Abstract
The inositol 1,4,5-trisphosphate receptor (IP3R) is a tetrameric intracellular Ca2+ channel, which mediates the release of Ca2+ from the endoplasmic reticulum in response to many different extracellular stimuli. We present a 3D structure of the type 1 IP3R obtained by electron microscopy and single-particle analysis that reveals its domain organization. The IP3R has a flower-like appearance with fourfold symmetry and is made up of three distinct domains connected by slender links. By relating the organization of the structural domains to secondary-structure predictions and biochemical data we develop a model in which structural domains are mapped onto the amino acid sequence to deduce the location of the channel region and the cytoplasmic inositol 1,4,5-trisphosphate-binding and modulatory subdomains. The structure of the IP3R is compared with that of other tetrameric cation channels. The channel domain is similar in size and shape to its counterparts in the ryanodine receptor and the Shaker voltage-gated K+ channel.
The receptors for the second messenger inositol 1,4,5-trisphosphate (IP3) are intracellular Ca2+ channels, the opening of which is controlled by both IP3 and Ca2+ (1, 2). This dual regulation is important in allowing IP3 receptors (IP3Rs) to generate spatially and temporally complex intracellular Ca2+ signals (3). In addition, IP3R may also be directly involved in regulating Ca2+ entry into cells by providing a functional association between intracellular Ca2+ stores and plasma-membrane channels responsible for store-regulated Ca2+ entry (4).
Functional IP3Rs are assemblies of four single-chain subunits. In mammals, three different genes encode closely related subtypes of IP3R subunits, and related genes are found in other species (5). Each of these subunits has about 2,700 amino acid residues and consists of a transmembrane region, which probably includes six transmembrane helices and a P loop (6, 7), a large cytoplasmic N-terminal region, and a short C-terminal cytoplasmic tail (see Fig. 2C).
Figure 2.
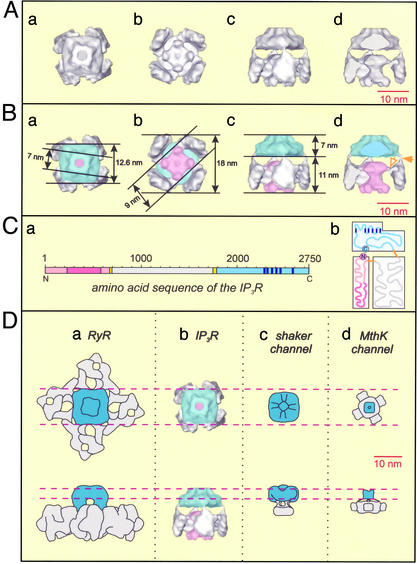
Structures of IP3R and other tetrameric cation channels. (A and B) Surface-rendered representations of the 3D map of IP3R viewed along the fourfold axis from the ER lumen (a); viewed along the fourfold axis from the cytoplasm (b); viewed orthogonal to the membrane plane (c); and viewed as in c but sectioned to remove one cytoplasmic petal domain to reveal the cytoplasmic stigma domain (d). In B the dimensions of the 3D map are indicated and the different domains of IP3R are identified by the following colors: blue, channel domain; pink, cytoplasmic stigma domain; and gray, cytoplasmic petal domains. In Bd the filled arrowhead marks one of the four slender links between the transmembrane domain and the petal subdomain, whereas a corresponding slender link between a petal subdomain and the stigma subdomain is marked with an open arrow. (C) Amino acid sequence of an IP3R subunit represented linearly (a) and drawn to reveal its overall domain distribution (b). Proposed domains are color-coded as follows: the cytoplasmic stigma domain (pink) containing the IP3-binding pocket (dark pink), cytoplasmic petal domain (gray), channel domain (blue), and its transmembrane segments (dark blue), and regions of low complexity corresponding to domain boundaries (orange). (D) Comparison of the 3D map of IP3R, viewed from the ER lumen (Upper) and orthogonal to the membrane plane (Lower), with other cation channels with known 3D structures. (a) Cartoon of the type 1 RyR (39). (b) IP3R 3D map. (c) Cartoon of the Shaker channel (40). (d) Cartoon of the MthK channel (41).
The large cytoplasmic region within each subunit of the IP3R binds IP3 and communicates this binding to the pore region of the protein. The IP3-binding site is formed by two regions lying between amino acids 226 and 576 (8), which include conserved basic residues that interact with the phosphate groups of IP3. Other regions interact with this IP3-binding core to modulate its affinity for IP3, notably the N-terminal region (9), sites that bind calmodulin and Ca2+ (10), and residues within the pore region and C-terminal tail (11, 12). The association of N and C termini in IP3R is a feature that may be shared with other tetrameric ion channels (11). For IP3R, this interaction appears to involve the N and C termini of different subunits, so that the IP3-binding domain of one subunit may influence the pore components of its neighbor (11). Each of a pair of sites that lie immediately downstream of the IP3-binding site of the type 3 IP3R has been shown to interact directly with trp3 channels, and perhaps thereby provide a link between the IP3R and activation of store-regulated Ca2+ entry (13).
