Abstract
Two substrates of insulin-degrading enzyme (IDE), amyloid β-protein (Aβ) and insulin, are critically important in the pathogenesis of Alzheimer's disease (AD) and type 2 diabetes mellitus (DM2), respectively. We previously identified IDE as a principal regulator of Aβ levels in neuronal and microglial cells. A small chromosomal region containing a mutant IDE allele has been associated with hyperinsulinemia and glucose intolerance in a rat model of DM2. Human genetic studies have implicated the IDE region of chromosome 10 in both AD and DM2. To establish whether IDE hypofunction decreases Aβ and insulin degradation in vivo and chronically increases their levels, we characterized mice with homozygous deletions of the IDE gene (IDE −/−). IDE deficiency resulted in a >50% decrease in Aβ degradation in both brain membrane fractions and primary neuronal cultures and a similar deficit in insulin degradation in liver. The IDE −/− mice showed increased cerebral accumulation of endogenous Aβ, a hallmark of AD, and had hyperinsulinemia and glucose intolerance, hallmarks of DM2. Moreover, the mice had elevated levels of the intracellular signaling domain of the β-amyloid precursor protein, which was recently found to be degraded by IDE in vitro. Together with emerging genetic evidence, our in vivo findings suggest that IDE hypofunction may underlie or contribute to some forms of AD and DM2 and provide a mechanism for the recently recognized association among hyperinsulinemia, diabetes, and AD.
Insulin-degrading enzyme (IDE, insulysin) is an ≈110-kDa thiol zinc-metalloendopeptidase located in cytosol, peroxisomes, endosomes, and on the cell surface (1–4) that cleaves small proteins of diverse sequence, many of which share a propensity to form β-pleated sheet-rich amyloid fibrils under certain conditions [e.g., amyloid β-protein (Aβ), insulin, glucagon, amylin, atrial natriuretic factor, and calcitonin] (5, 6). IDE is the major enzyme responsible for insulin degradation in vitro (1), but the extent to which it mediates insulin catabolism in vivo has been controversial, with doubts expressed that IDE has any physiological role in insulin catabolism (7). Insulin, which is critical for glucose, lipid, and protein metabolism, as well as for cell growth and differentiation, is cleared mainly by the liver and kidney, but most other tissues also degrade the hormone. It was recently shown that transferring an ≈3.7-cM chromosomal region containing the IDE gene from an inbred rat model of type 2 diabetes mellitus (DM2) (the GK rat) to a normoglycemic rat recapitulated several features of the diabetic phenotype, including hyperinsulinemia and postprandial hyperglycemia (8). The GK allele of IDE in this chromosomal region was found to bear two missense mutations that, when transfected into COS-1 cells, resulted in 31% less insulin degradation compared with cells transfected with the WT allele. Furthermore, the IDE region of chromosome 10q has been genetically linked to DM2 (9, 10) and to elevated fasting glucose levels [20-year means (11)].
In addition to its putative role in insulin catabolism, IDE has been found to degrade Aβ in neuronal and microglial cell cultures (4, 12–14), and to eliminate Aβ's neurotoxic effects (15). Although cerebral accumulation of Aβ is believed to play a central role in Alzheimer's disease (AD) pathogenesis, in the vast majority of cases the underlying causes for this elevation are unknown. Several lines of evidence demonstrate that newly generated Aβ is rapidly cleared from the brain (16, 17), suggesting that Aβ-degrading proteases could play a role in regulating cerebral levels of the peptide. Two proteases, neprilysin (NEP) and endothelin-converting enzyme, have recently been shown to degrade Aβ in vivo (18, 19), validating the role of proteolysis in regulating endogenous Aβ levels. In addition to its genetic linkage to DM2, the IDE region of chromosome 10q has also been linked to late-onset (conventional) AD (20) and to age of disease onset in AD families (21). Allelic association in or near the IDE gene has been reported in some AD populations (20, 22) but not others (23, 24). In two other reports, the occurrence of AD (25) and high plasma Aβ42 levels (26) were linked to a region of chromosome 10q that is ≈40 Mb proximal to the linkage peaks over the IDE region. It remains unclear whether these linkage peaks represent the same underlying locus or two separate loci.
It has recently been demonstrated in cell cytosol fractions that IDE also degrades the β-amyloid precursor protein (APP) intracellular domain (AICD) (27), which can otherwise reach the nucleus (28, 29) and participate in transcriptional regulation (30, 31). To establish whether hypofunction of IDE impairs Aβ, insulin, and AICD degradation in vivo, causing accumulation of these peptides and recapitulation of features of AD and DM2, we characterized mice in which IDE was genetically disrupted.
Materials and Methods
IDE −/− Mice.
IDE +/− founder mice were created at Lexicon Genetics (The Woodlands, TX) by using a gene-trapping method (32). We identified the integration site of the targeting cassette in intron 1 and developed a PCR-based assay that distinguishes the three possible mouse genotypes (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). IDE WT and −/− (homozygous deletion) mice were derived by breeding the IDE +/− founder mice, which are in a hybrid genetic background (50% C57BL/6 albino and 50% 129SvEvBrd strains). The −/− and WT animals were then bred independently of one another and fed a normal chow diet ad libitum (unless specified otherwise).
Preparation and Immunoblotting of Brain Fractions.
