Abstract
Tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), is a chemical carcinogen thought to be involved in the initiation of lung cancer in smokers. NNK is metabolically activated to methylating and pyridyloxobutylating species that form promutagenic adducts with DNA nucleobases, e.g. O6-[4-oxo-4-(3-pyridyl)butyl]guanine (O6-POB-dG). O6-POB-dG is a strongly mispairing DNA lesion capable of inducing both G→A and G→T base changes, suggesting its importance in NNK mutagenesis and carcinogenesis. Our earlier investigations have identified the ability of O6-POB-dG to hinder DNA digestion by snake venom phosphodiesterase (SVPDE), a 3′-exonuclease commonly used for DNA ladder sequencing and as a model enzyme to test nuclease sensitivity of anti-sense oligonucleotide drugs. We now extend our investigation to three other enzymes possessing 3′-exonuclease activity: bacteriophage T4 DNA polymerase, Escherichia coli DNA polymerase I, and E.coli exonuclease III. Our results indicate that, unlike SVPDE, 3′-exonuclease activities of these three enzymes are not blocked by O6-POB-dG lesion. Conformational analysis and molecular dynamics simulations of DNA containing O6-POB-dG suggest that the observed resistance of the O6-POB-dG lesion to SVPDE-catalyzed hydrolysis may result from the structural changes in the DNA strand induced by the O6-POB group, including C3′-endo sugar puckering and the loss of stacking interaction between the pyridyloxobutylated guanine and its flanking bases. In contrast, O6-methylguanine lesion used as a control does not induce similar structural changes in DNA and does not prevent its digestion by SVPDE.
INTRODUCTION
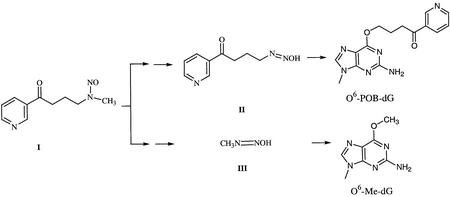
Cigarette smoking is a well known risk factor for lung cancer. The tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, (NNK; I in Scheme 1) is thought to be involved in the induction of lung cancer in smokers (1,2). Indeed, NNK has been reported to be metabolically activated to [4-(3-pyridyl)-4-oxobutyl]diazohydroxides, II and methyldiazohydroxide, III (Scheme 1), which pyridyloxobutylate and methylate guanines in DNA, respectively (2–4). If not repaired before DNA replication, NNK-induced DNA lesions can be misread by DNA polymerases, resulting in heritable mutations and the initiation of cancer (Scheme 1).
Scheme 1.
We and others have developed methods for mapping carcinogen-induced nucleobase lesions within DNA sequences by mass spectral analysis of controlled exonuclease digests (5,6). Exonuclease enzymes sequentially remove mononucleotides in either the 5′→3′ or the 3′→5′ direction. Since time controlled exonuclease digests contain a mixture of DNA fragments differing from each other by one or more mononucleotides (DNA ‘ladders’), mass differences between adjacent peaks in the resulting mass spectra correspond to individual nucleotides, making it possible to determine the complete DNA sequence (5,6). The 16mer d(AACAGC CATAT[O6-POB-dG]GCCC) was digested with 5′→3′ and 3′→5′ exonucleases in order to confirm the position of O6-[4-oxo-4-(3-pyridyl)butyl]guanine (O6-POB-dG) in the sequence (5). Interestingly, while bovine spleen phosphodiesterase (5′-exonuclease) was able to remove pyridyloxobutylated guanine nucleotide (O6-POB-dG, M = 477.2), snake venom phosphodiesterase (SVPDE, 3′-exonuclease) was incapable of bypassing the lesion (5).
The ability of O6-POB-dG to block SVPDE-catalyzed DNA hydrolysis is of potential interest because of the continuing effort to develop nuclease-resistant antisense oligodeoxynucleotides (ODNs). 3′-Exonucleases have been shown to be primarily responsible for enzymatic degradation of therapeutic ODNs in serum-containing medium and in various cell lines (7–9). The antisense activity of phosphodiester ODNs in cells is primarily determined by their resistance to exonucleolytic degradation (10). Monia et al. (10) have proposed that SVPDE can be used as a model nuclease in experiments that examine nuclease sensitivity of antisense cellular nuclease responsible for degradation of antisense oligonucleotides. Many chemically modified antisense ODNs have been designed to confer nuclease resistance while preserving the antisense activity, including the introduction of cationic groups (9,11), bulky tricyclic base moieties (12), phosphothiolate backbone linkages and 2′-substituted sugars (10), self-forming 3′ hairpins (13), 2′-O,4′-C-ethylene linked nucleosides (14), 3′-conjugates (8), ODNs of reversed polarity (2′,5′) (15), and α-anomeric oligonucleotides (16).
Although SVPDE alone is commonly used to assess the exonucleolytic stability of chemically modified ODNs, structural differences between the active site geometries of different nucleases may render O6-POB-dG-containing DNA less resistant towards physiologically relevant 3′-exonucleases. We thus examined the ability of three other common 3′-exonucleases [E.coli exonuclease III, E.coli DNA polymerase I (KF), and T4 DNA polymerase] to hydrolyze O6-POB-dG-containing DNA. Furthermore, torsional analysis and molecular modeling simulations of O6-POB-dG- containing DNA were performed in an attempt to uncover the structural basis for the observed exonuclease resistance of pyridyloxobutylated DNA.
MATERIALS AND METHODS
3-Hydroxypicolinic acid (HPA) was purchased from Bruker Daltonics (Billerica, MA). Bio-gel P-6 cartridges were obtained from Bio-Rad Laboratories (Hercules, CA). SVPDE and E.coli DNA polymerase I were purchased from Worthington Biochemicals (Lakewood, NJ). T4 DNA polymerase was from Invitrogen Life Technologies (Carlsbad, CA). Escherichia coli exonuclease III was obtained from Epicentre Technologies (Madison, WI), and the Klenow fragment of E.coli DNA polymerase I was purchased from Sigma (St Louis, MO). O6-Me-dG phosphoamidite was obtained from Glen Research (Sterling, VA). Parafilm™ was obtained from American National Can (Greenwich, CT). Cation exchange beads in ammonium form were purchased from Applied Biosystems (Foster City, CA). Water was purified with a Milli-Q ultrapure water filtration system from Millipore (Bedford, MA).
Oligodeoxynucleotides
The DNA oligomer sequences used in our study are listed in Table 1. The O6-POB-dG-containing ODN d(AACAGCCA TAT[O6-POB-G]GCCC) was kindly provided by Professor Lisa Peterson (University of Minnesota Cancer Center). The hexadecamer of the same sequence with O6-Me-dG in place of O6-POB-dG and the complemetary DNA strands were prepared by standard phosphoramidite chemistry at Microchemical Facility at University of Minnesota. The O6-Me-dG phosphoramidite was obtained from Glen Research. The synthetic DNA oligomer was deprotected by a manufacturer-supplied method, and HPLC purified as described elsewhere (17). The concentrations of pure oligonucleotides were determined by UV spectrophotometry using estimated values of molar extinction coefficients (Table 1). To obtain double-stranded DNA, equimolar amounts of the complementary DNA strands were combined, dried under vacuum and dissolved in the STE buffer (10 mM Tris, 50 mM NaCl, pH 8) to achieve a concentration of ∼10 OD units/100 µl buffer. The solution was briefly heated to 94°C and allowed to slowly cool to room temperature.
