Abstract
Ribosomal RNA gene transcription by RNA polymerase I (Pol I) is the driving force behind ribosome biogenesis, vital to cell growth and proliferation. The key activator of Pol I transcription, UBF, has been proposed to act by facilitating recruitment of Pol I and essential basal factor SL1 to rDNA promoters. However, we found no evidence that UBF could stimulate recruitment or stabilization of the pre-initiation complex (PIC) in reconstituted transcription assays. In this, UBF is fundamentally different from archetypal activators of transcription. Our data imply that UBF exerts its stimulatory effect on RNA synthesis, after PIC formation, promoter opening and first phosphodiester bond formation and before elongation. We provide evidence to suggest that UBF activates transcription in the transition between initiation and elongation, at promoter escape by Pol I. This novel role for UBF in promoter escape would allow control of rRNA synthesis at active rDNA repeats, independent of and complementary to the promoter-specific targeting of SL1 and Pol I during PIC assembly. We posit that stimulation of promoter escape could be a general mechanism of activator function.
Keywords: promoter escape, recruitment, RNA polymerase I, transcription activation, UBF
Introduction
More than half of the total RNA synthesis in a eukaryotic cell is committed to the production of ribosomal RNAs (Reeder, 1999) and, as rRNA constitutes the enzymatic and structural scaffold of ribosomes (Moore and Steitz, 2002), this drives the biogenesis of the ribosomes, required for normal cell growth and division (Warner, 1999). For transcription of the rRNA genes, eukaryotic cells have evolved a dedicated machinery, which includes RNA polymerase I (Pol I), confined to the nucleolar subcompartment of the nucleus (Hannan et al, 1998; Reeder, 1999; Moss and Stefanovsky, 2002; Grummt, 2003; Comai, 2004; Russell and Zomerdijk, 2005). To understand how cells accomplish the tight control of Pol I transcription, balancing the supply of rRNA with demand under different growth conditions, it is necessary to determine the exact functions of the transcription factors that act in conjunction with Pol I at the rRNA gene promoter (reviewed by: Grummt, 2003; Moss, 2004; Russell and Zomerdijk, 2005). One key aspect to understanding mammalian rRNA gene regulation is to identify the critical step(s) in the Pol I transcription cycle affected by the major activator of transcription, UBF (Jantzen et al, 1990). In general, models for activator function propose that they positively influence kinetically limiting steps in transcription and/or interact with components of the basal transcription machinery leading to cooperative recruitment and assembly of the pre-initiation complex (PIC) (Kingston and Green, 1994). Indeed, one of the predominant mechanisms by which activators enhance Pol II transcription involves sequence-specific binding at regulatory promoter elements and sequestering of components of the basal transcription machinery to the gene promoter, thereby facilitating PIC assembly (Ptashne and Gann, 1997). UBF was proposed to operate in such a manner (reviewed by: Zomerdijk and Tjian, 1998; Comai, 2004), although this had not been tested directly. We sought to determine the step in the Pol I transcription cycle activated by UBF.
Basal levels of transcription from the rDNA promoter can be produced with SL1, a TBP–TAFI complex (Comai et al, 1992, 1994; Zomerdijk et al, 1994), and Pol I in a reconstituted system (Schnapp and Grummt, 1991; Smith et al, 1993; Friedrich et al, 2005). To achieve activated levels of rDNA transcription, UBF is essential (Bell et al, 1988; Jantzen et al, 1990). In addition to this ‘true' activation in reconstituted transcription systems, UBF has a positive effect on Pol I transcription as an anti-repressor (Kuhn and Grummt, 1992; Brou et al, 1993; Pelletier et al, 2000). Additionally, in cells UBF can repress Pol I transcription elongation, an effect that can be reversed by growth factor-induced ERK phosphorylation of UBF (Stefanovsky et al, 2006).
UBF might function as an architectural protein (Jantzen et al, 1992; Reeder et al, 1995). It contains six HMG-box domains (Jantzen et al, 1990; Bachvarov and Moss, 1991), which are thought to interact with the minor groove of DNA, exhibiting a relaxed specificity of sequence binding, and display the ability to dramatically bend DNA (Jantzen et al, 1992; Leblanc et al, 1993; Bazett Jones et al, 1994; Copenhaver et al, 1994; Hu et al, 1994; Putnam et al, 1994; Stefanovsky et al, 2001a). The DNA-binding and transactivation domains of UBF overlap and dimerization through the amino-terminus is essential for the activation function of UBF (McStay et al, 1991; Jantzen et al, 1992). UBF activation requires the upstream control element (UCE; −156 to −107) of the rDNA promoter, whereas SL1 functions through the essential core element (−45 to +18), overlapping the start site (+1) of transcription (Zomerdijk and Tjian, 1998). DNase I footprinting showed UBF binding at the UCE and core. Human SL1 alone failed to produce a DNase I footprint, but SL1 and UBF in combination extended the footprint of UBF alone, suggesting a cooperative interaction at the promoter (Learned et al, 1986; Bell et al, 1988). UBF can interact with SL1 (Bodeker et al, 1996; Hempel et al, 1996), via its highly acidic carboxy-terminal domain (Jantzen et al, 1992; Kihm et al, 1998; Tuan et al, 1999), as well as with Pol I (Schnapp et al, 1994; Hanada et al, 1996). On the basis of these findings, it was proposed that UBF recruits SL1 and Pol I to the rDNA promoter, activating transcription by facilitating PIC assembly. However, binding of UBF occurs throughout the rDNA repeat (O'Sullivan et al, 2002; Mais et al, 2005), not easily reconcilable with a role for UBF in nucleating PIC assembly at the rDNA promoter. Moreover, SL1 can nucleate PIC formation (Schnapp and Grummt, 1991; Smith et al, 1993; Friedrich et al, 2005), directly contacting Pol I-associated factor hRRN3, also known as TIF-IA (Bodem et al, 2000), thereby recruiting initiation-competent Pol I to the core promoter (Miller et al, 2001). SL1 can also stabilize UBF binding at the rDNA promoter (Friedrich et al, 2005). Although we observed no detectable effect of UBF on SL1 binding to the promoter, a role for UBF in the recruitment of Pol I was not excluded.
Here, we provide evidence that activation of rDNA transcription by UBF in a reconstituted system occurs subsequent to PIC formation. We have defined a specific and novel function for UBF in activating the rate of RNA synthesis at promoter escape and clearance by Pol I. This mechanism enables UBF to activate transcription both from previously inactive promoters following PIC assembly and from SL1-engaged promoters at each successive round of transcription following re-initiation.
Results
UBF activates Pol I transcription subsequent to PIC formation
UBF had been suggested to activate Pol I transcription in vitro by facilitating recruitment of SL1 (Bell et al, 1988) and perhaps Pol I to the rDNA promoter (Schnapp et al, 1994; Hanada et al, 1996). We tested this hypothesis using an immobilized template in reconstituted transcription reactions with highly purified human SL1 and Pol Iβ (Miller et al, 2001; Panov et al, 2001) and insect cell (recombinant baculovirus) expressed human UBF1, which contains the necessary modifications for activity (Figure 1A). The human Pol I complex is over 1 MDa large and contains the core Pol I subunits and a number of associated factors, among which are RRN3 (Miller et al, 2001), topoisomerase IIα and CK2 (Panova et al, 2006). In the absence of UBF, SL1 efficiently directs recruitment of Pol Iβ to the rDNA promoter, via its interaction with Pol I-associated factor hRRN3 (Miller et al, 2001), supporting basal levels of transcription (Figure 1B, lanes 2, 7 and 12). Transcription was activated by addition of purified recombinant UBF during PIC assembly (Figure 1B, lanes 3–5, and C).
Figure 1.
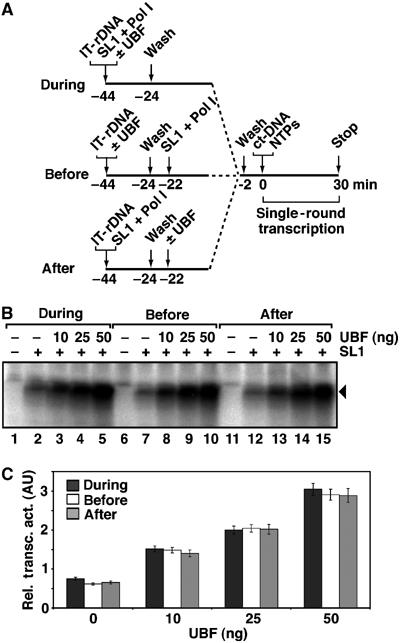
UBF activates transcription from a pre-assembled SL1–Pol I–rDNA promoter complex. (A) UBF (0, 10, 25 or 50 ng) was added before, during or after PIC assembly with SL1 and Pol Iβ on an immobilized rDNA template (IT-rDNA, Fr4), as outlined. Assembly of factors on the promoter was for 20 min on ice, with gentle mixing. Unbound factors were removed by washing templates in TM10/0.05. Missing factors were added, incubation was continued for a further 20 min and the templates were washed in TM10/0.05 before initiation of single-round transcription upon addition of NTPs and calf thymus DNA (0.5 μg, ct-DNA). Reactions were stopped after 30 min. (B) Transcripts produced in the reactions outlined in panel A were analysed in S1 nuclease protection assays. Protected radiolabelled fragments, reflecting accurately initiated transcripts of 40 nt, are indicated by an arrowhead. Signals in lanes 1, 6 and 11 are not transcription-related. (C) Mean and standard error of transcription levels (in arbitrary units) of single-round transcription with 0, 10, 25 and 50 ng UBF added before, during or after PIC assembly as outlined in panel A, for three independent experiments.
