Abstract
Telomeres, the protective caps of eukaryotic chromosomes, are maintained by the enzyme telomerase. This telomere-specific reverse transcriptase (RT) uses a small region of its RNA subunit as template to synthesize telomeric DNA, which is generally G/T rich in the strand that contains the 3′ end. To further our understanding of why telomeres are usually G/T rich, we screened Saccharomyces cerevisiae telomerase RNA (TLC1) libraries with randomized template sequences for complementation of a tlc1 deletion and decapping of existing telomeres. Surprisingly, the vast majority of the 60 000 different mutant telomerase templates tested showed no activity in vivo. This deficiency was not due to impaired assembly with the catalytic subunit (Est2p) nor could it be alleviated by enforced telomerase recruitment to the telomeres. Rather, the mutant templates reduced the nucleotide addition processivity of telomerase. The functional RNA template sequences recovered in our screens preferentially contained two or more consecutive rC nucleotides, reminiscent of the wild-type template. Thus, in contrast to retroviral RTs that can reverse transcribe any RNA sequence into DNA, the budding yeast telomerase RT is specialized for its C-rich RNA template.
INTRODUCTION
In most organisms, telomere sequences are rich in guanine and thymine nucleotides in the DNA strand that runs 5′ to 3′ towards the chromosome end (1). Furthermore, telomere repeats usually contain runs of several adjacent deoxyguanosine nucleotides. This sequence feature allows the formation of a stable DNA secondary structure, the G-quadruplex, in vitro [reviewed by Williamson (2)]. The high degree of telomere sequence conservation suggests some functional importance. However, the precise molecular events in which sequence-specific functions of the telomere are involved have not been identified so far.
The essential function of telomeres is to prevent chromosome end-to-end fusions and extensive nucleolytic degradation [reviewed by McEachern et al. (3)]. Loss of end protection could be provoked by changes in the telomeric DNA sequence (4–7). Presumably, the mutant telomere sequences interfere with the formation of an essential telomeric chromatin and DNA structures such as the G-quadruplex. G-rich DNA secondary structures have also been proposed to participate in the telomerase reaction cycle (8). Specifically, the folding of newly synthesized telomeric repeats into G–G hairpins or G-quadruplex structures may facilitate the product dissociation and translocation steps by lowering the energy difference between the extended, base-paired telomere–template hybrid and the dissociated individual strands (9,10).
Whether the telomerase RNA template participates in a sequence-specific manner in the reaction or serves only as a passive template has been discussed controversially in the past. Several completely non-telomeric RNA templates can be reverse transcribed by Tetrahymena thermophila telomerase (11), and limited incorporation of mutant telomere sequences specified by ectopically expressed mutant telomerase RNA templates occurred in human tumor cells (12,13). On the other hand, changes in the product dissociation pattern, reduced fidelity and lowered processivity have been described for template mutant Tetrahymena telomerases (14–16). A template mutant yeast telomerase was inactive in mutant enzyme homomultimers but active in wild-type/mutant heteromultimers (17,18). So far, no general rule for the RNA template’s role in the telomerase reaction has emerged from the published experiments.
In this study, we chose a genetic approach to define the template sequence requirements of budding yeast telomerase and gain insight into the reasons why telomeres are generally G/T rich. A telomerase RNA template library, in which 10 of the 16 templating nucleotides were randomized, was screened for complementation of a tlc1 deletion and, in a separate screen, for the induction of growth arrest. This unbiased analysis of a large number of template sequences revealed that telomerase can reverse transcribe only a minor fraction of all possible templates in vivo. Mutant template RNAs that complemented a tlc1 deletion preferentially contained at least two consecutive rC nucleotides, similar to the central part of wild-type TLC1. To verify the functional importance of this sequence, we constructed several telomerase RNA template libraries with five or six randomized template nucleotides, thus spanning the entire RNA template region. The number of complementing templates was especially low in a library where the central 477CCCAC475 template sequence was randomized, emphasizing the importance of this region. The deficiency conveyed by the template mutations was a reduced telomerase nucleotide addition processivity, indicating that the Saccharomyces cerevisiae TERT enzyme has a functional dependence on the C/A-rich RNA template sequence.
MATERIALS AND METHODS
Library construction
Mutagenesis of TLC1 was carried out as previously described (19) by ligating a PCR product obtained with oligonucleotide primers carrying random nucleotides at the desired positions (sequence of the mutagenesis primer: 5′-TAATTATCATGAGAAGCCTACCATCACCACCCACACACAAATGTTACAG-3′; the underlined sequence corresponds to the template region and was changed according to the desired library design) with a plasmid vector containing the rest of the TLC1 gene. The ligation products were transformed into competent Escherichia coli cells for amplification. For the short libraries (Library483–478, Library477–473 and Library472–468), the number of transformants was considerably higher than the theoretical complexity of these libraries. For Library480–471, approximately 200 000 bacterial transformants were obtained, which corresponds to one-fifth of the theoretical complexity. Individual clones from each library were sequenced to confirm randomization of each nucleotide position in the desired region.
Screen for complementation of tlc1-Δ
YKF19 [Mat a ade2 his3-11 can1-Δ leu2 trp1 ura3-52 DIA5-1 (ADE2 telomere VR) tlc1::HIS3 rad52::LEU2] was re-streaked progressively until senescence. The second from last streak was used to inoculate a liquid culture for transformation, and the cells were incubated until no further growth was detectable. The transformation efficiency was determined by counting the number of colonies obtained with wild-type TLC1 (pSD107). Control transformations with the empty vector (pRS314) gave rise to no or very few colonies. Colonies obtained upon transformation with the library plasmids were re-streaked at least twice. The tlc1 template region from plasmid library-harboring colonies was PCR amplified using the two oligonucleotides 5′-TLC1_long 5′-GGCCCGGGAATAAAACTAGAGAGGAAGATAGG-3′ and 3′-TLC1_short 5′-GGCCCGGGACAGTGTCAGAAAAAATACTAGG-3′. The PCR product was digested with NcoI to exclude contaminating wild-type plasmids, which were present in Library480–471 at a frequency of 0.5%. The template region was sequenced either directly from the PCR product or from the plasmid after recovery in E.coli.