Table 1. DNA oligomer sequences employed in this study.
| Name | Sequence | MW | ε260(103 M–1 cm–1) |
|---|---|---|---|
| O6-POB-dG 16mer | d(AACAGCCATATG[O6-POB-dG]CCC) | 4982.5 | 161.8a |
| O6-Me-dG 16mer | d(AACAGCCATATG[O6-Me-dG]CCC) | 4847.0 | 161.8a |
| (–) strand | d(GGGCCATATGGCTGTT) | 3879.0 | 166.9 |
aEstimated extinction coefficient based on unmodified strand AACAGCCATATGGCCC.
Exonuclease digestions
All DNA oligomers used in exonuclease experiments were carefully desalted with Bio-gel P-6 cartridges as described below to remove any salts that may potentially interfere with exonuclease activity. Pilot experiments have demonstrated that this desalting step was necessary for obtaining reproducible results.
Snake venom phosphodiesterase
Single-stranded DNA oligomers (100 pmol) were incubated at 37°C with 6 mU of enzyme in 30 mM ammonium acetate buffer containing 30 mM MgSO4 (pH 9.4, total volume = 20 µl). Aliquots (5 µl) were taken after 0, 10, 20 and 30 min and immediately frozen at –20°C. After the incubation, aliquots were combined and analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) as described below. To achieve full digestion, the enzyme amount was increased to 12 mU, with the incubation time of 2 h.
Klenow fragment of E.coli polymerase I
DNA oligomers (100 pmol) were incubated with 2 U of E.coli polymerase I in 20 mM Tris buffer (pH 8.6) containing 40 mM KCl and 10 mM MgCl2 at 37°C (total volume = 14 µl). Aliquots (3 µl) were removed at 0, 10, 20 and 30 min and immediately frozen at –20°C. At the end of incubation, aliquots were combined, and samples were prepared for analysis by MALDI-TOF MS as described below.
Exonuclease III
Double-stranded DNA oligomers (100 pmol) were incubated with 10 U of exonuclease III in a buffer containing 330 mM Tris-acetate, pH 7.8, 660 mM potassium acetate, 100 mM magnesium acetate and 5 mM dithiothreitol (total volume = 13 µl) at 37°C. Aliquots (3 µl) were removed at 0, 10, 20 and 30 min, and immediately frozen at –20°C. The aliquots were combined and analyzed by MALDI-TOF MS following desalting as described below.
T4 DNA polymerase
Single-stranded or double-stranded DNA oligomers (30 pmol) were combined with 2.5 U of T4 DNA polymerase in 82 mM Tris-acetate buffer (pH 7.9) containing 100 mM sodium acetate, 25 mM MgSO4, 25 µg/ml BSA and 1.5 mM DTT (total volume = 5 µl). Aliquots were removed after 1 and 2 h incubations and immediately frozen at –20°C. Combined aliquots were prepared for analysis by MALDI-TOF MS as described below.
DNA desalting
All exonuclease digestion mixtures were desalted prior to MALDI-TOF MS analysis by using Bio-gel P-6 polyacrylamide gel size exclusion cartridges. The cartridges were rinsed with 2 ml of deionized water to remove the buffer, and DNA samples (in 100 µl of deionized water) were loaded, followed by centrifugation at 1000 g for 5 min. Desalted DNA passed through the column, while the salts were retained on the cartridge. This desalting method has been previously demonstrated to be optimal for the removal of metal ions present in exonucleolytic digests (5). The samples were dried under reduced pressure and dissolved in 1 mM ammonium citrate.
MALDI-TOF MS analysis
A Bruker Reflex III MALDI-TOF MS (Bruker Daltonics) was equipped with a 337 nm N2 laser set to a repetition rate of 3 Hz and an energy of 175 microjoule/pulse. The instrument was operated in the linear positive ion mode at laser attenuation of 45–52 and a delay time of 150 ns. The mass range was m/z 500–18 375. Spectra were averaged over 192 laser shots. The instrument was internally calibrated using synthetic ODNs of known molecular weight bracketing the m/z range of interest. Small pieces of Parafilm were stretched into a thin film over a 10-position MultiPROBE target (Bruker Daltonics) to create a smooth lining over the disc. MALDI matrix solution was prepared by combining 33 µl of saturated aqueous solution of 3-HPA with 5 µl of 10 mM ammonium citrate, 8 µl of acetonitrile and 4 µl of water. MALDI matrix (1 µl) was applied to the surface and allowed to dry at room temperature. DNA samples were dissolved in 3 µl of water and incubated with cation exchange beads for at least 2 h at 4°C. For sample loading, 1 µl of the aqueous sample solution was placed on the top of a crystallized matrix spot and allowed to dry. The spots were checked under a light microscope to ensure proper crystallization.
Computational studies
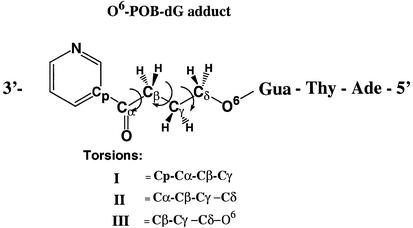
DNA trimers 5′-AT[O6-POB-dG]-3′ were model built using standard B-form geometrical parameters with the Insight II molecular modeling package (18). The force-field parameters for O6-POB-dG group were developed by following the procedure outlined by Cornell et al. (19). The charges for the modified guanine residue and the pyridyloxobutyl side chain were derived by using Gaussian 98 (20) to calculate the HF/6-31G* electrostatic potential, and then fitted to obtain the charges using the RESP approach outlined by Cornell et al. (19). (A listing of the POB side chain parameters and charges is given in Supplementary Material Fig. S1 and Table S1.) The system was neutralized by placement of sodium counterions using the addion option of the LEAP module of AMBER (21). The conformation of the O6-POB side chain was evaluated using a dihedral search algorithm. Structures were generated by varying the three most critical torsions of the pyridyloxobutyl group as defined in Table 2. These torsions were varied from 0 to 360°, with an increment of 60°, using the dihedral search option of the spasms module of AMBER 4.1 (21). The 10 lowest energy structures were selected for unrestrained energy minimization with the sander module of AMBER, version 6.00. Each optimization was started with 100 steps of steepest descent minimization, followed by 4900 steps using the conjugate gradient algorithm. Optimization was halted when the RMS in the gradient reached 0.0001 kcal/mol. The minimized structures were then overlaid and visualized using UCSF Midas Plus (22).
