 | 0.009 |
| 2 Mb | 22 | 18.4 | 0.123 | 46 | 39.1 | 0.162 |
| 5 Mb | 29 | 30.4 | 0.610 | 69 | 68.4 | 0.447 |
The occurrence of perturbed gene expression in two or more genes located within the distance shown defined a chromosomal cluster. Parameters are as indicated for Table 2.
Discussion
Whereas irreversible proliferative arrest is mediated by telomere shortening and is abrogated by adventitious expression of hTERT in both human fibroblasts and HMECs, the transcriptional changes occurring during such arrest were different in these cell types. Gene expression perturbations common to replicative senescence in three fibroblast lines derived from fetal skin, foreskin, and fetal lung were observed, whereas transcriptional changes in two breast-derived HMEC lines differed from each other and from the perturbations observed in fibroblasts. The commonality of gene expression changes during senescence in fibroblasts now provides a fingerprint for the senescence state in such cells and additionally identifies specific cellular functions likely to be affected by senescence. Whether the differences between the senescent fibroblasts and postselection senescent HMECs we studied are related to methylation/inactivation of p16INK4A in these HMECs is not known. The lack of commonality of senescence-specific gene expression changes among HMEC lines is consistent with evidence that senescent HMECs show extensive genetic instability in culture (16). Surprisingly, however, despite the different identities of genes up-regulated during senescence in fibroblasts and HMECs, and in HMEC lines of disparate origin, up-regulated genes in all of the senescent cell lines we examined showed the common property of chromosomal clustering.
Several mechanisms have been proposed to account for the irreversible proliferative arrest accompanying telomere shortening. One hypothesis is that telomere shortening causes uncapping or structural alteration of telomeres, yielding chromosome ends recognized as DNA breaks and inducing a DNA damage response (9–12). Whereas certain DNA damaging agents and other stresses have in fact been found to induce a senescence-like proliferative arrest phenotype (28, 34, 35), it is unclear whether this phenotype represents true senescence (36, 37). If senescence is indeed a response to DNA damage, the extensive differences in senescence-specific gene expression observed between human fibroblasts and HMECs would imply that the effects of DNA damage must vary according to cell type and line.
It also has been proposed that telomere shortening may activate ordinarily silent human “senescence genes” located in subtelomeric regions (38). Although subtelomeric silencing is a well demonstrated phenomenon (39, 40), the genes we found to be up-regulated during senescence were not located disproportionately in subtelomeric regions, and we did not detect altered expression of known subtelomeric genes (e.g., some olfactory receptor family genes, IL9 receptor, and RABL2B) during senescence.
Chromosomal clustering of genes whose expression was up-regulated during senescence was a prominent and unexpected finding that contrasted with the generally random distribution of genes we observed to be down-regulated during senescence, or genes whose up-regulation was limited to quiescence. Earlier evidence that histone deacetylase inhibitors, which decondense chromatins, can induce a senescence-like state in human fibroblasts (41) has led to the suggestion that the opening of certain chromatin domains (i.e., conversion of heterochromatins to euchromatins) may be a feature of replicative senescence (42, 43). Possibly related to chromatin alteration during senescence, over-expression of a histone deacetylase, Sir2, can extend life span in Saccharomyces cerevisiae and Caenorhabditis elegans (44, 45), and its mammalian homolog SIRT1 can antagonize PML/p53-induced senescence (46). Our results support the view that processes occurring during senescence may lead to localized alterations in chromatin structure and the consequent up-regulation of groups of genes within “opened” domains.
Chromosomal clustering of genes showing congruent expression recently has been observed for yeast (47), for certain highly expressed human genes (48, 49), in C. elegans, where genes expressed in muscle are clustered in groups of 2–5 (50), and in Drosophila (51, 52). Although it has been suggested that chromosomal clustering may be a general concomitant of concurrent up-regulation of gene expression (51), our finding that genes up-regulated specifically during replicative senescence but not genes up-regulated uniquely in quiescence show chromosomal clustering does not support this notion.
Supplementary Material
Acknowledgments
We thank P. Yaswen and M. Stampfer for kindly providing HMECs; J. Shay, E. Blackburn, and R. Tibshirani for comments on the manuscript; Y. Teng and J. Lam for technical assistance; and members of the Cohen laboratory for helpful discussions and assistance. We thank P. Brown and D. Botstein for providing the IMAGE clone DNA we amplified for use in cDNA microarrays. This work was supported by Postdoctoral Fellowship 5FB-0067 from the California Breast Cancer Research Program (to H.Z.), by a Stanford Graduate Fellowship (to K.-H.P.), and by grants from National Foundation for Cancer Research and the California Breast Cancer Research Program (to S.N.C.).
