Abstract
A variety of genetic alterations and gene expression changes are involved in the pathogenesis of bladder tumor. To explore these changes, oligonucleotide array analysis was performed on RNA obtained from carcinogen-induced mouse bladder tumors and normal mouse bladder epithelia using Affymetrix (Santa Clara, CA) MGU74Av2 GeneChips. Analysis yielded 1164 known genes that were changed in the tumors. Certain of the upregulated genes included EGFR-Ras signaling genes, transcription factors, cell cycle-related genes, and intracellular signaling cascade genes. However, downregulated genes include mitogen-activated protein kinases, cell cycle checkpoint genes, Rab subfamily genes, Rho subfamily genes, and SH2 and SH3 domains-related genes. These genes are involved in a broad range of different pathways including control of cell proliferation, differentiation, cell cycle, signal transduction, and apoptosis. Using the pathway visualization tool GenMAPP, we found that several genes, including TbR-I, STAT1, Smad1, Smad2, Jun, NFκB, and so on, in the TGF-β signaling pathway and p115 RhoGEF, RhoGDI3, MEKK4A/MEKK4B, PI3KA, and JNK in the G13 signaling pathway were differentially expressed in the tumors. In summary, we have determined the expression profiles of genes differentially expressed during mouse bladder tumorigenesis. Our results suggest that activation of the EGFR-Ras pathway, uncontrolled cell cycle, aberrant transcription factors, and G13 and TGF-β pathways are involved, and the cross-talk between these pathways seems to play important roles in mouse bladder tumorigenesis.
Keywords: Mouse bladder cancer, expression profile, Affymetrix microarray, differential expression, signaling pathway
Introduction
Bladder cancer is the fifth most common cancer in the United States and is associated with exposure to cigarette smoke; it is predicted to account for 57,400 new cases and 12,500 cancer-related deaths in 2003. Approximately 15% of bladder tumors evolve into invasive tumors after infiltration through the basement membrane. Patients with muscle invasive disease are at high risk for recurrence, progression, and metastases. The incidence of bladder cancer has been steadily increasing and, despite improvements in treatment, the majority of the patients will not survive for 5 years [1].
Ras, erb-B2, and epidermal growth factor receptor (EGFR) are the most important oncogenes in urinary bladder cancer. The transforming potential of ras is due to a mutation, whereas EGFR and erb-B2 are commonly overexpressed in transformed cells. Reported frequencies of H-ras point mutations with a glycine-to-valine substitution in codon 12 in bladder neoplasms varied widely between studies from 0% to 45% [2–5]. Recently, several ways to suppress Ras activities, including inhibitors of Ras signal transduction and a ras suppressor mutant, have been reported [6]. Overexpression of EGFR or erb-B2 and ras mutation could result in constitutive MAPK activation [7] and correlates with muscular invasion and extent of tumor invasion [8]. Almost all advanced bladder carcinomas lack either pRb or p16INK4a, with cyclin D1 overexpression preferentially occurring in earlier stages [9,10].
There are two primary chemically induced models of urinary bladder cancers in rodents. Both employ repeated intragastric administration of 4-hydroxybutyl(butyl)nitrosamine (OH-BBN) to induce bladder cancers in either mice or rats [11,12]. The bladder cancers typically have a mixed histology showing elements of both transitional and squamous cells. Investigators have found a relatively low frequency of Ras mutation in these cancers [13] and roughly 50% of these tumors develop p53 mutations [14], which are similar to those found in humans. Complete loss of p53 is a prerequisite for collaborating with activated Ha-ras to promote bladder tumorigenesis [15]. Inactivation of p53 and pRb induced carcinoma in situ and invasive and metastatic bladder cancer, whereas activation of Ha-ras in transgenic mice caused urothelial hyperplasia and superficial papillary noninvasive bladder tumors. These results provide strong, direct experimental evidence that the two phenotypic pathways of bladder tumorigenesis are caused by distinctive genetic defects [16]. There has been further characterization of these tumors for various gene products of the EGFR loop [17]. Similar to human bladder tumors, these tumors tend to show overexpression of EGFR and amphiregulin.
Significant progress has been made in understanding the underlying molecular and genetic events in bladder cancer. Numerous markers have been described to correlate to some extent with tumor stage and prognosis of patients with bladder cancer. However, the power of many of these markers is limited; there remains a great need to develop reliable alternative markers that can provide more useful information regarding diagnosis and prognosis, and to facilitate the selection of appropriate therapy in the individual patient. Expression profiling with high-throughput DNA microarrays has the potential of providing critical clues. In this study, we employed Affymetrix (Santa Clara, CA) microarrays representing over 12,000 genes and expressed sequence tags (ESTs) to identify differentially expressed genes in mouse bladder tumors. The purposes of the present study were: 1) to detect and identify differential gene expression profiles in mouse bladder tumors; and 2) to elucidate the underlying mechanisms of mouse bladder tumorigenesis. The genes identified in this study can be employed in a variety of applications: 1) for use as early detection markers for bladder lesions in the mouse model; 2) for comparison of gene expression changes observed in mouse to human bladder cancers; 3) for basic understanding of the bladder cancer process; 4) for help in defining potential molecular targets, which can be tested in therapeutic or prevention studies in bladder tumor models; and 5) for use as potential modulatable biomarkers, which can be employed in screening for potential agents, or in determining the efficacy of those agents.