Sequence analysis and more direct evidence (6) suggest the presence of six transmembrane helices near the C terminus of each IP3R subunit. These helices both mediate oligomerization of the receptor (14) and contribute to the pore structure. The two helices closest to the C terminus (helices 5 and 6) together with the intervening loop (“P-loop”) form the pore itself (7).
How IP3 binding at one end of the IP3R causes the opening of a pore at the opposite end of the primary sequence is unknown. IP3 binding is certainly accompanied by a large conformational change in the receptor (15), but the linkage between IP3 binding and channel opening is complex. It is likely that several of the receptor subunits must first bind IP3 and then Ca2+ before the pore opens (16). Detailed structural information is required to test the predictions arising from biochemical and physiological studies to understand the gating, modulation, and ion selectivity of the IP3R. Here we present the 3D map of this membrane receptor, which we discuss in terms of domain distribution and its relation with other known ion-channel structures.
Materials and Methods
Purification of IP3R.
A membrane fraction was prepared from rat cerebella (17), resuspended (50 mg/ml in 50 mM Tris/1 mM EDTA, pH 8.3), and solubilized by the addition of 1% Triton X-100. After centrifugation (100,000 × g for 60 min), the supernatant, supplemented with 250 mM NaCl, was loaded onto an Econopac heparin column (1 ml, Bio-Rad), washed with 25 ml of medium A (50 mM Tris/250 mM NaCl/10% glycerol, pH 8.3), supplemented with 1 mM EDTA, 1% Surfact-Amps X-100 (Pierce), and then with medium A supplemented with 0.1% Surfact-Amps X-100 (50 ml). IP3R was eluted from the heparin column directly onto a 2-ml Con A-agarose column (Sigma) by using medium A supplemented with 0.1% Surfact-Amps X-100 and 50 μM decavanadate (18). After washing with 150 ml of medium B (50 mM Tris/100 mM NaCl/10% glycerol/0.1% Surfact-Amps X-100, pH 8.3), IP3R was eluted by overnight incubation with 4 ml of medium B containing 800 mM methyl mannopyranoside (17). To assess the quaternary structure of the purified receptor, 0.2-ml fractions of the final eluate were loaded onto linear 10–30% sucrose gradients (10 ml) in 50 mM Tris/1 mM EDTA/0.1% Surfact-Amps X-100, pH 8.3, and centrifuged (134,000 × g for 60 min). Gradients were calibrated by using a high-molecular-weight gel filtration standards kit (Amersham Biosciences). Size-exclusion HPLC was preformed by using a Zorbax GF-450 column (Jones Chromatography, Lakewood, CO) coupled to a Kontron system (Kontron Instruments, Watford, U.K.). All media included the following mixture of protease inhibitors: 150 nM aprotinin, 1 μM pepstatin, 20 μg/ml soybean trypsin inhibitor, 1 mM benzamidine, 250 μM PMSF, 250 μM captopril, and 1 μM bestatin.
Electron Microscopy and Image Analysis.
Purified IP3R, in 50 mM Tris/1 mM EDTA/0.1% Surfact-Amps X-100, pH 8.3, was loaded onto glow-discharged carbon-coated copper electron microscope grids and negatively stained with 2% (wt/vol) uranyl acetate. Micrographs were collected on a Philips (Eindhoven, the Netherlands) CM200 FEG electron microscope under low-dose conditions, at an acceleration voltage of 200 kV and a calibrated magnification of ×37,600. Focal pairs were recorded at defocus levels of ≈1.5 and 5 μm. The micrographs selected for image processing were digitized by using a Leaf Scan 45 densitometer (Scitex, Tel Aviv, Israel) at a step size of 10 μm and then 2 × 2 pixel areas were averaged, resulting in a pixel size of 0.53 nm. Individual molecular images of IP3R were identified interactively by using the program dsload (an X-windows display program for Medical Research Council (MRC) images, available on request from E.P.M.), windowed into 100 × 100 pixel boxes and converted for processing by using imagic programs (19). The defocus value for each micrograph was estimated by using dsload, in amplitude displays of Fourier transforms of representative regions, on the basis of the location of minima and maxima associated with the Thon rings. Particle images were corrected for the effect of the contrast transfer function (CTF) by phase reversal (20). Surface-rendered views of the final 3D map were visualized by using avs software (21).