Screen for growth arrest
Yeast strain YKF20 [Mat a ade2 his3-11 can-1Δ leu2 trp1 ura3-52 DIA5-1 (ADE2 telomere VR) tlc1::HIS3 pTLC1-URA3] was transformed with Library480–471 and grown under conditions that selected for both the wild-type pTLC1-URA3 and the mutant tlc1 from Library480–471. The colonies were replica-plated onto 5-fluoro-orotic acid (FOA)/–Trp medium to select for loss of the plasmid containing wild-type TLC1 but retention of the mutant tlc1 plasmid. The library plasmids were recovered from candidate TLC1/tlc1 colonies and re-transformed into a tlc1-Δ yeast strain well before the onset of senescence. Growth arrest-inducing tlc1 alleles were identified by reduced transformant colony size and number relative to pRS314.
IP–RT–PCR experiments
Yeast strain YKF103 (Mat a ura3-52 ade2-101 lys2-Δ1 trp1-Δ1 his3-Δ200/CF+ ProteinA-EST2) (20) was transformed with either pKF5 (19) or Library480–471. Individual colonies were picked and 10 ml cultures were grown to an OD600 of 0.3–0.5. The cells were harvested, washed and lysed by bead bashing in 300 µl of immunoprecipitation (IP) buffer high salt [10 mM Tris–HCl pH 7.5, 150 mM NaCl, 150 mM KCl, 1 mM MgCl2, 1 mM phenylmethylsulfonylfluoride, 0.1 mM dithiothreitol (DTT), 10% glycerol, 0.1% Tween-20, 0.1% NP-40, 0.1 U/µl of RNase inhibitor]. The extracts were cleared by centrifugation, and 500 µg of total protein were incubated with 50 µl of a 50% slurry of rabbit serum agarose beads (Sigma) to bind the protein A-tagged Est2p and 20 U of DNase I (Roche Molecular Biochemicals) for 120 min at 4°C. The beads were washed twice with 1 ml of IP buffer high salt, once with 1 ml of IP buffer low salt (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.1 mM DTT, 10% glycerol) and resuspended in 30 µl of IP buffer low salt. The reverse transcriptase reactions were carried out with Superscript II RT (Life Technologies) according to the manufacturer’s instructions using 10 µl of the resuspended beads as template and oligo 3′-TLC1_short (see above) to prime cDNA synthesis. PCR amplification and NcoI digestion were performed as described above.
Telomere length analysis
Telomere length was analyzed by telomere-PCR (21) or Southern blotting. Telomere PCR products were separated on 3% agarose gels and the size was determined using AIDA image quantification software (Fuji). For Southern blotting, restriction fragments were separated on 0.7% agarose gels, transferred to nylon membrane and subsequently hybridized with a Y′-probe (22) and a probe for the 1 kb DNA size standard (Life Technologies). Autoradiographs were acquired with a Fuji BAS PhosphorImager and analyzed with AIDA software (Fuji).
Telomerase assays
Telomerase extracts were prepared and reactions performed as described (19,23). The amount of TLC1 RNA in each preparation was determined by northern hybridization, and equal amounts were used in the reactions. For the preparation without TLC1 RNA (empty vector), the same amount of protein as for the wild-type preparation was employed.
RESULTS
Functional telomerase RNA templates are highly enriched in C and A nucleotides
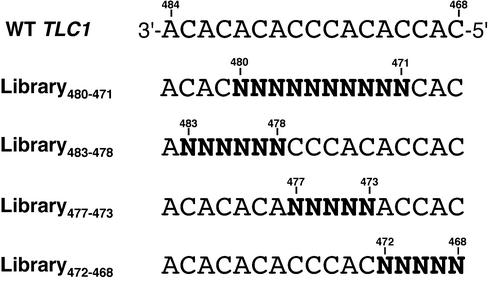
To increase the likelihood of mutant telomerases adding multiple repeats to the telomeres, we randomized only the central 10 nt of the wild-type TLC1 template region (Library480–471, Fig. 1). This library design should enable all mutant telomerases to base pair to the existing wild-type telomeres. In addition, the conservation of 3 nt at either end of the template should allow re-alignment of the telomeres after reverse transcription of a mutant template up to the template 5′ boundary.
Figure 1.
Schematic representation of the different TLC1 template libraries used in this study. All telomerase RNA template sequences in the figures and text of this manuscript are written in the 3′ to 5′ direction, reflecting the order in which the template nucleotides are reverse transcribed by the telomerase reverse transcriptase.
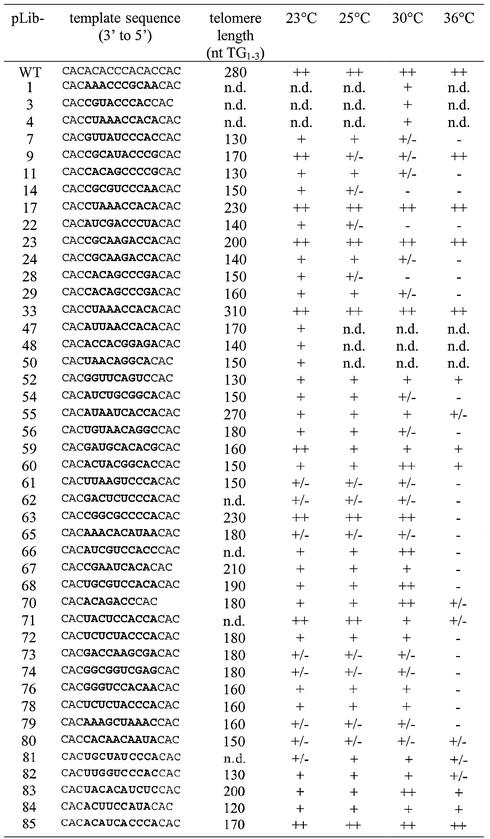
To select for templates that complemented a deletion of the TLC1 gene, we transformed YKF19 (tlc1-Δ rad52-Δ) with Library480–471 as the cells underwent senescence. The colonies obtained were re-streaked at least twice before further analysis. Out of an estimated 60 000 transformants (see Materials and Methods), 40 different mutant templates complemented the tlc1 deletion (Table 1). Two template mutations were isolated twice (23 + 24 and 72 + 78), and one template mutation was isolated three times independently (4 + 17 + 33). Five mutant templates were shorter than the wild-type TLC1 template region and were not taken into account in deriving the consensus sequence. Most telomerase RNA template mutations that complemented the tlc1 deletion nevertheless resulted in slow growth, various degrees of temperature sensitivity and short telomeres. Sequence comparison of complementing full-length mutant templates revealed little similarity in positions close to the template 3′ boundary, while the region close to the template 5′ boundary showed a strong bias for C and A as templating nucleotides (Fig. 2, nucleotide composition over mutant region different from 25% each: P > 99.9, χ2 analysis). This sequence bias was not present in the library before the screen (data not shown).