Materials and Methods
Mouse Bladder Tumors
Male B6D2F1 (C57Bl/6 x DBA/2 F1) mice were obtained from Harlan Sprague-Dawley, Inc. (Indianapolis, IN) at 28 days of age and were housed in polycarbonate cages (five per cage). The animals were kept in a lighted room 12 hours each day and maintained at 22 ± 0.5°C. Teklad 4% mash diet (Harlan Teklad, Madison, WI) and tap water were provided ad libitum. At 56 days of age, mice received the first of 12 weekly gavage treatments with OH-BBN (TCI America, Portland, OR). Each 7.5-mg dose was dissolved in 0.1 ml of ethanol:- water (25:75). Mice (unless sacrificed early because of a large palpable bladder mass) were sacrificed 8 months following the first OH-BBN treatment. Bladder tumors were removed and frozen for subsequent molecular assays. A portion of each tumor was fixed and processed for routine paraffin embedding, cut into 5-µm sections, and mounted for hematoxylin and eosin (H&E) staining for histopathology. All bladder tumors used in this study were diagnosed as bladder cancers with a mixed histology showing elements of both transitional and squamous cells. Both bladder tissues and normal bladder epithelia come from age-matched controls.
RNA Isolation and Amplification
To isolate bladder epithelia, we conducted microdissection, under a dissecting microscope employing control mice who were at least 8 months old, by separating the epithelia from the stroma and muscle tissues using surgical blade and forceps. A 5-µm frozen section was made and H&E-stained to examine the purity of the isolated epithelia. Total RNA from normal bladder epithelia, normal bladder tissues, and bladder cancers were isolated by Trizol (Invitrogen, Carlsbad, CA) and purified using the RNeasy Mini Kit and RNase-free DNase Set (QIAGEN, Valencia, CA) according to the manufacturer's protocol. In vitro transcription-based RNA amplification was then performed on each sample. cDNA for each sample was synthesized using a Superscript cDNA Synthesis Kit (Invitrogen) and a T7-(dT)24 primer: 5′-GGCCAGTGAATTGTAATACGACT-CACTATAGGGAGGCGG-(dT)24-3′. The cDNA was cleaned using phase-lock gel (Fisher Cat ID E0032005101) phenol/chloroform extraction. Then, the biotin-labeled cRNA was transcribed in vitro from cDNA using a BioArray High Yield RNA Transcript Labeling Kit (ENZO Biochemistry, New York, NY) and purified, again using the RNeasy Mini Kit.
Affymetrix GeneChip Probe Array and Semiquantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR) Confirmation
The labeled cRNA was applied to the Affymetrix Mu74Av2 GeneChips, which contain >12,000 genes and ESTs on one array according to the manufacturer's recommendations. Every gene or EST is represented by a probe set consisting of approximately 16 probe pairs (oligonucleotides) of 25-mer oligonucleotides. One sequence of a probe pair represents the complementary strand of the target sequence, whereas the other has a 1-bp mismatch at the central base pair position. This mismatch sequence serves as an internal control for specificity of hybridization. To evaluate the reliability of the array results, 10 genes were randomly selected from the genes detected in the microarray assay for further confirmation by semiquantitative RT-PCR as previously described [18]. The large number of differentially expressed genes led us to take a further quality control step in which the distribution of fold changes was examined.
Grouping Gene
Genes were functionally annotated using the GO-Biologic Process annotations as provided by Affymetrix. To organize the differentially expressed genes into a small number of mutually exclusive categories, each GO category represented in the data set was mapped to 1 of 14 categories of Table 1. This mapping resulted in genes that were categorized either unambiguously, ambiguously, or not at all. Genes with no or ambiguous categorization were examined and manually placed in 1 of 14 categories.
Table 1.
Classification of 1164 Known Genes Found to Be Differentially Expressed in Mouse Bladder Cancers by Microarray Analysis (Fold Change ≥ 2 and P < .05) Into 14 Subgroups Using the GO-Biological Process Annotations as Provided by Affymetrix.
| Group | Description | Number of Genes Changed in Tumors | |
| Up | Down | ||
| 1 | Cell cycle | 42 | 10 |
| 2 | Immune response | 99 | 7 |
| 3 | Transcription | 54 | 47 |
| 4 | G-protein signaling | 23 | 8 |
| 5 | Cell adhesion | 42 | 9 |
| 6 | Small GTPase signaling | 13 | 16 |
| 7 | Other signaling effectors | 43 | 24 |
| 8 | Transport | 40 | 36 |
| 9 | Metabolism | 112 | 129 |
| 10 | Apoptosis | 13 | 8 |
| 11 | Development/differentiation | 20 | 25 |
| 12 | Cell proliferation | 23 | 12 |
| 13 | Cytoskeleton | 20 | 22 |
| 14 | Others | 15 | 6 |
| Total annotated | 559 | 359 | |
| Total unannotated | 144 | 102 | |
Cluster and GenMAPP
Array normalization and gene expression estimates were obtained using Affymetrix Microarray Suite 5.0 software (MAS5). The array mean intensities were scaled to 1500. These estimates formed the basis for statistical testing. To eliminate the false calls of the gene expression, the raw data values below 300 were excluded from the data set. Differential expression was determined using the combined basis of t-test with P < .05 and fold changes (either up or down) greater than two-fold. Thus, for a gene to be included in our list, it had to be expressed at moderate to high levels and display at least a two-fold alteration in expression, and that difference had to be statistically significant. Genes meeting all these criteria were called positive for differential expression. Hierarchical clustering was performed as follows. For the selected genes, expression indexes were transformed across samples to an N(0,1) distribution using a standard statistical Z-transform. These values were inputted to the GeneCluster program of Eisen et al. [19] and genes were clustered using average linkage and correlation dissimilarity. Signal transduction pathways, metabolic pathways, and other functional groupings of genes were evaluated for differential regulation using the visualization tool GenMAPP [20]. We imported the statistical results of our data set into the program and used GenMAPP to illustrate pathways containing differentially expressed genes.