Secondary Structure Prediction.
IP3R secondary structure predictions were performed online with 3D-PSSM (ref. 22; www.sbg.bio.ic.ac.uk/servers/3dpssm/), SEG screening for low-complexity regions (23), and PROF (24) (SEG and PROF are available at http://cubic.bioc.columbia.edu/predictprotein/). Our assignment of low-complexity regions in the amino acid sequence of IP3R was made by comparing the results obtained by using these programs.
Results and Discussion
Characterization of Purified IP3R.
From sucrose gradient analysis it is clear that most IP3R migrated as tetramers (≈1200 kDa) and that the subunit mobility (≈260 kDa on SDS/PAGE) is indistinguishable from that of native receptors. The homogeneity of purified IP3R was further assessed by size-exclusion HPLC, which showed a single protein peak consistent with a homogenous tetrameric assembly. The maximal IP3-binding capacity of fractions eluted from the Con A column (Bmax = 0.93 IP3/receptor subunit) confirmed the purity of the IP3R preparation. From equilibrium-competition binding assays using [3H]IP3 (25), the values of the affinity for IP3 of the receptor in its native membrane (equilibrium dissociation constant, Kd, = 11.5 ± 1.6 nM, n = 6) and after purification (Kd = 3.6 ± 0.6 nM) were similar. Under the conditions used to load IP3R onto electron microscope grids (medium lacking both IP3 and Ca2+), we expect IP3R to be in a closed state. The characterization of the purified IP3R is supplied as Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org.
Image Processing.
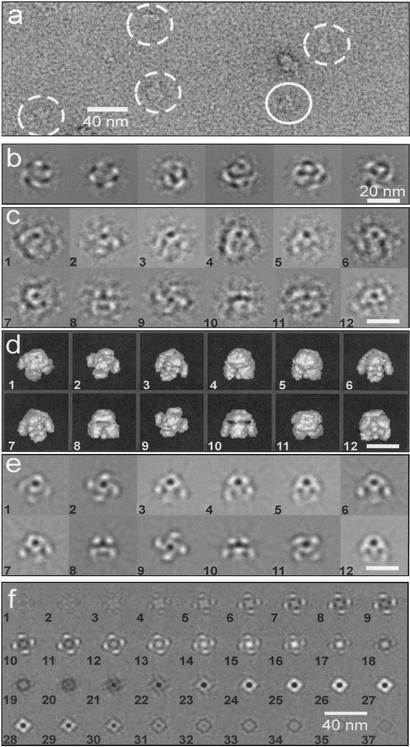
Negatively stained IP3R is shown in Fig. 1a. The data set was restricted to images recorded at ≈5 μm underfocus, which at a 200-kV accelerating voltage gives substantial phase contrast in the resolution range of 10–4 nm, accentuating the features in projection images of IP3R that allow different views of the protein to be distinguished (see below). From an initial set of ≈4,500 projection images of IP3R, 3,435 were selected by visual inspection rejecting clearly damaged, aggregated, or otherwise defective images. The selected images were corrected for the effect of the contrast transfer function, reference-free aligned, and subjected to multivariate statistical analysis, i.e., eigenvector/eigenvalue data compression (19). The resulting eigenvectors correspond to eigenimages, examples of which are shown in Fig. 1b. Strong fourfold modulation is shown by two of the eigenimages obtained from this first multivariate statistical analysis. This modulation is characteristic of the C4 point group, i.e., fourfold symmetry, in a similar manner to the observation of thirteenfold symmetry in eigenimages of the portal protein of SPP1 bacteriophage (26).
Figure 1.
Electron microscopy and image analysis. (a) Electron micrograph of negatively stained IP3R. Images selected for analysis are identified by circles, with one top view of the complex marked by a continuous line, whereas tilted views are indicated by dashed lines. (b) Sequence of eigenimages obtained during classification by multivariate statistical analysis. The first two eigenimages show fourfold modulation, characteristic of C4 point-group symmetry. (c) Examples of class averages used for angular reconstitution. (d) Surface-rendered views of the 3D map with orientations inferred for the class averages shown in c. (e) Reprojections of the 3D map corresponding to the class averages shown in c. (f) Sections through the 3D map of IP3R. Sections are 0.524 nm thick and cut normal to the fourfold axis starting from the cytoplasmic end (image 1) and running to the ER end (image 36). (Scale bars: 40 nm in a and f; 20 nm in b–e.)