Table 1. Complementing templates from Library480–471.
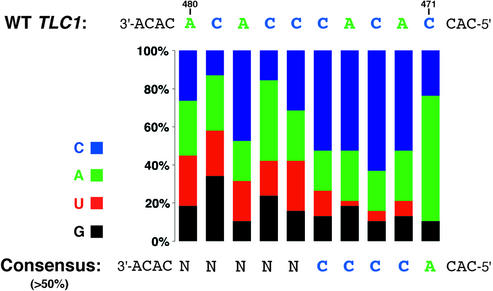
Figure 2.
Nucleotide frequencies at the randomized positions in Library480–471 of those alleles that complemented a tlc1 deletion. The color-coded bars show the frequency of the four nucleotides found at the respective position. The consensus indicated at the bottom represents nucleotides that are present in more than half of the complementing template mutations at the indicated positions.
The sequence 3′-CCCCA-5′ (indicated below the bar graph) is proposed as a consensus since these nucleotides are present in more than half of the template sequences at the respective positions. Together with the invariant positions due to the library design, this suggested that the sequence 3′-CCC CACAC-5′, reminiscent of the central portion in the wild-type TLC1 template sequence, can fulfill the sequence-specific requirements for S.cerevisiae telomere maintenance. Since this consensus represents the most frequent nucleotide at each position but not the most frequent template sequence, the full consensus is found in only one of the mutant templates. The slightly shorter sequence 3′-CCCACAC-5′ is present in six mutant templates, and the relaxed consensus C2–4(AC)1–3 is found in 18 of the full-length and two of the non full-length mutant templates (see Table 1). It should be noted, however, that at least the short versions of this consensus (e.g. 3′-CCAC-5′) are not sufficient for telomere maintenance; they presumably require additional sequence features within the template (see, for example, the results with Library477–473 below).
The template positions 3′ of 477C make no essential contributions to the sequence-dependent function of telomerase
In wild-type telomerase, >70% of the alignment events take place between positions 484A and 479C (19). However, no sequence conservation was apparent for positions 480–476 in complementing clones from Library480–471. To directly test the sequence requirements in this region, we designed a library in which the nucleotides 3′ of 477C were mutagenized (Library483–478, Fig. 1). The majority of the templates from Library483–478 rescued the cells from senescence (95 ± 11%, average ± SD, n = 3), arguing that this region either does not contribute significantly to sequence-dependent telomerase functions or that substrate annealing 5′ of 477C can be sufficient for in vivo telomere maintenance.
Consecutive rC template nucleotides mediate a sequence-dependent function of TLC1
The consensus template sequence derived from the complementing clones of Library480–471 resembles the central 477CCCAC473 sequence of wild-type TLC1 but is found closer to the template 5′ boundary. To further test whether the region at the template 5′ boundary contributes to the sequence-dependent function, we designed a library in which only the five template positions adjacent to the 5′ template boundary were randomized (Library472–468, Fig. 1). Only about half of the template sequences from this library could complement the tlc1 deletion (40 ± 12%, n = 3). Since Library472–468 contained fewer complementing template RNAs than Library483–478, even though the complexity of Library472–468 is lower, we propose that part of the sequence-dependent function resides within positions 472–468 of TLC1. We cannot exclude, however, that some templates of Library472–468 are non-functional because mutant nucleotides, once incorporated into the telomere, impair the translocation or re-annealing step during the next round of telomere extension.
We next analyzed whether the central 477CCCAC473 is important for TLC1 function. Templates from Library477–473 could complement the tlc1 deletion to roughly the same extent as those from Library472–468 (32 ± 3%, n = 4). Thirty-two complementing templates were recovered and sequenced (Table 2). Four templates were recovered twice (4 and 45, 17 and 33, 18 and 47, and 20 and 23), and all of these contained at least a CC dinucleotide in the mutant region. Considering the entire set of the recovered templates, 22 out of the 28 distinct sequences contained a CC dinucleotide or a CCC trinucleotide in the mutagenized region (78% of the complementing templates). This bias is significantly higher than the random frequency of a CC dinucleotide in a 5mer sequence [25% expected: P = (0.25)2 × 4 = 0.25] and was not found in the library before selection (data not shown). Furthermore, the consensus 477NNNCC473 could be derived. Taken together with the results from Library480–471, this indicates that at least two consecutive rC template nucleotides are important for efficient telomerase function. Two separate blocks of consecutive rC nucleotides, as found in the wild-type template sequence, were clearly selected for in the context of Library477–473 and therefore appear to enhance telomerase efficiency further.
Table 2. Complementing templates from Library477–473.
Few mutant telomerase RNA templates induce a growth arrest in S.cerevisiae
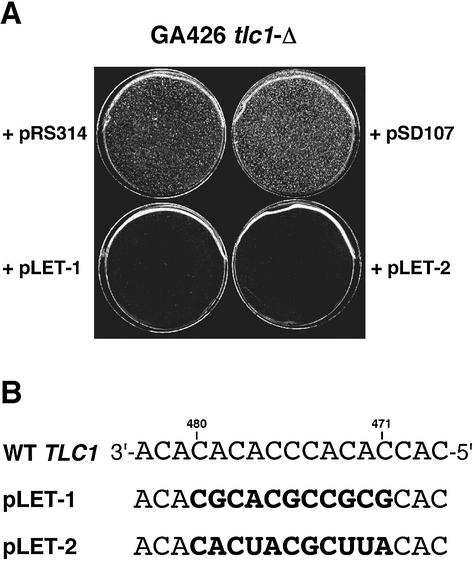
The incorporation of mutant sequences at the 3′ end of a telomere can interfere with its capping function and can lead to cell death (5,6,12,13,24). Since an inducible expression system for mutant TLC1 genes using the GAL1–10 promotor did not give satisfactory results (data not shown), we employed a plasmid shuffling technique to recover mutant library plasmids. This approach allowed us to screen for tlc1 alleles that were recessive to wild-type TLC1 but lethal when expressed on their own. From a total of 60 000 transformants with Library480–471, we obtained only two candidates that reproducibly induced growth arrest in the context of equilibrium-length telomeres (Fig. 3). This corresponds to a frequency of 0.003%, even lower than that determined in the complementation screen with Library480–471 (0.07%).