Table 2. Torsional analysis of the 4-oxo-4-(3-pyridyl)butyl substituent.
| Structure no. | Torsion I | Torsion II | Torsion III | AMBER energy (kcal/mol) | Adduct orientation |
|---|---|---|---|---|---|
| 1 | –174 | 66 | 64 | –174.0 | 3′ End |
| 2 | –78 | –63 | 74 | –184.2 | Extended |
| 3 | 76 | 55 | 64 | –184.5 | 3′ End |
| 4 | –110 | 180 | 65 | –184.5 | Extended |
| 5 | 90 | 66 | 176 | –187.5 | 5′ End |
| 6 | –120 | 70 | –66 | –187.6 | 3′ End |
| 7 | –80 | –175 | 71 | –188.3 | Extended |
| 8 | 179 | –178 | 69 | –190.3 | Extended |
| 9 | 99 | 69 | –81 | –191.3 | 5′ End |
| 10 | 73 | 60 | –65 | –197.1 | 3′ End |
The optimized adduct side chain torsions I, II and III, along with their AMBER minimized energies. Orientations of the O6-POB-group are indicated.
Molecular dynamics simulations
Molecular dynamics (MD) simulations of the DNA duplexes 5′-(CATAT[O6-POB-dG]GCCC)-3′ and 5′-(CATAT[O6-Me-dG]GCCC)-3′ were performed using the SANDER module of AMBER version 6.0 (21). An updated version of the Cornell et al. (19) force-field (PARM99), including the newly developed POB-dG parameters, was applied in all simulations (23). Counterions were placed using the addion option of the LEAP module of AMBER. The neutralized system was placed in a periodic box of TIP3P water molecules using a 12 Å layer. This yielded a simulation cell ∼52 × 56 × 66 Å in dimensions, containing ∼4500 water molecules. A constant temperature of 300 K and pressure of 1 atm was effected using the Berendsen coupling algorithm (24) with coupling constants of 2 and 0.2 ps, respectively. All simulations were carried out with periodic boundary conditions, and SHAKE (25) was used to constrain all hydrogen atoms. A non-bonded cut-off of 9 Å and an integration time step of 2.0 fs were applied. The particle mesh Ewald (PME) method (26) was used to treat Coulombic long-range interactions, using a grid spacing of 1 Å.
The equilibration of the system was carried out similar to a method reported by Miller and Kollman (27). In brief, a 25 kcal/mol restraint was initially placed on all solute atoms and counter-ions, and the water molecules were first minimized for 1000 steps. Next, the solvent was allowed to relax during a 3 ps MD simulation of the solvent around the solute, maintaining the temperature at 300 K. The entire system was then minimized to remove any high energy contacts using 500 steps of constrained energy minimization with a force constant of 5 kcal/mol applied to all solute heavy atom positions. Finally, the system was heated to 300 K over 10 ps, while the restraints were removed from the solute atoms. Pre-equilibration of the entire system was carried out for a time period of 50 ps to completely relax the system and to evaluate the structure for stability. Subsequent productions runs were performed for 2 ns from which physical properties were monitored over the final 1 ns. An analysis of the structural drift of the system (via RMS in structure) and drift in total energy, temperature and pressure (density) indicated that the system had reached equilibrium at ∼400 ps. (A plot of the RMS difference in structure over time is given in Fig. S2)
RESULTS
Two sets of experimental conditions were used to evaluate the effects of O6-POB-dG on 3′-exonuclease activity: (i) controlled digest designed to produce a complete set of DNA ladders and (ii) incubation with an excess of exonuclease to achieve complete digestion of DNA to 2′-deoxynucleotides. Optimal enzyme amounts and incubation times for each experiment were established in pilot studies using unmodified DNA 16mers (data not shown).
Snake venom phosphodiesterase
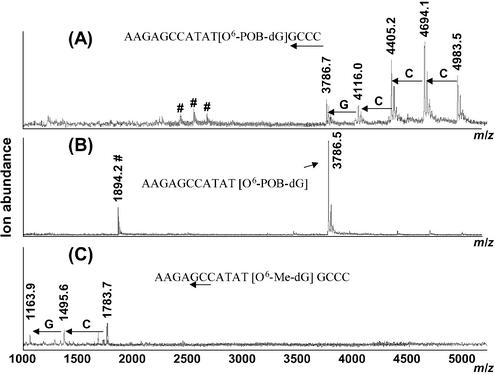
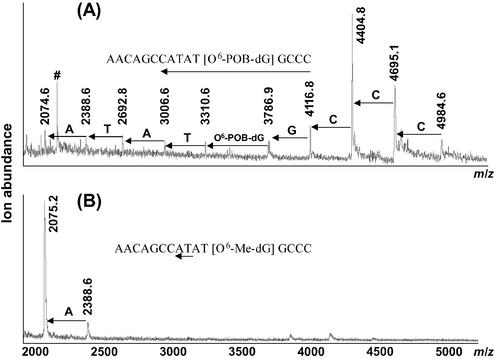
The results of MALDI-TOF MS analysis of O6-POB-dG-containing DNA hexadecamer d(AACAGCCATATG[O6-POB-dG]CCC) following SVPDE digestion are illustrated in Figure 1. Under controlled conditions (Fig. 1A), five peaks observed in the MALDI-TOF mass spectra correspond to the intact ODN (m/z = 4983.5, [M+H]+) and to the DNA fragments resulting from the removal of the first four residues: –C (m/z = 4694.1), –CC (m/z = 4405.2), –CCC (m/z = 4116.0), and –CCCG (m/z = 3786.7). No fragments below m/z = 3786.7 are observed (a series of small peaks marked with # in the area m/z 2300–2800 corresponds to doubly charged ions). The accumulation of the fragment at m/z = 3786.7 (AACAGCCATAT[O6-POB-dG]) in the exonucleolytic digest (Fig. 1A) suggests that O6-POB-dG hinders the 3′-exonuclease activity of SVPDE, preventing further hydrolysis. Indeed, further incubation of this DNA sample with an excess of SVPDE (complete digestion conditions) leads to disappearance of the peaks at higher m/z values, with the incomplete digestion product AACAGCCATAT[O6-POB-dG] becoming the only species observed [m/z = 3786.5, (M+H)+and m/z = 1894.2, (M+2H)2+] (Fig. 1B). In contrast, a control DNA 16mer containing O6-Me-dG instead of O6-POB-dG is nearly completely digested at the same conditions (Fig. 1C). These results indicate that O6-POB-dG blocks SVPDE-catalyzed hydrolysis of the phosphodiester bond between the modified guanosine nucleotide and the rest of the DNA chain, preventing exonucleolytic degradation beyond the adduct site.
Figure 1.
MALDI-TOF mass spectra of SVPDE digests of modified DNA 16mers d(AACAGCCATATGXCCC): (A) X = O6-POB-dG, time-controlled digest; (B) X = O6-POB-dG, complete digest conditions; (C) O6-Me-dG-containing oligomers, controlled digest conditions. Arrows indicate the portion of the sequence represented in the spectra, and doubly charged ions are marked with #.