Strikingly, UBF efficiently activated transcription from templates with a pre-formed and functional SL1–Pol I–promoter DNA complex (Figure 1B, lanes 13–15, and C). This level of activation by UBF added after PIC assembly was comparable to that observed from templates to which UBF was pre-bound (Figure 1B, lanes 8–10, and C) and to which UBF was added during PIC assembly (Figure 1B, lanes 3–5, and C). Note that where UBF was added after PIC assembly, the bulk of unbound factors, SL1 and Pol I, were removed in a wash step before the addition of UBF (Figure 1B, lanes 13–15, and C) and that the amount of SL1 surviving this wash step that might be bound nonspecifically to the beads is negligible (Friedrich et al, 2005) and therefore cannot contribute significantly to formation of PICs following the addition of UBF. UBF thus appears to activate transcription by a mechanism distinct from facilitated recruitment of SL1 and Pol I to the rDNA. Furthermore, as competitor DNA was included in these reactions to limit transcription to a single round (Panov et al, 2001), the activation by UBF is not owing to stimulation of recycling and re-initiation of Pol I during multiple rounds of transcription.
The rate of PIC assembly is not affected by UBF
We predicted that UBF would not accelerate PIC formation, given that it did not appear to stimulate recruitment of basal factors to the rDNA promoter template. This was confirmed by measuring the time-dependent formation of transcriptionally active PICs for UBF-activated versus basal transcription. PIC assembly was initiated in the presence or absence of UBF and allowed to proceed for various periods of time. The level of transcriptionally active PICs formed at each time point was then determined by measuring the amount of transcripts produced from these PICs in a 30 min transcription reaction. A comparison of the relative increase in transcript levels for UBF-activated and basal transcription revealed that the rate of PIC formation was not increased in the presence of UBF (Figure 2A). Indeed, no difference was found in the calculated rate constants (Panov et al, 2001) for PIC formation in the absence or presence of UBF (1.5±0.3 × 10−2 s−1).
Figure 2.
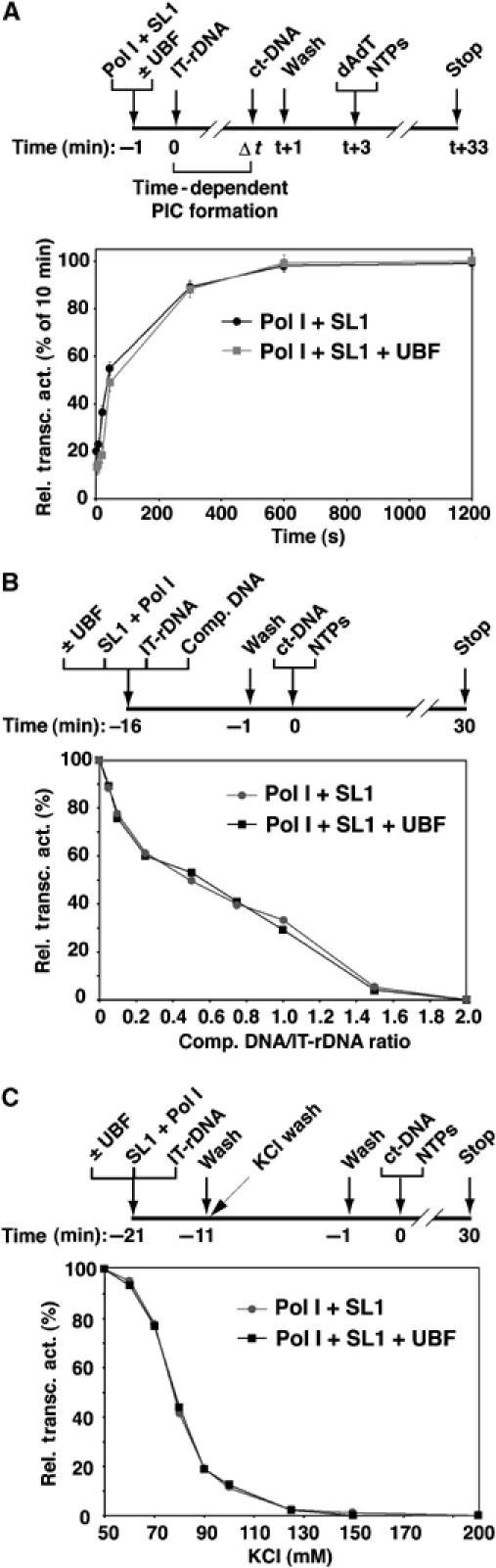
Analysis of the assembly and stability of PICs in the presence or absence of UBF. (A) UBF does not increase the rate of PIC assembly. Immobilized template (2.5 μl of IT-rDNA; Fr3) was incubated in a final volume of 20 μl with Pol Iβ and SL1, without and with 100 ng UBF on ice for varying periods (t, seconds) as indicated. Ct-DNA (0.5 μg) was added to stop PIC formation, and templates were washed in TM10i/0.05 buffer. Transcription was initiated by the addition of NTPs and 2 μg poly(dA.dT). Transcripts were detected in S1 nuclease protection assays. Transcript levels of three independent experiments were quantitated by phosphorimaging, expressed (in percentage), separately for basal and UBF-activated transcription, relative to their maximal levels at 20 min (set at 100%) and plotted against time. Basal transcription by SL1 and Pol Iβ is depicted in black and UBF-activated transcription in grey. (B) The affinity of SL1–Pol I for promoter DNA is not affected by UBF. In a competition experiment for DNA binding by SL1 and Pol I between nonspecific and promoter DNA, IT-rDNA (Fr3, 125 ng) and increasing amounts of a 423 bp nonspecific DNA fragment (comp. DNA, derived from pBR322) were incubated with Pol Iβ and SL1 on ice for 15 min in a final volume of 20 μl with or without 50 ng UBF. Beads were washed in TM10i/0.05 buffer, and single-round transcription was initiated by adding NTPs and 0.5 μg ct-DNA. Transcript levels were determined in an S1 nuclease protection assay and quantitated by phosphorimaging. The signals were expressed as a % of activity (100% set for no competitor DNA) and plotted against the molar ratio between nonspecific (comp.) and promoter DNA (IT-rDNA). (C) The salt stability of Pol I in the PIC is not affected by UBF. IT-rDNA (Fr3, 125 ng) was incubated in a final volume of 20 μl with Pol Iβ and SL1 on ice for 15 min with or without 50 ng UBF. Beaded templates were washed in TM10i/0.05 and incubated for 10 min at the indicated KCl concentrations (50–200 mM) in TM10. Then, the beaded templates were re-equilibrated to TM10i/0.05 and single-round transcription was initiated by adding NTPs and 0.5 μg ct-DNA. Transcript levels were determined by S1 nuclease protection assay and quantitated by phosphorimaging. The signals were expressed as a % of activity (100% set at 50 mM KCl) and plotted against the salt concentrations (mM KCl).
Affinity of SL1–Pol I for promoter DNA is unaltered by UBF
We analysed the effect of UBF on highly purified SL1 and Pol I in the assembly of functional PICs in the presence of competing nonspecific DNA. Interestingly, the relative decrease in transcriptional activity for PICs assembled with or without UBF and challenged with increasing amounts of competitor DNA was similar over the entire range of competitor to promoter template ratios (Figure 2B). These results suggest that UBF does not significantly alter the affinity and specificity of interaction of SL1–Pol I for promoter DNA, consistent with an inability of UBF to stimulate the rate of PIC formation.
UBF does not increase the stability of Pol I in the PIC
To test whether UBF stabilizes Pol I in the PIC, we pre-assembled PICs with or without UBF, and then subjected these complexes to increasing salt concentrations known to affect the stability of Pol I in the PIC (Panov et al, 2001). The relative amounts of functional PICs remaining on the rDNA after salt treatment were determined by transcription assays under salt conditions established to be optimal for transcription in this human reconstituted transcription system (Figure 2C). A drop in the stability of functional PICs occurred with increasing salt concentrations. Initiation was effectively precluded by KCl concentrations in excess of 150 mM, consistent with findings in extracts from murine lymphoma cells (Gokal et al, 1990) and in HeLa nuclear extracts (Panov et al, 2001). No significant difference in salt stability was observed between PICs of SL1–Pol I and PICs assembled in the presence of UBF, suggesting that UBF does not activate transcription by increasing PIC stability.