Figure 3.
(A) Growth arrest can be induced by template mutant telomerase. tlc1-Δ cells were transformed with the indicated plasmids well before the onset of replicative senescence. (B) Two template sequences of death- inducing template RNAs. The tetranucleotide 3′-ACGC-5′ is present in both lethal templates, but also in complementing RNA templates from Library477–473 (21, identical with 29) and Library480–471 (59). It is therefore not solely responsible for the lethal effect.
Since the applied selection scheme depends on a recessive phenotype of the tlc1 template mutation with respect to the wild-type TLC1 plasmid, we also determined the proportion of Library480–471 plasmids that show a dominant phenotype. We observed no reduced viability upon transformation of 12 randomly chosen template library plasmids into TLC1 wild-type cells. Thus, the level of mutant tlc1 genes in Library480–471 with a dominant lethal growth phenotype is likely to be <10% and can therefore not explain the low frequency of plasmids in this library that complemented a tlc1 deletion.
Template mutant telomerase RNAs are efficiently assembled into telomerase enzymes
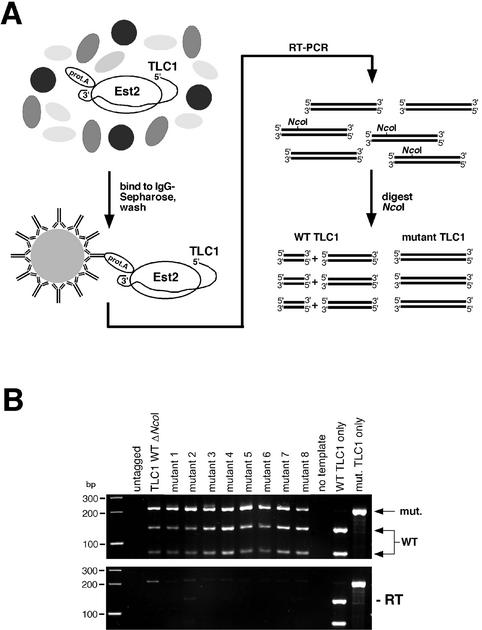
We examined whether mutant telomerase RNAs were efficiently assembled into telomerase ribonucleoprotein complexes with an IP–RT–PCR strategy. Since in all template library plasmids an NcoI restriction site close to the TLC1 template region is deleted, the mutant template RNAs can be distinguished from wild-type TLC1 RNA in a mixed population by digestion with NcoI. We transformed a strain carrying a fusion gene of the telomerase catalytic subunit (EST2) and protein A with Library480–471. The protein A-tagged telomerase was immunoprecipitated from protein extracts with IgG-coated beads (20,25) and the associated RNA was reverse transcribed. A fragment around the TLC1 template region was amplified by PCR, digested with NcoI and analyzed by gel electrophoresis (Fig. 4A). The wild-type TLC1 RNA present in this strain served as a positive control for the efficiency of the IP. We found that eight randomly chosen template mutants were incorporated into telomerase enzymes as efficiently as wild-type TLC1 (Fig. 4B). Thus, the assembly of telomerase does not show sequence specificity with regard to the template region. This is consistent with the finding that the TLC1 template region is dispensable for assembly with the catalytic subunit, Est2p (26).
Figure 4.
Template mutant RNAs are associated with Est2p. (A) Outline of the IP–RT–PCR experiment to assess the association of template mutant telomerase RNAs with the telomerase catalytic subunit, Est2p. All yeast strains contained a wild-type TLC1 gene, which serves as internal control for IP efficiency and as competitor for the assembly of the template mutant RNAs with Est2p in vivo. (B) All eight randomly chosen template mutants (mutants 1–8) from Library480–471 are efficiently assembled into telomerase enzymes. Upper panel, PCR products can be obtained for both the endogenous wild-type TLC1 RNA (cut with NcoI) and the template mutant TLC1 RNAs (larger, uncut fragment). TLC1 WT ΔNcoI refers to a control mutation where the NcoI site was abolished but the template sequence was left unchanged. Lower panel, PCR amplification of the immunopurified RNA without prior RT treatment to reveal products due to DNA contamination.
Enforced interaction of telomerase with the telomeres does not increase the frequency of functional template RNAs
Telomerase is recruited to or activated at telomeres through the interaction of Est1p with Cdc13p (27–31). Fusions of the open reading frames of telomerase components and CDC13 or its DNA-binding domain (CDC13DBD) force the interaction of telomerase with the telomere and lead to vigorous telomere elongation (27,32,33). We included two such fusion proteins, Cdc13–Est2p (27) and Cdc13DBD–Est3p (32), in our screen for growth arrest. If the mutant telomerases were inactive due to an access defect, the CDC13 fusion proteins should have alleviated this problem.
In the presence of either Cdc13 fusion protein, the frequency of colonies that did not grow after counterselection of the wild-type TLC1 plasmid varied only slightly between the empty vector (3.1 ± 0.75%, n = 2), the wild-type plasmid (2.3 ± 0.3%, n = 2) and the Library480–471 (5.3 ± 1.6%, n = 2). Twenty-four colonies without an apparent growth defect after counterselection of the wild-type TLC1 plasmid were re-streaked successively, and all showed senescence at the same time as control clones with the empty vector (data not shown). In addition, the overall transformation efficiency of the library was not reduced by the introduction of the fusion proteins, indicating that the frequency of dominant lethal template mutations also did not increase.
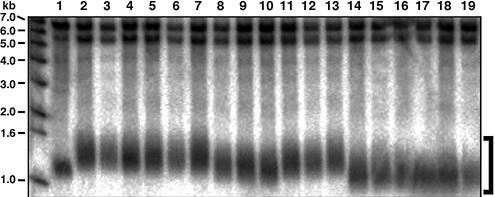
The presence of the Cdc13–telomerase fusion proteins therefore does not rescue the in vivo incorporation defect seen with the majority of the mutant templates in Library480–471. Telomere elongation in the presence of the Cdc13–telomerase fusion proteins did occur in the presence of a wild-type TLC1 gene (Fig. 5, compare lane 1 with lanes 2–10). However, the elongated telomeres became shortened in cells with the empty vector or library plasmids after the wild-type TLC1 plasmid had been shuffled out (Fig. 5, lanes 14–19).
Figure 5.