Fresh-frozen mouse brain was homogenized in 8 vol (wt/vol) of 250 mM sucrose in 50 mM Tris⋅HCl (pH 7.4) in a Potter-Elvehjem homogenizer. After pelleting nuclei and unbroken cells, the supernatant was spun again at 100,000 × g for 1 h. The resulting supernatant was saved as the soluble fraction, and the membrane pellet was resuspended in 100 mM Na2CO3 (pH 11.3) to open microsomes and remove proteins loosely associated with the membranes (33). The membranes were pelleted again, resuspended in 50 mM Tris⋅HCl (pH 7.4), and sonicated. Protein concentrations were determined by using a bicinchoninic acid-based protein assay (Pierce). For immunoblotting, membranes were probed with antibodies raised to IDE [IDE-1 (4)]; NEP (56C6, Novocastra, Newcastle, U.K.); APP and its proteolytic derivatives C83, C99 [antibody C8 (34)], and sAPP (22C11, Chemicon); presenilin N-terminal (Ab14; a gift from S. Gandy, Thomas Jefferson University, Philadelphia) and C-terminal [4627 (35)] fragments; BACE (Ab-2, Oncogene Science); and ADAM10 (AB19026, Chemicon).
Primary Neuronal Cultures.
Cerebral cortices were collected from embryonic day 17 litters, incubated in 0.025% trypsin in dissection buffer [Hanks' balanced salt solution (HBSS)/10 mM Hepes/10 mM glucose/10 mM sucrose], mechanically dissociated, and plated at 40,000 cells per cm2 onto poly(l)-lysine coated dishes. Cells were maintained in a neuronal-selecting medium [Neurobasal medium with the growth supplement B27 (Invitrogen)].
Trichloroacetic Acid (TCA) Precipitation Degradation Assays.
To measure proteolysis of Aβ by brain fractions, 100 pM synthetic human 125I-Aβ1-40 (Amersham Pharmacia) was incubated at 37°C with 100 μg/ml protein for various intervals. To measure proteolysis of insulin by liver fractions, 100 pM recombinant human 125I-insulin (Amersham Pharmacia) was incubated at 37°C with 50 μg/ml protein. Neuronal cultures, after being washed twice with medium (Neurobasal) containing 0.1% BSA, were incubated in the same medium with 40 pM of either 125I-insulin or 125I-Aβ1-40. At each degradation assay time point, an aliquot of the sample was added to an equal volume of 15% TCA to allow the uncleaved peptides to precipitate. Following centrifugation, cpm of γ radiation in the TCA-insoluble pellet (undegraded peptide) and TCA supernatant (degraded peptide fragments) were determined. For each sample, the cpm in the supernatant at time 0 (background) was subtracted from the cpm in the supernatants at the other time points to obtain the “adjusted supernatant cpm.” The percent degradation for each time point was determined by dividing the adjusted supernatant cpm by the “adjusted total cpm” (sum of the cpm in the pellet and the adjusted supernatant cpm).
Quantification of Endogenous Aβ.
After primary neurons conditioned Neurobasal media with B27 supplement, the media were removed, 1,10-phenanthroline and a protease inhibitor mixture (Sigma) were added, and the samples were centrifuged to remove any cellular material. Aβ X-40 and Aβ X-42 ELISAs were performed by using the well characterized BNT-77/BA-27 (Aβ X-40) and BNT-77/BC-05 (Aβ X-42) systems (36). To measure brain Aβ, each cerebral hemisphere was homogenized in 9 vol (wt/vol) of 0.2% diethylamine in 50 mM NaCl (16, 37) by using a Potter-Elvehjem homogenizer. After centrifugation at 100,000 × g for 1 h, the supernatant was neutralized with 1/10 vol of 500 mM Tris⋅HCl (pH 6.8), then Aβ ELISAs were performed.
Detection and Dephosphorylation of AICD.
Fresh mouse brains were homogenized in 9 vol (wt/vol) of buffer (50 mM Tris, pH 7.4/150 mM NaCl/1% SDS/5 mM EDTA/2 mM 1,10-phenanthroline/Sigma protease inhibitor mixture), boiled, sonicated, and then boiled again. The post-16,000 × g supernatant was diluted 10-fold with 50 mM Tris⋅HCl, and AICD was immunoprecipitated with antibody C8 attached to protein A Sepharose beads (Sigma). After washes, SDS sample buffer was added and the samples were boiled, run on 10–20% tricine gels, and transferred to nitrocellulose membranes. Membranes were boiled in PBS and blotted with antibodies raised against the APP C terminus [C8 or anti-0443 (Calbiochem)]. For dephosphorylation, samples were prepared as above until after the final wash following immunoprecipitation, when they were incubated at 37°C for 1 h with either calf intestinal phosphatase in Buffer 3 (New England Biolabs) or Buffer 3 alone.
Quantification of Endogenous Insulin, Glucagon, and Glucose.
Serum insulin and plasma glucagon levels were determined by using sensitive rodent radioimmunoassays kits (Linco Research, St. Charles, MO). Blood glucose levels were measured with Glucometer Elite (Bayer, Elkhart, IN). For the glucose tolerance test, 2 g/kg d-glucose was injected i.p. and blood glucose levels were measured at the indicated time points.
Results
IDE −/− mice are viable and fertile. The absence of the characteristic 110-kDa IDE protein in brain fractions (see Fig. 4, which is published as supporting information on the PNAS web site) and liver fractions was confirmed by immunoblotting. Immunoblotting for NEP in IDE −/− and WT brain membranes revealed equal amounts of this protease. There was no significant difference in mean body weights between IDE −/− and age-matched WT animals ranging from 8 to 42 weeks of age. Routine histological examination of all major tissues of IDE −/− mice at age 20 weeks was normal.