IP3R images were averaged into 340 classes, examples of which are shown in Fig. 1c. A selection was made of well-preserved classes with good signal-to-noise ratios, and these were assigned Euler angles and back-projected to produce 3D maps using C4 point-group symmetry. Further selection of classes was based on agreement with reprojections of the 3D map. Reprojections were also used as references for further refinement of classes and angular assignment. Three independent analyses of the data were conducted, giving rise to similar 3D maps. In the final analysis the refinement cycle was repeated 15 times. During the final four cycles no significant changes in density distribution were observed.
The final 3D map, which was calculated from 96 classes containing a total of 1,153 images, is shown as a set of sections (0.524 nm thick and cut normal to the fourfold axis) in Fig. 1f and as surface views in Figs. 1d and 2. Reprojections from the 3D map calculated in directions inferred for the original classes are shown in Fig. 1e. The density distribution of the reprojections agrees well with the corresponding classes (Fig. 1c), demonstrating the reliability of the 3D analysis.
Orientation of Molecular Images.
A typical characteristic of macromolecules loaded onto carbon-coated electron microscope grids is the restricted number of molecular views, arising from preferred orientation of the molecules on the carbon surface. IP3R appears to be unusual in this regard, in that although the analysis was made by using negatively stained particles on carbon-coated grids, a wide distribution of molecular views was obtained. This wide range of orientations, which is advantageous for angular reconstitution, is revealed by the distribution of the three angles that define the direction of view (Euler angles) assigned to the 96 classes used in the final 3D map (see Fig. 4, which is published as supporting information on the PNAS web site), and by the diversity of molecular appearances of the raw images (Fig. 1a). The classes used for the final 3D reconstruction were well represented, with 77% of them containing 10 or more molecular images (the distribution of molecular images within these classes is shown in Fig. 4b).
Three-Dimensional Map of the IP3R.
The 3D map of the IP3R is represented as sections normal to the fourfold symmetry axis (Fig. 1f) and as surface-rendered views (Fig. 2 A and B), the latter contoured to give a molecular mass of ≈1,200 kDa for the tetrameric complex assuming a protein density of 83 Da/nm3 (27). The resolution of this map is ≈3 nm according to the Fourier shell correlation criterion (Fig. 4), where the 3σ threshold is corrected for C4 symmetry (28).
The receptor has a flower-like appearance with overall dimensions of about 18 × 18 × 18 nm (Fig. 2B). It is made up of a large domain (Fig. 1f, sections 1–20) representing the petals and stigma of the flower (gray and pink, respectively, in Fig. 2B) and a small domain (Fig. 1f, sections 21–36) representing the stalk (blue in Fig. 2B). Consideration of the amino acid sequence of IP3R suggests that the small and large domains correspond to the channel and cytoplasmic domains, respectively.
The channel domain (blue in Fig. 2B) has a square outline ≈12.6 nm wide when viewed from the lumen of the endoplasmic reticulum (ER) (Fig. 2Ba), it is ≈7 nm deep (Fig. 2Bc), and it maintains a square profile that twists and tapers toward the lumen of the ER to a width of ≈7 nm (Fig. 2Ba). Within this domain there is a central region of low density, ≈2.5 nm wide, running along the fourfold axis.
The cytoplasmic domain (gray and pink in Fig. 2B) is ≈18 nm wide as viewed from the cytoplasm (Fig. 2Bb) and ≈11 nm deep (Fig. 2Bc). It contains four petal-like subdomains (gray in Fig. 2B), located at a radius of ≈7 nm from the fourfold axis (Fig. 2Bb), which extend from the junction of the cytoplasmic and channel domains to the cytoplasmic end of IP3R (Fig. 1f, sections 1–18; Fig. 2Bc). A slender density connects each petal to the corners of the channel domain (filled arrowhead in Fig. 2Bd). The cytoplasmic domain also contains a subdomain closer to the fourfold axis representing the stigma of the flower (pink in Fig. 2B). The cytoplasmic end of this subdomain has a square profile ≈9 nm wide when viewed along the fourfold axis (Fig. 1f, sections 4–9; Fig. 2 A and Bb) and it terminates with four lobes in each corner of the square that protrude toward the cytoplasmic end of IP3R. The opposite end of the stigma subdomain is located close to the junction between the channel and the cytoplasmic domains and has an approximately circular profile ≈7 nm in diameter with four slender links (open arrowhead in Fig. 2Bd) running radially to each petal subdomain. The two ends of the stigma subdomain are connected by a region running along the fourfold axis that narrows to a waist with a diameter of ≈3.5 nm (Fig. 2 A and Bd).
Comparison with Previous Structural Data on the IP3R.