Telomere length analysis by Southern blotting of cells expressing a Cdc13DBD–Est3 fusion protein and various template mutant telomerase RNAs. Except for the wild-type strain, three independent colonies were analyzed for each genotype. Genomic DNA was prepared 50–75 generations after introduction of the Cdc13DBD–Est3 fusion protein. The library candidates were chosen at random. Lane 1, GA426 WT; lanes 2–4, YKF20 + pVL1292 + pTLC1-URA3 + pSD107; lanes 5–7, YKF20 + pVL1292 + pTLC1-URA3 + pRS314; lanes 8–10, YKF20 + pVL1292 + pTLC1-URA3 + candidates from Library480–471; lanes 11–13, YKF20 + pVL1292 + pSD107, pTLC1-URA3 shuffled out; lanes 14–16, YKF20 + pVL1292 + pRS314, pTLC1-URA3 shuffled out; lanes 17–19, YKF20 + pVL1292 + candidates from Library480–471, pTLC1-URA3 shuffled out. Terminal restriction fragments of the Y′-telomeres are indicated by the bracket on the right.
Most templates in Library480–471 do not lead to incorporation of mutant sequences into the telomeres
The frequency at which candidate templates were obtained in either of the two screens was <0.1% of Library480–471. Since the mutant tlc1 RNAs were efficiently assembled into telomerase enzymes and most likely not limited in their access to the telomeres, we examined whether the telomeres had acquired mutant sequences. We employed telomere-PCR (21) to compare the telomere length of cells that contained either a wild-type TLC1 gene, an empty vector or a plasmid from Library480–471. While the presence of wild-type TLC1 allowed normal telomere length maintenance (266 ± 18 nt, n = 20), cells that had received an empty vector lacked telomerase activity and consequently had significantly shorter telomeres (179 ± 34 nt, n = 20, shorter than wild-type TLC1 P < 0.001, t-test) 25 generations after a plasmid containing wild-type TLC1 had been shuffled out. The telomere length of cells that contained a plasmid from Library480–471 showed the same extent of telomere shortening (188 ± 34 nt, n = 20, shorter than wild-type TLC1 P < 0.001, and shorter than empty vector P > 0.4, t-test), and these mutant yeast cells senesced upon further re-streaking. To rule out the possibility of very low levels of mutant sequence incorporation, 12 telomeres from four different template mutants were cloned but no mutant sequences were recovered.
Non-functional template mutations affect the nucleotide addition processivity of telomerase
We compared the in vitro telomerase activities of complementing and non-complementing template mutants obtained from Library477–473. The candidates were taken from this library because the same DNA oligonucleotide substrate, d(TG)7, can be used for all mutants. It presumably anneals within the sequence 484ACACACA476 which is present in all mutant templates [note that in the context of a wild-type template sequence, the nucleotides 479CAC477 and 473CAC471 are excluded for initial substrate annealing in vivo (19)]. Telomerase was prepared from cells carrying either the wild-type TLC1 gene, an empty vector, one of three non-complementing telomerase RNA template mutants (chosen from a random collection of individual mutant templates from Library477–473) or one of three complementing telomerase RNA template mutants. Strikingly, all three non-complementing template mutants showed a reduced activity compared with wild-type and complementing mutant telomerases (Fig. 6).
Figure 6.
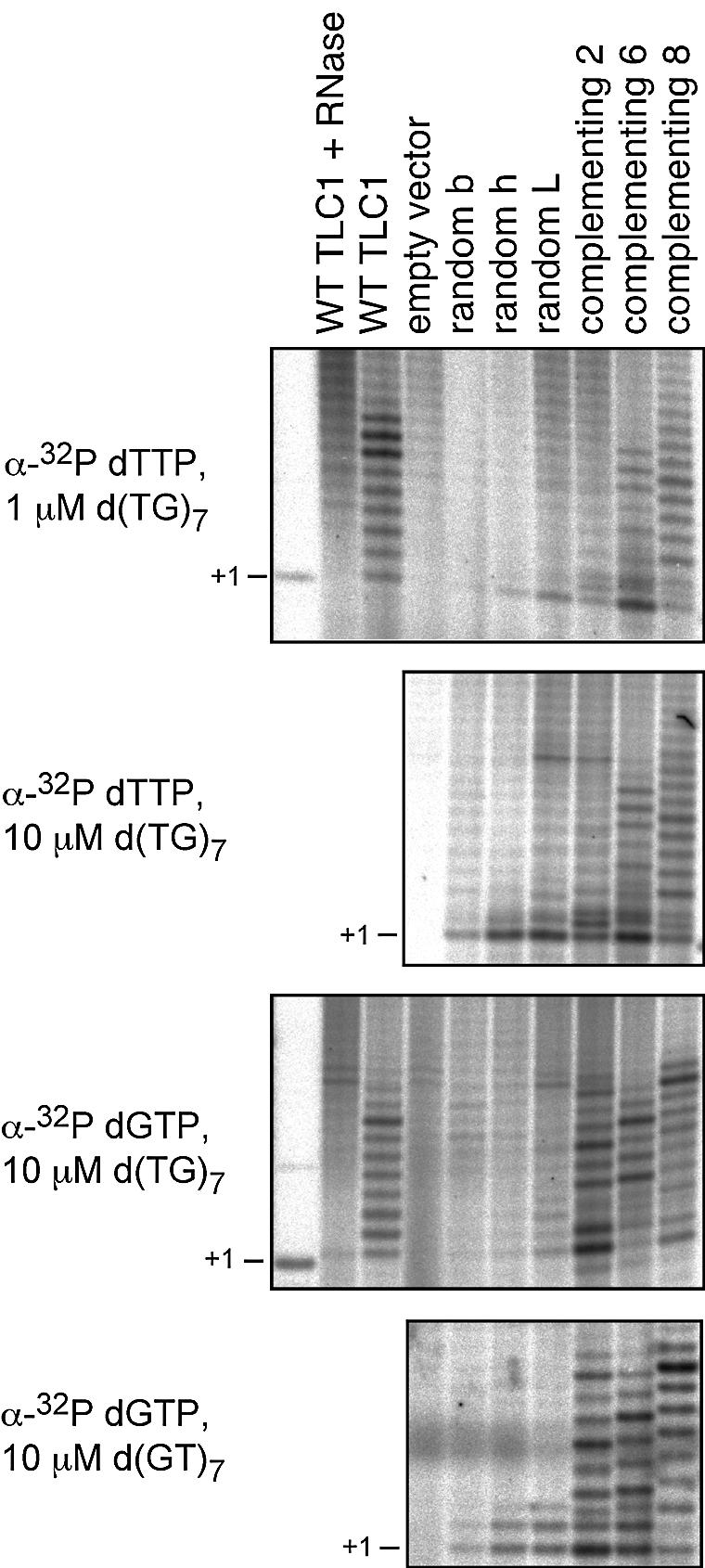
In vitro telomerase activity assays. The DNA oligonucleotide substrate and the labeled dNTP are indicated on the left. TLC1 template mutants were selected from Library477–473. The reactions contained all unlabeled dNTP substrates at 50 µM and labeled dNTP substrates at 5 µM. The template sequences were (mutant stretches underlined): random b 484ACACACAUGGAGACCAC468, random h 484CACACAUAGUGACCAC468, random L 484CACACAUCUAGACCAC468, complementing 2 484CACACAGUGCCACCAC468, complementing 6 484CACACACAUGCACCAC468 and complementing 8 484CACACAGACCCACCAC468.