Escherichia coli DNA polymerase I
The 3′-exonuclease activity of E.coli DNA polymerase I and the Klenow fragment of E.coli DNA polymerase I (KF) towards pyridyloxobutylated DNA was tested using single- and double-stranded DNA 16mers containing O6-POB-dG. The DNA ladders derived from the O6-POB-dG-containing single-stranded DNA include the mass shift corresponding to pyridyloxobutylated guanine nucleotide [M = 477.2 (theory), and mass shift between m/z = 3786.9 and 3310.6 is 476.3 (Fig. 2A)]. Control incubations in the absence of enzyme provide no evidence for adduct degradation (data not shown), ruling out the possibility of O6-POB-dG decomposition to normal guanine. At the same conditions, the control DNA 16mer containing O6-Me-dG instead of O6-POB-dG is digested far beyond the modified guanine nucleotide, with only small DNA fragments remaining in the hydrolytic mixture (Fig. 2B). Similar results were obtained for double-stranded DNA (data not shown). Therefore, the 3′-exonuclease activity of E.coli DNA polymerase I is hindered, but not completely blocked by O6-POB-dG. No detectable 5′-exonuclease activity was observed at the conditions of our experiments.
Figure 2.
Overlaid MALDI-TOF mass spectra of E.coli polymerase I digests of double-stranded DNA 16mers containing O6-guanine lesions d(AACAGCCATATGXCCC): (A) X = O6-POB-dG, (B) X = O6-Me-dG incubated at identical conditions. Arrows indicate the portion of the sequence represented in the spectra.
Escherichia coli exonuclease III
Since exonuclease III digests only double-stranded DNA, an O6-POB-dG-containing 16mer was annealed to the complementary strand to prepare a suitable substrate (Table 1). Exonuclease III digest of double-stranded DNA is expected to contain two series of DNA ladders corresponding to each of the complementary strands (28). MALDI spectrum of the digest shows a DNA ladder corresponding to the strand d(AGCCATAT[O6-POB-dG]GCCC) including the pyridyloxobutylated nucleotide [M = 477.2 (theory), mass difference between m/z = 3786.5 and 3311.2 is 475.3 (Fig. S3)]. This result demonstrates the ability of exonuclease III to digest past O6-POB-dG lesion. Exonuclease III incubation of an analogous DNA duplex containing O6-Me-dG in place of O6-POB-dG at the same conditions results in extensive hydrolysis of both strands (Fig. S3). Interestingly, exonuclease III digestion of the pyridyloxobutylated (+) strand appears to proceed two nucleotides beyond the predicted mid point in this duplex (m/z 2389.0), while the corresponding (–) strand digestion product (GGGCCATA, theoretical m/z 2435.7) is not observed (Fig. S3). The reasons for this result are presently unknown. Despite these discrepancies, MALDI-TOF MS results clearly indicate that exonuclease III-catalyzed hydrolysis of the O6-POB-dG-containing DNA strand is slowed down, but not completely blocked by O6-POB-dG nucleotide.
T4 DNA polymerase
MALDI-TOF MS analysis of T4 DNA polymerase digests of the O6-POB-dG-containing 16mer resulted in a nearly complete set of DNA ladders (Fig. S4), including the O6-POB-dG nucleotide (M = 477.2). The corresponding control O6-Me-dG-containing oligonucleotide was hydrolyzed to a similar extent. These results indicate that, unlike SVPDE, 3′-exonuclease activity of T4 DNA polymerase is capable of bypassing O6-POB-dG. A similar result was obtained when using double-stranded DNA substrates (data not shown), with no significant differences between the DNA ladders generated from exonucleolytic degradation of O6-POB-dG- and O6-Me-dG-containing hexadecamers.
Molecular modeling studies of O6-POB-dG-containing DNA
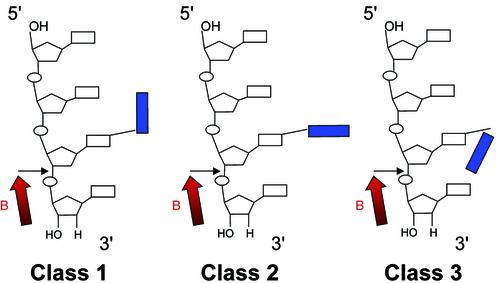
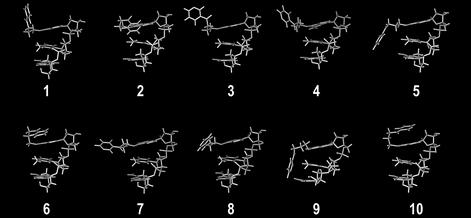
The experimental results described above demonstrate the ability of O6-POB-dG lesions to block 3′-exonuclease digestion catalyzed by SVPDE (Fig. 1). In contrast, the other three 3′-exonucleases tested were capable of bypassing the lesion (Fig. 2; Figs S3 and S4). To help understand the origins of the blocking effect of O6-POB-dG on 3′-exonuclease activity of SVPDE, DNA trinucleotides containing the O6-POB-dG adduct at the 3′ end were used to model the pyridyloxobutylated DNA substrate. Mao et al. (29) have proposed that the exonuclease resistance of DNA containing N2-guanine adducts of benzo[a]pyrene diolepoxide (BPDE) is defined by the orientation of the BPDE group relative to the rest of the DNA chain. Only N2-BPDE-dG stereoisomers with the pyrenyl group pointing towards the 3′ terminus of the DNA chain were resistant to SVPDE digestion (29). We thus performed a torsional study to establish the preferred orientation of the POB substituent relative to the DNA strand. Multiple conformers of the adduct side chain were generated using the SPASMS module of AMBER by systematically varying the torsional angles I–III (Table 2), followed by energy minimization. Extensive conformational searches have been previously applied to study the preferred orientation of BPDE-DNA adducts (30,31). The final structures obtained from our analyses are shown in Figure 3, and the energy values for each structure are reported in Table 2. The POB side chain was found to adopt three general orientations (Scheme 2): (i) the POB group pointing towards the 5′ end of the DNA (e.g. structure 9 in Fig. 3); (ii) the POB group parallel to the neighboring bases (e.g. structures 8 and 3 in Fig. 3); and (iii) the POB group pointing towards the 3′ end of the DNA chain (e.g. structure 10 in Fig. 3). These results (Table 2 and Fig. 3) indicate that, unlike the pyrenyl group of BPDE-DNA adducts, the POB side chain is quite flexible and can assume a variety of conformations. Given the inherent rigidity of the cyclized side chain of a BPDE adduct, this result is not surprising and may be partly responsible for the differences in nuclease activity against POB-dG- and BPDE-dG-containing oligomers.
Figure 3.
Wireframe representation of the 10 lowest energy conformations of O6-[4-oxo-4-(3-pyridyl)butyl]guanine.
Scheme 2.