Results
Different Expression Patterns between Epithelia and Whole Tissues
The experiment design for this study includes the use of mouse bladder epithelium, bladder tissue, and bladder tumor to test the usefulness of whole bladder tissues versus purified epithelium as controls, and to profile the gene differential expression during the mouse bladder tumorigenesis. Untutored cluster diagrams of whole mouse bladder tissues, epithelia, and tumors, and dendrograms were created from hierarchical clustering of the gene expression profiles of each sample. The whole bladder tissues, epithelia, and tumors were clustered in groups by tissue type (Figure 1). Comparing the epithelium with the tumors, 1554 genes were found to be differentially expressed in the tumors. When comparing whole bladder tissues and tumors, 805 genes were found to be differentially expressed in tumors. About 51.8% of 1554 genes found in tumors with epithelium controls had the same results as with whole bladder controls (Figure 1). There were also another 456 genes found to be differentially expressed when using whole bladder as controls, which did not show changes when using epithelium controls (data not shown). Further comparisons are made with epithelium controls, unless otherwise noted.
Figure 1.
Comparison of bladder epithelia and whole bladder tissues as controls to bladder cancers. Untutored clusters of whole mouse bladder tissues, epithelia, and tumors were created from hierarchical clustering of the gene expression profiles of each sample. Among the 1554 genes found differentially expressed in mouse cancers compared with epithelia, 51.8% genes were consistent between epithelium and whole tissues as controls. E, bladder epithelium; N, whole bladder tissue; T, bladder tumor.
Gene Expression Profile in Bladder Tumors
In this study, microarray data were available from five mouse bladder tumors and four mouse bladder normal epithelia samples; fold changes of gene expression were based on the ratios of mean values between tumors and epithelium controls. Among 1554 differentially expressed genes, 867 genes were overexpressed and 687 genes were underexpressed in bladder tumors, and 1164 are known genes. We categorized these genes into 14 subgroups, as given in Table 1. Many of the upregulated genes were Ras family genes, transcription factors, cell cycle-related genes, and intracellular signaling cascades (Table 2). Downregulated genes include the mitogen-activated protein kinase genes, cell cycle checkpoint genes, Rab subfamily genes, Rho subfamily genes, and SH2 and SH3 domains-related genes (Table 3).
Table 2.
Selected Genes Whose Expression Is Upregulated in Mouse Bladder Tumors Compared with Normal Bladder Epithelia Identified by Microarray.
| Gene | Access | Description | Fold Change* | Incidence |
| Cell cycle-related genes | ||||
| Clk4 | AF005423 | CDC-like kinase 4 | 2.3 | 3/5 |
| Cks1 | AB025409 | CDC28 protein kinase 1 | 4.0 | 4/5 |
| CDC2a | M38724 | CDC2 homolog A | 16.0 | 5/5 |
| CDC20 | AW061324 | CDC20 homolog | 4.6 | 4/5 |
| CDC25c | L16926 | CDC25 homolog C | 5.3 | 4/5 |
| Ccna2 | X75483 | Cyclin A2 | 2.6 | 3/5 |
| Ccnb1 | X64713 | Cyclin B1 | 16.0 | 4/5 |
| Ccnb2 | X66032 | Cyclin B2 | 2.5 | 3/5 |
| Ccnd1 | AI849928 | Cyclin D1 | 2.6 | 4/5 |
| Ccne1 | X75888 | Cyclin E1 | 2.5 | 4/5 |
| Mad2l1 | U83902 | MAD2-like 1 | 2.6 | 4/5 |
| Plk | U01063 | Polo-like kinase homolog | 9.1 | 5/5 |
| Plk-ps1 | U73170 | Polo-like kinase, pseudogene 1 | 2.6 | 4/5 |
| Dp1 | AF043939 | DP1 gene | 2.0 | 3/5 |
| Gadd45b | AV138783 | GADD45 β- | 4.3 | 5/5 |
| Bub1 | AF002823 | mitotic checkpoint protein kinase Bub1 | 9.6 | 4/5 |
| Ras pathway effectors | ||||
| Racgap1 | AW122347 | Rac GTPase-activating protein 1 | 3.5 | 4/5 |
| Rad51 | D13803 | RAD51 homolog (Saccharomyces cerevisiae) | 2.3 | 3/5 |
| Rad9 | AF045663 | RAD9 homolog (S. pombe) | 8.6 | 5/5 |
| Ranbp1 | X56045 | RAN-binding protein 1 | 2.1 | 3/5 |
| Rap2ip | U73941 | Rap2-interacting protein | 2.8 | 4/5 |
| Rin2 | AI835968 | Ras and Rab interactor 2 | 2.9 | 3/5 |
| Rassf1 | AW049415 | RalGDS/AF-6 domain family 1 | 2.8 | 4/5 |
| Rasgrp1 | AF106070 | RAS guanyl releasing protein 1 | 4.0 | 5/5 |
| Arhg | AB025943 | Ras homolog gene family, member G | 2.5 | 5/5 |
| Arhh | AA739233 | Ras homolog gene family, member H | 2.9 | 5/5 |
| Arhj | AW121127 | Ras homolog gene family, member J | 2.0 | 4/5 |
| Rasl2-9 | L32752 | RAS-like, family 2, locus 9 | 2.8 | 3/5 |
| Rac3 | AA967636 | RAS-related C3 Botulinum substrate 3 | 10.5 | 4/5 |
| Arhgef1 | U58203 | Rho GEF 1 | 2.5 | 4/5 |
| Rhoip3 | AV277546 | Rho-interacting protein 3 | 6.5 | 4/5 |
| Transcription regulators | ||||
| Atf3 | U19118 | Activating transcription factor 3 | 5.3 | 5/5 |
| Elk4 | Z36885 | ELK4, member of ETS oncogene family | 3.3 | 3/5 |
| Etv1 | L10426 | Ets variant gene 1 | 2.1 | 3/5 |
| Etv4 | X63190 | Ets variant gene 4 (E1AF) | 4.9 | 5/5 |
| Etv6 | AI845538 | Ets variant gene 6 (TEL oncogene) | 2.1 | 3/5 |
| Foxc2 | AV251191 | Forkhead box C2 | 4.0 | 5/5 |
| Foxm1 | Y11245 | Forkhead box M1 | 2.0 | 4/5 |
| Fosl1 | AF017128 | Fos-like antigen 1 | 9.8 | 5/5 |
| Jun | X12761 | Jun oncogene | 7.5 | 5/5 |
| Nfkb1 | M57999 | NF-κB1, p105 | 2.2 | 4/5 |
| Nfkbie | AF030896 | IκB epsilon | 3.9 | 4/5 |
| Mybbp1a | U63648 | MYB-binding protein (P160) 1a | 4.3 | 4/5 |
| Nmi | AF019249 | N-myc (and STAT) interactor | 2.3 | 3/5 |
| EGF/EGFR pathway | ||||
| Egfr | AW049716 | Epidermal growth factor receptor | 2.6 | 4/5 |
| Mapk10 | L35236 | Mitogen-activated protein kinase 10 | 2.3 | 3/5 |
| Map4k4 | U88984 | MAP kinase kinase kinase kinase 4 | 2.0 | 3/5 |
| Mknk1 | Y11091 | MAPK-interacting serine/threonine kinase 1 | 4.3 | 5/5 |
| Scap2 | AB014485 | Src family-associated phosphoprotein 2 | 2.3 | 4/5 |
| Shd | AB018423 | Src homology 2-transforming protein D | 3.0 | 4/5 |
| Sla | U29056 | Src-like adaptor | 4.6 | 4/5 |
| Pik3c2a | U52193 | PI3 kinase, C2 domain containing α | 7.5 | 4/5 |
| Pik3ca | U03279 | PI3 kinase, catalytic, α-polypeptide | 2.3 | 3/5 |
Most of the upregulated genes were Ras family genes, transcription factors, cell cycle-related genes, and intracellular signaling cascade factors.
Fold change is the ratio of mean gene expression values of the tumors to the mean gene expression values of the epithelia from the microarray.
Table 3.
Selected Genes Whose Expression Is Downregulated in Mouse Bladder Tumors Compared with Normal Bladder Epithelia Identified by Microarray.
| Downregulated Genes in Mouse Bladder Tumors Identified by Microarray | |||||||||
| Gene | Access | Description | Fold Change* | Incidence | |||||
| Cell cycle-related genes | |||||||||
| Ccng1 | L49507 | Cyclin G1 | 2.0 | 3/5 | |||||
| Gas1 | X65128 | Growth arrest-specific 1 | 2.8 | 4/5 | |||||
| Madh2 | U60530 | MAD homolog 2 | 2.9 | 5/5 | |||||
| Atm | U43678 | Ataxia telangiectasia-mutated homolog | 4.9 | 5/5 | |||||
| Rbbp7 | U35142 | Retinoblastoma - binding protein 7 | 2.1 | 3/5 | |||||
| Rbl2 | U36799 | Retinoblastoma-like 2 | 2.0 | 3/5 | |||||
| Ras pathway effectors | |||||||||
| Rab11a | AI853996 | RAB11a, member of RAS oncogene family | 2.5 | 3/5 | |||||
| Rab33b | AW208630 | RAB33B, member of RAS oncogene family | 2.6 | 5/5 | |||||
| Rab3d | AI835706 | RAB3D, member of RAS oncogene family | 2.1 | 4/5 | |||||
| Rab9 | AB027290 | RAB9, member of RAS oncogene family | 2.1 | 5/5 | |||||
| Arhgdia | AI836322 | Rho GDP dissociation inhibitor (GDI) α | 7.0 | 5/5 | |||||
| Arhgdig | U73198 | Rho GDP dissociation inhibitor (GDI) gamma | 2.8 | 4/5 | |||||
| Rhob | X99963 | rhoB gene | 2.1 | 4/5 | |||||
| Arhq | D50264 | Ras homolog gene family, member Q | 2.6 | 4/5 | |||||
| Transcription regulators | Tbx2 | U15566 | T-box 2 | 4.0 | 5/5 | ||||
| Ndr2 | AB033921 | N-myc downstream-regulated 2 | 21.1 | 5/5 | |||||
| Lasp1 | AW122780 | LIM and SH3 protein 1 | 2.3 | 4/5 | |||||
| Lmo1 | AW124311 | LIM domain only 1 | 6.5 | 5/5 | |||||
| Lmo4 | AF074600 | LIM domain only 4 | 2.3 | 4/5 | |||||
| Pdlim3 | AF002283 | PDZ and LIM domain 3 | 4.9 | 5/5 | |||||
| Tcf2 | AB008174 | Transcription factor 2 | 2.0 | 3/5 | |||||
| Tcf21 | AF035717 | Transcription factor 21 | 3.7 | 4/5 | |||||
| Tcf3 | AJ223069 | Transcription factor 3 | 2.5 | 3/5 | |||||
| Gata2 | AB000096 | GATA-binding protein 2 | 3.5 | 5/5 | |||||
| Gata3 | X55123 | GATA-binding protein 3 | 9.2 | 5/5 | |||||
| Gata4 | M98339 | GATA-binding protein 4 | 2.3 | 3/5 | |||||
| Cri1 | AI844939 | CREBBP/EP300-inhibitory protein 1 | 2.3 | 5/5 | |||||
| MAPK/Src homolog | |||||||||
| Erk2 | D87271 | ERK2 | 2.6 | 4/5 | |||||
| Map2k6 | U39066 | Mitogen-activated protein kinase kinase 6 | 2.5 | 3/5 | |||||
| Map3k4 | AV270901 | Mitogen-activated protein kinase kinase kinase 4 | 3.0 | 4/5 | |||||
| Map3k5 | AB006787 | Mitogen-activated protein kinase kinase kinase 5 | 2.5 | 4/5 | |||||
| Map3k8 | AV341985 | Mitogen-activated protein kinase kinase kinase 8 | 3.0 | 3/5 | |||||
| Sh2bpsm1 | AF020526 | SH2-B PH domain containing signaling mediator 1 | 2.1 | 3/5 | |||||
| Sh3bgr | AW048272 | SH3-binding domain, glutamic acid-rich protein | 4.9 | 5/5 | |||||
| Sh3bp1 | X87671 | SH3 domain-binding protein 1 | 2.8 | 4/5 | |||||
| Sh3gl2 | U58886 | SH3 domain GRB2-like 2 | 13.0 | 5/5 | |||||
| Sh3gl3 | U58887 | SH3 domain GRB2-like 3 | 2.5 | 4/5 | |||||
Downregulated genes include the mitogen-activated protein kinase genes, cell cycle checkpoint genes, Rab subfamily genes, Rho subfamily genes, and SH2 and SH3 domains-related genes.
Fold change is the ratio of mean gene expression values of the tumors to the mean gene expression values of the epithelia from the microarray.
Gene Distribution and RT-PCR Confirmation
With such a large number of differentially expressed genes, we examined the distribution of fold changes to detect if any large skew could account for the results. The distribution of fold changes for the differentially expressed genes is shown in Figure 2A, and its symmetry suggests that no skew artifact is present. We validated the differential expression of 10 genes by semiquantitative RT-PCR. Nine of 10 genes were confirmed by RT-PCR. The confirmation rate is 90% at the cutoff of two-fold change and P < .05. The RTPCR results of these nine genes agreed well with the microarray data (Figure 2B).
Figure 2.
Distribution of the 1164 differentially expressed known genes by microarray analysis and semiquantitative RT-PCR confirmation for selected genes. (A) Overview of the number of genes has different fold changes compared with normal bladder epithelia. (B) Comparison of fold change produced by microarray with relative expression ratio obtained from RT-PCR; the concordance is good.
Ras-Related Genes in Bladder Tumors
Tables 2 and 3 list selected genes that were upregulated or downregulated, respectively. The Ras superfamily is a diverse group of small G proteins participating in many cellular processes and also widely involved in tumorigenesis. In this study, many Ras superfamily members were found to be abnormally expressed in bladder tumors. Except for the Rab subfamily, including Rab3D, Rab9, Rab11A, and Rab33B, which were underexpressed, almost all other rasrelated genes, such as Ras, Rap, Rin, Rac, Ran, and Rad, were overexpressed in bladder tumors. For Rho-related genes, Rho-GEF1 and RhoIP3 were overexpressed, and Rho-GDIα, Rho-GDIβ, and RhoB were underexpressed in mouse bladder tumors.
Cell Cycle-Related Genes and Transcription Regulators in Bladder Tumors
Many of the overexpressed genes were cell cycle-related genes that promote the entry into cell cycle and mitosis, including cyclin B1, B2, D1, E1, CDK2, CDC2, CDC20, CDC25, and CDC28 protein kinase 1. Cyclin G1, retinoblastoma (Rb)-like 2, ATM, Gas1, and Rb-binding protein 7 were found to be downregulated in mouse bladder tumors; these genes play important roles in the cell cycle arrest and G1/S and G2 checkpoints. Several genes that function in cell cycle as transcription regulators, and which are associated with carcinogenesis in various cancers, were also overexpressed in mouse bladder tumors. These genes included ets, fos, Jun, myb, N-myc, NF-κB1, and IκB-ε. In mouse bladder tumors, we also found some transcription regulators that function in normal development and differentiation to be downregulated, including LMO1 and LMO4, GATA-BP2, GATA-BP3, and GATA-BP4 (Tables 2 and 3).
Differentially Expressed Genes Interpreted by GenMAPP
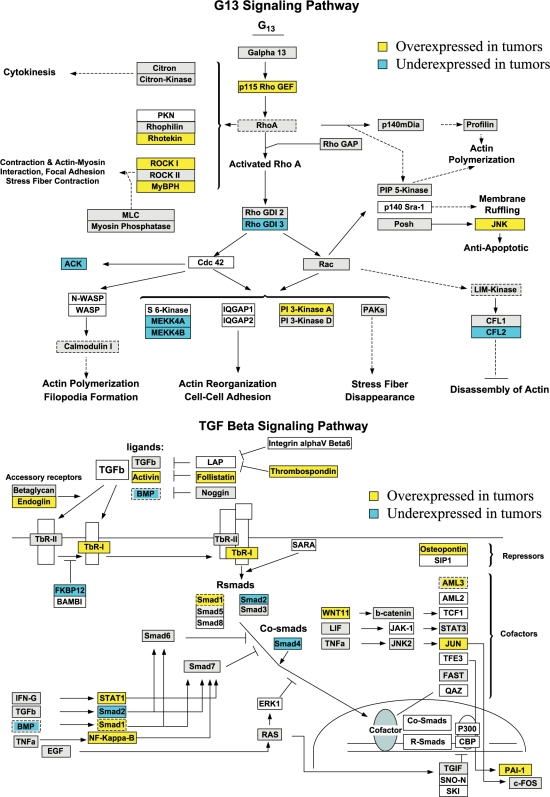
GenMAPP is a tool for visualizing expression data in the context of biologic pathways [20]. Using the GenMAPP, we found MAPK cascade, G protein signaling pathway, apoptosis, Wnt signaling pathway, and TGF-4β signaling, each of which may be involved in bladder tumorigenesis. Figure 3 represents the genes differentially expressed in the mouse bladder tumors that are involved in G13 and TGF-β signaling pathways.
Figure 3.
GenMAPP G13 and TGF-β signaling pathways integrated in the mouse bladder tumorigenesis with cutoff fold change ≥1.5 and P < .05. Yellow and blue indicate overexpressed and underexpressed genes in the tumor samples, respectively. Grey indicates that the selection criteria were not met but the gene is represented on the array. White boxes indicate that the gene was not present on the chip.
Discussion
One question for gene expression analysis both in human and animal studies is the type of normal tissues to use as controls. This problem is acute in complex tissues, such as lung, prostate, and mammary tissues, in which the stroma is mixed with the epithelium cells. For the whole organ, the epithelium may only account for less than 20%. In these situations, is it reasonable to use the whole tissue as the control? Our results reveal that when the whole bladder tissues rather than the epithelia are used as controls, only 51.8% of the 1554 genes that changed in tumors compared with the epithelia were found to be differentially expressed. Another 456 genes were also found to be differentially expressed when using whole bladder as controls (data not shown). However, these 456 genes did not show any changes when comparing tumors versus partially purified epithelia. Our results indicate that numerous genes accounting for cellular diversity would also be interpreted as tumorigenesis genes when using whole bladder tissues as controls in the study. Thus, it would appear that by preferentially examining genes whose expression was altered both when comparing tumors versus normal bladder and tumors versus isolated bladder epithelia, we may achieve a subset of genes that might be particularly useful as biomarkers or modulatable surrogate endpoints.
The transformation of normal cell into malignant cell is a multistep process that involves mutations or chromosomal aberrations. Like most types of cancer, the generation of bladder cancer is caused by the accumulation of various molecular changes, which can be categorized into 1) chromosomal alterations; 2) loss of cell cycle regulation, resulting in altered cellular proliferation; 3) growth control events such as angiogenesis, resulting in metastasis; and 4) decreases in cellular apoptosis. It is becoming apparent that the accumulation of genetic and epigenetic changes ultimately determines a tumor's phenotype and subsequent clinical behavior.
Ras, erb-B2, and EGFR are the most important oncogenes in bladder cancer. Ras superfamily regulates many cellular processes, such as cell cycle progression, actin cytoskeletal dynamics, and membrane traffic. The transforming potential of ras is due to a mutation, which, in bladder tumors, occurs in H-ras [21]. Overexpressions of H-ras, K-ras, and N-ras transcripts have also been associated with bladder tumor transition [22,23]. Guanine nucleotide exchange factors (GEFs) stimulate Ras superfamily members to exchange bound GDP for GTP, thereby increasing the amount of active form [24]. EGFR is known to signal, at least in part, through H-ras activation. A potential role for either a normal or a mutated overexpressed H-ras in upregulating EGFR during the progression of human bladder cancer to invasive phenotype has been demonstrated in the human papillary TCC cell line [25]. Rho family gene mutations in tumors are quite rare, but overexpression is more common [26]. Dysregulation of Rho family member activity probably also contributes to human cancer, in that some RhoGEFs act as oncogenes [27], whereas RhoGAPs [28] act as tumor suppressors. Reduced expression of RhoGDIs has recently been shown to correlate with increasing invasive and metastatic ability in human bladder carcinoma cell lines [29,30]. Increased activity of another Ras effector, PI3 kinase, is associated with many types of human cancer. Because PI3 kinase is an immediate downstream effector of Ras and EGFR, multiple pathways may contribute to an increase in PI3 kinase activity in bladder cancer. PI3 kinase consistently prevents apoptosis in many cell systems through activation of the Rac GTPase, possibly through activation of NF-κB [31]. Thus, the activation of PI3 kinase associated with excessive Ras activity may promote oncogenesis by blunting the apoptosis-inducing stimuli associated with oncogenic transformation.
In our present study, we found that Ras superfamily members significantly changed in mouse bladder tumorigenesis with several GEFs overexpressed, such as RhoGEF1 and RasGRP1, and GDIs underexpressed, including RhoGDIα and RhoGDIγ, in mouse bladder tumors, respectively. Several EGFR-Ras pathway effectors were also found to be overexpressed in mouse bladder tumors, including EGFR, Ras superfamily members, Src, PI3 kinase, and downstream transcription factors, such as Fos, Jun, NF-κB, and Myc. Our data suggest that bladder tumors can most likely develop through the EGFR-Ras pathway.
Another group of genes found to be differentially expressed in bladder tumors are cell cycle-related genes. Tumor proliferation depends on the derangement of normal cell cycle progression and control. Cell cycle-associated protein complexes composed of cyclins and cyclin-dependent kinases (CDKs) regulate normal cellular proliferation. Different CDK-cyclin complexes cooperate to drive cells through different phases of the cell cycle. Activation of CDK4 and CDK6 by D-type cyclins is thought to be involved in progression through early G1. CDK2 is sequentially activated by E-type cyclins during the G1/S transition, and the A-type cyclins during S phase [32]. CDK1/cyclin B is critical for the onset of mitosis. Proper regulation of CDK1 (CDC2 and CDC28 in fission and budding yeast, respectively) requires both activating activating and inhibitory phosphorylation [33]. Activation of tyrosine phosphatase CDC25 results in activation of CDK1 by dephosphorylation on Tyr15, triggering the onset of mitosis. The activity of CDK1/cyclin B is also regulated through proteolysis. Anaphase-promoting complex (APC), a gatekeeper of the spindle assemble checkpoint, can be activated by CDC20 and activated APC can mediate the cyclin B proteolysis, resulting in rapid decline in CDK1 activity [34]. APC activity is primarily regulated by MAD2, which has implicated the BUB family of kinase. Polo kinases regulate several stages of mitotic progression. Some of their proposed substrates are CDC25C, β-tubulin, APC/C subunits, and the kinesin-related protein MKLP-1 [35]. Cks1 promotes mitosis by modulating the transcriptional activation of the APC/C protein ubiquitin ligase activator CDC20. The essential role of Cks1 is to recruit the proteasome to, and/or dissociate the CDC28 kinase from, the CDC20 promoter, thus facilitating transcription by remodeling transcriptional complexes or chromatin associated with the CDC20 gene [36].
Several tumor-suppressor genes and their protein products (p53, pRb, p27Kip1, p16INK4A, and p14ARF) act at the G0/G1 checkpoint of the cell cycle to prevent loss of cell cycle control, and, ultimately, tumor progression. RBL2/p130 is a member of the Rb family of proteins, which are structurally and functionally similar to the pRb. Overexpression RBL2/p130 can induce growth arrest in certain cell types [37] and can bind to and inhibit the transcriptional activity of E2F transcription factors [38]. RBBP7 was initially identified as a Rb-binding protein [39] and was shown to repress E2F-regulated promoters together with HDAC proteins and BRG1 in a Rb-containing complex [40,41]. RBBP7 is located on the X chromosome and it is interesting to note that rates of bladder cancer in males exceed that in females by approximately four-fold by the age of 60 years, with an increasing sex difference throughout life. RBBP7 is a potent suppressor of cell growth in transformed cell lines and inhibits tumorigenesis in nude mice [42,43]. Expression of this gene was decreased in tumors relative to controls. Thus, RBBP7 may have an essential role in cell cycle control and may act as a tumor suppressor.
In this study, the cell cycle commitment genes, such as cyclins and CDKs, were found to be overexpressed. RBL2/p130 and RBBP7, which act at the checkpoint and suppress cell growth, were underexpressed in bladder tumorigenesis, respectively. This result is in agreement with our finding of a relatively high proliferative index in larger lesions derived from this model. Ligand binding to EGFR may activate the Ras pathway, resulting in induction of the cell cycle and causing an uncontrolled cell growth.
In addition to the involvement of EGFR, Ras pathways, and cell cycle, G13 and TGF-β signal pathways are also involved in mouse bladder tumorigenesis (Figure 3). G13 directly interacts with and activates a GEF for the GTPase Rho, p115RhoGEF, and thus activates Rho, leading to a variety of effects such as the regulation of actin cytoskeleton. G13 may also engage the PI3K pathway to activate the protein kinase Akt and regulate NF-κB [44]. The TGF-β pathways regulate many processes, including cellular proliferation, differentiation, apoptosis, inflammation, hematopoiesis, wound repair, and specification of development. Disruption of these pathways can lead to a range of diseases, including cancer. TGF-β binding type I and type II receptors on the cell surface allow receptor II to phosphorylate the receptor I kinase domain, which then propagates the signal through phosphorylation of the Smad proteins. The activated Smad complexes are translocated into the nucleus and, in conjunction with other nuclear cofactors, regulate the transcription of target genes [45]. TGF-β switches from tumor suppressor in the premalignant stages of tumorigenesis to proto-oncogene at a later stage, leading to cancer progression, survival, and metastasis [46,47]. Biphasic roles of TGF-β in signal transduction are associated with the cross-talk between TGF-β and other signaling pathways, such as inhibition of early EGF-induced p42/p44 MAPK, PKA-Raf1 interaction in delayed EGF-induced cell cycle [48], and Rholike GTPase in activation of TGF-β downstream pathways [49,50].