Previous analyses of IP3R by using electron microscopy have reported a variety of structures. Chadwick et al. (29) described a “pinwheel”-like structure with fourfold symmetry, 25 × 25 nm in size. Katayama et al. (30) described a native 2D lattice on the surface of the intracellular membrane of freeze-etched and platinum-shadowed Purkinje neurons, measuring ≈14 × 16 nm. Packing considerations suggest that our IP3R 3D structure may be accommodated in such an array, particularly if potential flexibility in the petal subdomain is taken into account. Particles of 12-nm width observed in these freeze-etch studies may well represent channel domains. More recently, IP3R has been studied by negative-staining electron microscopy as a function of Ca2+ concentration (31). A “windmill”-shaped structure measuring 31 nm diagonally predominated at elevated Ca2+ concentration, whereas a more compact square structure with sides measuring 19 nm predominated in the absence of Ca2+. In each case, labeling studies with gold-heparin appeared to locate the IP3-binding site to the peripheral domains (i.e., the tips of the sails of the windmill). Our 3D structure (determined in the absence of Ca2+), which measures about 18 × 18 × 18 nm and is approximately square in both top and side projections, corresponds more closely with the square structure observed by Hamada et al. (31). However, the peripheral location of the IP3-binding domain suggested by heparin-gold labeling (31) is not consistent with recent studies (32) suggesting that the IP3-binding sites within a tetrameric receptor are no more than 2 nm apart. It is possible that the large size of the heparin, gold, and linker may have exaggerated the separation of the IP3-binding sites in the labeling study (31).
During the preparation of this manuscript, another 3D map of the type 1 IP3R was reported based on cryo-electron microscopy (cryo-EM) and single-particle image analysis at a reported resolution of 2.4 nm (33). The overall size (≈18 × 18 × 17 nm) and shape of this 3D structure are broadly consistent with our results but there is little detailed agreement between the two studies. It might be expected that an analysis of cryo-EM data at a 2.4-nm resolution would produce a more detailed and accurate 3D map than would an analysis of negatively stained images at 3 nm. We argue, however, that our analysis is of higher quality and reliability because of the increased contrast in our negatively stained images, and our use of imagic (19), a well established set of programs that provide very effective image classification and model-free angular assignment. Different views of IP3R have superficially similar projection appearances (Fig. 1 c and e), which at the early stages of analysis may be confused with respect to classification and angular assignment. This potential problem is accentuated if the signal-to-noise ratio is low, as frequently occurs in cryo-EM images, and no prior knowledge of the 3D structure exists. Our IP3R structure shows a level of detail consistent with the 3-nm resolution estimated, whereas less detail is observed by Jiang et al. (33). Furthermore, our map has allowed us to assign specific functional domains, which are consistent with secondary structure predictions and biochemical studies, and show similarities to related cation channels (see below).
Identifying Domains in the Structure of the IP3R.
The 3D map of IP3R is made up of the channel domain and petal and stigma subdomains as described above. Within each monomeric subunit, these domains and subdomains are linked together consecutively and in this order by single, slender connections. Based on this observation, we suggest that the different domains may be linearly mapped onto the amino acid sequence. Individual domains may then be assigned to regions of the sequence by estimating their relative volumes in the 3D map.
Channel and Cytoplasmic Domains.
According to our 3D map, the channel domain of IP3R represents ≈35% of the total volume of the protein. This volume corresponds to an estimated molecular mass of ≈110 kDa (35% of 313 kDa, the subunit molecular mass) and to ≈960 amino acid residues. The channel domain should contain the region where six transmembrane helices and their interconnecting loops have been identified, located close to the C terminus between residues 2275 and 2590. It thus seems likely that this domain corresponds approximately to the 960 amino acid residues at the C terminus (residues 1790–2750). In this light, it is interesting to note that in previous studies of type 1 IP3R (8, 11, 34), limited trypsin digestion released a C-terminal fragment with a molecular mass (91–94 kDa; residues 1932–2749) similar to the size of the channel domain suggested by our structural analysis (≈110 kDa). This assignment is further supported by secondary structure analysis of the type 1 IP3R sequence, which clearly reveals a glycine-rich low-complexity region immediately upstream of residue 1790, between residues 1750 and 1788. Corresponding low-complexity regions are also present in types 2 and 3 IP3R. In multidomain proteins, adjacent domains are often found to be linked by low-complexity regions (35), suggesting that this region is indeed likely to separate the channel and cytoplasmic domains of IP3R. The channel domain, therefore, appears to be composed of the transmembrane region closely integrated with two cytoplasmic regions corresponding to residues 1790–2274 and the C-terminal tail (residues 2591–2750).