Decreased Aβ Catabolism and in Vivo Accumulation of Aβ in IDE −/− Mice.
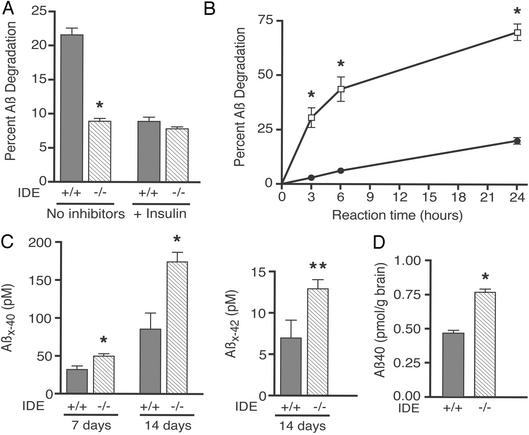
To characterize the role of IDE in Aβ metabolism in brain, the principal site of Aβ accumulation, we compared degradation of 100 pM 125I-Aβ40 in both soluble and Na2CO3-washed membrane fractions of brain from WT and IDE −/− mice. TCA precipitation assays (see Materials and Methods) were performed at reaction times 0, 3, and 8 h for membranes and at 0, 30, and 60 min for soluble fractions. IDE −/− brain membranes showed highly significant ≈59% and ≈47% deficits in Aβ degradation at 3 and 8 h, respectively, compared with identically prepared membranes from WT mice (P < 0.00001 for each time point; Fig. 1A, 3-h time point shown). Brain membrane fractions are particularly relevant, because a membrane-associated form of IDE occurs in intracellular vesicles‖ and on the neuronal cell surface (4), where Aβ is produced (38). The brain soluble fraction of the IDE −/− mice had an even greater Aβ-degradation deficit, with ≈80% less degradation at 30 min and ≈72% less at 60 min compared with WT (P < 0.00001 for each time point; data not shown). In both the membrane and soluble fractions, insulin (10 μM), a potent competitive inhibitor of IDE, had little effect on Aβ degradation in the IDE −/− mice but inhibited the IDE WT activity to a level of residual Aβ proteolysis similar to that of the IDE −/− mice (Fig. 1A and data not shown). The latter result supports the conclusion that the difference in Aβ-degradative capacity between the two mouse genotypes is due to the deficiency of IDE. Degradation assays were also performed in brain fractions by using rodent Aβ (differs from the human peptide at three amino acids) that was iodinated at His-6 or -14, as opposed to the human Aβ labeled at Tyr-10 that was used above. The IDE −/− Aβ-degrading deficits in both brain membrane and soluble fractions were the same for the rodent 125I-Aβ as for the human peptide. This result indicates that the IDE −/− degradation deficit extends to rodent Aβ, and that the site of iodination does not affect the results of our Aβ degradation assays.
Figure 1.
Aβ degradation and accumulation in IDE gene-disrupted mice. (A) Aβ degradation in brain membrane fractions. 125I-Aβ1-40 proteolysis in Na2CO3-washed brain membranes from two IDE −/− and two WT (+/+) mice was measured by TCA precipitation assay in the presence or absence of unlabeled insulin (3-h time point shown). Bars represent means ± SEM of 4–10 determinations from two to three independent assays (*, P < 0.00001 by Student's t test for IDE −/− compared with the IDE +/+ without inhibitors). (B) Aβ degradation in primary neurons. Age-matched cortical neurons (7–22 days in vitro) from two IDE −/− (filled circles) and two +/+ (open squares) embryonic day 17 litters were incubated with 40 pM 125I-Aβ1-40, and TCA precipitation assays were performed at the indicated time points. Graph points represent the mean ± SEM of 9–11 determinations from five to six independent assays (*, P < 0.00001). Some error bars are obscured by the symbols for data points. (C) Endogenous Aβ levels in IDE −/− neuronal conditioned media. Cortical neurons from two IDE −/− and two +/+ embryonic day 17 litters were allowed to condition media for 7 (4–11 days in vitro) or 14 days (4–18 days in vitro) before being analyzed by Aβ X-40 and Aβ X-42 ELISA. X-40 and X-42 bars represent mean Aβ concentrations ± SEM of six to eight and four to six determinations, respectively, and each determination is a mean of duplicate ELISA values (*, P < 0.005; **, P = 0.01). (D) Endogenous Aβ levels in IDE −/− brains. Represented are the mean pmol of Aβ per g of brain ± SEM of six WT and seven −/− brains from 12-week-old animals determined by Aβ X-40 ELISA (*, P < 0.00001).
To extend our finding of the IDE-mediated Aβ-degradation deficit to intact, living neurons, the principal producers of Aβ, we added 125I-Aβ40 (40 pM) to primary cortical neuronal cultures and performed TCA degradation assays. IDE −/− neurons had ≈70–90% less degradation of this extracellular Aβ than did WT neurons (P < 0.00001 for each time point; Fig. 1B). Next, quantification of endogenously produced neuronal Aβ by ELISA revealed significant elevations of the peptide in the IDE −/− cultures. Compared with WT neuronal media, steady-state Aβ X-40 levels in IDE −/− media were increased by 56% (P < 0.005) and 104% (P = 0.001) after 7 and 14 days of conditioning, respectively (Fig. 1C). Aβ X-42 levels were undetectable at 7 days, but at 14 days were 86% higher in the IDE −/− neuronal media than the WT media (P = 0.01; Fig. 1C).