Abbreviations
- HMECs
human mammary epithelial cells
- FPR
false positive rate
- FNR
false negative rate
References
- 1.Hayflick L, Moorhead P S. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Sager R. Environ Health Perspect. 1991;93:59–62. doi: 10.1289/ehp.919359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright W E, Shay J W. Curr Opin Genet Dev. 2001;11:98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 4.Faragher R G, Kipling D. BioEssays. 1998;20:985–991. doi: 10.1002/(SICI)1521-1878(199812)20:12<985::AID-BIES4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 5.Smith J R, Pereira-Smith O M. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 6.Hayflick L. N Engl J Med. 1976;295:1302–1308. doi: 10.1056/NEJM197612022952308. [DOI] [PubMed] [Google Scholar]
- 7.Greider C W, Blackburn E H. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 8.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 9.Harley C B. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 10.Holt S E, Shay J W, Wright W E. Nat Biotechnol. 1996;14:836–839. doi: 10.1038/nbt0796-836. [DOI] [PubMed] [Google Scholar]
- 11.Karlseder J, Smogorzewska A, de Lange T. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn E H. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 14.Hammond S L, Ham R G, Stampfer M R. Proc Natl Acad Sci USA. 1984;81:5435–5439. doi: 10.1073/pnas.81.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster S A, Galloway D A. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 16.Romanov S R, Kozakiewicz B K, Holst C R, Stampfer M R, Haupt L M, Tisty T D. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 17.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 18.Brenner A J, Stampfer M R, Aldaz C M. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- 19.Huschtscha L I, Noble J R, Neumann A A, Moy E L, Barry P, Melki J R, Clark S J, Reddel R R. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- 20.Ramirez R D, Morales C P, Herbert B S, Rohde J M, Passons C, Shay J W, Wright W E. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbert B S, Wright W E, Shay J W. Oncogene. 2002;21:7897–7900. doi: 10.1038/sj.onc.1205902. [DOI] [PubMed] [Google Scholar]
- 22.Vojta P J, Barrett J C. Biochim Biophys Acta. 1995;1242:29–41. doi: 10.1016/0304-419x(95)00002-w. [DOI] [PubMed] [Google Scholar]
- 23.Lundberg A S, Hahn W C, Gupta P, Weinberg R A. Curr Opin Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 24.Cristofalo V J, Volker C, Francis M K, Tresini M. Crit Rev Eukaryot Gene Expr. 1998;8:43–80. doi: 10.1615/critreveukargeneexpr.v8.i1.30. [DOI] [PubMed] [Google Scholar]
- 25.Shelton D N, Chang E, Whittier P S, Choi D, Funk W D. Curr Biol. 1999;9:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 26.Schwarze S R, DePrimo S E, Grabert L M, Fu V X, Brooks J D, Jarrard D F. J Biol Chem. 2002;277:14877–14883. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- 27.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O, et al. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robles S J, Adami G R. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- 29.Severino J, Allen R G, Balin S, Balin A, Cristofalo V J. Exp Cell Res. 2000;257:162–171. doi: 10.1006/excr.2000.4875. [DOI] [PubMed] [Google Scholar]
- 30.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 31.Pan K H, Lih C J, Cohen S N. Proc Natl Acad Sci USA. 2002;99:2118–2123. doi: 10.1073/pnas.251687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pignolo R J, Martin B G, Horton J H, Kalbach A N, Cristofalo V J. Exp Gerontol. 1998;33:67–80. doi: 10.1016/s0531-5565(97)00090-9. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Ames B N. Proc Natl Acad Sci USA. 1994;91:4130–4134. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K, Mori I, Nakayama Y, Miyakoda M, Kodama S, Watanabe M. Radiat Res. 2001;155:248–253. doi: 10.1667/0033-7587(2001)155[0248:rislga]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Sherr C J, DePinho R A. Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 37.Wright W E, Shay J W. Nat Biotechnol. 2002;20:682–688. doi: 10.1038/nbt0702-682. [DOI] [PubMed] [Google Scholar]
- 38.Wright W E, Shay J W. Trends Genet. 1992;8:193–197. doi: 10.1016/0168-9525(92)90232-s. [DOI] [PubMed] [Google Scholar]
- 39.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 40.Baur J A, Zou Y, Shay J W, Wright W E. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko V V, Hirai T H, Russanova V R, Barbie D A, Howard B H. Mol Cell Biol. 1996;16:5210–5218. doi: 10.1128/mcb.16.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howard B H. Exp Gerontol. 1996;31:281–293. doi: 10.1016/0531-5565(95)00022-4. [DOI] [PubMed] [Google Scholar]
- 43.Villeponteau B. Exp Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 44.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 45.Tissenbaum H A, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 46.Langley E, Pearson M, Faretta M, Bauer U M, Frye R A, Minucci S, Pelicci P G, Kouzarides T. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen B A, Mitra R D, Hughes J D, Church G M. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 48.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus M C, van Asperen R, Boon K, Voute P A, et al. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 49.Lercher M J, Urrutia A O, Hurst L D. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 50.Roy P J, Stuart J M, Lund J, Kim S K. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 51.Spellman P T, Rubin G M. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutanaev A M, Kalmykova A I, Shevelyov Y Y, Nurminsky D I. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.