Cytoplasmic Subdomains.
The cytoplasmic domain of IP3R is further divided into the petal and stigma subdomains that make up ≈45% and 20% of the total volume of IP3R, corresponding approximately to 1,240 (141 kDa) and 550 (63 kDa) amino acid residues, respectively. Results from both limited proteolysis and measurements of the location and properties of the IP3-binding sites can be used as guides to assign these domains. The tetrameric IP3R has a single IP3-binding site on each subunit, located between residues 226 and 576 (9), and recent studies with dimeric molecules of IP3 suggest that the sites are likely to be separated by no more than ≈2 nm (32). On this basis, the petal subdomains in our 3D map are unlikely to contain the IP3-binding sites, because equivalent sites within these subdomains are at least 8 nm apart (Fig. 2Bb). On the other hand, the stigma subdomain lies close to the fourfold axis and could readily accommodate IP3 molecules bound in equivalent positions and spaced 2 nm or less apart. According to secondary structure prediction, there is a low-complexity region in the sequence of IP3R downstream of the IP3-binding pocket, which is between residues 670 and 695. Thus, it seems reasonable to propose that the stigma subdomain corresponds approximately to residues 1–670, which includes the IP3-binding pocket. These 670 residues account for 24% of the 2,750 amino acid residues of IP3R, which is reasonably close to the value of 20% estimated for the relative volume of the stigma subdomain in our 3D map. Accordingly, the low-complexity region between residues 670 and 695 would represent the link between stigma and petal subdomains. These results are broadly consistent with those obtained from limited proteolytic cleavage of type 1 receptors, which released a soluble protein that bound IP3 and had a mass (68 kDa) (34) similar to that (≈63 kDa) predicted for the stigma subdomain.
The remaining part of the cytoplasmic region of IP3R, lying approximately between residues 695 and 1750, should correspond to the petal domain. Again, results from trypsin digestion lend support to this conclusion, because mild digestion of type 1 IP3R released a fragment (116 kDa) that includes residues 943-1731 (8). This region of IP3R is part of what has been referred to as the “modulatory domain,” which contains binding sites for Ca2+, calmodulin and FKBP12, and the FK506-binding protein, as well as phosphorylation sites (36). In type 3 IP3R, this region also includes sites thought to interact with Ca2+ entry-channel proteins (13). The peripheral location of the petal domains appears well suited to these roles. From these considerations, we propose that the structural domains of the IP3R are mapped onto the primary sequence in a linear manner, as illustrated in Fig. 2Ca.
Location of N and C Termini.
The 3D arrangement described above places the N and C termini of IP3R in separate domains (Fig. 2Ca), yet crosslinking and immunoprecipitation studies suggest that they are close together (11). It is possible that an interaction between N and C termini may occur at the interface between the stigma and channel domains (Fig. 2Cb). In the 3D map of IP3R, these domains appear to be ≈1.5 nm apart, although it is possible that regions of closer approach were not identified at the current resolution. Nevertheless, a spacing of ≈1.5 nm is compatible with the length of the crosslinking agent (1.2 nm) used to identify the interaction between the N and C termini (11). It is also noteworthy that a very small fragment of the extreme C terminus of the type 1 receptor appears to be uniquely sensitive to trypsin cleavage (8), suggesting that it may be exposed and, perhaps, thereby free to interact with the stigma domain.
Comparison with Other Tetrameric Cation Channels.
The 3D structure of IP3R can be compared with the structures of other tetrameric cation channels. The ryanodine receptor (RyR), like IP3R, is an intracellular Ca2+ channel. These two channels are partly homologous in amino acid sequence, particularly in the transmembrane pore-forming region, although the RyR is made up of ≈5,000 residues per subunit, and thus has nearly twice the mass of the IP3R. Three-dimensional structures at a 2- to 3-nm resolution have been determined for types I, II ,and III RyR by cryo-EM and single-particle image analysis (reviewed in refs. 37 and 38). At this resolution, the different isoforms of RyR are substantially similar. The 3D structure of type I RyR (39) is illustrated as a cartoon in Fig. 2Da for comparison with the IP3R (Fig. 2Db). The channel domain of RyR is ≈6.5 nm deep and has a square profile viewed along the fourfold axis with sides measuring ≈12.5 nm and it tapers toward the luminal end. These features correspond quite closely to the channel domain of IP3R. The tapered ends of the channel domains of both the IP3R (Fig. 2Db) and RyR (Fig. 2Da) are twisted ≈7° clockwise with respect to their wider bases. We use this feature to select the 3D map of IP3R presented here over its mirror-related alternative, which is not excluded by the current analysis. Aside from comparisons with RyR, other interpretations of our 3D map are not affected by this selection. The similar sizes and shapes of the channel domains of RyR and IP3R suggest that they share a similar organization. It seems possible, therefore, that the RyR channel contains a substantial cytoplasmic component closely integrated with the transmembrane region similar to the arrangement we propose for the channel domain of IP3R. On the other hand, the cytoplasmic domain of the RyR is considerably larger than its counterpart in the IP3R and quite different in appearance, presumably reflecting the larger size of RyR and differences in organization associated with regulation and ligand binding.