To determine whether the IDE-mediated Aβ-degradation deficit results in cerebral accumulation of naturally produced, endogenous Aβ, as occurs in AD, we quantified levels of the peptide in IDE −/− and +/+ brains by ELISA. In a preliminary experiment, we compared Aβ X-40 levels in a small group of 12-week-old IDE −/− and +/+ mice (n = 3 for each genotype). The IDE −/− animals had a significant 19% increase in brain Aβ X-40 levels over WT (1.71 ± 0.08 vs. 1.43 ± 0.06 pmol/g brain; P < 0.05). Next, we quantified cerebral Aβ X-40 levels in a larger group of 12-week-old IDE −/− and WT mice (n = 7 −/− and 6 WT) and found a 64% elevation of Aβ X-40 in the IDE −/− animals (0.767 ± 0.022 vs. 0.469 ± 0.018 pmol/g; P < 0.00001; Fig. 1D). Cerebral Aβ X-42 levels were also examined in the 12-week-old animals, but in the WT brains most of the values were below the reliable range of the ELISA. As a positive control, we quantified Aβ X-40 in 12-week-old NEP −/− mouse brains (39) and their WT controls (n = 3 −/− and 2 WT mice), and found an elevation of Aβ X-40 in the NEP −/− animals (64%; 2.13 ± 0.34 vs. 1.30 ± 0.13 pmol/g; P ≤ 0.05) that was identical to that in the larger set of IDE −/− animals of the same age. The magnitude of this increase is similar to that previously reported in NEP −/− mouse cerebral cortex (18). Comparison of endogenous cerebral Aβ levels in 6-month-old IDE −/− and WT mice (n = 7 −/− and 10 WT) revealed significant elevations in both Aβ X-40 (13%; 2.24 ± 0.05 vs. 1.98 ± 0.02 pmol/g; P < 0.0001) and Aβ X-42 (7%; 0.70 ± 0.01 vs. 0.65 ± 0.02 pmol/g; P < 0.05) (data not shown).
AICD Accumulation in IDE −/− Mice.
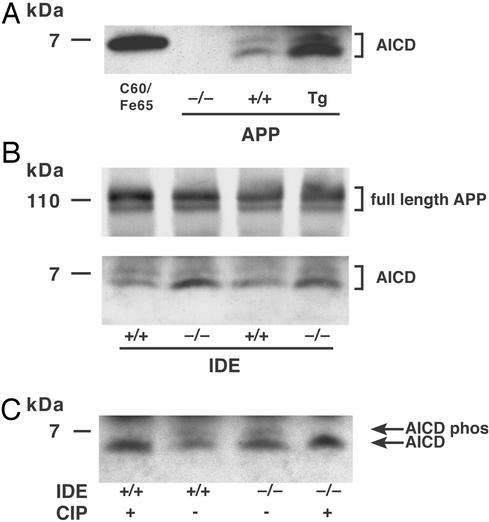
We next determined whether IDE mediates AICD catabolism in vivo. Endogenous brain AICD levels were quantified in six IDE −/− and six +/+ mice by immunoprecipitation with antibody C8 (raised against the APP C terminus), followed by immunoblotting with either C8 or another APP C-terminal antibody, anti-0443. Two specifically immunoreactive AICD proteins, a major band of ≈5 kDa and a minor band of ≈7 kDa, were routinely detected in freshly prepared brain homogenates with either antibody. Both these proteins were absent in the brains of mice lacking APP expression (40) and increased in the brains of transgenic mice overexpressing human APP (41) (Fig. 2A). There was a clear and consistent increase in the lower AICD band in the IDE-deficient compared with WT brains, but no increase in the upper band (Fig. 2B). In contrast, levels of APP (Fig. 2B) and its other proteolytic derivatives (C99, C89, C83, and sAPP) were all indistinguishable by immunoblotting in the two genotypes, as were the levels of presenilin NTF and CTF, BACE, and ADAM10. Dephosphorylation of the homogenates with calf intestinal phosphatase eliminated the upper band in both mouse genotypes and increased the density of the lower band without changing its migration, indicating that the former is a phosphorylated form and the latter a nonphosphorylated form of AICD (Fig. 2C). Thus, IDE appears to selectively regulate the levels of unphosphorylated AICD, but not its minor phosphorylated form, in vivo.
Figure 2.
Cerebral levels of APP and AICD in IDE-deficient mice. Shown are immunoprecipitation immunoblots of brain lysate with antibody C8. Identical results were obtained with C8 immunoprecipitation followed by anti-0443 immunoblotting. (A) AICD in APP gene-disrupted (−/−), WT (+/+), and APP transgenic (Tg) mouse brains. Lane C60/Fe65, the positive control, is lysate from transfected COS cells coexpressing a 59-aa AICD construct, which runs slightly higher than the endogenous 50-aa AICD, and Fe65, which stabilizes AICD (28). (B) Full-length APP and AICD in IDE −/− vs. +/+ brain. The APP and AICD bands are from identical lanes of the same blot, but the APP bands are from a lighter exposure. (C) Dephosphorylation of AICD. After the final wash following immunoprecipitation, samples were incubated for 1 h with either calf intestinal phosphatase (CIP) or buffer alone.
Decreased Insulin Catabolism and Glucose Intolerance in IDE −/− Mice.