In conclusion, we show in this study that microarrays can be used to significantly enhance the search for the molecular pathogenesis of tumors. We found that inappropriate regulation of Ras, cell cycle, and TGF-β pathways may be the three major steps in the tumorigenesis of mouse bladder malignancy. In addition, we were able to identify a variety of genes whose expression was highly increased, independent of whether they are directly involved in the mechanism of tumorigenesis in this model. These highly modulated genes—should they prove to be changed at the protein level—may prove highly useful in identifying early lesions as well as in identifying tumors in samples from urine or serum. In addition, both these highly overexpressed genes as well as many of the genes, which are along the mechanistic pathway, may prove to be modulated by effective preventive or therapeutic agents. Finally, with regards to our initial question as to what is the proper control for these studies, we may not be able to reach a definitive conclusion. It would appear that combined use of both normal bladder and bladder epithelia might be most useful. These results support the relevance of OHBBN-induced bladder cancer in mice as an in situ model of bladder cancer.
References
- 1.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer Statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Czerniak B, Cohen GL, Etkind P, Deitch D, Simmons H, Herz F, Koss LG. Concurrent mutations of coding and regulatory sequences of the Ha-ras gene in urinary bladder carcinomas. Hum Pathol. 1992;23:1199–1204. doi: 10.1016/0046-8177(92)90285-b. [DOI] [PubMed] [Google Scholar]
- 3.Olderoy G, Daehlin L, Ogreid D. Low-frequency mutation of Ha-ras and Ki-ras oncogenes in transitional cell carcinoma of the bladder. Anticancer Res. 1998;18:2675–2678. [PubMed] [Google Scholar]
- 4.Buyru N, Tigli H, Ozcan F, Dalay N. Ras oncogene mutations in urine sediments of patients with bladder cancer. J Biochem Mol Biol. 2003;36:399–402. doi: 10.5483/bmbrep.2003.36.4.399. [DOI] [PubMed] [Google Scholar]
- 5.Knowles MA. Molecular genetics of bladder cancer. Br J Urol. 1995;1:57–66. [PubMed] [Google Scholar]
- 6.Shinohara N, Koyanagi T. Ras signal transduction in carcinogenesis and progression of bladder cancer: molecular target for treatment? Urol Res. 2002;30:273–281. doi: 10.1007/s00240-002-0275-0. [DOI] [PubMed] [Google Scholar]
- 7.Swiatkowski S, Seifert HH, Steinhoff C, Prior A, Thievessen I, Schliess F, Schulz WA. Activities of MAP-kinase pathways in normal uroepithelial cells and urothelial carcinoma cell lines. Exp Cell Res. 2003;282:48–57. doi: 10.1006/excr.2002.5647. [DOI] [PubMed] [Google Scholar]
- 8.Brandau S, Bohle A. Bladder cancer: I. Molecular and genetic basis of carcinogenesis. Eur Urol. 2001;39:491–497. doi: 10.1159/000052494. [DOI] [PubMed] [Google Scholar]
- 9.Orntoft TF, Wolf H. Molecular alterations in bladder cancer. Urol Res. 1998;26:223–233. doi: 10.1007/s002400050050. [DOI] [PubMed] [Google Scholar]
- 10.Knowles MA. The genetics of transitional cell carcinoma: progress and potential clinical application. BJU Int. 1999;84:412–427. doi: 10.1046/j.1464-410x.1999.00217.x. [DOI] [PubMed] [Google Scholar]
- 11.Grubbs CJ, Lubet RA, Koki AT, Leahy KM, Masferrer JL, Steele VE, Kelloff GJ, Hill DL, Seibert K. Celecoxib inhibits N-butyl-N-(4-hydroxybutyl)-nitrosamine-induced urinary bladder cancers in male B6D2F1 mice and female Fischer-344 rats. Cancer Res. 2000;60:5599–5602. [PubMed] [Google Scholar]
- 12.Grubbs CJ, Moon RC, Squire RA, Farrow GM, Stinson SF, Goodman DG, Brown CC, Sporn MB. 13-cis-Retinoic acid: inhibition of bladder carcinogenesis induced in rats by N-butyl-N-(4-hydroxybutyl)-nitrosamine. Science. 1977;198:743–744. doi: 10.1126/science.910158. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa K, Uzvolgyi E, St John MK, de Oliveira ML, Arnold L, Cohen SM. Frequent p53 mutations and occasional loss of chromosome 4 in invasive bladder carcinoma induced by N-butyl-N-(4-hydroxybutyl)nitrosamine in B6D2F1 mice. Mol Carcinog. 1998;21:70–79. doi: 10.1002/(sici)1098-2744(199801)21:1<70::aid-mc9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto S, Chen T, Murai T, Mori S, Morimura K, Oohara T, Makino S, Tatematsu M, Wanibuchi H, Fukushima S. Genetic instability and p53 mutations in metastatic foci of mouse urinary bladder carcinomas induced by N-butyl-N-(4-hydroxybutyl)nitrosamine. Carcinogenesis. 1997;18:1877–1882. doi: 10.1093/carcin/18.10.1877. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Huang HY, Pak J, Cheng J, Zhang ZT, