Abstract
Helicobacter pylori infection of the human stomach is common and typically benign, although a subset of hosts develops severe pathology. Infection occurs in an organ with distinct microenvironments characterized by pronounced differences in the composition of acid-producing parietal cells. In this study, we examine determinants of bacterial tropism to various gastric niches by using germ-free normal and transgenic mice with an engineered parietal cell ablation. Mice were colonized for 8 weeks with a clinical isolate (Hp1) that expresses adhesins recognized by epithelial NeuAcα2,3Galβ1,4 glycan receptors. In normal mice, Hp1 has tropism for a parietal cell-deficient niche where sialylated glycans are expressed by a narrow band of pit cells positioned at the boundary between the squamous epithelium (forestomach) and the proximal glandular epithelium. Lymphoid aggregates that develop in this niche, but not elsewhere in the stomach, were analyzed by GeneChip and quantitative RT-PCR studies of laser capture microdissected mucosa and yielded a series of biomarkers indicative of immune cell activation and maturation. Genetic ablation of parietal cells produced a new source of NeuAcα2,3Galβ1,4 glycans in amplified gastric epithelial lineage progenitors, with accompanying expansion of Hp1 within the glandular epithelium. Lymphoid aggregates that develop in this formerly acid-protected epithelium have molecular features similar to those observed at the forestomach/glandular junction. These findings demonstrate the important roles played by parietal cells and glycan receptors in determining the positioning of H. pylori within the gastric ecosystem, and emphasize the need to consider the evolution of pathology within a given host in a niche-specific context.
Keywords: gastric acid‖adhesin receptors‖immune response‖germ-free animals
At least half of all humans harbor Helicobacter pylori in their stomachs. This microaerophilic bacterium is usually acquired in childhood and remains in the stomach of its host for decades (1). In most cases, the host–microbial relationship is benign, marked only by mild mucosal inflammation. However, in a subset of individuals, this relationship evolves to produce gastric or duodenal ulcers, adenocarcinoma, or mucosal lymphoma (2).
Lee and coworkers (3) have emphasized that severe pathology more often develops in regions of the gastric epithelium where there is a marked transition in the census of acid-producing parietal cells. They and others proposed that local environmental pH gradients at these transitions provide the organism with an opportunity to sample and occupy niches where growth conditions are optimal (3, 4). Once the organism becomes entrenched, pH and other niche-associated factors (e.g., redox state, nutrient availability) then presumably shape the nature of the host–microbial interaction (5).
These speculations emphasize the importance of viewing the nature of the host–microbial cross-talk, the genetic microevolution of this bacterium (6, 7), and the structure of the pathogenic cascade in the context of the niche occupied by the colonizing strain in a given host's gastric ecosystem. At present, it is not feasible to analyze one or more of these aspects of infection in different human gastric niches: we are genetically diverse, and our macroenvironment (e.g., living conditions, diet) cannot be rigorously controlled over the time scale during which H. pylori infection evolves. However, germ-free (GF) mice provide an attractive model for deciphering how the interplay of microbial, host, and environmental factors shapes the destiny of an infection.
The proximal third of the adult mouse stomach (forestomach) is lined with a squamous epithelium, whereas its distal two thirds are covered with a glandular epithelium punctuated by thousands of tubular-shaped invaginations known as gastric units. The glandular epithelium can be subdivided into three geographically distinct regions based on the cell lineages that populate these units (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Gastric units in the zymogenic region contain three principal cell types: pit (produce mucus), parietal, and zymogenic (export digestive enzymes). Units in the mucoparietal region contain pit cells, parietal cells and the precursor to zymogenic cells (neck cells), whereas units in the most distal region (antrum) only have mucus-producing pit and neck cells. Thus, the junctions between forestomach and zymogenic regions (FS/Z), and the mucoparietal and antral regions, represent two well-demarcated areas of abrupt change in parietal cell census.
H. pylori is the only documented bacterial species that is able to survive in the normal human stomach for prolonged periods of time. Because mice are coprophagic, conventionally raised animals develop an entrenched gastric microbiota, especially in regions where acid-production is reduced. Introducing clinical isolates of H. pylori into GF animals allows ready definition of the pattern of bacterial tropism to various gastric niches. Moreover, any ensuing host responses can be directly ascribed to the colonizing strain.
In this report, we examine the role of parietal cells and bacterial attachment to epithelial cell-associated glycan receptors in H. pylori pathogenesis by colonizing GF normal mice, and transgenic animals that lack parietal cells (8), with a clinical isolate recovered from a patient with gastritis (Hp1; refs. 8 and 9). Whole genome genotyping of Hp1 indicated that it contains the 93 of 95 H. pylori genes reported to be regulated by acid, suggesting that it is equipped to respond to the presence or absence of acid in its microenvironments (9–11). Our results demonstrate that this bacterium seeks out niches with sparse parietal cell populations and underscore the point that understanding the forces that shape the destiny of an infection requires a commitment to defining host and microbial responses as a function of the various gastric milieus that coexist within a single host.
Materials and Methods
Animals.
A pedigree of GF FVB/N transgenic mice that express an attenuated diphtheria toxin A fragment (tox176) under the control of nucleotides −1,035 to +24 of the mouse gene encoding the β-subunit of H+/K+ ATPase, and their normal littermates, were maintained in plastic gnotobiotic isolators (12).
Colonization of GF Mice.
Hp1 was cultured on selective medium under microaerophilic conditions (13), concentrated to 108 colony-forming units (CFU)/ml PBS, brought into the gnotobiotic isolator (12), and inoculated into the stomachs of 6- week-old mice. Animals were killed 8 weeks later, and their stomachs were dissected along the cephalocaudal axis. One half was homogenized in 0.5 ml of brain–heart infusion broth and plated on selective medium to assay for CFU. The other half was immediately frozen in OCT compound (Sakura) for laser capture microdissection and/or immunohistochemical analysis.
Navigated Laser Capture Microdissection (n-LCM).
n-LCM was performed with a PixCell II system by using 7.5-μm-diameter laser spot and CapSure HS LCM Caps (Arcturus) (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for further details). RNA was isolated from each patch of captured mucosa (PicoPure RNA Isolation kit, Arcturus; ref. 14).
GeneChip Analysis.
cRNA targets were generated by using two rounds of amplification (RiboAmp RNA Amplification kit, Arcturus; ENZO Bioarray High Yield transcription kit, Affymetrix). Each cRNA (25 μg) was hybridized to a U74A GeneChip (Affymetrix). The overall fluorescence intensity across each chip was scaled to 1,500 and pair-wise comparisons of the levels of transcripts were performed with genechip software (version 4.0).
Real-Time Quantitative RT-PCR (qRT-PCR).
RNA from n-LCM cells was used for random hexanucleotide-primed cDNA synthesis (14). cDNA was added to a qRT-PCR reaction that contained 1× SYBR Green PCR master mix (Applied Biosystems), 0.25 units of UDP-N-glycosidase (Invitrogen), and 900 nM gene-specific primers (see Table 1, which is published as supporting information on the PNAS web site for a list of these primers). Assays were performed in triplicate with an ABI Prism 7700 Sequence Detector (Applied Biosystems). Data were normalized to 18S ribosomal RNA (ΔΔCT analysis).
Immunohistochemistry.
Methods and reagents used for these studies are described in Supporting Materials and Methods and in the legends to Figs. 1, 2, and 4.
Figure 1.
The tropism of H. pylori strain Hp1 for a band of NeuAcα2,3Galβ1,4 glycan-positive pit cells located at the FS/Z of normal FVB/N mice. Multilabel immunohistochemical studies were performed on GF mice killed 8 weeks after monocontamination with Hp1. (A) FS/Z showing Hp1 (red, arrows) in close proximity to neutrophils (green, marked with antibodies to Gr-1). Distally, the epithelium contains a few scattered parietal cells (magenta, arrowhead; tagged with Dolichos biflorus agglutinin, DBA). (B) NeuAcα2,3Galβ1,4-glycans, identified with MAA (red), are expressed in a band of pit cells located in the upper portion of gastric units positioned within the FS/Z (e.g., arrows). Arrowheads highlight the receding pattern of sialylated glycan expression in pit cells positioned at the periphery of this band: MAA-positive cells decrease, whereas Anguilla anguilla agglutinin (AAA)-positive cells (blue) increase. Parietal cells are marked green with DBA. (C) Higher-power view of the FS/Z showing the association of Hp1 (red) with MAA-positive epithelial cells (green). Nuclei are stained blue with bis-benzamide. (Bars = 25 μm.)
Figure 2.
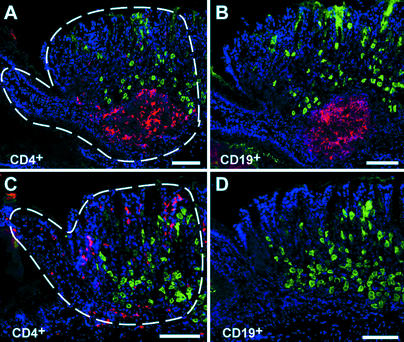
Patterns of inflammation encountered at the FS/Z of an Hp1-infected normal mouse. Serial sections were prepared 8 weeks after infection of a GF mouse and stained with reagents to visualize CD4+ T cells or CD19+ B cells as red. (A and B) Patch of FS/Z mucosa with a lymphoid aggregate containing CD4+ and CD19+ cells. Parietal cells are marked green with DBA lectin. Nuclei are blue (bis-benzamide). The dashed lines in A illustrate what is defined as a patch of mucosa targeted for n-LCM. (C and D) Neighboring patch of diffuse gastritis containing a sparse population of CD4+ T cells. CD19+ B cells are absent. Dashed lines in C outline a typical targeted population for n-LCM. (Bars = 100 μm.)
Figure 4.
Hp1 tropism extends to the zymogenic region of parietal-cell deficient tox176 transgenic mice. (A and B) Gastric units from the zymogenic region of age- and gender-matched normal and tox176 mice (A and B, respectively). Ablation of parietal cells (stained magenta in A with DBA) produces an increase in proliferating isthmal GEPs (green; marked in S-phase with BrdU administered 90 min before death). GEPs produce NeuAcα2,3Galβ1,4 glycans (red with MAA). Pit cells in the upper portion of gastric units are marked blue with A. anguilla agglutinin. (C) Hp1 (red) shows tropism for MAA-positive cells (green) located in the expanded isthmal domain of tox176 gastric units. (D) Transmission electron microscopy demonstrating direct interaction between Hp1 and a granule-free GEP in a gastric unit positioned within the zymogenic region of an ex-GF tox176 mouse stomach. (E) Hematoxylin and eosin-stained section of the zymogenic region. (Inset) A cluster of Hp1 (visualized as red by staining a serial section with antibodies to Hp surface proteins). (F and G) Serial sections prepared from an ex-GF tox176 mouse colonized for 8 weeks with Hp1. A prominent lymphoid aggregate containing CD4+ T cells and CD19+ B cells (red) is apparent. Nuclei are counterstained with bis-benzimide. (Bars, 25 μm in A–C; 0.5 μm in D; 100 μm in E–G.)
Results
Hp1 Shows Tropism for the Junction Between the Forestomach and the Glandular Epithelium.
Six week-old GF normal FVB/N mice were inoculated once with 107 CFU of H. pylori strain Hp1 and killed 8 weeks later. Viable organisms were recovered in 31/34 mice (91%; 104 to 108 CFU per stomach).
We began our analysis of the host response to Hp1 in different regions of the stomach by staining serial sections with antibodies to bacterial surface proteins. The results showed that this clinical isolate had tropism for the glandular epithelium positioned at the junction between the forestomach and zymogenic region (FS/Z; Fig. 1 A and C). This region resembles the area of the human stomach (cardia) positioned immediately adjacent to the esophagus: it encompasses the first few rows of gastric units in the proximal glandular epithelium and contains few if any parietal cells. In contrast, bacteria were not seen in the parietal cell-rich zymogenic region, at the junction between the mucoparietal and antral regions, or in the antrum (n = 16 mice).
The Hp1 strain expresses adhesins that bind to an oncofetal carbohydrate epitope (NeuAcα2,3Galβ1,4) found in the stomachs of humans with H. pylori-associated chronic atrophic gastritis (8, 15). We had shown previously that these sialylated glycans are also synthesized in the small population of gastric epithelial lineage progenitors (GEPs) that reside in the isthmal domains of gastric units located within the zymogenic region of the normal FVB/N mouse stomach (8).
We discovered that these epitopes, which can be detected by Maackia amurensis agglutinin (MAA), were also expressed in pit cells positioned in a narrow band of gastric units located at the FS/Z of GF (and conventionally raised) FVB/N mice (Fig. 1B). In contrast, MAA-positive pit cells were not evident in the other area of the stomach where there is abrupt drop-off in parietal cell density, the mucoparietal/antral transition. Hp1 attachment and colonization occurred in this band of MAA-positive pit cells at the FS/Z (Fig. 1C).
Histological analysis of serial sections prepared from the stomachs of 14-week-old ex-GF normal mice disclosed that the severity of the inflammatory response correlated with bacterial tropism. Large clusters of mucosal CD4+ T cells (lymphoid aggregates) were present within the FS/Z of each stomach studied (16 of 16) and were absent in the stomachs of noninfected, age-matched controls (n = 8). These foci also contained aggregates of CD19+ B cells (Fig. 2 A and B). The remainder of the FS/Z mucosa was infiltrated with scattered CD4+ T cells and lacked B cells (Fig. 2 C and D). This nonfocal, less intense inflammatory response (“mild diffuse gastritis”) was the only histological pattern detected in the parietal cell-rich zymogenic region, at the mucoparietal/antral junction, and in the antrum (data not shown).
Together, these results suggest that bacterial tropism for, and subsequent development of lymphoid aggregates within the FS/Z reflects a combination of at least two host factors: (i) the availability of host glycan receptors for bacterial adhesins in this niche, and (ii) a favorable microenvironment arising from parietal cell deficiency.
Molecular Characterization of the Host Response at the FS/Z in Normal Mice.
A navigated form of laser capture microdissection (16) was combined with GeneChip-based expression profiling to characterize host responses at the principal site of colonization. Equivalent-sized neighboring patches of mucosa (one with a lymphoid aggregate and the other with the diffuse pattern of mild gastritis) were harvested from the stomach of a single mouse that had been infected for 8 weeks with Hp1. GeneChips representing ≈12,000 mouse genes and ESTs were used to compare the response in these juxtaposed areas (see Fig. 6, which is published as supporting information on the PNAS web site). Two cRNA targets were independently generated from each RNA preparation and each cRNA hybridized to a GeneChip. Chips probed with cRNAs from the area of mild diffuse gastritis were designated as “baseline,” whereas chips probed with cRNAs from the lymphoid aggregate were designated as “experimental.” Four chip-to-chip comparisons were performed with each experimental chip compared with the two baseline chips. A total of 361 genes, plus 171 ESTs satisfied our selection criteria: (i) the difference in their expression between experimental and baseline chips was ≥2-fold (increased or decreased) and (ii) the difference was evident in the comparison of a given experimental chip with each of the two baseline chips (see Table 2, which is published as supporting information on the PNAS web site).
The data set contained a collection of transcripts indicating more robust representation of T cells (CD53, CD84), B cells (CD19, CD20, CD84), natural killer (NK) cells (CD53, CD56), and macrophages (MARCO; macrophage receptor with collagenous structure) in the lymphoid aggregate. Expression of chemokine (C-C) receptor 6 (CCR6) and CD83 (average fold difference in mRNA levels = +8.2 and +3.6, respectively) suggested the presence of specialized antigen-presenting dendritic cells, a notion supported by the increased level of ephrin A4 mRNA (mediates interactions between dendritic cells and B cells, +2.5) and by immunostaining with antibodies to CD11b and CD11c (data not shown). Increased expression of lymphotoxin β (LTβ, +2.4), a gene involved in lymphoid aggregate development (17), was also documented.
Blood vessel formation in the area surrounding lymphoid aggregates should provide entry and exit points for immune cells. Increases in vascular endothelial growth factor (VEGF-A, +2.4) and its receptor, fms-like tyrosine kinase-1 (Flt-1, +5.3) likely reflect this process. EphrinB2 arrests endothelial cell proliferation and migration through suppression of VEGF activities (18): its down-regulation in the lymphoid aggregate (-3.9) should help promote VEGF activity. The data set also included two other genes involved in angiogenesis: endothelin-2 (+6.5) and adenosine A2A receptor (+3.4).
In lymphoid structures such as Peyer's patches, there is immune cell activation and maturation so that cells are equipped to respond to antigenic stimuli. The data set provided evidence that these processes were occurring in the FS/Z lymphoid aggregate. CD45 (+3.7) is tyrosine phosphatase required for lymphocyte activation. Zap70 (T cell receptor ζ-chain associated protein kinase, 70 kDa, +3.1) plays a vital signaling role in the interaction between antigen presenting cells and T cells that leads to T cell activation (19). T cell factor 1 (+8.4) is critical for T cell maturation (20), whereas B lymphoid kinase (+2.2) is important for B cell proliferation and developmental progression (21). Wiskott–Aldrich syndrome homolog (WAS, +3.0) and its effector, actin related protein 2/3 complex (Arp2/3, +2.4), contribute to lymphocyte development, activation, and function; WAS deficiency restricts progression to the CD4+CD8+ double positive stage during thymocyte development, and impairs chemotactic, phagocytic, and adhesive functions (22).
The data set provided evidence for increased inflammatory activity in the lymphoid aggregate compared with the adjacent region of mild diffuse gastritis. There was elevated expression of NF-κB1 (+4.6), various immunoglobulins, members of the complement cascade (complement component 3, +2.4; complement component 1 r subunit, +2.2), IFN-γ inducible protein 47 kDa (Irg-47, +3.2; plays an important role in defense against bacterial infections; ref. 23), retinoic acid early transcript γ (Rae-1, +2.2; interacts with the NK2GD glycoprotein receptor to activate NK cells at sites of inflammation; ref. 24), and chitinase-3 like 1 (+3.9; released by macrophages to stimulate turnover of extracellular matrix; ref. 25).
The reduced expression of three genes also indicated local activation of inflammatory mediators. Tritetraprolin (-3.0) is a TNF-α-induced product that blocks TNF-α production by facilitating degradation of TNF-α mRNA (26). PKC interacting cousin of thioredoxin 2 (−2.5) is an inhibitor of two factors important for T cell activation: NF-κB and AP1 (27). Oct1 (−2.4) represses the transcription of proinflammatory genes such as IL-8 and vascular cell adhesion molecule 1 (28).
Our data set contained several genes involved in cholesterol biosynthesis (farnesyl diphosphate synthetase, +2.9; mevalonate decarboxylase, +3) that were also identified in a recent DNA microarray analysis of the responses of a cultured gastric adenocarcinoma-derived epithelial cell line (AGS) to H. pylori (29). In addition, we identified two other transcripts encoding proteins involved in modulating cholesterol efflux in macrophages that were increased in the lymphoid aggregate [ATP binding cassette (ABC) subfamily A1 (ABCA1), +2.7; ABCG1, +4.2]. Modulating cholesterol synthesis and fluxes could play an important role in host defense by altering membrane fluidity, forming lipid rafts for signaling, increasing the levels of endotoxin neutralizing lipoprotein, and recruiting macrophages (29, 30).
n-LCM/qRT-PCR Validation of the GeneChip Data Set.
A subset of five genes was culled from the data set for follow-up qRT-PCR analysis to determine whether the GeneChip results were representative of immune responses that occur in the FS/Z of other Hp1-infected mice. The selected genes encode proteins that represent biomarkers for (i) formation of lymphoid aggregates (lymphotoxin β); (ii) the presence of specialized antigen-presenting dendritic cells [chemokine (C-C) receptor 6]; (iii) active inflammation (complement component 3, an acute phase protein that triggers the complement cascade and promotes bacterial opsonization); and (iv) epithelial barrier function/renewal [serine protease inhibitor 1–2; and regenerating gene (Reg) 3γ, a secreted mitogen; ref. 31].
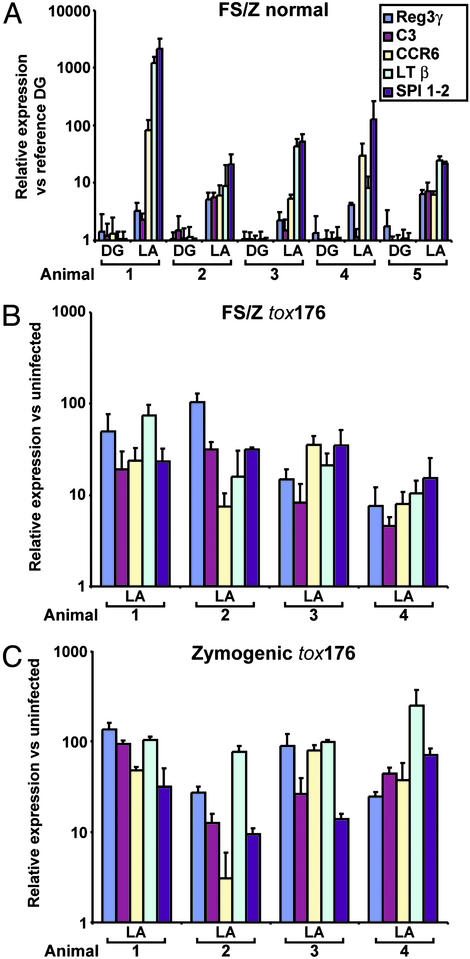
n-LCM was used to procure cells from equivalent-sized neighboring patches of FS/Z mucosa, one with a lymphoid aggregate, the other with a diffuse pattern of mild gastritis, from five 14-week-old ex-GF normal mice. The results confirm that these genes provide a robust set of biomarkers of the host response to Hp1 within the FS/Z niche (Fig. 3A and Fig. 7, which is published as supporting information on the PNAS web site).
Figure 3.
qRT-PCR studies of LCM mucosa harvested from different niches in Hp1-infected normal and tox176 mice. (A) Comparison of the levels of five mRNAs in a patch of FS/Z mucosa containing a lymphoid aggregate (LA), versus an adjacent equivalent-sized patch with mild diffuse gastritis (DG). The analysis was performed using mucosal patches dissected from five Hp1-infected normal mice. The level of each mRNA in each LA-containing and each DG-containing patch is expressed relative to a reference RNA prepared from a DG patch from one of the colonized animals. (B) Patches of mucosa with LA recovered by n-LCM from the FS/Z of four Hp1-infected tox176 mice. mRNA levels were defined by using FS/Z mucosa retrieved from age-matched GF tox176 mice as a baseline. (C) n-LCM/qRT-PCR analysis of LA-containing mucosa from the zymogenic region of tox176 stomachs. Each RNA preparation was assayed in triplicate. Mean values ± SD are plotted.
Hp1 Is More Broadly Distributed in the Gastric Ecosystem of Mice with a Genetically Engineered Parietal Cell Ablation.
The absence of lymphoid aggregates in the parietal-cell rich zymogenic region of the normal mouse stomach is consistent with the hypothesis that acid-producing parietal cells and sialylated NeuAcα2,3Galβ1,4 epithelial glycans play critical roles in defining the location of Hp1 infection and subsequent development of an adaptive host immune response. To further test this hypothesis, we introduced Hp1 into GF transgenic mice with an engineered parietal cell deficiency.
Expressing an attenuated diphtheria toxin A fragment (tox176) under the control of parietal-cell specific transcriptional regulatory elements from the gene encoding the noncatalytic β-subunit of H+/K+ ATPase results in ablation of this lineage and achlorhydria (pH 6–7). Loss of parietal cells is accompanied by increased proliferation of GEPs located in the isthmal stem cell niches of gastric units. This leads to progressive expansion of GEPs, so that by postnatal weeks 14–18, they represent ≈20% of the total epithelial census in units positioned within the zymogenic region (see ref. 16 for a discussion of molecular mechanisms that underlie this expansion). Like the band of MAA-positive pit cells positioned at the FS/Z of normal adult mice, the amplified GEPs in tox176 littermates express NeuAcα2,3Galβ1,4 glycans (Fig. 4 A and B). Previous in situ binding assays established that Hp1, as well as strains recovered from patients with H. pylori-associated chronic atrophic gastritis or adenocarcinoma, produce adhesins that recognize these sialylated GEP-associated epitopes (8, 9).
GF tox176 littermates received a single gavage of 107 CFU of Hp1 at 6 weeks of age. 100% of the mice killed 8 weeks later were colonized (n = 18). There were no statistically significant differences in the number of viable bacteria recovered in these compared with normal FVB/N mice (Student's t test). However, immunohistochemical studies revealed that Hp1 was distributed throughout the parietal cell-deficient zymogenic region. Binding to GEPs was documented by multilabel immunohistochemistry as well as by transmission electron microscopy (Fig. 4 C and D).
The expanded distribution of Hp1 was accompanied by an expanded distribution of lymphoid aggregates. In each of six tox176 mice studied, there was a uniformly high density of CD4+ cells in the FS/Z epithelium, leaving essentially no intervening patches of mild diffuse gastritis. In contrast to normal mice, there were also aggregates of CD4+ and CD19+ lymphocytes distributed throughout the parietal cell-deficient zymogenic region (Fig. 4 E–G).
n-LCM was used to harvest a patch of mucosa containing a CD4+/CD19+ lymphoid aggregate from the FS/Z mucosa, and a patch containing a lymphoid aggregate from the mid-zymogenic region from each of four 14-week-old tox176 mice. Comparably sized and positioned patches of mucosa were microdissected from the stomachs of uninfected tox176 mice to serve as baseline controls for qRT-PCR analysis. Each of the five genes previously analyzed in the FS/Z mucosa of normal mice was markedly induced in both the FS/Z and zymogenic lymphoid aggregates of tox176 animals (Fig. 3 B and C).
Discussion
H. pylori infection occurs in an organ with distinct microenvironments. Some of these microenvironments are established before colonization and reflect normal regional differences in the cellular composition of the gastric mucosa. Other microenvironments are created as a consequence of infection, such as H. pylori-induced chronic atrophic gastritis with loss of acid-producing parietal cells (32). In the present study, we used gnotobiotic mice to study the impact of manipulating the preexisting parietal cell content of the stomach on H. pylori tropism and the subsequent development of host pathology. We have found that normal FVB/N mice express NeuAcα2,3Galβ1,4 glycans, recognized by at least one known H. pylori adhesin (SabA; ref. 33), in a sharply demarcated, narrow band of pit cells positioned at the boundary between the squamous epithelium of the forestomach and the glandular epithelium. These sialylated glycan receptors are located in gastric units that are largely devoid of parietal cells. A clinical isolate of H. pylori (Hp1), expressing adhesins recognized by these glycan receptors, homes to this niche, resulting in formation of lymphoid aggregates that express biomarkers indicating the presence of T cells, B cells, dendritic cells, NK cells, and macrophages. This restriction of bacterial tropism and lymphoid aggregate formation to the FS/Z is relieved by ablation of parietal cells from the glandular epithelium. Loss of this acid-producing lineage produces a new source of NeuAcα2,3Galβ1,4 glycans: amplified GEPs. There is accompanying expansion of Hp1 colonization from the FS/Z to the now achlorhydric glandular epithelium, with lymphoid aggregate formation in what was formerly an acid-protected environment. The molecular features of these lymphoid aggregates mimic those in the FS/Z, indicating that with removal of parietal cells, the glandular epithelium is fully capable of hosting a robust immune response to this microbe.
The normal human stomach does not contain organized lymphoid structures. However, H. pylori infection often results in MALT formation (34, 35). Studies in both humans and animal models indicate that the primary adaptive immune response to H. pylori is Th1-polarized (IFNγ-driven with strong activation of macrophages) (36–39). This type of response can increase the risk for cell-mediated mucosal atrophy, with reductions in parietal census and proliferation of GEPs (38). If the colonizing strain expresses adhesins recognized by progenitor-associated glycan receptors, then attachment to multi- or oligopotential GEPs with high proliferative potential may increase the risk for initiating tumorigenesis. In contrast, a Th2-balanced response (IL-4-mediated with B cell activation) may be protective (40) by specifically targeting and removing H. pylori without endangering parietal cell census.
Together, these observations support the notion that local environmental pH gradients that form in regions of the stomach where there are abrupt transitions in parietal cell census provide H. pylori with an initial favorable niche. The likelihood for entrenchment is increased if epithelial receptors are available for expressed bacterial adhesins. Persistence is favored by metabolic and genetic adaptations made by this microorganism which has a remarkably plastic genome (9, 41). If the adaptive immune response to colonization is strongly Th1-polarized, the host is put at risk for parietal cell loss in the neighboring epithelium: such loss may be promoted if there are structural similarities between bacterial cell surface immunodeterminants and epitopes normally expressed by parietal cells (e.g., Lewisx in the case of Hp1 and FVB/N mice; ref. 13). By reshaping the parietal cell landscape in chronic atrophic gastritis, the bacterium enlarges its preferred niche. However, the ensuing territorial expansion of H. pylori into the parietal cell-depleted, GEP-enriched environment can put the host at risk for development of more severe pathology.
Our results emphasize the gatekeeper role played by parietal cells, and by extension acid production, in H. pylori pathogenesis. We have recently used GeneChips and qRT-PCR to show that Hp1 colonization of GF mice has no demonstrable effect on the expression profile of parietal cells (42). The present study raises the question of what effects parietal cells and acid have on bacterial gene expression, and what bacterial genes are critical for entry into and persistence within host niches that are relatively devoid of acid. For example, the impact of acid on phase variation of genes involved in production of outer membrane proteins (43) and surface carbohydrate epitopes (44) could influence attachment (persistence), nutrient acquisition (growth), and the degree to which synthesized bacterial surface immunodeterminants are recognized as foreign by the host. In vitro studies have shown that acid regulates expression of both acid-resistance and virulence genes, including members of the organism's 31 gene cag pathogenicity island (cagA and cagE), vacuolating cytotoxin (vacA), wbcJ (homologous to bacterial O-antigen biosynthetic proteins; links lipopolysaccharide expression and acid survival), and urease (buffers the organism from acid) (5). Colonizing GF normal and tox176 mice with isogenic strains of H. pylori containing null alleles of these genes and observing the effects on tropism, persistence, and the evolution of host pathology should be informative.
The pathogenicity island (cag PAI) of H. pylori encodes components of a type IV secretion system that can deliver (inject) bacterial products into epithelial cells, including products linked to greater host pathology (29, 45). It is remarkable that Hp1 can elicit formation of a robust immune response in our gnotobiotic mouse model given that its cag PAI has only retained ORFs 1–8 (e.g., it lacks cagA and cagE). Clinical isolates have been identified recently that have an intact cag PAI and bind in vitro to sections of tox176 gastric epithelium (9). They may be a useful starting point for mutagenesis studies, and for assessing the long-term impact of attachment to GEPs on development of adenocarcinoma.
Supplementary Material
Acknowledgments
We thank Thad Stappenbeck for helpful suggestions about laser capture microdissection, Sabrina Wagoner and Lisa Roberts for superb technical assistance, and Sherif Karam and Per Falk for their support. This work was funded by National Institutes of Health Grant DK58529.
Abbreviations
- GF
germ-free
- CFU
colony-forming unit
- n-LCM
navigated laser capture microdissection
- qRT-PCR
quantitative RT-PCR
- GEP
gastric epithelial lineage progenitor
- MAA
Maackia amurensis agglutinin
- NK
natural killer
References
- 1.Mitchell H M, Li Y Y, Hu P J, Liu Q, Chen M, Du G G, Wang Z J, Lee A, Hazell S L. J Infect Dis. 1992;166:149–153. doi: 10.1093/infdis/166.1.149. [DOI] [PubMed] [Google Scholar]
- 2.Peek R M, Jr, Blaser M J. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 3.Van Zanten S J, Dixon M F, Lee A. Gastroenterology. 1999;116:1217–1229. doi: 10.1016/s0016-5085(99)70025-9. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Rosberg K, Scott D R, Rex D, Melchers K, Sachs G. Gastroenterology. 1996;111:886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 5.de Vries N, van Vliet A H M, Kusters K G. In: Helicobacter pylori: Physiology and Genetics. Mobley H L T, Mendz G L, Hazell S L, editors. Washington, DC: Am. Soc. Microbiol. Press; 2001. pp. 321–334. [Google Scholar]
- 6.Kuipers E J, Israel D A, Kusters J G, Gerrits M M, Weel J, van Der Ende A, van Der Hulst R W, Wirth H P, Hook-Nikanne J, Thompson S A, et al. J Infect Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb G F, Blaser M J. Proc Natl Acad Sci USA. 2002;99:3135–3140. doi: 10.1073/pnas.042685799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syder A J, Guruge J L, Li Q, Hu Y, Oleksiewicz C M, Lorenz R G, Karam S M, Falk P G, Gordon J I. Mol Cell. 1999;3:263–274. doi: 10.1016/s1097-2765(00)80454-2. [DOI] [PubMed] [Google Scholar]
- 9.Bjorkholm B M, Guruge J L, Oh J D, Syder A J, Salama N, Guillemin K, Falkow S, Nilsson C, Falk P G, Engstrand L, et al. J Biol Chem. 2002;277:34191–34197. doi: 10.1074/jbc.M203613200. [DOI] [PubMed] [Google Scholar]
- 10.Ang S, Lee C Z, Peck K, Sindici M, Matrubutham U, Gleeson M A, Wang J T. Infect Immun. 2001;69:1679–1686. doi: 10.1128/IAI.69.3.1679-1686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allan E, Clayton C L, McLaren A, Wallace D M, Wren B W. Microbiology. 2001;147:2285–2292. doi: 10.1099/00221287-147-8-2285. [DOI] [PubMed] [Google Scholar]
- 12.Hooper L V, Mills J C, Roth K A, Stappenbeck T S, Wong M H, Gordon J I. Methods Microbiol. 2002;31:559–589. [Google Scholar]
- 13.Guruge J L, Falk P G, Lorenz R G, Dans M, Wirth H P, Blaser M J, Berg D E, Gordon J I. Proc Natl Acad Sci USA. 1998;95:3925–3930. doi: 10.1073/pnas.95.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stappenbeck T S, Hooper L V, Manchester J K, Wong M H, Gordon J I. Methods Enzymol. 2002;356:168–196. doi: 10.1016/s0076-6879(02)56932-9. [DOI] [PubMed] [Google Scholar]
- 15.Amado M, Carneiro F, Seixas M, Clausen H, Sobrinho-Simoes M. Gastroenterology. 1998;114:462–470. doi: 10.1016/s0016-5085(98)70529-3. [DOI] [PubMed] [Google Scholar]
- 16.Mills J C, Andersson N, Hong C V, Stappenbeck T S, Gordon J I. Proc Natl Acad Sci USA. 2002;99:14819–14824. doi: 10.1073/pnas.192574799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koni P A, Sacca R, Lawton P, Browning J L, Ruddle N H, Flavell R A. Immunity. 1997;6:491–500. doi: 10.1016/s1074-7613(00)80292-7. [DOI] [PubMed] [Google Scholar]
- 18.Kim I, Ryu Y S, Kwak H J, Ahn S Y, Oh J L, Yancopoulos G D, Gale N W, Koh G Y. FASEB J. 2002;16:1126–1128. doi: 10.1096/fj.01-0805fje. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard N, Di Bartolo V, Hivroz C. Immunity. 2002;17:389–399. doi: 10.1016/s1074-7613(02)00421-1. [DOI] [PubMed] [Google Scholar]
- 20.Staal F J, Meeldijk J, Moerer P, Jay P, van de Weerdt B C, Vainio S, Nolan G P, Clevers H. Eur J Immunol. 2001;31:285–293. doi: 10.1002/1521-4141(200101)31:1<285::AID-IMMU285>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Malek S N, Dordai D I, Reim J, Dintzis H, Desiderio S. Proc Natl Acad Sci USA. 1998;95:7351–7356. doi: 10.1073/pnas.95.13.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thrasher A J. Nat Rev Immunol. 2002;2:635–646. doi: 10.1038/nri884. [DOI] [PubMed] [Google Scholar]
- 23.Collazo C M, Yap G S, Sempowski G D, Lusby K C, Tessarollo L, Woude G F, Sher A, Taylor G A. J Exp Med. 2001;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diefenbach A, Jensen E R, Jamieson A M, Raulet D H. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recklies A D, White C, Ling H. Biochem J. 2002;365:119–126. doi: 10.1042/BJ20020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson B A, Blackwell T K. Oncogene. 2002;21:4237–4246. doi: 10.1038/sj.onc.1205526. [DOI] [PubMed] [Google Scholar]
- 27.Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 28.Ortego M, Hernandez A G, Bustos C, Blanco-Colio L M, Hernandez-Presa M A, Tunon J, Egido J. Eur J Pharmacol. 2002;448:113–121. doi: 10.1016/s0014-2999(02)01938-6. [DOI] [PubMed] [Google Scholar]
- 29.Guillemin K, Salama N R, Tompkins L S, Falkow S. Proc Natl Acad Sci USA. 2002;99:15136–15141. doi: 10.1073/pnas.182558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Eck M, Bos I S, Kaminski W E, Orso E, Rothe G, Twisk J, Bottcher A, Van Amersfoort E S, Christiansen-Weber T A, Fung-Leung W P, et al. Proc Natl Acad Sci USA. 2002;99:6298–6303. doi: 10.1073/pnas.092327399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazumori H, Ishihara S, Hoshino E, Kawashima K, Moriyama N, Suetsugu H, Sato H, Adachi K, Fukuda R, Watanabe M, et al. Gastroenterology. 2000;119:1610–1622. doi: 10.1053/gast.2000.20262. [DOI] [PubMed] [Google Scholar]
- 32.Correa P. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 33.Mahdavi J, Sonden B, Hurtig M, Olfat F O, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson K A, et al. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du M Q, Isaccson P G. Lancet Oncol. 2002;3:97–104. doi: 10.1016/s1470-2045(02)00651-4. [DOI] [PubMed] [Google Scholar]
- 35.Genta R M, Hamner H W, Graham D Y. Hum Pathol. 1993;24:577–583. doi: 10.1016/0046-8177(93)90235-9. [DOI] [PubMed] [Google Scholar]
- 36.Bamford K B, Fan X, Crowe S E, Leary J F, Gourley W K, Luthra G K, Brooks E G, Graham D Y, Reyes V E, Ernst P B. Gastroenterology. 1998;114:482–492. doi: 10.1016/s0016-5085(98)70531-1. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi M, Redline R, Nedrud J, Czinn S. Infect Immun. 1996;64:238–245. doi: 10.1128/iai.64.1.238-245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth K A, Kapadia S B, Martin S M, Lorenz R G. J Immunol. 1999;163:1490–1497. [PubMed] [Google Scholar]
- 39.Eaton K A, Mefford M, Thevenot T. J Immunol. 2001;166:7456–7461. doi: 10.4049/jimmunol.166.12.7456. [DOI] [PubMed] [Google Scholar]
- 40.Fox J G, Beck P, Dangler C A, Whary M T, Wang T C, Shi N H, Nagler-Anderson C. Nat Med. 2000;6:536–542. doi: 10.1038/75015. [DOI] [PubMed] [Google Scholar]
- 41.Israel D A, Salama N, Krishna U, Rieger U M, Atherton J C, Falkow S, Peek R M., Jr Proc Natl Acad Sci USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills J C, Syder A J, Hong C V, Guruge J L, Raaii F, Gordon J I. Proc Natl Acad Sci USA. 2001;98:13687–13692. doi: 10.1073/pnas.231332398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tannaes T, Dekker N, Bukholm G, Bijlsma J J, Appelmelk B J. Infect Immun. 2001;69:7334–7340. doi: 10.1128/IAI.69.12.7334-7340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran A P, Knirel Y A, Senchenkova S N, Widmalm G, Hynes S O, Jansson P E. J Biol Chem. 2002;277:5785–5795. doi: 10.1074/jbc.M108574200. [DOI] [PubMed] [Google Scholar]
- 45.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.