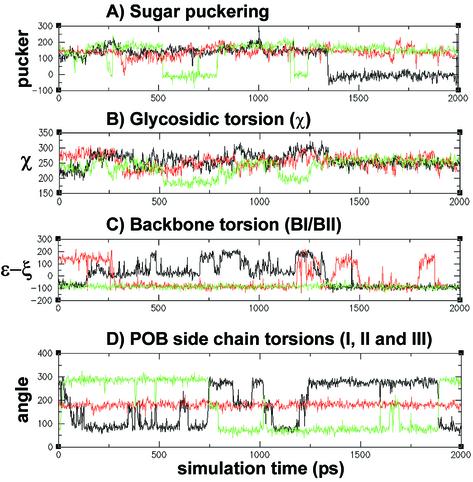
To investigate the effects of the O6-POB-dG adduct on the structural parameters of DNA, duplexes of the sequence d(CATATXGCCC) (X = O6-POB-dG or O6-Me-dG) were evaluated using MD simulations. A comparison of the sugar pucker phase angles, glycosidic torsions, and backbone conformations for O6-POB-dG, O6-Me-dG and unmodified dG over the entire 2000 ps of the simulation time is given in Figure 4. The sugar pucker phase angle of O6-POB-dG nucleotide shows a sharp C2′-endo to C3′-endo transition at ∼1.35 ns of MD simulation (Fig. 4A). This event occurs at approximately the same time as a shift in the backbone conformation about the modified nucleotide phosphate (Fig. 4C). The value of [ε–ξ], which is commonly used to describe the phosphate conformation in nucleic acid structure (32), initially shows a significant fluctuation at the pyridyloxobutylated nucleotide, but settles in at a value of –100° at ∼1.35 ns (Fig. 4C). The O6-POB side chain, which orients along the major groove of the DNA duplex, appears to induce a shift from C2′-endo to C3′-endo sugar puckering of the modified deoxyguanosine, which in turn drives a conformational change of the phosphate backbone (Fig. 4C). The control O6-Me-dG-containing oligomer does not show a similar event, with fluctuations in the [ε–ξ] values occurring throughout the entire simulation time (Fig. 4C). Examination of the glycosidic torsion angle of the O6-POB-dG nucleotide (χ) over the course of our MD simulation (Fig. 4B) indicates that the base adopts a glycosidic torsion angle typical for B-form geometries (32). Although some fluctuation of χ is observed during the initial stages of the simulation, it eventually equilibrates to a value within the anti conformation range (∼–113°) (Fig. 4B).
Figure 4.
Plot of (A) sugar puckering, (B) glycosidic torsion (χ) and (C) backbone phosphate conformation, represented as (ε–ξ), versus time for POB-, Me-guanine containing DNA duplexes. Residue POB-dG6 values are in black, Me-dG6 values are in red and the values for a control guanine (dG7), from the POB-dG containing duplex, are in green. (D) POB side chain torsion plots for torsions: I (black), II (red) and III (green) are also shown. All y-axis units are in degrees.
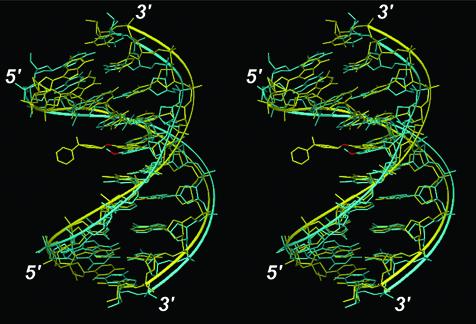
The least-squares fit of the final structures of O6-POB-dG- and O6-Me-dG-containing DNA duplexes is presented in Figure 5. Apart from the obvious steric consequences of POB-adduct formation, the two structures are very similar, with an overall RMS difference of 2.6 Å. One noticeable structural difference appears to be a slight tilting of the modified base, resulting in a partial loss of stacking interactions between O6-POB-dG and its neighboring nucleotides. The H-bond distances between the corresponding atoms in O6-POB-dG (G6 in Tables S2–S5) and dC in the opposite strand provide evidence for the loss of base pairing between the modified nucleobase and its partner cytosine (Tables S2–S5). A plot of the side chain torsion angles over the time course of the simulation is also given in Figure 4D. The side chain prefers an extended structure as indicated by the persistence of the trans central torsion conformation throughout the simulation. In fact, extended conformers (e.g. similar to conformers 4, 7 and 8 in Fig. 3 and Table 2) were observed during the MD simulations. This preference, however, is most likely due to steric effects of flanking bases in the duplex that may not be present in single-stranded DNA. A complete listing of the structural parameters comparing the O6-POB-dG- and O6-Me-dG-containing oligomers is given in Tables S2–S5.
Figure 5.
Stereoview showing the overlay of the average structures of POB- (yellow) and Me- (cyan) guanine containing DNA duplexes over the last 1.0 ns of MD simulations. The O6- of the adducted guanines are highlighted in red, and the 3′ and 5′ terminals of the duplex are labeled. Hydrogen atoms are not shown.
DISCUSSION
The main purpose of this study was to evaluate the ability of O6-POB-dG adducts to block 3′-exonucleases and to establish possible structural reasons for this effect. We found that O6-POB-dG is capable of blocking DNA hydrolysis mediated by SVPDE, but does not have the same effect on the corresponding 3′-exonuclease activities of E.coli DNA polymerase I, exonuclease III and T4 DNA polymerase. SVPDE digests DNA strands all the way up to the pyridyloxobutylated base, but is unable to hydrolyze the phosphodiester bond between the O6-POB-dG nucleotide and its 5′-neighbouring nucleotide, even following prolonged incubation with an excess of exonuclease (Fig. 1B). Although the rates of exonucleolytic hydrolysis of O6-POB-dG-containing DNA catalyzed by the 3′-exonuclease activity of E.coli polymerase I and E.coli DNA exonuclease III are significantly reduced as compared with the control O6-Me-dG-containing strand, these enzymes are capable of bypassing the pyridyloxobutylated base (Fig. 2; Figs S3 and S4). Our observation of the differential processing of pyridyloxobutylated DNA by various 3′-exonucleases suggests that mammalian 3′-exonucleases may exhibit substrate preferences dissimilar to that of SVPDE. In an earlier study, Xu and Kool (33) have reported that 5′-bridging of phosphothioesters blocks T4 DNA polymerase, but not SVPDE, suggesting differences in their substrate specificity. Our results lend further support to differences between the substrate requirements of different 3′-exonucleases.
The observed differences between the 3′-exonuclease activities of E.coli DNA polymerase I, exonuclease III, T4 DNA polymerase and SVPDE towards O6-POB-dG-containing DNA may result from their distinct active site geometries and/or different mechanisms of phosphoryl transfer employed by different exonucleases. The 3′-exonuclease activities of E.coli DNA polymerase I and T4 DNA polymerase reside in their C-terminal domains and serve the proofreading function, excising incorrectly added nucleotides (34,35). Both single- and double-stranded DNA can be hydrolyzed. Crystal structures of the 3′-exonuclease domains of the KF and T4 DNA polymerase (34,36–38) indicate that, despite low sequence homology, the 3′-exonuclease active sites of the two enzymes have similar structures. Both active sites contain two divalent metal ions (A and B) coordinated to four conserved carboxylate residues (37,39). Mutations of these residues lead to a significant loss of 3′-exonuclease activity (40). The role of the divalent metal ion A is to coordinate a water molecule, resulting in the production of a metal-hydroxide anion that participates in nucleophilic attack and hydrolyzes the phosphodiester (38). Metal ion B interacts with the 3′-hydroxyl of the scissile phosphate, stabilizing the leaving group (38).