The low activity levels detected might have been caused by the changed template sequence, resulting in a lower incorporation rate of the labeled nucleotide [compare, for example, complementing mutant 2 in Figure 6 with labeled dTTP (second panel) and labeled dGTP (third panel)]. We tested this hypothesis by including labeled dATP in the reaction. The non-complementing mutant templates tested in our assay all contain an rU nucleotide at the first mutant position, which should direct the incorporation of the labeled dATP. No increased activity for the non-complementing mutants was detected in these reactions (Fig. 7A), confirming an impaired enzymatic activity.
Figure 7.
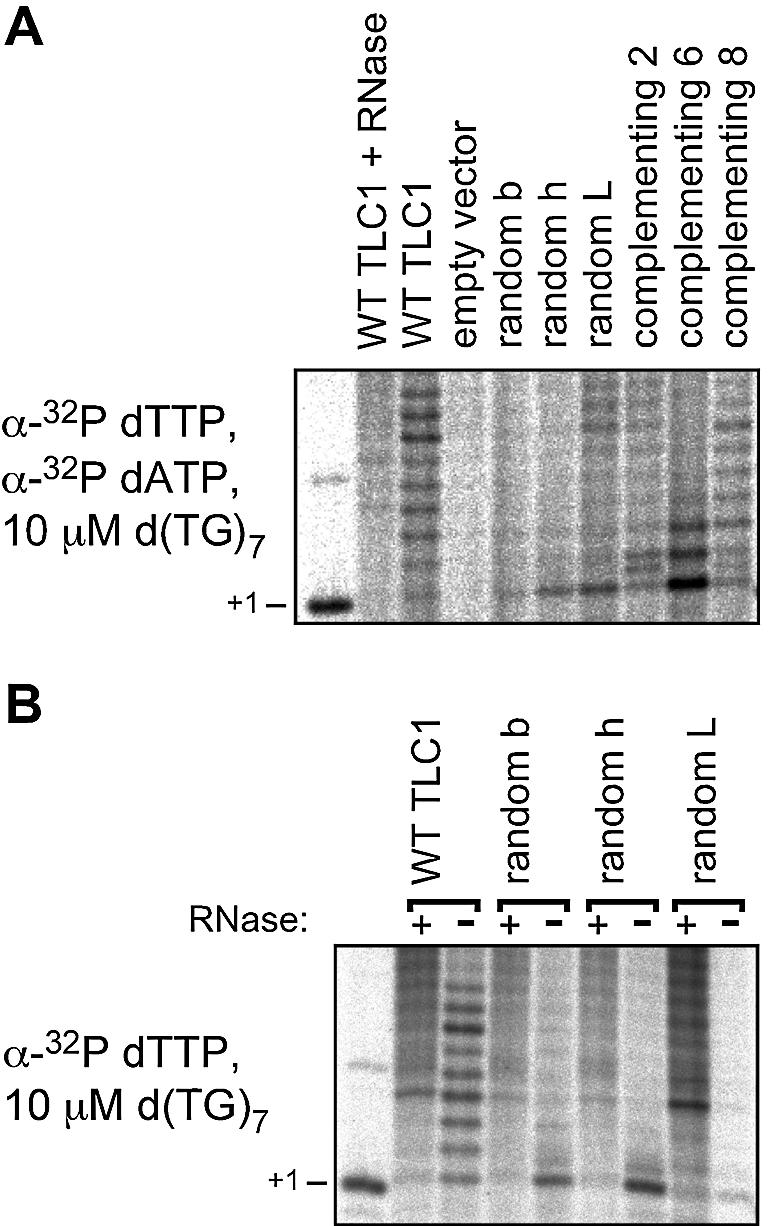
In vitro telomerase activity assays. (A) Control reactions containing both labeled dTTP and labeled dATP. No increased activity could be detected for the non-complementing telomerase RNA template mutations (random b, random h and random L). (B) The +1 band obtained in the reactions with the non-complementing telomerase RNA template mutations is RNase sensitive.
Despite their reduced activity, the non-complementing template mutant telomerases could add a single nucleotide to the substrate DNA oligonucleotide (TG)7 (e.g. see Fig. 6, second panel) and up to 3 nt to the substrate oligonucleotide (GT)7 (Fig. 6, bottom panel). This activity was RNase sensitive (Fig. 7B), absent in extracts from cells that had received an empty vector instead of a TLC1 gene (e.g. Fig. 6, second panel, first lane) and weak when labeled dGTP was used in the reaction with the substrate (TG)7 (Fig. 6, third panel). We therefore conclude that it represents bona fide template-directed telomerase activity, strongly suggesting that the processivity is perturbed in the mutant enzymes. This may result either from an inefficient incorporation of dATP and dCTP by telomerase or from an inability of the mutant enzymes to perform structural transitions necessary to advance to the next template position. It is unlikely that the templating nucleotide 477U is solely responsible for the enzymatic defect, as many complementing mutants from Library477–473 also contain the mutation 477U (Table 2). Furthermore, we have shown previously that a 469A→U mutation leads to the incorporation of dA into telomeres in vivo (19), arguing that dATP can be a substrate for yeast telomerase. The low frequency of functional template RNAs contained in Library477–473 and, most probably, Library480–471 therefore appears to be due to an impact of mutant template sequences on telomerase nucleotide addition processivity. While this reduced processivity is the most striking phenotype of the template mutant telomerases, we cannot exclude additional effects on, for example, the DNA substrate affinity or the fidelity of the telomerase enzyme.