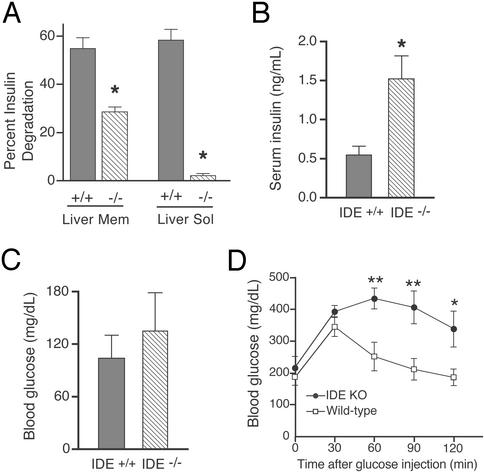
The majority of insulin degradation occurs in endosomes and at the plasma membrane, but there is some evidence that it can also occur in the cytosol (1). Therefore, we performed 125I-insulin TCA degradation assays on both Na2CO3-washed membrane fractions and cytosolic fractions of liver, the primary site of insulin clearance (1). IDE −/− liver membranes showed highly significant 58% and 48% decreases in insulin degradation compared with WT at 1 and 3 h, respectively (P < 0.00001 at each time point; Fig. 3A). The liver cytosol had >96% less insulin degradation at both 15 and 30 min in the IDE −/− vs. WT mice (P < 0.00001 at each time point; Fig. 3A). Next, we assayed insulin degradation in brain membrane and soluble fractions and found an ≈72% (P < 0.0001) and ≈98% (P < 0.0001) decrease in insulin proteolysis, respectively, in the IDE −/− mice (data not shown). To determine whether the insulin degradation deficit chronically elevated endogenous insulin levels, we quantified fasting levels in IDE −/− and WT sera. A marked ≈2.8-fold increase in insulin levels (P < 0.01) was detected in the IDE-deficient mice aged 17–20 weeks (Fig. 3B). We then analyzed blood glucose levels to assess the physiological effect of this chronic hyperinsulinemia. Although fasting glucose levels did not differ significantly between IDE −/− and WT mice (Fig. 3C), an i.p. glucose tolerance test revealed significant hyperglycemia at 60 (P < 0.01), 90 (P < 0.01), and 120 (P < 0.05) min after glucose injection in the IDE −/− mice (Fig. 3D). Endogenous levels of plasma glucagon, another reported substrate of IDE in vitro (42), were not different between IDE −/− (114.6 pg/ml, n = 7) and WT animals (120.2 pg/ml, n = 8) aged 28–34 weeks. Thus, IDE −/− mice have both hyperinsulinemia and glucose intolerance, two classic features of DM2.
Figure 3.
Insulin degradation, hyperinsulinemia, and glucose intolerance in IDE-deficient mice. (A) Insulin degradation in liver fractions. TCA degradation assays were performed on liver Na2CO3-washed membranes (3 h shown) and soluble fractions (30 min shown) from three IDE −/− and three WT mice. Bars represent means ± SEM of 8–12 determinations from two independent assays (*, P < 0.00001). Serum insulin (B) and blood glucose (C) levels after an overnight fast. Bars represent the means ± SEM of measurements from eight 17- to 20-week-old mice of each genotype (*, P < 0.05). (D) Glucose tolerance test. Graph points represent the mean blood glucose ± SEM of six IDE −/− (filled circles) and four WT (open squares) 28- to 30-week-old male mice after i.p. injection of d-glucose at time 0 (*, P < 0.05; **, P < 0.01).
Discussion
Taken together, our findings demonstrate that two proteins, insulin and Aβ, which each play a central role in a common and devastating human disease, are regulated in vivo by IDE. In addition to defining a key role for IDE in both Aβ and insulin metabolism in vivo, selective deletion of the IDE gene recapitulates some of the hallmark phenotypic characteristics of AD and DM2, namely chronic elevation of cerebral Aβ, as in AD, and hyperinsulinemia and glucose intolerance, as in DM2. Although the elevation of cerebral Aβ in the IDE −/− animals (≈10–65%) validates a role for IDE in Aβ proteolysis in vivo, there are probably additional mechanisms of Aβ clearance in the intact brain, especially for Aβ42. Other proteases (e.g., NEP, endothelin-converting enzyme), microglia, and/or transport into the circulation may each play a significant role in Aβ clearance and partially compensate for the lack of IDE function. Future experiments will determine whether the smaller Aβ elevation seen in the 6-month-old IDE −/− brains compared with the 12-week-old brains represents biological compensation or experimental variation. Supporting the latter explanation is the difference in Aβ elevation measured in the two sets of 12-week-old IDE −/− animals (19% and 64% in the smaller and larger sets, respectively). In any event, our data make clear that IDE hypofunction in vivo leads to a highly significant, chronic accumulation of cerebral Aβ. Considering that IDE was the major Aβ-degrading protease in the neuronal and brain Aβ degradation assays, the similar elevation (64%) of endogenous Aβ40 in the NEP −/− and the larger set of IDE −/− brains was not expected. One possible explanation offered by recent evidence is that IDE principally degrades soluble Aβ (such as the 125I-Aβ40 used in our degradation assays), whereas NEP does not; its Aβ proteolytic activity is mainly in the insoluble fraction (14). In our study, we quantified cerebral Aβ levels in DEA-extracted brain fractions, which represent a combination of the soluble and insoluble pools of endogenous Aβ (unpublished observations). Thus, the insoluble Aβ pool was analyzed in the DEA-extracted brains but not in the degradation assays. It is not yet fully established which pool of Aβ is more pathogenic in AD, although several studies suggest that cognitive dysfunction correlates more tightly with soluble than insoluble levels of cortical Aβ (43, 44).