Unlike KF and T4 polymerase, exonuclease III is specific for double-stranded DNA. It hydrolyzes DNA duplex by removing nucleotides from both 3′ termini in a synchronous fashion, until the point where no double-stranded substrate is available (28). The rate of exonuclease III-mediated DNA hydrolysis varies 3-fold depending on the nature of the excised nucleotide, following the order: dC > dA, dT > dG. Although it is relatively non-sensitive to chemical modifications at nucleobases, introduction of a 5-alkyl substituent on a cytosine has been reported to induce resistance (41).
SVPDE (venom exonuclease, phosphodiestarase I) is found in the venom of rattlesnake species Crotalus terrificus terrificus and Crotalus adamanteus (42,43). SVPDE successively removes 5′-mononucleotides from the free hydroxy-terminated 3′ end of single-stranded DNA and RNA. Kinetic analyses indicate that the rate of SVPDE-catalyzed hydrolysis is only slightly affected by nucleic acid sequence (44), with digestion rates changing in the following order: G,T > A > C (45). SVPDE is a complex glycoprotein containing neutral sugars, amino sugars, sialic acid residues, triacylglycerols and cholesterol esters (46). Each molecule of the enzyme contains one Zn2+, six Mg2+ and 35 Ca2+ atoms. While Ca2+ and Zn2+ affect the affinity of the enzyme towards DNA substrate, Mg2+ is required for catalysis (46). Since the hydrolysis reaction proceeds with the retention of configuration at the phosphorus, the formation of a covalent intermediate has been proposed (47). Interestingly, several groups have reported that SVPDE has an additional endonuclease activity which enables it to completely hydrolyze chemically altered DNA by ‘skipping’ the modified base (45,48,49). It should be noted, however, that endonuclease activity was only observed following prolonged incubations with an excess of SVPDE (48,49) and may be a result of a contaminating endonuclease activity.
Previous studies have noted SVPDE blockage by certain nucleobase modifications, including abasic sites (45,50), deoxyadenosine adducts of polycyclic aromatic hydrocarbons (51–53), 9-(2-aminoethoxy)-phenoxazine (12) and tandem base lesions (49,54–56). The resistance of modified nucleotides towards SVPDE-catalyzed hydrolysis has been attributed to a variety of factors, including steric hindrance, interference with the metal ion binding in the exonuclease active site and altered conformation of the phosphodiester backbone.
Steric factors have been proposed to induce the 3′-exonuclease resistance of bulky DNA lesions, e.g. benzo[a]pyrene diolepoxide (BPDE) adducts (29,51–53), cholesterol adducts (57) and N6-adenine lesions formed by 4-nitroquinoline 1-oxide (58). Interestingly, only the (–) trans-anti-N2-BPDE-dG slowed down SVPDE digestion, while the (+) trans-anti-N2-BPDE-dG stereoisomer was readily bypassed (29). Mao et al. (29) have proposed that the differences in exonuclease resistance of the (+) and (–) BPDE-modified oligonucleotide results from the opposite spatial orientations of the pyrenyl residues in these adducts. While the lesion pointing towards the 3′ end of the DNA chain [(–) trans-anti-N2-BPDE-dG] interfered with 3′-exonuclease activity of SVPDE, the 5′ oriented adduct [(+) trans-anti-N2-BPDE-dG] was readily bypassed (29). This mechanism, however, is unlikely to explain the effect of O6-POB-dG on SVPDE, since the POB side chain is quite flexible and is capable of assuming multiple low energy conformations (Table 2, Fig. 3 and Scheme 2). Furthermore, while ODNs containing bulky BPDE adducts can be completely hydrolyzed at higher enzyme concentrations and longer incubation times, O6-POB-dG represents a block to SVPDE-catalyzed DNA hydrolysis that cannot be bypassed even in the presence of a large excess of the enzyme (Fig. 1B).
Teplova et al. (11) have demonstrated that the 3′-exonuclease activity of SVPDE and E.coli polymerase I is blocked by 2′-O-aminopropyl RNA modification. The crystal structure of a complex between a DNA fragment containing the 2′-O-aminopropyl modification and the KF has demonstrated that the positively charged 2′-O-substituent interferes with metal binding site B of the exonuclease, slowing down the hydrolysis (11). A similar mechanism can be proposed for O6-POB-dG, with the pyridyl group disrupting the coordination of the metal ions in the active site of SVPDE. However, unlike the 2′-aminopropyl substituent (pKa = 9.2), which is protonated at physiological pH, the pyridyl group of POB (pKa ∼ 5) is expected to be largely uncharged at physiological conditions.
Our 2 ns MD simulations of a DNA decamer containing O6-POB-dG (Fig. 4) demonstrate that the presence of the O6-POB substituent affects the sugar puckering of the modified guanine, which is shifted from the normal C2′-endo conformation to the C3′-endo range characteristic for RNA and the A-form of DNA (Fig. 4A). This conformational change may affect SVPDE processing of O6-POB-dG-containing DNA, especially if the sugar portion of the molecule is involved in substrate recognition. RNA is known to be a worse SVPDE substrate than DNA (44). Structural changes induced by DNA modifications have been previously reported to interfere with exonucleolytic cleavage. For example, SVPDE is unable to recognize nucleosides in syn conformation (59). An SVPDE-resistant diastereomer of 5′-O-phosphorothiolate O-nitrophenyl ester has been shown to have a C2′-endo ribose pucker, with the heterocycle in anti position and the C5′–O5′ bond in the gauche, gauche conformation (47). Therefore, the change of the sugar pucker observed upon pyridyloxobutylation of the O6-guanine position may play a role in processing by SVPDE. Furthermore, POB adduct formation may have an entropic effect on nuclease activity. Our simulations (Fig. 4) show that the O6-POB side chain induces a phosphodiester backbone motion localized primarily at the site of adduct formation. SVPDE activity and substrate specificity are likely to be medicated by a formation of a unique enzyme–substrate complex (leading to a reduction in the number of degrees of freedom). The presence of the O6-POB group may result in a loss of configurational entropy, destabilizing the enzyme– substrate complex. It is important to point out, however, that the minor structural changes induced by the O6-POB-dG group uncovered by our simulations are unlikely to fully explain the structural basis to its blocking effect on SVPDE. Studies are currently in progress at our laboratories to establish the structure–activity relationship between the O6-guanine substituent and the nuclease resistance of the chemically modified DNA, which should help explain the mechanism of O6-POB-dG-induced exonuclease blockage.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor Lisa Peterson (University of Minnesota Cancer Center) for the generous gift of O6-POB-dG-containing DNA oligomers. This research was partially supported by a seed grant from the Academic Health Center (University of Minnesota).