DISCUSSION
Functional requirements on the telomerase RNA template sequence
Our template library screens reveal for the first time that only a minor fraction of all possible template sequences will reconstitute active telomerase in S.cerevisiae. Functional templates from Library480–471 often contained the sequence 3′-C2–4(AC)1–3-5′ positioned at or near the template 5′ boundary. This motif resembles the central 477CCCAC473 and the 5′ template boundary 471CCAC468 of wild-type TLC1, indicating that these sequences play a role in telomerase biochemistry that goes beyond their function as a passive template for nucleotide addition. Analysis of template mutant telomerases in vitro revealed that non-complementing template mutations lead to reduced nucleotide addition processivity. Similar results were reported previously for T.thermophila telomerase based on a series of site-specific RNA template mutants (14–16). The present study not only extends this notion to budding yeast, but also explores 60 000 different template sequences in parallel.
In wild-type TLC1, the nucleotides 479CAC477 and 473CAC471 are not available for telomere alignment but become available for base pairing during reverse transcription. In addition, the central 477CCC475 trinucleotide is reverse transcribed in a processive manner, while product dissociation can occur 3′ or 5′ of this motif (19). Thus, the biochemical properties of telomerase vary with the template position that is being reverse transcribed. Also, defined changes in the sequence of the RNA template could influence telomerase activity. For example, the mutation 476CCA474→GUG in S.cerevisiae telomerase RNA gave rise to an inactive enzyme (18) when it was the only telomerase RNA species in the cell. However, telomerase activity was restored even for the mutant template in the context of wild-type/mutant enzyme heteromultimers. It is unclear at this point which enzymatic defect is conveyed by this mutation. Whatever the rules governing the properties of yeast telomerase may be, the reduced in vitro processivity of the three template mutant telomerases tested in our study indicates that the threading of subsequent template nucleotides through the active site is very sensitive to changes in the RNA template sequence. A previous study has shown that a mutant yeast telomerase RNA specifying human telomeric repeats is functional in vivo and leads to the incorporation of human telomere repeats onto yeast chromosome ends (34). This mutation corresponds roughly to our derived consensus as it contains several CCC trinucleotides. However, since the putative total length of the template region is longer, this mutant template sequence cannot be superimposed without prior assumptions onto the results obtained in our study. Several point mutations in the budding yeast telomerase catalytic subunit (Est2p) resulted in reduced nucleotide addition processivity in vitro (35,36). It is not clear whether these est2 mutations affect the same mechanism as our RNA template mutations, especially since the deficiency conferred by RNA template sequence changes is far more pronounced.
Our screen also demonstrates that a stretch of successive rC template nucleotides is not absolutely required for telomere maintenance (see, for example, template 65 in Table 1) and that the sequence 472CCAC468 is not sufficient for telomerase activity in vivo (complementation <100% with Library477–473). The sequence requirements on the budding yeast RNA template therefore must be more complex than the minimal consensus found in our study. It is noteworthy that in the context of the very long telomerase RNA template regions of certain yeasts, the telomerase RNA templates do not need to be particularly C/A rich (4,37).
The active templates identified in our screen may have been subject to a further selection for telomerases that synthesize DNA with binding sites for essential telomere-binding proteins such as Rap1p, Cdc13p and Est1p. While Cdc13p provides essential end-protecting functions and recruits or activates telomerase via its interaction with Est1p (28,29,31,38–40), the binding of Rap1p to the telomeres negatively regulates extension (41–43). Thus, a fully functional telomerase that does not incorporate Rap1p-binding sites into the telomere is predicted to lead to strong telomere elongation. Mutant telomerase enzymes with this phenotype can be obtained through single nucleotide substitutions in the telomerase RNA template region (4,7,44), leading to rapid telomere elongation by >2 kb within 50 generations (7,44). In contrast, almost all of the complementing template mutations identified in our screen of Library480–471 resulted in shortened telomeres. Since it is unlikely that all the corresponding mutant telomere sequences result in increased Rap1p binding, we propose that the mutant telomerases do not obtain the full activity of the wild-type enzyme. This hypothesis is corroborated by the reduced in vitro nucleotide addition processivity of non-complementing template mutant telomerases. On the other hand, incorporated mutant telomere sequences that lead to reduced binding of Cdc13p (a complete lack of Cdc13p binding should be lethal) may decrease the recruitment or activation of telomerase. Even in the context of a Cdc13DBD–telomerase fusion protein, this could have prevented the generation of long stretches of mutant telomeric DNA and thus may have limited the number of complementing RNA template sequences recovered in our screens.
Has the need for C/A-rich template RNAs contributed to the conservation of the telomeric repeat sequences?
The relatively strong conservation of the telomeric repeat sequence during evolution is an unusual feature for non-coding DNA. The propensity of single-stranded telomeric DNA from most species to form stable secondary structures based on G–G pairing has been proposed to play a protective role at the chromosome end. Loss of the telomeric 3′ single-stranded extensions correlates with loss of end protection (45), and experimentally induced chromosome end-to-end fusions occurred preferentially at telomeres replicated by the leading strand machinery (46), which leaves blunt ends after replication. This putative protective function of the telomere sequence could certainly explain its conservation.
On the other hand, the template sequence dependence of telomerase activity described here may also limit the divergence of telomeric sequences during evolution. Consistent with this hypothesis, the telomeres of Drosophila melanogaster, which are maintained by retrotransposition rather than telomerase, are not G/T rich [reviewed by Pardue et al. (47) and Louis (48)]. Their chromosome ends nonetheless are specifically recognized and protected since mutations in a telomere-binding protein lead to chromosome end-to-end fusions (49). This indicates that telomere capping can be achieved without the help of G-rich DNA secondary structures and that telomere sequences can, in principle, deviate from the T/G-rich consensus. Our template library screen has revealed that extensive changes of the budding yeast telomerase RNA template sequence most probably result in non-functional telomerase enzymes. If this specialization of the TERT enzyme for a C/A-rich RNA template is conserved in other telomerases, it may give an alternative explanation for the conservation of the telomere sequence.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr V. Lundblad for plasmids encoding the Cdc13-Est2 (pVL1107) and Cdc13DBD-Est3 (pVL1292) fusion proteins. This work was supported by a Human Frontier Science Program grant (laboratories of J.L. and T.R.C.), the Swiss National Science Foundation (J.L.) and the National Center of Competence in Research ‘Frontiers in Genetics’ program (J.L.).