It is of particular interest that in vivo deficiency of a protease responsible for degrading insulin actually results in hyperglycemia in response to a glucose load (i.e., glucose intolerance). Although it is generally accepted that the hyperinsulinemia in DM2 is a compensatory response to insulin resistance of target tissues, there is increasing evidence that, at least in some populations, basal hyperinsulinemia itself can have a primary role in the pathogenesis of DM2 (45–47). Our findings suggest that elevated insulin levels secondary to chronically decreased clearance are sufficient to induce glucose intolerance, supporting the hypothesis that hyperinsulinemia can play a primary pathogenic role in DM2. The mechanism of this effect is presently unclear, but it is possible that IDE −/− mice develop peripheral insulin resistance, perhaps by (i) absence of IDE-mediated release of insulin from the receptor (48) with a resulting decrease in the number of insulin receptors available for activation; (ii) compensatory down-regulation of insulin receptors or factors in the postreceptor insulin signaling pathways; or (iii) an abnormal (i.e., nonphasic) pattern of insulin elevation in response to a glucose challenge (49).
The etiologies of most cases of AD and DM2 are probably multifactorial, consisting of significant genetic predilections onto which are imposed environmental factors. Combined with emerging evidence from human genetics, our findings recommend IDE as a disease susceptibility gene in both AD and DM2. By recapitulating major features of both the AD and DM2 phenotypes through disruption of a single gene, our model is also relevant to the growing evidence that DM2 and, in particular, hyperinsulinemia, are associated with an increased risk of developing AD. Although DM2 has long been known as a risk factor for all vascular diseases, including vascular dementia, its epidemiological relationship to AD has only recently been explored in large-scale studies. Prospective and historical population-based cohort studies (6,370 and 1,455 subjects, respectively) have shown that patients with DM2 are approximately twice as likely to develop AD, independent of vascular factors (50, 51). A third study limited to Japanese-American men (3,774 subjects) reported no association between AD and diabetes (52). Hyperinsulinemia (both fasting and after a glucose load) has been correlated with dementia in nondiabetic patients, with epidemiological evidence suggesting that insulin effects the brain directly rather than through vascular factors (53, 54). In addition, abnormally high levels of insulin were reported in the cerebrospinal fluid of AD patients after fasting compared with those in patients with vascular dementia or normal controls (55). Supporting a role for IDE in the hyperinsulinemia–AD correlation is evidence that an exaggerated serum insulin response to administered glucose seen in AD patients is secondary to an insulin clearance defect rather than to overproduction of the hormone (56). There is work suggesting that the advanced glycation end products that occur in DM2 can be found in amyloid plaques and could lead to increased brain Aβ aggregation in AD (57, 58). However, this mechanism probably does not explain the above epidemiological data, as one would expect to see an association between AD and hyperglycemia, not hyperinsulinemia, and particularly not hyperinsulinemia in the absence of DM2.
Combining the genetic and epidemiological evidence discussed above with our findings in an in vivo model, we offer two hypotheses to explain the association between DM2, hyperinsulinemia, and AD. (i) A primary decrease in IDE function can put an individual at risk for these conditions, as suggested by the genetic evidence for an AD and a DM2 gene in the same IDE-containing region of chromosome 10q. (ii) Chronic hyperinsulinemia, whether caused by IDE dysfunction or another mechanism, could provide an avid competitive substrate to IDE, resulting in decreased degradation and resultant gradual accumulation of Aβ. Regardless of whether IDE has a role in the pathogenesis of AD, our results have potential therapeutic implications in that compounds which up-regulate or disinhibit IDE would be expected to lower Aβ levels in vivo.
We have demonstrated that, in addition to its role in endogenous insulin and Aβ economy, IDE is also involved in regulating the cerebral levels of unphosphorylated (but not phosphorylated) AICD in vivo. Perhaps phosphorylation protects AICD from cleavage by IDE, either directly or via interaction with other proteins, but this question has yet to be answered, and the physiological significance of elevated AICD in the IDE −/− animals remains to be determined.
In conclusion, our findings demonstrate that IDE regulates Aβ, insulin, and AICD levels in vivo, and suggest, in light of the emerging genetic data, that defects in the protease could underlie or contribute to the etiologies of AD and DM2.
Supplementary Material
Acknowledgments
We thank Laura Rosen for advice on neuronal cultures, Taylor Kimberly and Bing Zheng for the C60/Fe65 lysate and useful discussions about AICD, and Bao Lu and Norma Gerard for the NEP −/− mice. This work was supported by National Institutes of Health Grants AG1247 (to W.F., S.M., and D.J.S.), AG15903 (to S.G., Y.C., and L.L.), and AG15379 (to M.P.F.); the Mayo Alzheimer's Disease Research Center (C.B.E. and E.A.E.); the Alzheimer's Association (C.B.E. and E.A.E.); a Smith Fellowship (to E.A.E.); and a Paul Beeson Physician Faculty Scholar in Aging award (to M.P.F.).
Abbreviations
- Aβ
amyloid β-protein
- AD
Alzheimer's disease
- AICD
APP intracellular domain
- APP
β-amyloid precursor protein
- DM2
type 2 diabetes mellitus
- IDE
insulin-degrading enzyme
- NEP
neprilysin
- TCA
trichloroacetic acid
Footnotes
Farris, R. W., Vekrellis, K., Mansourian, S., Chiu, S., Condron, M. & Selkoe, D. J. (2001) Soc. Neurosci. Abstr. 27, 508.