REFERENCES
- 1.Hecht S.S., Chen,C.B., Ornaf,R.M., Hoffmann,D. and Tso,T.C. (1978) Chemical studies on tobacco smoke LVI. Tobacco specific nitrosamines: origins, carcinogenicity and metabolism. IARC Sci. Publ., 395–413. [PubMed] [Google Scholar]
- 2.Hecht S.S. (1999) DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res., 424, 127–142. [DOI] [PubMed] [Google Scholar]
- 3.Hecht S.S. (1998) Biochemistry, biology and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol., 11, 559–603. [DOI] [PubMed] [Google Scholar]
- 4.Castonguay A., Stoner,G.D., Schut,H.A. and Hecht,S.S. (1983) Metabolism of tobacco-specific N-nitrosamines by cultured human tissues. Proc. Natl Acad. Sci. USA, 80, 6694–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tretyakova N., Matter,B., Ogdie,A., Wishnok,J.S. and Tannenbaum,S.R. (2001) Locating nucleobase lesions within DNA sequences by MALDI-TOF mass spectral analysis of exonuclease ladders. Chem. Res. Toxicol., 14, 1058–1070. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L.-K. and Gross,M.L. (2000) Matrix-assisted laser desorption/ionization mass spectrometry methods for oligodeoxynucleotides: improvements in matrix, detection limits, quantification and sequencing. J. Am. Soc. Mass Spectrom., 11, 854–865. [DOI] [PubMed] [Google Scholar]
- 7.Shaw J.P., Kent,K., Bird,J., Fishback,J. and Froehler,B. (1991) Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res., 19, 747–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamper H.B., Reed,M.W., Cox,T., Virosco,J.S., Adams,A.D., Gall,A.A., Scholler,J.K. and Meyer,R.B.,Jr (1993) Facile preparation of nuclease resistant 3′ modified oligodeoxynucleotides. Nucleic Acids Res., 21, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zendegui J.G., Vasquez,K.M., Tinsley,J.H., Kessler,D.J. and Hogan,M.E. (1992) In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic Acids Res., 20, 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monia B.P., Johnston,J.F., Sasmor,H. and Cummins,L.L. (1996) Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J. Biol. Chem., 271, 14533–14540. [DOI] [PubMed] [Google Scholar]
- 11.Teplova M., Wallace,S.T., Tereshko,V., Minasov,G., Symons,A.M., Cook,P.D., Manoharan,M. and Egli,M. (1999) Structural origins of the exonuclease resistance of a zwitterionic RNA. Proc. Natl Acad. Sci. USA, 96, 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier M.A., Leeds,J.M., Balow,G., Springer,R.H., Bharadwaj,R. and Manoharan,M. (2002) Nuclease resistance of oligonucleotides containing the tricyclic cytosine analogues phenoxazine and 9-(2-aminoethoxy)-phenoxazine (‘G- clamp’) and origins of their nuclease resistance properties. Biochemistry, 41, 1323–1327. [DOI] [PubMed] [Google Scholar]
- 13.Samani T.D., Jolles,B. and Laigle,A. (2001) Best minimally modified antisense oligonucleotides according to cell nuclease activity. Antisense Nucleic Acid Drug Dev., 11, 129–136. [DOI] [PubMed] [Google Scholar]
- 14.Morita K., Hasegawa,C., Kaneko,M., Tsutsumi,S., Sone,J., Ishikawa,T., Imanishi,T. and Koizumi,M. (2002) 2′-O,4′-C-ethylene-bridged nucleic acids (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett., 12, 73–76. [DOI] [PubMed] [Google Scholar]
- 15.Xiao W., Li,G., Player,M.R., Maitra,R.K., Waller,C.F., Silverman,R.H. and Torrence,P.F. (1998) Nuclease-resistant composite 2′,5′-oligoadenylate-3′, 5′-oligonucleotides for the targeted destruction of RNA: 2–5A-iso-antisense. J. Med. Chem., 41, 1531–1539. [DOI] [PubMed] [Google Scholar]
- 16.Vichier-Guerre S., Pompon,A., Lefebvre,I. and Imbach,J.L. (1994) New insights into the resistance of alpha-oligonucleotides to nucleases. Antisense Res. Dev., 4, 9–18. [DOI] [PubMed] [Google Scholar]
- 17.Tretyakova N., Matter,B., Jones,R. and Shallop,A. (2002) Formation of benzo[a]pyrene diol epoxide-DNA adducts at specific guanines within K-oras and p53 gene sequences: stable isotope-labeling mass spectrometry approach. Biochemistry, 41, 9535–9544. [DOI] [PubMed] [Google Scholar]
- 18.MSI (2002) InsightII User Guide. MSI, San Diego.
- 19.Cornell W.D., Cieplak,P., Bayly,C.L., Gould,L.R., Merz,K.M.J., Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins and nucleic acids. J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 20.Frisch M.J., Trucks,G.W., Schlegel,H.B., Scuseria,G.E., Robb,M.A., Cheeseman,J.R., Zakrzewski,V.G., Montgomery,J.A.,Jr, Stratmann,R.E. and Burant,J.C. (1998) Gaussian 98, Rev. A.6. Gaussian, Inc., Pittsburgh, PA.
- 21.Pearlman D.A., Case,D.A., Caldwell,J.W., Ross,W.S., Cheatham,T.E., DeBolt,S., Ferguson,D.M., Seibel,G. and Kollman,P.A. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun., 91, 1–41. [Google Scholar]
- 22.Ferrin T.E., Huang,C.C., Jarvis,L.E. and Langridge,R. (1988) The MIDAS display system. J. Mol. Graph., 6, 13–27. [Google Scholar]
- 23.Wang M., Cieplak,P. and Kollman,P.A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem., 21, 1049–1074. [Google Scholar]
- 24.Berendsen H.J.C., Postma,J.P.M., Gunsteren,W.F., DiNola,A. and Haak,J.R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys., 81, 3684–3690. [Google Scholar]
- 25.Ryckaert J.P., Ciccotti,G. and Berendsen,H.J.C. (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys., 23, 327–342. [Google Scholar]
- 26.Essmann U., Perera,L., Berkowitz,M.L., Darden,T. and Pedersen,L.G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys., 103, 8577–8593. [Google Scholar]
- 27.Miller J.L. and Kollman,P.A. (1997) Theoretical studies of an exceptionally stable RNA tetraloop: observation of convergence from an incorrect NMR structure to the correct one using unrestrained molecular dynamics. J. Mol. Biol., 270, 436–450. [DOI] [PubMed] [Google Scholar]
- 28.Linxweiler W. and Horz,W. (1982) Sequence specificity of exonuclease III from E. coli. Nucleic Acids Res., 10, 4845–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao B., Li,B., Amin,S., Cosman,M. and Geacintov,N.E. (1993) Opposite stereoselective resistance to digestion by phosphodiesterases I and II of benzo[a]pyrene diol epoxide-modified oligonucleotide adducts. Biochemistry, 32, 11785–11793. [DOI] [PubMed] [Google Scholar]
- 30.Tan J., Geacintov,N.E. and Broyde,S. (2000) Conformational determinants of structures in stereoisomeric cis-opened anti-benzo[a]pyrene diol epoxide adducts to adenine in DNA. Chem. Res. Toxicol., 13, 811–822. [DOI] [PubMed] [Google Scholar]
- 31.Xie X.M., Geacintov,N.E. and Broyde,S. (1999) Origins of conformational differences between cis and trans DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides. Chem. Res. Toxicol., 12, 597–609. [DOI] [PubMed] [Google Scholar]
- 32.Saenger W. (1984) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY.