Shaker is a voltage-gated K+ channel which, like IP3R, is believed to have six transmembrane helices and a P loop. Its 3D structure was recently calculated by single-particle analysis of negatively stained electron micrographs at a resolution of ≈2.5 nm (40) and is illustrated as a cartoon in Fig. 2Dc. The channel domain of Shaker has a square profile similar to its counterpart in IP3R when viewed along the fourfold axis, although it is somewhat narrower; 10 nm compared with 12.6 nm for IP3R. The Shaker channel domain is joined to its cytoplasmic domain by four thin connectors reminiscent of the slender connections between the IP3R channel and cytoplasmic domains. Slender connections between channel and cytoplasmic domains have also been identified in the x-ray crystallographic structure of MthK, a prokaryotic Ca2+-gated K+ channel (ref. 41; Fig. 2Dd) in which the channel-domain subunits are made up of two transmembrane helices and a P loop. These connections correspond to low-complexity regions in the MthK amino acid sequence analogous to the low-complexity regions that we suggest form the slender connections in the IP3R. Slender linkages between channel and cytoplasmic domains appear to be a common theme in these different cation channels. They may allow substantial conformational changes associated with regulation or ligand binding to occur in the cytoplasmic domains alongside more subtle rearrangements in the conformation of the channel domains related to channel opening as has been proposed for K+ channels (42).
Our model for the domain organization of the IP3R is consistent with our 3D structure, secondary-structure predictions, and biochemical measurements (Fig. 2C). It predicts the 3D location of key functional sites and reveals interesting parallels with other tetrameric cation channels.
Supplementary Material
Acknowledgments
We thank Elena Orlova for support and advice on image processing; Marin van Heel for use of electron microscopy facilities; Claudia Büchel for help with HPLC; and John Squire for encouragement and advice. This work was supported by grants from The Wellcome Trust.
Abbreviations
- ER
endoplasmic reticulum
- IP3
inositol 1,4,5-trisphosphate
- IP3R
IP3 receptor
- RyR
ryanodine receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mak D-O D, McBride S, Foskett J K. Proc Natl Acad Sci USA. 1998;95:15821–15825. doi: 10.1073/pnas.95.26.15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins C E, Taylor C W. Curr Biol. 1999;9:1115–1118. doi: 10.1016/s0960-9822(99)80481-3. [DOI] [PubMed] [Google Scholar]
- 3.Berridge M J, Lipp P, Bootman M D. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Tang J, Tikunova S, Johnson J D, Chen Z, Qin N, Dietrich A, Stefani E, Birnbaumer L, Zhu M X. Proc Natl Acad Sci USA. 2001;98:3168–3173. doi: 10.1073/pnas.051632698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor C W, Genazzani A A, Morris S A. Cell Calcium. 1999;26:237–251. doi: 10.1054/ceca.1999.0090. [DOI] [PubMed] [Google Scholar]
- 6.Michikawa T, Hamanaka H, Otsu H, Yamamoto A, Miyawaki A, Furuichi T, Tashiro Y, Mikoshiba K. J Biol Chem. 1994;269:9184–9189. [PubMed] [Google Scholar]
- 7.Ramos-Franco J, Galvan D, Mignery G A, Fill M. J Gen Physiol. 1999;114:243–250. doi: 10.1085/jgp.114.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshikawa F, Iwasaki H, Michikawa T, Furuichi T, Mikoshiba K. J Biol Chem. 1999;274:328–334. doi: 10.1074/jbc.274.1.328. [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa F, Morita M, Monkawa T, Michikawa T, Furuichi T, Mikoshiba K. J Biol Chem. 1996;271:18277–18284. doi: 10.1074/jbc.271.30.18277. [DOI] [PubMed] [Google Scholar]
- 10.Sienaert I, Kasri N N, Vanlingen S, Parys J, Callewaert G, Missiaen L, De Smedt H. Biochem J. 2002;365:269–277. doi: 10.1042/BJ20020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehning D, Joseph S K. EMBO J. 2000;19:5450–5459. doi: 10.1093/emboj/19.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakade S, Maeda N, Mikoshiba K. Biochem J. 1991;277:125–131. doi: 10.1042/bj2770125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulay G, Brown D M, Qin N, Jiang M, Dietrich A, Zhu M X, Chen Z, Birnbaumer M, Mikoshiba K, Birnbaumer L. Proc Natl Acad Sci USA. 1999;96:14955–14960. doi: 10.1073/pnas.96.26.14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joseph S K, Boehning D, Pierson S, Nicchitta C V. J Biol Chem. 1997;272:1579–1588. doi: 10.1074/jbc.272.3.1579. [DOI] [PubMed] [Google Scholar]