References
- 1.Duckworth W C, Bennett R G, Hamel F G. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 2.Goldfine I D, Williams J A, Bailey A C, Wong K Y, Iwamoto Y, Yokono K, Baba S, Roth R A. Diabetes. 1984;33:64–72. doi: 10.2337/diab.33.1.64. [DOI] [PubMed] [Google Scholar]
- 3.Seta K A, Roth R A. Biochem Biophys Res Commun. 1997;231:167–171. doi: 10.1006/bbrc.1997.6066. [DOI] [PubMed] [Google Scholar]
- 4.Vekrellis K, Ye Z, Qiu W Q, Walsh D, Hartley D, Chesneau V, Rosner M R, Selkoe D J. J Neurosci. 2000;20:1657–1665. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett R G, Duckworth W C, Hamel F G. J Biol Chem. 2000;275:36621–36625. doi: 10.1074/jbc.M006170200. [DOI] [PubMed] [Google Scholar]
- 6.Kurochkin I V. Trends Biochem Sci. 2001;26:421–425. doi: 10.1016/s0968-0004(01)01876-x. [DOI] [PubMed] [Google Scholar]
- 7.Authier F, Posner B I, Bergeron J J M. Clin Invest Med. 1996;19:149–160. [PubMed] [Google Scholar]
- 8.Fakhrai-Rad H, Nikoshkov A, Kamel A, Fernstrom M, Zierath J R, Norgren S, Luthman H, Galli J. Hum Mol Genet. 2000;9:2149–2158. doi: 10.1093/hmg/9.14.2149. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S, Watanabe R M, Valle T T, Hauser E R, Magnuson V L, Langefeld C D, Ally D S, Mohlke K L, Silander K, Kohtamaki K, et al. Am J Hum Genet. 2000;67:1174–1185. [PMC free article] [PubMed] [Google Scholar]
- 10.Wiltshire S, Hattersley A T, Hitman G A, Walker M, Levy J C, Sampson M, O'Rahilly S, Frayling T M, Bell J I, Lathrop G M, et al. Am J Hum Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meigs J B, Panhuysen C I, Myers R H, Wilson P W, Cupples L A. Diabetes. 2002;51:833–840. doi: 10.2337/diabetes.51.3.833. [DOI] [PubMed] [Google Scholar]
- 12.Qiu W Q, Ye Z, Kholodenko D, Seubert P, Selkoe D J. J Biol Chem. 1997;272:6641–6646. doi: 10.1074/jbc.272.10.6641. [DOI] [PubMed] [Google Scholar]
- 13.Qiu W Q, Walsh D M, Ye Z, Vekrellis K, Zhang J, Podlisny M, Rosner M R, Safavi A, Hersh L B, Selkoe D J. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 14.Sudoh S, Frosch M P, Wolf B A. Biochemistry. 2002;41:1091–1099. doi: 10.1021/bi011193l. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee A, Song E, Kihiko-Ehmann M, Goodman J P, Jr, Pyrek J S, Estus S, Hersh L B. J Neurosci. 2000;20:8745–8749. doi: 10.1523/JNEUROSCI.20-23-08745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage M J, Trusko S P, Howland D S, Pinsker L R, Mistretta S, Reaume A G, Greenberg B D, Siman R, Scott R W. J Neurosci. 1998;18:1743–1752. doi: 10.1523/JNEUROSCI.18-05-01743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dovey H F, John V, Anderson J P, Chen L Z, de Saint Andrieu P, Fang L Y, Freedman S B, Folmer B, Goldbach E, Holsztynska E J, et al. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 18.Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard N P, Gerard C, Hama E, Lee H J, Saido T C. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 19.Eckman E A, Watson M, Marlow L, Sambamurti K, Eckman C B. J Biol Chem. 2003;278:2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- 20.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis M G, Go R C, Vekrellis K, et al. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 21.Li Y J, Scott W K, Hedges D J, Zhang F, Gaskell P C, Nance M A, Watts R L, Hubble J P, Koller W C, Pahwa R, et al. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ait-Ghezala G, Abdullah L, Crescentini R, Crawford F, Town T, Singh S, Richards D, Duara R, Mullan M. Neurosci Lett. 2002;325:87–90. doi: 10.1016/s0304-3940(02)00243-4. [DOI] [PubMed] [Google Scholar]
- 23.Abraham R, Myers A, Wavrant-DeVrieze F, Hamshere M L, Thomas H V, Marshall H, Compton D, Spurlock G, Turic D, Hoogendoorn B, et al. Hum Genet. 2001;109:646–652. doi: 10.1007/s00439-001-0614-1. [DOI] [PubMed] [Google Scholar]
- 24.Boussaha M, Hannequin D, Verpillat P, Brice A, Frebourg T, Campion D. Neurosci Lett. 2002;329:121–123. doi: 10.1016/s0304-3940(02)00586-4. [DOI] [PubMed] [Google Scholar]
- 25.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze F W, Crook R, et al. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 26.Ertekin-Taner N, Graff-Radford N, Younkin L H, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin S G. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 27.Edbauer D, Willem M, Lammich S, Steiner H, Haass C. J Biol Chem. 2002;277:13389–13393. doi: 10.1074/jbc.M111571200. [DOI] [PubMed] [Google Scholar]
- 28.Kimberly W T, Zheng J B, Guenette S Y, Selkoe D J. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 29.Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B. J Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 30.Cao X, Sudhof T C. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 31.Baek S H, Ohgi K A, Rose D W, Koo E H, Glass C K, Rosenfeld M G. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 32.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 33.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podlisny M B, Tolan D, Selkoe D J. Am J Pathol. 1991;138:1423–1435. [PMC free article] [PubMed] [Google Scholar]
- 35.Podlisny M B, Citron M, Amarante P, Sherrington R, Xia W, Zhang J, Diehl T, Levesque G, Fraser P, Haass C, et al. Neurobiol Dis. 1997;3:325–337. doi: 10.1006/nbdi.1997.0129. [DOI] [PubMed] [Google Scholar]
- 36.Duff K, Eckman C, Zehr C, Yu X, Prada C-M, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 37.Petanceska S S, Nagy V, Frail D, Gandy S. Exp Gerontol. 2000;35:1317–1325. doi: 10.1016/s0531-5565(00)00157-1. [DOI] [PubMed] [Google Scholar]
- 38.Xia W, Ray W J, Ostaszewski B L, Rahmati T, Kimberly W T, Wolfe M S, Zhange J, Goate A M, Selkoe D J. Proc Natl Acad Sci USA. 2000;97:9299–9304. doi: 10.1073/pnas.97.16.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu B, Gerard N P, Kolakowski L F, Jr, Bozza M, Zurakowski D, Finco O, Carroll M C, Gerard C. J Exp Med. 1995;181:2271–2275. doi: 10.1084/jem.181.6.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Jiang M, Trumbauer M E, Sirinathsinghji D J S, Hopkins R, Smith D W, Heavesn R P, Dawson G R, Boyce S, Conner M W, et al. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- 41.Mucke L, Masliah E, Yu G Q, Mallory M, Rockenstein E M, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duckworth W C, Kitabchi A E. Diabetes. 1974;23:536–543. doi: 10.2337/diab.23.6.536. [DOI] [PubMed] [Google Scholar]
- 43.Lue L F, Kuo Y M, Roher A E, Brachova L, Shen Y, Sue L, Beach T, Kurth J H, Rydel R E, Rogers J. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean C A, Cherny R A, Fraser F W, Fuller S J, Smith M J, Beyreuther K, Bush A I, Masters C L. Ann Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 45.Weyer C, Hanson R L, Tataranni P A, Bogardus C, Pratley R E. Diabetes. 2000;49:2094–2101. doi: 10.2337/diabetes.49.12.2094. [DOI] [PubMed] [Google Scholar]
- 46.Ferrannini E, Natali A, Bell P, Cavallo-Perin P, Lalic N, Mingrone G. J Clin Invest. 1997;100:1166–1173. doi: 10.1172/JCI119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weyer C, Salbe A D, Lindsay R S, Pratley R E, Bogardus C, Tataranni P A. Metabolism. 2001;50:223–230. doi: 10.1053/meta.2001.20170. [DOI] [PubMed] [Google Scholar]
- 48.Backer J M, Kahn C R, White M F. J Biol Chem. 1990;265:14828–14835. [PubMed] [Google Scholar]
- 49.Pratley R E, Weyer C. Diabetologia. 2001;44:929–945. doi: 10.1007/s001250100580. [DOI] [PubMed] [Google Scholar]
- 50.Ott A, Stolk R P, van Harskamp F, Pols H A, Hofman A, Breteler M M. Neurology. 1999;53:1937–1942. doi: 10.1212/wnl.53.9.1937. [DOI] [PubMed] [Google Scholar]
- 51.Leibson C L, Rocca W A, Hanson V A, Cha R, Kokmen E, O'Brien P C, Palumbo P J. Am J Epidemiol. 1997;145:301–308. doi: 10.1093/oxfordjournals.aje.a009106. [DOI] [PubMed] [Google Scholar]
- 52.Curb J D, Rodriguez B L, Abbott R D, Petrovitch H, Ross G W, Masaki K H, Foley D, Blanchette P L, Harris T, Chen R, et al. Neurology. 1999;52:971–975. doi: 10.1212/wnl.52.5.971. [DOI] [PubMed] [Google Scholar]
- 53.Stolk R P, Breteler M M, Ott A, Pols H A, Lamberts S W, Grobbee D E, Hofman A. Diabetes Care. 1997;20:792–795. doi: 10.2337/diacare.20.5.792. [DOI] [PubMed] [Google Scholar]
- 54.Razay G, Wilcock G K. Age Ageing. 1994;23:396–399. doi: 10.1093/ageing/23.5.396. [DOI] [PubMed] [Google Scholar]
- 55.Fujisawa Y, Sasaki K, Akiyama K. Biol Psychiatry. 1991;30:1219–1228. doi: 10.1016/0006-3223(91)90158-i. [DOI] [PubMed] [Google Scholar]
- 56.Craft S, Newcomer J, Kanne S, Dagogo-Jack S, Cryer P, Sheline Y, Luby J, Dagogo-Jack A, Alderson A. Neurobiol Aging. 1996;17:123–130. doi: 10.1016/0197-4580(95)02002-0. [DOI] [PubMed] [Google Scholar]
- 57.Sasaki N, Fukatsu R, Tsuzuki K, Hayashi Y, Yoshida T, Fujii N, Koike T, Wakayama I, Yanagihara R, Garruto R, et al. Am J Pathol. 1998;153:1149–1155. doi: 10.1016/S0002-9440(10)65659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitek M P, Bhattacharya K, Glendening J M, Stopa E, Vlassara H, Bucala R, Manogue K, Cerami A. Proc Natl Acad Sci USA. 1994;91:4766–4770. doi: 10.1073/pnas.91.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.