UBF increases the rate of RNA synthesis
We next asked whether UBF stimulates the rate of RNA synthesis following PIC formation. We measured the rate of RNA synthesis (by S1 nuclease protection assay) in single-round transcription from pre-assembled PICs that did or did not include UBF (Figure 3). In this reconstituted transcription system with purified factors, transcripts continued to accumulate for about 10–20 min. This likely reflects asynchronous firing of PICs and single-round transcription rather than multi-round transcription following re-initiation events because the level and kinetics of transcription were not altered by inclusion of competitor DNA (Supplementary Figure S1A, lanes 1 and 2 and S1B) at a concentration that prevents re-initiation and multi-round transcription in a nuclear extract (Supplementary Figure S1A, lanes 4 and 5). Calculated rate constants for productive RNA transcript synthesis (Panov et al, 2001) reflect the combined rate constants of all steps following PIC formation, including promoter opening, initiation, promoter escape and elongation. The rate constant of UBF-activated RNA synthesis was three times that of basal transcription (3.4±0.3 × 10−3 and 1.1±0.1 × 10−3 s−1, respectively). It follows that UBF can stimulate transcription at a step subsequent to PIC assembly, and, therefore, at promoter opening, initiation, promoter escape or elongation.
Figure 3.
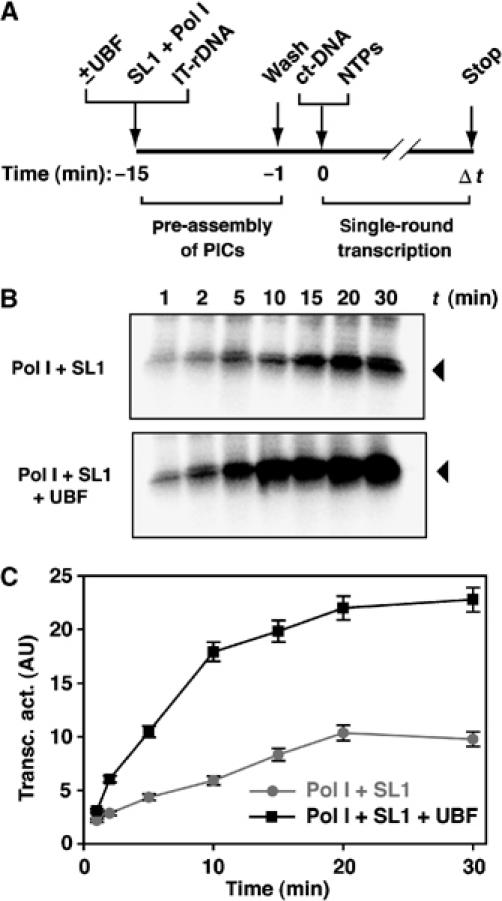
UBF increases the rate of RNA synthesis. (A) To determine the rate constant for RNA synthesis in the presence or absence of UBF, 50 μl of immobilized template (50 ng Fr3 per 1 μl of beads) was incubated for 15 min on ice in a final volume of 200 μl with 20 μl Pol Iβ, 5 μl SL1 and with or without 750 ng UBF. Templates were washed in TM10i/0.05 and single-round RNA synthesis was initiated by adding NTPs and 10 μg of ct-DNA. Samples (20 μl) were taken at varying time points (Δt, min) and the RNA from each time point was isolated and annealed to the 5′- labelled oligonucleotide (−20 to +40) in S1 nuclease protection assay. (B) Transcript levels were assessed in the S1 nuclease protection assay (arrowheads). (C) Transcript levels were quantitated by phosphorimaging. Transcriptional activity is expressed in arbitrary units and plotted against time. The grey line represents basal transcription supported by SL1 and Pol Iβ at the rDNA promoter, whereas the black line describes activated levels of transcription in the presence of UBF. The calculated rate constants are 3.4±0.3 × 10−3 s−1 for UBF-activated RNA synthesis and 1.1±0.1 × 10−3 s−1 for basal transcription.
A stimulatory role for UBF in transcription distinct from initial promoter opening and abortive initiation
We investigated the relationship between promoter opening and the requirement for Pol I transcription factors in efficient initiation of transcription with engineered heteroduplex promoter templates (Figure 4A). Despite the open configuration at the start site, Pol I alone did not initiate specifically from these ‘bubble' templates (data not shown). First, we assessed the levels of α-amanitin-resistant transcription supported by these pre-melted templates in nuclear extracts (Figure 4B). Overall, the heteroduplex promoters showed increased levels of transcription initiation compared to the wild-type (WT) homoduplex promoter and, thus, pre-opening facilitated transcription initiation from these linear promoter fragments (Figure 4B, compare lanes 2–5 with lane 1). The rDNA promoter with mutations introduced in the non-template strand to create a heteroduplex with three unpaired bases (HD3-t) supported the highest level of transcription. Further opening of the promoter, achieved using HD4-t and HD5-t, resulted in a relative lower level of transcription compared to that seen with HDt-3 (Figure 4B, compare lanes 4 and 5 with lane 3). Homoduplex variants of the rDNA promoter containing the M4 and M5 mutations supported reduced levels of transcription compared to the WT template (Supplementary Figure S2), suggesting that the M4 and M5 promoter mutations were responsible for the lower levels of transcription from the heteroduplexes HD4-t and HD5-t compared to the HDt-3 heteroduplex. HDt-3 was selected for analysis of the effect of UBF on promoter opening as transcription from this template was dramatically higher than that from the WT template (Figure 4), owing to its artificially opened promoter, and because the M3 promoter mutations did not adversely affect transcription levels in the context of the homoduplex template (Supplementary Figure S2). We asked whether there was an increase in transcription from HDt-3, compared to WT, in a reconstituted transcription system with Pol I and SL1, and whether or not UBF could activate this transcription. We observed a dramatic increase in SL1–Pol I-directed transcription from this heteroduplex containing three unpaired bases (Figure 4C, lane 3 compared to lane 1). Crucially, transcription from this template was stimulated still further by UBF (Figure 4C, lane 4 compared to lane 3). Moreover, the fold activation of transcription by UBF was comparable to that seen on the WT homoduplex promoter (Figure 4C). These results therefore uncouple elevated levels of transcription initiation as a consequence of facilitated initial promoter opening from stimulation of transcription initiation by UBF.
Figure 4.
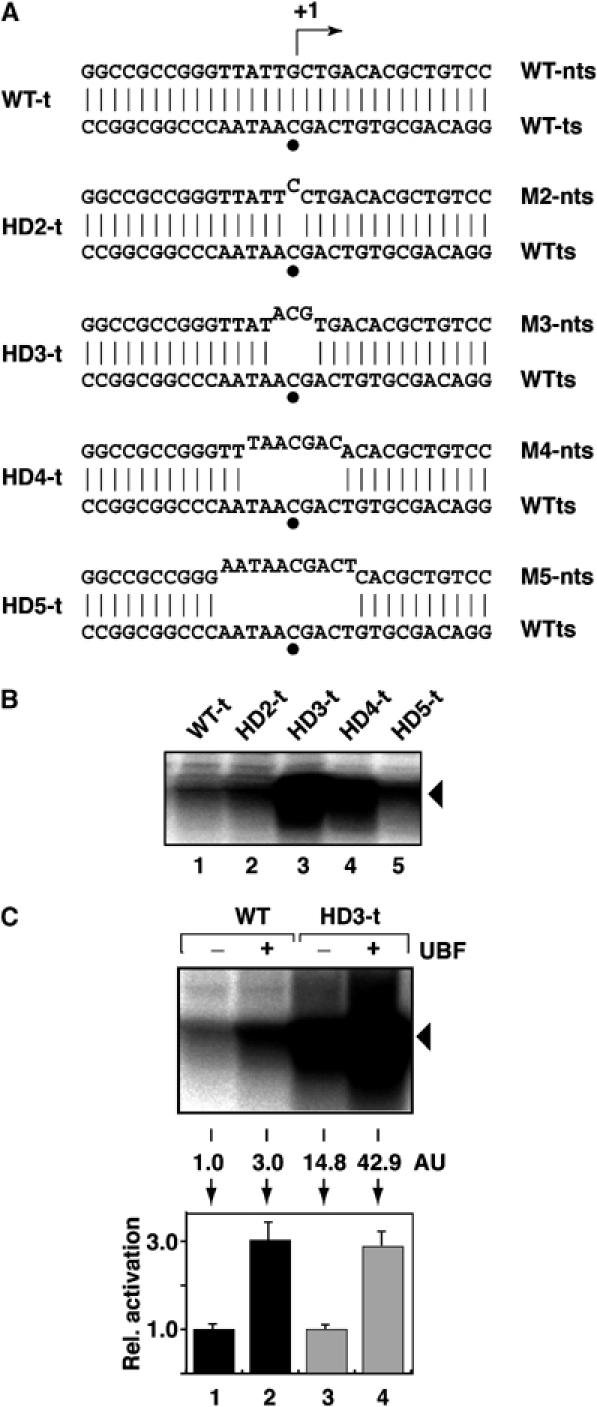
UBF activates Pol I transcription from pre-opened promoter templates. (A) Schematic of promoter sequences around the human rDNA transcription start site (+1 and black dot) for heteroduplex templates (HD2-t to HD5-t) between WT template strand (ts) and mutant non-template strands (M2- to M5-nts), with one, three, seven and 10 unpaired bases, respectively. (B) In vitro transcription assays contained 2 μl of HeLa cell nuclear extract (NE) and 120 ng of the indicated templates (see panel A). Transcripts were analysed by the S1 nuclease protection assay. (C) Reconstituted transcription reactions contained 120 ng of the WT (lanes 1 and 2) or artificial bubble template HD3-t (lanes 3 and 4), Pol Iβ and SL1 (lanes 1–4) and 75 ng UBF (lanes 2 and 4). Transcripts were analysed by S1 nuclease protection and quantitated by phosphorimaging. Relative activation was expressed as a ratio between basal and UBF-activated transcription for the WT (lanes 1 and 2, respectively; black bars) and pre-opened template (lanes 3 and 4, respectively; grey bars).