REFERENCES
- 1.Wellinger R.J. and Sen,D. (1997) The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer, 33, 735–749. [DOI] [PubMed] [Google Scholar]
- 2.Williamson J. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 3.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 4.McEachern M.J. and Blackburn,E.H. (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature, 376, 403–409. [DOI] [PubMed] [Google Scholar]
- 5.Kirk K.E., Harmon,B.P., Reichardt,I.K., Sedat,J.W. and Blackburn,E.H. (1997) Block in anaphase chromosome separation caused by a telomerase template mutation. Science, 275, 1478–1481. [DOI] [PubMed] [Google Scholar]
- 6.Smith C.D. and Blackburn,E.H. (1999) Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J. Cell Biol., 145, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prescott J.C. and Blackburn,E.H. (2000) Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol. Cell. Biol., 20, 2941–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shippen-Lentz D. and Blackburn,E.H. (1990) Functional evidence for an RNA template in telomerase. Science, 247, 546–552. [DOI] [PubMed] [Google Scholar]
- 9.Salazar M., Thompson,B.D., Kerwin,S.M. and Hurley,L.H. (1996) Thermally induced DNA–RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry, 35, 16110–16115. [DOI] [PubMed] [Google Scholar]
- 10.Jarstfer M. and Cech,T. (2002) Effects of nucleotide analogues on Euplotes aedicluatus telomerase processivity: evidence for product-assisted translocation. Biochemistry, 41, 151–161. [DOI] [PubMed] [Google Scholar]
- 11.Ware T.L., Wang,H. and Blackburn,E.H. (2000) Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J., 19, 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marusic L., Anton,M., Tidy,A., Wang,P., Villeponteau,B. and Bacchetti,S. (1997) Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol. Cell. Biol., 17, 6394–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M.M., Rivera,M.A., Botchkina,I.L., Shalaby,R., Thor,A.D. and Blackburn,E.L. (2001) A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl Acad. Sci. USA, 98, 7982–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilley D., Lee,M.S. and Blackburn,E.H. (1995) Altering specific telomerase RNA template residues affects active site function. Genes Dev., 9, 2214–2226. [DOI] [PubMed] [Google Scholar]
- 15.Autexier C. and Greider,C.W. (1995) Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev., 9, 2227–2239. [DOI] [PubMed] [Google Scholar]
- 16.Gilley D. and Blackburn,E.H. (1996) Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol., 16, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prescott J. and Blackburn,E.H. (1997) Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- 18.Prescott J. and Blackburn,E.H. (1997) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev., 11, 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forstemann K. and Lingner,J. (2001) Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol., 21, 7277–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman K.L. and Cech,T.R. (1999) Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev., 13, 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forstemann K., Hoss,M. and Lingner,J. (2000) Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res., 28, 2690–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie K.B., Mallory,J.C. and Petes,T.D. (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1 and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohn M. and Blackburn,E.H. (1995) Telomerase in yeast. Science, 269, 396–400. [DOI] [PubMed] [Google Scholar]
- 24.McEachern M.J., Iyer,S., Fulton,T.B. and Blackburn,E.H. (2000) Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl Acad. Sci. USA, 97, 11409–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto A.G., Zaug,A.J., Sobel,S.G., Wolin,S.L. and Cech,T.R. (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature, 401, 177–180. [DOI] [PubMed] [Google Scholar]
- 26.Livengood A.J., Zaug,A.J. and Cech,T.R. (2002) Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol., 22, 2366–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- 28.Pennock E., Buckley,K. and Lundblad,V. (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell, 104, 387–396. [DOI] [PubMed] [Google Scholar]
- 29.Chandra A., Hughes,T.R., Nugent,C.I. and Lundblad,V. (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev., 15, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi H. and Zakian,V.A. (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev., 14, 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 31.Taggart A.K., Teng,S.C. and Zakian,V.A. (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science, 297, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 32.Hughes T.R., Evans,S.K., Weilbaecher,R.G. and Lundblad,V. (2000) The est3 protein is a subunit of yeast telomerase. Curr. Biol., 10, 809–812. [DOI] [PubMed] [Google Scholar]
- 33.Grandin N., Damon,C. and Charbonneau,M. (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol., 20, 8397–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henning K.A., Moskowitz,N., Ashlock,M.A. and Liu,P.P. (1998) Humanizing the yeast telomerase template. Proc. Natl Acad. Sci. USA, 95, 5667–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosoy D. and Lue,N.F. (2001) Functional analysis of conserved residues in the putative ‘finger’ domain of telomerase reverse transcriptase. J. Biol. Chem., 276, 46305–46312. [DOI] [PubMed] [Google Scholar]
- 36.Peng Y., Mian,I.S. and Lue,N.F. (2001) Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell, 7, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 37.McEachern M.J. and Hicks,J.B. (1993) Unusually large telomeric repeats in the yeast Candida albicans. Mol. Cell. Biol., 13, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garvik B., Carson,M. and Hartwell,L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J.J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- 41.Lustig A., Kurtz,S. and Shore,D. (1990) Involvement of the silencer and UAS binding protein Rap1 in regulation of telomere length. Science, 250, 549–553. [DOI] [PubMed] [Google Scholar]
- 42.Kyrion G., Boake,K. and Lustig,J. (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcand S., Gilson,E. and Shore,D. (1997) A protein-counting mechanism for telomere length regulation in yeast. Science, 275, 986–990. [DOI] [PubMed] [Google Scholar]
- 44.Chan S., Chang,J., Prescott,J. and Blackburn,E. (2001) Altering telomere structure allows telomerase to act in yeasts lacking ATM kinases. Curr. Biol., 11, 1240–1250. [DOI] [PubMed] [Google Scholar]
- 45.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 46.Bailey S., Cornforth,M., Kurimasa,A., Chen,D. and Goodwin,E. (2001) Strand-specific postreplicative processing of mammalian telomeres. Science, 293, 2462–2465. [DOI] [PubMed] [Google Scholar]
- 47.Pardue M., Danilevskaya,O., Lowenhaupt,K., Slot,F. and Traverse,K. (1996) Drosophila telomeres: new views on chromosome evolution. Trends Genet., 12, 48–52. [DOI] [PubMed] [Google Scholar]
- 48.Louis E.J. (2002) Are Drosophila telomeres an exception or the rule? Genome Biol., 3, REVIEWS0007.1–0007.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fanti L., Giovinazzo,G., Berloco,M. and Pimpinelli,S. (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell, 2, 527–538. [DOI] [PubMed] [Google Scholar]