- 15.Mignery G A, Südhof T C. EMBO J. 1990;9:3893–3898. doi: 10.1002/j.1460-2075.1990.tb07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchant J S, Taylor C W. Curr Biol. 1997;7:510–518. doi: 10.1016/s0960-9822(06)00222-3. [DOI] [PubMed] [Google Scholar]
- 17.Richardson A, Taylor C W. J Biol Chem. 1993;268:11528–11533. [PubMed] [Google Scholar]
- 18.Föhr K J, Scott J, Ahnert-Hilger G, Gratzl M. Biochem J. 1989;262:83–89. doi: 10.1042/bj2620083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Heel M, Harauz G, Orlova E V, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Shaw A, Bates P A, Newman R H, Gowen B, Orlova E, Gorman M A, Kondo H, Dokurno P, Lally J, et al. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 21.Sheehan B, Fuller S D, Pique M E, Yeager N. J Struct Biol. 1996;116:99–106. doi: 10.1006/jsbi.1996.0017. [DOI] [PubMed] [Google Scholar]
- 22.Kelley L A, MacCallum R M, Sternberg M J E. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 23.Wootton J C, Federhen S. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 24.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 25.Nerou E P, Riley A M, Potter B V L, Taylor C W. Biochem J. 2001;355:59–69. doi: 10.1042/0264-6021:3550059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dube P, Tavares P, Lurz R, van Heel M. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harpaz Y, Gerstein M, Chothia C. Structure (London) 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 28.Orlova E V, Dube P, Harris J R, Beckman E, Zemlin F, Markl J, van Heel M. J Mol Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- 29.Chadwick C C, Saito A, Fleischer S. Proc Natl Acad Sci USA. 1990;87:2132–2136. doi: 10.1073/pnas.87.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama E, Funahashi H, Michikawa T, Shiraishi T, Ikemoto T, Iino M, Hirosawa K, Mikoshiba K. EMBO J. 1996;15:4844–4851. [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada K, Miyata T, Mayanagi K, Hirota J, Mikoshiba K. J Biol Chem. 2002;277:21115–21118. doi: 10.1074/jbc.C200244200. [DOI] [PubMed] [Google Scholar]
- 32.Riley A M, Morris S A, Nerou E P, Correa V, Potter B V L, Taylor C W. J Biol Chem. 2002;277:40290–40295. doi: 10.1074/jbc.M206925200. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Q X, Thrower E C, Chester D W, Ehrlich B E, Sigworth F J. EMBO J. 2002;21:3575–3581. doi: 10.1093/emboj/cdf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph S K, Pierson S, Samanta S. Biochem J. 1995;307:859–865. doi: 10.1042/bj3070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnhammer E L L, Wootton J C. Proteins. 2001;45:262–273. doi: 10.1002/prot.1146. [DOI] [PubMed] [Google Scholar]
- 36.Patel S, Joseph S K, Thomas A P. Cell Calcium. 1999;25:247–264. doi: 10.1054/ceca.1999.0021. [DOI] [PubMed] [Google Scholar]
- 37.Samso M, Wagenknecht T. J Struct Biol. 1998;121:172–180. doi: 10.1006/jsbi.1997.3955. [DOI] [PubMed] [Google Scholar]
- 38.Orlova E V, Serysheva I I, Hamilton S L, Chiu W, van Heel M. In: The Structure and Function of Ryanodine Receptors. Sitsapesan R, Williams A J, editors. London: Imperial College Press; 1998. pp. 23–46. [Google Scholar]
- 39.Orlova E V, Serysheva I I, van Heel M, Hamilton S L, Chiu W. Nat Struct Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 40.Sokolova O, Kolmakova-Partensky L, Grigorieff N. Structure (London) 2001;9:215–220. doi: 10.1016/s0969-2126(01)00578-0. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y X, Lee A, Chen J Y, Cadene M, Chait B T, MacKinnon R. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 42.Jiang Y X, Lee A, Chen J Y, Cadene M, Chait B T, MacKinnon R. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.