- 33.Xu Y. and Kool,E.T. (1998) Chemical and enzymatic properties of bridging 5′-S-phosphorothioester linkages in DNA. Nucleic Acids Res., 26, 3159–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beese L.S., Derbyshire,V. and Steitz,T.A. (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science, 260, 352–355. [DOI] [PubMed] [Google Scholar]
- 35.Abdus Sattar A.K., Lin,T.C., Jones,C. and Konigsberg,W.H. (1996) Functional consequences and exonuclease kinetic parameters of point mutations in bacteriophage T4 DNA polymerase. Biochemistry, 35, 16621–16629. [DOI] [PubMed] [Google Scholar]
- 36.Brautigam C.A., Sun,S., Piccirilli,J.A. and Steitz,T.A. (1999) Structures of normal single-stranded DNA and deoxyribo-3′-S- phosphorothiolates bound to the 3′-5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry, 38, 696–704. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Yu,P., Lin,T.C., Konigsberg,W.H. and Steitz,T.A. (1996) Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry, 35, 8110–8119. [DOI] [PubMed] [Google Scholar]
- 38.Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beese L.S., Friedman,J.M. and Steitz,T.A. (1993) Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry, 32, 14095–14101. [DOI] [PubMed] [Google Scholar]
- 40.Elisseeva E., Mandal,S.S. and Reha-Krantz,L.J. (1999) Mutational and pH studies of the 3′→5′ exonuclease activity of bacteriophage T4 DNA polymerase. J. Biol. Chem., 274, 25151–25158. [DOI] [PubMed] [Google Scholar]
- 41.He K., Porter,K.W., Hasan,A., Briley,J.D. and Shaw,B.R. (1999) Synthesis of 5-substituted 2′-deoxycytidine 5′-(alpha-P- borano)triphosphates, their incorporation into DNA and effects on exonuclease. Nucleic Acids Res., 27, 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipps G.R. (1975) Purification and characterization of phosphodiesterase from Crotalus venom. Hoppe Seylers Z. Physiol Chem., 356, 1085–1096. [DOI] [PubMed] [Google Scholar]
- 43.Dolapchiev L.B., Sulkowski,E. and Laskowski,M.,Sr (1974) Purification of exonuclease (phosphodiesterase) from the venom of Crotalus adamanteus. Biochem. Biophys. Res. Commun., 61, 273–281. [DOI] [PubMed] [Google Scholar]
- 44.Razzell W.E. and Khorana,H.G. (1959) Studies on polynucleotides. J. Biol. Chem., 234, 2105–2113. [PubMed] [Google Scholar]
- 45.Zhang L.K., Rempel,D. and Gross,M.L. (2001) Matrix-assisted laser desorption/ionization mass spectrometry for locating abasic sites and determining the rates of enzymatic hydrolysis of model oligodeoxynucleotides. Anal. Chem., 73, 3263–3273. [DOI] [PubMed] [Google Scholar]
- 46.Dolapchiev L.B., Vassileva,R.A. and Koumanov,K.S. (1980) Venom exonuclease. II. Amino acid composition and carbohydrate, metal ion and lipid content in the Crotalus adamanteus venom exonuclease. Biochim. Biophys. Acta, 622, 331–336. [DOI] [PubMed] [Google Scholar]
- 47.Burgers P.M., Eckstein,F. and Hunneman,D.H. (1979) Stereochemistry of hydrolysis by snake venom phosphodiesterase. J. Biol. Chem., 254, 7476–7478. [PubMed] [Google Scholar]
- 48.Ilankumaran P., Pannell,L.K., Gebreselassie,P., Pilcher,A.S., Yagi,H., Sayer,J.M. and Jerina,D.M. (2001) Patterns of resistance to exonuclease digestion of oligonucleotides containing polycyclic aromatic hydrocarbon diol epoxide adducts at N6 of deoxyadenosine. Chem. Res. Toxicol., 14, 1330–1338. [DOI] [PubMed] [Google Scholar]
- 49.Bourdat A.G., Gasparutto,D. and Cadet,J. (1999) Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res., 27, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart G.R. and Chambers,R.W. (1987) Synthesis and properties of oligodeoxynucleotides with an AP site at a preselected position. Nucleic Acids Res., 15, 7451–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dipple A. and Pigott,M.A. (1987) Resistance of 7,12-dimethylbenz[a]anthracene-deoxyadenosine adducts in DNA to hydrolysis by snake venom phosphodiesterase. Carcinogenesis, 8, 491–493. [DOI] [PubMed] [Google Scholar]
- 52.Cheh A.M., Yagi,H. and Jerina,D.M. (1990) Stereoselective release of polycyclic aromatic hydrocarbon-deoxyadenosine adducts from DNA by the 32P postlabeling and deoxyribonuclease I/snake venom phosphodiesterase digestion methods. Chem. Res. Toxicol., 3, 545–550. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs R. and Daune,M. (1973) Physical basis of chemical carcinogenesis by N-2-fluorenylacetamide derivatives and analogs. FEBS Lett., 34, 295–298. [DOI] [PubMed] [Google Scholar]
- 54.Weinfeld M. and Soderlind,K.J. (1991) 32P-postlabeling detection of radiation-induced DNA damage: identification and estimation of thymine glycols and phosphoglycolate termini. Biochemistry, 30, 1091–1097. [DOI] [PubMed] [Google Scholar]
- 55.Buchko G.W., Cadet,J., Morin,B. and Weinfeld,M. (1995) Photooxidation of d(TpG) by riboflavin and methylene blue. Isolation and characterization of thymidylyl-(3′,5′)-2-amino-5-[(2-deoxy-beta-d-erythro-pentofuranosyl)amino]-4H-imidazol-4-one and its primary decomposition product thymidylyl-(3′,5′)-2,2-diamino-4-[(2-deoxy-beta-d-erythro-pentofuranosyl)amino]-5(2H)-oxazolone. Nucleic Acids Res., 23, 3954–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman K.J., Pla,R.L., Guichard,Y., Farmer,P.B. and Jones,G.D. (2001) Evaluation of phosphodiesterase I-based protocols for the detection of multiply damaged sites in DNA: the detection of abasic, oxidative and alkylative tandem damage in DNA oligonucleotides. Nucleic Acids Res., 29, E101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Machwe A., Ganunis,R., Bohr,V.A. and Orren,D.K. (2000) Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res., 28, 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panigrahi G.B. and Walker,I.G. (1990) The N2-guanine adduct but not the C8-guanine or N6-adenine adducts formed by 4-nitroquinoline 1-oxide blocks the 3′-5′ exonuclease action of T4 DNA polymerase. Biochemistry, 29, 2122–2126. [DOI] [PubMed] [Google Scholar]
- 59.Ogilvie K.K. and Hruska,F.H. (1976) Affect of spleen and snake venom phosphodiesterases on nucleotides containing nucleosides in the syn conformation. Biochem. Biophys. Res. Commun., 68, 375–378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.