Next, we assessed whether UBF activates Pol I transcription by promoting the conversion of PICs into complexes that initiate transcription more efficiently. Promoter-specific initiation was measured by synthesis of abortive dinucleotide-primed trinucleotide transcripts (Figure 5A). There is a considerable level of GpCpU synthesis by Pol I primed with GpC (Figure 5B, lane 1), yet a significantly (∼2-fold) higher level of SL1-dependent specific abortive initiation by Pol I was detected, which was template-dependent (Figure 5B, lanes 2 and 3). UBF neither stimulated this SL1-dependent abortive initiation (Figure 5C, lanes 6–8) nor affected nonspecific abortive initiation by Pol I (Figure 5C, lanes 2–4). The finding that no increase in abortive initiation was observed in the presence of UBF suggests that UBF does not stimulate initiation of transcription and provides further evidence against a role for UBF in PIC assembly. Collectively, the data from experiments using the pre-opened promoter template and the abortive initiation assay argue that UBF stimulates Pol I transcription at a step after initial rDNA promoter opening and phosphodiester bond formation, implicating promoter escape or elongation as possible targets of UBF in activation of transcription.
Figure 5.
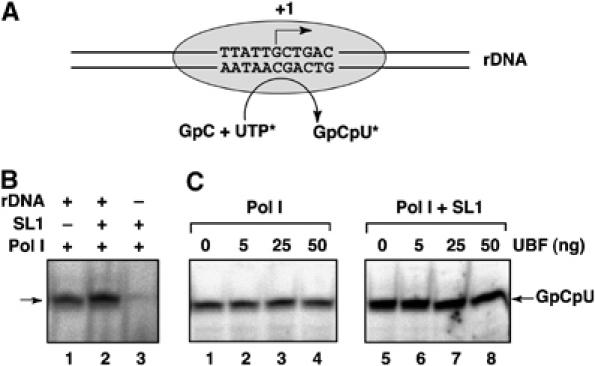
UBF does not stimulate SL1-dependent abortive initiation by Pol I. (A) Schematic representation of the start site sequence of the human rRNA gene promoter. In the abortive initiation assay, pre-assembled PICs (with and without UBF) were provided with the dinucleotide GpC (corresponding to the first 2 nt in the pre-rRNA) and [α-32P]UTP. Abortive initiation by Pol I yields 32P-labelled trinucleotide products, GpCpU. (B) rDNA (Fr3; lanes 1 and 2), Pol Iβ (lanes 1–3) and SL1 (lanes 2 and 3) were incubated on ice for 15 min. GpC and [α32P]UTP were then added to initiate the abortive RNA synthesis. The reaction products (GpCpU; arrow) were resolved on a denaturing 30% polyacrylamide gel. (C) rDNA (Fr3, 10 ng), Pol Iβ (lanes 1–8), SL1 (lanes 5–8) and various amounts of UBF (as indicated, lanes 1–8) were incubated in a final volume of 20 μl on ice for 15 min. Abortive RNA synthesis was initiated and analysed as in panel B.
Elongation of Pol I transcription in vitro is not a target for UBF activation
So, does UBF affect elongation of transcription in our reconstituted transcription system? We assessed the effect of UBF on both nonspecific end-to-end transcription by Pol I of an rDNA fragment (in the absence of SL1) and on specific transcription (in the presence of SL1) in a run-off assay (Figure 6A and B). End-to-end transcription elongation (producing a transcript of 432 nt) was not detectably stimulated by UBF (Figure 6B, lanes 1–3 and 5–7) under conditions where UBF stimulated specific transcription (producing a 239 nt transcript) in the presence of SL1 from the same promoter DNA fragment (Figure 6B, lanes 9–11 and 13–15). At relatively high concentrations of UBF, we consistently observed inhibition of specific transcription rather than activation, for example, at 1 μg of UBF (molar ratio of UBF to DNA of ∼10) there was a decrease in specific transcription (Figure 6B, lanes 12 and 16). As nonspecific transcription in this run-off assay was also inhibited by this amount of UBF (Figure 6B, lanes 4 and 8), we suggest that the repressive effect of UBF on specific transcription was the result of UBF interfering with elongation of transcription, consistent with recent findings (Stefanovsky et al, 2006). UBF also failed to activate nonspecific transcription by Pol I of random (calf thymus) DNA fragments (yielding transcript of ∼500 nt; data not shown). Importantly, the fold activation by UBF of transcription over the first 40 nt, as determined in an S1 nuclease protection assay, was similar to the fold activation by UBF of synthesis of a run-off transcript of 239 nt (Figure 6C). Collectively, these data suggest that UBF activation of reconstituted transcription is not the result of stimulated elongation of transcription.
Figure 6.
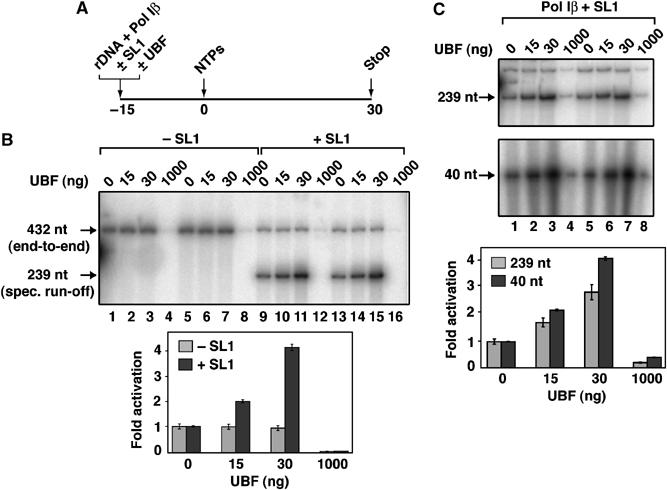
Elongation of transcription by Pol I is not activated by UBF. (A) The influence of UBF on the run-off transcription by Pol I in the presence or absence of SL1 was assessed as outlined schematically. UBF (0, 15, 30 and 1000 ng) was added to rDNA (Fr4, 150 ng) promoter fragment with Pol Iβ, with or without SL1. Reactions of 20 μl (final volume) were incubated on ice for 15 min. NTPs and [α-32P]CTP were then added to initiate the RNA synthesis. Radiolabelled transcripts were resolved on a denaturing 11% polyacrylamide gel. (B) Transcription reactions were performed as outlined in panel A. End-to-end transcription by Pol I (in the absence of SL1) yielded run-off transcripts of 432 nt, whereas specific initiation of transcription (in the presence of SL1) from the same promoter template yielded run-off transcripts of 239 nt. The experiment has been repeated three times and two representative experiments are shown. Transcript levels (432 nt for the ‘−SL1' and 239 nt for the ‘+ SL1' transcription reactions), determined with a phosphorimager, were expressed relative to the sample that did not contain UBF, set at 1.0 (lanes 1, 5, 9 and 13). (C) UBF activates synthesis of long (239 nt) and short (40 nt) transcripts with similar efficiency. UBF (0, 15, 30 and 1000 ng; lanes 1 and 5, 2 and 6, 3 and 7, and 4 and 8, respectively) was added to specific transcription reactions with immobilized rDNA promoter template (Fr4), Pol Iβ and SL1. Reactions (20 μl), in duplicate, were incubated for 20 min on ice and templates were washed in TM10i/0.05 to remove unbound factors. The single-round transcription was initiated upon addition of NTPs (including [α-32P]CTP) and 1 μg ct-DNA. After 45 min at 30°C, reactions were divided in two. Half of the reaction was analysed in a run-off assay, which yielded a full-length transcript of 239 nt (upper panel), and the other half by S1 nuclease protection, which yielded a protected fragment of the first 40 nt (lower panel). Products were quantitated by phosphorimaging. The experiment has been repeated three times and two representative experiments are shown. Fold-activation of transcription was expressed as a ratio between UBF-activated and basal transcription, for the 239 and 40 nt transcripts at the different UBF concentrations.
UBF stimulates transcription during promoter escape by Pol I
Taken together, the data suggest that UBF can activate Pol I transcription in vitro by stimulating an early stage of transcript synthesis following initiation and before elongation, at the step of promoter escape/clearance. Should UBF act to stimulate promoter escape, it would facilitate conversion of a Pol I complex that produces abortive transcripts of up to ∼10–15 nt to a stable elongating complex that produces full-length transcripts. However, demonstration of this was precluded owing to the sensitivity of the assay; no promoter proximal transcripts were detectable under any of our run-off transcription conditions with SL1 and Pol Iβ, without or with UBF (Supplementary Figure S3). Therefore, we designed an alternative assay to assess the effect of UBF on promoter escape by Pol I.
We reasoned that if UBF stimulates promoter escape, UBF-dependent activation would be detectable only at the very early stages of transcription and there would be no activation following completion of promoter escape and clearance by Pol I. Therefore, we constructed T-less templates (Figure 7A), allowing us to stall Pol I at defined distances from the transcription start site by omission of UTP (no run-off transcripts were detectable in the absence of UTP; Supplementary Figure S4) and then resume transcription to yield full-length transcripts by inclusion of all four NTPs (Figure 7C and Supplementary Figure S4). We tested whether or not UBF could activate this resumed transcription (Figure 7B–D). Control transcription reactions determined that all T-less templates have equal promoter strength, as they yielded similar levels of basal transcription (minus UBF) and of activated transcription when UBF was added during PIC formation and before stalling (bs) of Pol I (Figure 7C and D). Immobilized templates were used for these studies because we could stringently ‘wash' the templates (TM10i/0.25 M KCl; Figure 7B), an essential step in the procedure, as this step removed relatively unstably associated Pol I in (not yet active) PICs from the promoter (Panov et al, 2001), and therefore allowed us to analyse the effect of UBF on transcription specifically from stalled ‘stable' Pol I ternary complexes on the T-less templates. Crucially, UBF added after stalling (as) of Pol I activated transcription from each T-less template to a different extent (Figure 7C and D). Specifically, the results indicate that UBF can activate transcription when added after Pol I synthesis of the first 10 nt to the same extent as when added during PIC formation (Figure 7C, lanes 7 and 8 compared to lanes 9 and 10, and Figure 7D). This demonstrates that UBF activates transcription at a step following both PIC assembly and incorporation of the first few nucleotides, consistent with data presented above. UBF activates transcription with decreasing efficiency after Pol I has synthesized 15 nt, with little or no effect of UBF on run-off transcription after Pol I has synthesized 31 nt (Figure 7C and D), supporting our conclusion that UBF does not activate transcription in vitro by stimulating elongation following promoter clearance by Pol I.
Figure 7.
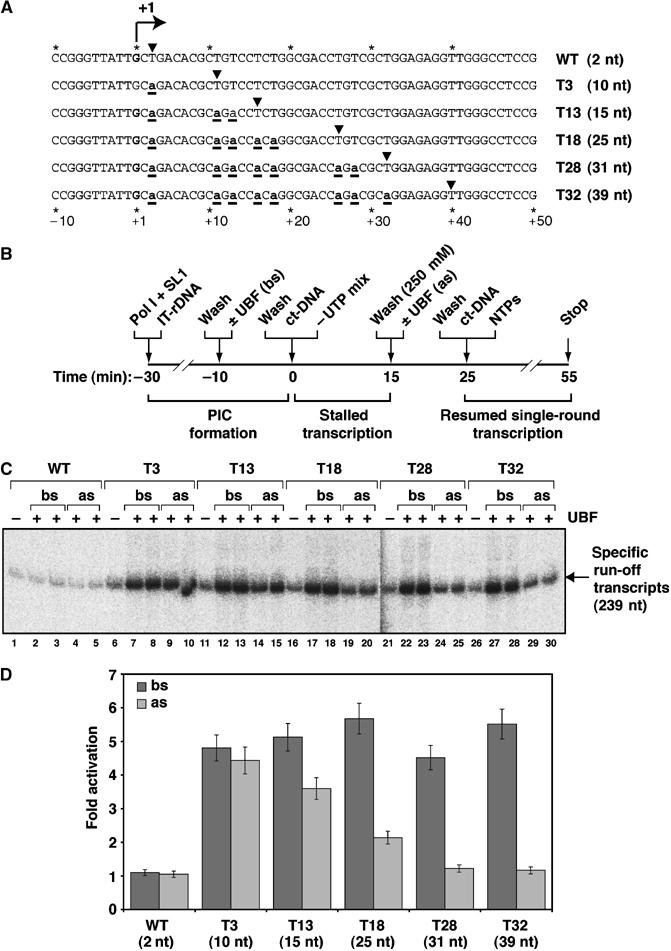
UBF activates transcription from stalled Pol I complexes only when Pol I is present in the vicinity of the transcription start site. (A) Schematic of part of the rDNA template strand sequences (−10 to +50) of WT and T-less (T3–T32) templates. The transcription start site (+1) is in bold type. Positions of T to A substitutions are underlined. The first thymidine residue in each transcribed region, indicated by arrowheads, corresponds to the predicted stall site of Pol I (following transcription in the absence of UTP). The distance from the start site of transcription to the site of stalled Pol I equals the length of the RNA transcript, shown in brackets. (B) Outline of the analysis of the effect of UBF on transcription when added before (bs) or after (as) stalling of Pol I at defined locations downstream of the transcription start site. Immobilized templates (IT-rDNA; WT or T-less Fr4 derivatives; 500 ng) were incubated in a final volume of 15 μl with Pol Iβ and SL1 for 20 min on ice. Templates were washed in TM10i/0.05 and incubation was continued for another 10 min without or with (32 or 53 ng) UBF (the combined period is referred to as ‘PIC formation'). Templates were then washed in TM10i/0.05, and transcription was initiated by adding the ‘−UTP mix', containing CTP, ATP and GTP, and 1 μg ct-DNA. Reactions were incubated for 15 min at 30°C (‘stalled transcription'). Pol I–template complexes stalled at various distances downstream of the start site, were then washed with TM10i/0.25, to remove Pol I not active in transcription and select for stable ternary complexes, and re-equilibrated in 15 μl TM10i/0.05. Incubation was continued for another 10 min on ice, without or with UBF (32 or 53 ng). Templates were washed in TM10i/0.05, and transcription was resumed upon addition of all NTPs (including [α-32P]CTP) and 1 μg ct-DNA for 15 min at 30°C (‘single-round transcription'). (C) Run-off transcripts (239 nt) from WT and T-less templates T3–T32 were analysed on a 6% denaturing polyacrylamide gel and quantitated by phosphorimaging. Transcription in the absence of UBF (basal, −) was compared to transcription in the presence of UBF (activated), where UBF was added before Pol I was stalled (bs) or when UBF was added after Pol I was stalled (as). Reactions with UBF ‘bs' and ‘as' contained 32 ng (left lane of pairs) or 53 ng (right lane of pairs). (D) Fold activation of transcription, expressed as a ratio between UBF-activated and basal transcription, for different stalled complexes when UBF was added before Pol I was stalled (bs, dark grey bars) or when UBF was added after Pol I was stalled (as, light grey bars). Note that as the transcript levels in the paired reactions with the two UBF amounts were similar (see panel C), the average is shown.
The inability of UBF added before or after stalling of Pol I at +3 to activate transcription from the WT template (Figure 7C, lanes 2–5, and D) is due to instability of Pol I complexes at the initial phase of transcription (Supplementary Figure S5), during which they probably undergo abortive initiation, and this is reflected in the lower amount of basal transcription from this template (Figure 7C, lane 1 compared to lanes 6, 11, 16, 21 and 26). This instability may be related to that seen for other RNA polymerases, for example, Pol II is unstable at the very early stages following initiation and only synthesis of a 4-nt RNA commits Pol II to promoter escape (Kugel and Goodrich, 2002), and similar distinct transitions have also been observed for T7 and Escherichia coli RNA polymerases after synthesis of a 4-nt RNA (Bowser and Hanna, 1991; Cheetham and Steitz, 1999, 2000).
The results suggest that UBF activates transcription following initiation of transcription and while the polymerase is in proximity to the transcription start site and that, once Pol I escapes and clears the promoter, elongation of transcription in this reconstituted system is unaffected by UBF. Therefore, we propose that UBF activates Pol I transcription at the step of promoter escape by Pol I.
Discussion
We provide evidence for a novel mode of activation by UBF, discrete from that of facilitated recruitment and stabilization of SL1 and/or Pol I at the rDNA promoter. The data presented here argue against UBF activation via recruitment of these factors, at least in vitro, for the following reasons. First, UBF activates transcription from pre-assembled SL1–Pol I–rDNA complexes and, importantly, the level of transcription supported is no different from that in which the PICs were assembled from free components in the presence of UBF (Figure 1). The order of addition experiments does not entirely exclude the possibility that activation by UBF following pre-assembly of an SL1–Pol I–rDNA complex might partly result from recruitment by UBF of a fraction of nonspecifically DNA-bound SL1 and/or Pol I to the core promoter DNA fragment to yield more PICs. However, the lack of stimulation by UBF of both abortive initiation (Figure 5) and resumed transcription by Pol I stalled at +31 or +39 nt (Figure 7) argues strongly against this possibility. Therefore, the number of functional PICs formed is independent of the order of PIC assembly, specifically whether UBF is present before, during or after the assembly reaction. Secondly, the rate constant for formation of an SL1–Pol I complex at the immobilized ribosomal promoter is indistinguishable from that for a PIC assembled in the presence of UBF, under conditions where we observe activation of transcription. Thirdly, UBF does not modulate the affinity and selectivity of SL1–Pol I for promoter DNA, does not detectably alter the stability of SL1 binding to DNA (Friedrich et al, 2005) and does not influence the salt stability of Pol I in the PIC, consistent with the failure of UBF to change the rate of productive PIC formation. We conclude that activation of Pol I transcription by UBF occurs at a stage subsequent to PIC formation.
Significantly, UBF increases the rate of RNA synthesis from pre-assembled PICs (Figure 3), implying a positive effect of UBF on initiation, promoter escape/clearance and/or elongation. UBF fails to stimulate abortive initiation (Figure 5), and, furthermore, while pre-opening of the promoter facilitates transcription initiation, UBF activates the heteroduplex ‘pre-opened' promoter to the same extent as the WT homoduplex template (Figure 4). Therefore, we infer that UBF does not affect the initiation frequency of the PICs and acts at a step subsequent to promoter opening and first phosphodiester bond formation. Furthermore, our combined data from Figures 6 and 7 indicate that transcription elongation by Pol I, whether specifically or randomly initiated in vitro, is not stimulated by UBF. Our analysis of the ability of UBF to activate resumed transcription following stalling of Pol I at defined distances close to the transcription start site demonstrates that UBF can activate transcription following first phosphodiester bond formation and at the early stages of transcript synthesis, most efficiently during transcription of the first 10–15 nt and through incorporation of up to 30 nt, but not thereafter (Figure 7). We propose that UBF can activate transcription at the step of promoter escape, operationally defined here as the short phase following initiation of transcription that includes formation of the first ∼10–15 phosphodiester bonds of the nascent transcripts and conversion of the transcribing polymerase from the unstable initiation mode to elongation mode. Thus, UBF can activate transcription by tipping the balance in favour of productive versus non-productive initiated transcription complexes (Carpousis and Gralla, 1980; McClure, 1985; Dvir, 2002). The reconstituted Pol I transcription system used here does not support multi-round transcription and therefore this might have precluded the direct observation of the conversion of small abortive transcripts into longer products in the presence of UBF as Pol I escapes the promoter (see Supplementary data).
Our study demonstrates the specific role of UBF in the ‘net' (or ‘true') activation of transcription at the rDNA promoters in vitro and this involves stimulated promoter escape by Pol I rather than stimulated PIC assembly or stability. This activation function for UBF in promoter escape does not exclude additional roles for UBF in the regulation of rRNA gene expression. The distribution of UBF throughout transcribed and non-transcribed regions of the entire rDNA repeat in cells, and at transcriptionally silent artificial arrays of enhancer sequences (UBF binding sites) in genomic loci other than rDNA, is likely to influence the organization of chromatin (O'Sullivan et al, 2002; Mais et al, 2005). UBF bound at those sites could sequester SL1 and Pol I (Mais et al, 2005) and increase the local concentration of these factors on DNA, but there is no strict correlation between the regions transcribed and those to which UBF binds. UBF bound to transcribed regions of the rDNA has been suggested to repress, rather than activate, transcription elongation and such repression can be partly alleviated following growth factor-induced ERK phosphorylation of UBF (Stefanovsky et al, 2006). At concentrations at which UBF activates transcription in vitro, UBF does not affect elongation (Figure 6B), whereas at significantly higher concentrations than those at which UBF activation of transcription is observed, UBF represses transcription elongation by Pol I in vitro (Figure 6B; Stefanovsky et al, 2006). In addition to this ‘dampening' function of UBF on elongation, there is also evidence to suggest a role for UBF as an anti-repressor (Kuhn and Grummt, 1992; Brou et al, 1993; Pelletier et al, 2000). Thus, it is likely that UBF influences rRNA gene expression at multiple levels, functioning as an activator at promoter escape by Pol I, as a regulator of elongation and as an anti-repressor in the context of chromatin in vivo.
The activity of UBF and its interactions with other components of the Pol I transcription machinery are modulated by post-translational modifications. The insect cell (recombinant baculovirus) expressed and purified human UBF used in this study contains modifications of residues known to be important for UBF activation function in vitro. UBF is phosphorylated by cyclin-dependent kinases (Klein and Grummt, 1999; Voit et al, 1999), and one site in particular is critical for interaction of UBF with Pol I and in the activation of transcription in vitro (Voit and Grummt, 2001). The interaction of UBF with Pol I-specific subunits CAST/hPAF49 and PAF53 is important for stimulated transcription at a step following PIC formation (Panov et al, 2006), and hence might be critical to promoter escape by Pol I. Furthermore, the growth factor-induced cyclic ERK1/2 phosphorylation of sites in HMG boxes 1 and 2 of UBF (Stefanovsky et al, 2001b) has been suggested to alter the interactions of UBF with DNA qualitatively, perhaps to facilitate passage of Pol I through UBF molecules bound throughout the transcribed region and so modulate elongation of transcription by Pol I (Stefanovsky et al, 2006). It is unlikely that UBF in our reconstituted transcription system undergoes such cyclic ERK-mediated phosphorylation events, yet it activates transcription robustly. Additionally, the carboxy-terminus of UBF, which contributes to its activation function (Jantzen et al, 1992), is prominently and differentially phosphorylated in vivo in response to cell growth conditions, and appears to be at the end of several signalling pathways. For example, recombinant CK2 in vitro can phosphorylate this domain of UBF (O'Mahony et al, 1992a, 1992b; Voit et al, 1992), and mutation of CK2-phosphorylated serine residues in this domain impairs the ability of UBF to activate transcription (Voit et al, 1995). We have recently obtained evidence to suggest that CK2 phosphorylation of UBF stimulates the ability of UBF to activate Pol I transcription through enhanced stabilization by SL1 of CK2-phosphorylated UBF at the rDNA promoter (Panova et al, 2006). The carboxy-terminus of UBF can also be phosphorylated by nuclear PI3-kinase p110 subunit, in response to insulin-like growth factor I (Drakas et al, 2004), and inhibition of mTOR signalling with rapamycin leads to a rapid dephosphorylation of the carboxy-terminus of UBF, which significantly reduces its ability to associate with SL1, and a loss of serum-induced activation of rDNA transcription by Pol I (Hannan et al, 2003). UBF activity, and Pol I transcription, can be repressed by tumour suppressor retinoblastoma protein (Rb) binding to UBF (Cavanaugh et al, 1995), and this interaction is mutually exclusive to CBP recruitment and acetylation of UBF, which enhances Pol I transcription (Pelletier et al, 2000). Acetylation of UBF enhances the interaction of UBF with Pol I (Meraner et al, 2006). The interaction of Rb with UBF interferes with the binding of UBF to SL1 (Hannan et al, 2000). Collectively, these data stress the importance of the UBF–SL1 and UBF–Pol I interactions in activation of transcription by UBF.
What could be the functional significance of interactions between UBF and SL1 (Bell et al, 1988; Jantzen et al, 1992; Kwon and Green, 1994; Beckmann et al, 1995; Kihm et al, 1998; Tuan et al, 1999) and between UBF and Pol I (Schnapp et al, 1994; Hanada et al, 1996; Voit and Grummt, 2001) in promoter escape? The interaction of UBF with SL1 is important in stabilization of UBF at the rDNA promoter (Friedrich et al, 2005) and the kinetic stability (increased lifetime) of this activator–DNA complex is likely to be a major determinant in the ability of UBF to activate transcription at promoter escape. The interaction of UBF with Pol I might be of direct relevance in promoting the transition and conformational changes in Pol I associated with the escape of Pol I from the promoter. Additionally, the interactions of UBF with SL1 and Pol I in cells might increase the proportion of UBF-containing Pol I holoenzymes (Seither et al, 1998; Hannan et al, 1999; Miller et al, 2001) and/or the local concentration of SL1 and Pol I on DNA (Mais et al, 2005) before the specific targeting of Pol I to the rDNA promoter, which is directed by SL1 (Miller et al, 2001; Friedrich et al, 2005).
In the transition of Pol I from initiation to elongation, UBF might function to weaken polymerase–promoter interactions by influencing protein–protein interactions of the PIC, facilitating conformational changes to yield productive polymerases, altering the local DNA topology, or a combination of these, and we present possible, not mutually exclusive, models for UBF activation by stimulation of promoter escape, with the aim to provide a framework for future experimentation. The first proposes a role for UBF in disrupting protein–protein interactions of the PIC, leading to promoter escape. The composition of the Pol I enzyme complex changes during the early stages of transcription, most notably by the dissociation of RRN3 from Pol I, potentially via disruption of its interaction with the Pol I subunit RPA43 (Brun et al, 1994; Milkereit and Tschochner, 1998; Peyroche et al, 2000; Hirschler-Laszkiewicz et al, 2003). This compositional change could occur as a consequence of promoter escape of Pol I, or instigate, or at least be intimately associated with, the process. One possibility is that UBF interacts with and facilitates changes in Pol I, following transcription initiation, which could promote release of hRRN3 and, concomitantly, assist in disruption of the interaction between Pol I and SL1. As UBF interacts with SL1, UBF might also convert SL1 from a complex that recruits Pol I to one that efficiently releases the enzyme by interfering with interactions between SL1 subunits TAFI110 and TAFI63 and hRRN3 (Miller et al, 2001). Events such as these may be critical for efficient promoter escape by Pol I.
In the second model, UBF converts Pol I to a processive enzyme complex. UBF might function to maintain the PIC intact until the polymerase has undergone this conversion, although a more active role for UBF in the conversion is suggested by the following. UBF interacts with at least two Pol I-specific subunits of the enzyme complex, CAST/hPAF49, the human orthologue of yeast RPA34.5 (Panov et al, 2006), and PAF53 (Hanada et al, 1996), the mammalian orthologue of yeast RPA49, and in so doing could perhaps facilitate conformational changes of Pol I (De Carlo et al, 2003) and its interactions with the template DNA in the critical transition of the enzyme between initiation and elongation (Panov et al, 2006).
In the third model, alterations in local DNA topology by UBF might facilitate promoter escape by Pol I. We have shown that UBF activation is independent of initial promoter opening and formation of the first few phosphodiester bonds, but it is possible that further opening during formation of the transcription bubble is stimulated by UBF. Another link between UBF influencing DNA topology and promoter escape is proposed in our ‘spring-load model'. It is likely that in the early stages of transcription Pol I draws the DNA through itself, while maintaining its interactions with the transcription factors at the promoter, such that the DNA would gather into a loop. In this model, UBF functions as an anchor at the promoter, through its ability to produce a structure with DNA resembling a nucleosome in DNA content and mass (Bazett Jones et al, 1994), thereby constraining the DNA. Torsional strain accumulates in the DNA loop as transcription proceeds, until the energy of DNA resilience disrupts the protein–protein interactions between the polymerase and transcription factors of the PIC, allowing Pol I to escape the promoter efficiently and continue synthesis of the pre-rRNA.
The novel role defined for UBF in Pol I transcription in activating the kinetics (rate) of RNA synthesis, stimulating promoter escape, is fundamentally different from that of recruitment of basal transcription factors and polymerase to the promoter, a defining feature of archetypal activators of transcription (Ptashne and Gann, 1997). Crucially, the ability of UBF to activate transcription at the step of promoter escape would enable stimulation of transcription in response to growth factors and nutrients both at previously inactive promoters following PIC assembly and also at SL1-engaged promoters at each successive round of transcription in re-initiation. The high levels of rRNA synthesis required for cell growth and division likely necessitate a high frequency of loading of Pol I at the rDNA promoter. This is directly affected by mechanisms that regulate the ability of SL1 to recruit Pol I; PIC assembly is instigated by SL1 core promoter binding and controlled in part by the availability of initiation-competent RRN3–Pol I complexes (Pol Iβ) (reviewed by Russell and Zomerdijk, 2005). However, a high frequency of loading of Pol I is only possible under conditions where polymerases are highly processive, efficiently escaping and clearing the promoter to allow SL1–promoter complexes to recruit the next Pol I. Therefore, UBF might also contribute indirectly to recruitment of Pol I to SL1-engaged promoters by stimulating promoter escape.
Our previous kinetic analyses had defined promoter escape and clearance as rate-limiting in reconstituted transcription by Pol I (Panov et al, 2001), yet it was not known whether there were critical regulators of this key step in the Pol I transcription cycle. Here, we have identified UBF as an important regulator of this rate-limiting step in Pol I transcription. Promoter escape has also been reported to be a rate-limiting step in Pol II-mediated transcription (Kugel and Goodrich, 1998). Examples have emerged of activators that regulate Pol II transcription at both initiation and promoter escape (Liu et al, 2001; Fukuda et al, 2004). Hence, stimulation of promoter escape by the nuclear RNA polymerases could be an important and more general mechanism by which transcription activators function.
Materials and methods
Protein purification
Pol Iβ and SL1 (free of UBF) were purified as described (Miller et al, 2001). Recombinant human UBF1, free of nucleic acid, was purified as outlined in Supplementary data (Supplementary Figure S6).
Preparation of rDNA promoter templates
Biotinylated human rDNA promoter templates, WT or T-less mutants generated by site-directed mutagenesis, were synthesized by PCR (Fr4, −193 to +239 bp; Fr3, −324 to +239) and immobilized on streptavidin-coated paramagnetic beads as described (Panov et al, 2001). Generation of the heteroduplex ‘pre-opened' promoter templates (HD2-t to HD5-t) is detailed in Supplementary data.
Assembly and isolation of Pol I PICs
Purified transcription factors UBF, SL1 and Pol Iβ were gently agitated and incubated for 5–40 min on ice with immobilized template (IT-rDNA), typically 5–20 μl (50 ng DNA/μl of M280 Dynabeads) in 20–200 μl total reaction volume of 50 mM KCl (final concentration) and TM10i (50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM sodium metabisulphite, 1 mM DTT, 50 ng/μl BSA, 0.015% NP-40, EDTA-free protease inhibitor (‘i') cocktail (Roche)). After separation using a magnetic stand, beads were washed twice with two reaction volumes of TM10/0.05 M KCl to remove unbound factors, and washed templates were used to initiate transcription reactions, as outlined schematically in the figures. Competitor DNA (calf thymus (ct-) DNA or poly(dA.dT)) was used to limit transcription to a single round, as established previously (Panov et al, 2001).
Reconstituted transcription reactions
Reconstituted transcription reactions and S1 nuclease protection (−20 to +40 5′-labelled template strand probe) were performed as described previously (Miller et al, 2001; Panov et al, 2001). Rate constants were calculated as described (Panov et al, 2001). In the experiments with pre-opened promoters, the WT sequence was retained for the template strand (see Supplementary data), so that RNA synthesis from the different ‘bubble' templates could be measured by S1 nuclease protection with the same WT S1 oligonucleotide.
Run-off transcriptions were performed as follows. Pol I transcription components were pre-incubated for 20 min on ice in 25 μl with 0.25–1.5 μg immobilized rDNA promoter template in TM10i/0.05. Transcriptions were initiated with 500 μM each UTP, GTP and ATP, 25 μM CTP and 2.5 μCi [α-32P]CTP (3000 Ci/mmol), and 2 U RNasin, 0.1 mg/ml α-amanitin, 10 mM creatine phosphate and 40 ng/μl ct-DNA were added. After 30 min at 30°C, 10 U RNase-free DNase I (Roche) was added and incubated at 37°C for 5 min. Reactions were terminated at 37°C for 5 min with 200 μl of 20 mM EDTA, 200 mM NaCl, 1% (w/v) SDS, 0.25 μg/μl tRNA and 20 mg/ml proteinase K. Nucleic acids were phenol–chloroform extracted, ethanol precipitated, dissolved in formamide loading buffer and analysed on denaturing (8 M urea) polyacrylamide gels.
For abortive initiation of transcription, pre-assembled PICs on immobilized rDNA promoter template were provided with the dinucleotide GpC (1.6 mM) and [α-32P]UTP (2 μCi at 3000 Ci/mmol) to yield the 32P-labelled 3-nt product GpCpU, which was resolved on a denaturing 30% polyacrylamide gel.
Supplementary Material
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Acknowledgments
We thank Steven Renwick, Anastasia Kotsoni, Marcus Lyall and Hannah Carney for technical assistance and Dr Jim Goodrich for advice on abortive initiation assays. We thank the National Cell Culture Center (Minneapolis, MN, USA) for growing HeLa cells. We thank our colleagues in the Zomerdijk laboratory and Professor Angus Lamond and Dr Tom Owen-Hughes for advice and critical reading of the manuscript. JKF received an MRC PhD studentship. JCBMZ is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
References
- Bachvarov D, Moss T (1991) The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res 19: 2331–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett Jones DP, Leblanc B, Herfort M, Moss T (1994) Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Beckmann H, Chen JL, O'Brien T, Tjian R (1995) Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 270: 1506–1509 [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241: 1192–1197 [DOI] [PubMed] [Google Scholar]
- Bodeker M, Cairns C, McStay B (1996) Upstream binding factor stabilizes Rib 1, the TATA-binding-protein-containing Xenopus laevis RNA polymerase I transcription factor, by multiple protein interactions in a DNA-independent manner. Mol Cell Biol 16: 5572–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem J, Dobreva G, Hoffmann-Rohrer U, Iben S, Zentgraf1 H, Delius1 H, Vingron M, Grummt I (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep 1: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser CA, Hanna MM (1991) Sigma subunit of Escherichia coli RNA polymerase loses contacts with the 3′ end of the nascent RNA after synthesis of a tetranucleotide. J Mol Biol 220: 227–239 [DOI] [PubMed] [Google Scholar]
- Brou C, Kuhn A, Staub A, Chaudhary S, Grummt I, Davidson I, Tora L (1993) Sequence-specific transactivators counteract topoisomerase II-mediated inhibition of in vitro transcription by RNA polymerases I and II. Nucleic Acids Res 21: 4011–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun RP, Ryan K, Sollner-Webb B (1994) Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol Cell Biol 14: 5010–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ, Gralla JD (1980) Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry 19: 3245–3253 [DOI] [PubMed] [Google Scholar]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI (1995) Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374: 177–180 [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Steitz TA (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science 286: 2305–2309 [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Steitz TA (2000) Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr Opin Struct Biol 10: 117–123 [DOI] [PubMed] [Google Scholar]
- Comai L (2004) Mechanism of RNA polymerase I transcription. Adv Protein Chem 67: 123–155 [DOI] [PubMed] [Google Scholar]
- Comai L, Tanese N, Tjian R (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68: 965–976 [DOI] [PubMed] [Google Scholar]
- Comai L, Zomerdijk JCBM, Beckmann H, Zhou S, Admon A, Tjian R (1994) Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266: 1966–1972 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Putnam CD, Denton ML, Pikaard CS (1994) The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res 22: 2651–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlo S, Carles C, Riva M, Schultz P (2003) Cryo-negative staining reveals conformational flexibility within yeast RNA polymerase I. J Mol Biol 329: 891–902 [DOI] [PubMed] [Google Scholar]
- Drakas R, Tu X, Baserga R (2004) Control of cell size through phosphorylation of upstream binding factor 1 by nuclear phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 101: 9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A (2002) Promoter escape by RNA polymerase II. Biochim Biophys Acta 1577: 208–223 [DOI] [PubMed] [Google Scholar]
- Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JCBM (2005) TBP–TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J Biol Chem 280: 29551–29558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Nakadai T, Shimada M, Tsukui T, Matsumoto M, Nogi Y, Meisterernst M, Hisatake K (2004) Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol Cell Biol 24: 6525–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokal PK, Mahajan PB, Thompson EA (1990) Hormonal regulation of transcription of rDNA. Formation of initiated complexes by RNA polymerase I in vitro. J Biol Chem 265: 16234–16243 [PubMed] [Google Scholar]
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Hanada K, Song CZ, Yamamoto K, Yano K, Maeda Y, Yamaguchi K, Muramatsu M (1996) RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J 15: 2217–2226 [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Rothblum LI (1998) Transcription by RNA polymerase I. Front Biosci 3: d376–d398 [DOI] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI (2000) Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 19: 4988–4999 [DOI] [PubMed] [Google Scholar]
- Hannan RD, Cavanaugh A, Hempel WM, Moss T, Rothblum L (1999) Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res 27: 3720–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel WM, Cavanaugh AH, Hannan RD, Taylor L, Rothblum LI (1996) The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol Cell Biol 16: 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschler-Laszkiewicz I, Cavanaugh AH, Mirza A, Lun M, Hu Q, Smink T, Rothblum LI (2003) Rrn3 becomes inactivated in the process of ribosomal DNA transcription. J Biol Chem 278: 18953–18959 [DOI] [PubMed] [Google Scholar]
- Hu CH, McStay B, Jeong SW, Reeder RH (1994) xUBF, an RNA polymerase I transcription factor, binds crossover DNA with low sequence specificity. Mol Cell Biol 14: 2871–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344: 830–836 [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Chow AM, King DS, Tjian R (1992) Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev 6: 1950–1963 [DOI] [PubMed] [Google Scholar]
- Kihm AJ, Hershey JC, Haystead TA, Madsen CS, Owens GK (1998) Phosphorylation of the rRNA transcription factor upstream binding factor promotes its association with TATA binding protein. Proc Natl Acad Sci USA 95: 14816–14820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Green MR (1994) Modeling eukaryotic transcriptional activation. Curr Biol 4: 325–332 [DOI] [PubMed] [Google Scholar]
- Klein J, Grummt I (1999) Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci USA 96: 6096–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA (1998) Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH, and ATP on negatively supercoiled DNA. Proc Natl Acad Sci USA 95: 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA (2002) Translocation after synthesis of a four-nucleotide RNA commits RNA polymerase II to promoter escape. Mol Cell Biol 22: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I (1992) Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc Natl Acad Sci USA 89: 7340–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Green MR (1994) The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box-binding protein. J Biol Chem 269: 30140–30146 [PubMed] [Google Scholar]
- Learned RM, Learned TK, Haltiner MM, Tjian RT (1986) Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 45: 847–857 [DOI] [PubMed] [Google Scholar]
- Leblanc B, Read C, Moss T (1993) Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J 12: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, Reinberg D, Levens D (2001) Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104: 353–363 [DOI] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B (2005) UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure WR (1985) Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54: 171–204 [DOI] [PubMed] [Google Scholar]
- McStay B, Frazier MW, Reeder RH (1991) xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev 5: 1957–1968 [DOI] [PubMed] [Google Scholar]
- Meraner J, Lechner M, Loidl A, Goralik-Schramel M, Voit R, Grummt I, Loidl P (2006) Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res 34: 1798–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Tschochner H (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J 17: 3692–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JCBM (2001) hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J 20: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB, Steitz TA (2002) The involvement of RNA in ribosome function. Nature 418: 229–235 [DOI] [PubMed] [Google Scholar]
- Moss T (2004) At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev 14: 210–217 [DOI] [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY (2002) At the center of eukaryotic life. Cell 109: 545–548 [DOI] [PubMed] [Google Scholar]
- O'Mahony DJ, Smith SD, Xie W, Rothblum LI (1992a) Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res 20: 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony DJ, Xie WQ, Smith SD, Singer HA, Rothblum LI (1992b) Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J Biol Chem 267: 35–38 [PubMed] [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B (2002) UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Zomerdijk JC (2001) A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol 21: 2641–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Panova TB, Gadal O, Nishiyama K, Saito T, Russell J, Zomerdijk JCBM (2006) RNA polymerase I-specific subunit CAST/hPAF49 has a role in the activation of transcription by UBF. Mol Cell Biol 26 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panova TB, Panov Ki, Russell J, Zomerdijk JCBM (2006) CK2 associates with initiation-competent RNA polymerase I and has multiple roles in rDNA transcription. Mol Cell Biol 26 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Cote J, Moss T (2000) Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell 6: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J 19: 5473–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A (1997) Transcriptional activation by recruitment. Nature 386: 569–577 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Copenhaver GP, Denton ML, Pikaard CS (1994) The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol 14: 6476–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH (1999) Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol 62: 293–327 [DOI] [PubMed] [Google Scholar]
- Reeder RH, Pikaard CS, McStay B (1995) UBF, an architectural element for RNA polymerase I promoters. In Nucleic Acids and Molecular Biology, Eckstein F, Lilley DMJ (eds) Vol. 9, pp 251–263. Berlin, Heidelberg: Springer-Verlag [Google Scholar]
- Russell J, Zomerdijk JCBM (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A, Grummt I (1991) Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem 266: 24588–24595 [PubMed] [Google Scholar]
- Schnapp G, Santori F, Carles C, Riva M, Grummt I (1994) The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J 13: 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seither P, Iben S, Grummt I (1998) Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol 275: 43–53 [DOI] [PubMed] [Google Scholar]
- Smith SD, O'Mahony DJ, Kinsella BT, Rothblum LI (1993) Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expr 3: 229–236 [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T (2006) Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell 21: 629–639 [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T (2001a) DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res 29: 3241–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T (2001b) An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell 8: 1063–1073 [DOI] [PubMed] [Google Scholar]
- Tuan JC, Zhai W, Comai L (1999) Recruitment of TATA-binding protein–TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol Cell Biol 19: 2872–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Grummt I (2001) Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci USA 98: 13631–13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Hoffmann M, Grummt I (1999) Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J 18: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Kuhn A, Sander EE, Grummt I (1995) Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res 23: 2593–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg HG, Grummt I (1992) The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J 11: 2211–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Zomerdijk JCBM, Beckmann H, Comai L, Tjian R (1994) Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 266: 2015–2018 [DOI] [PubMed] [Google Scholar]
- Zomerdijk JCBM, Tjian R (1998) Initiation of transcription on human rRNA genes. In Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Paule MR (ed) New York, Austin, TX: Springer-Verlag [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6


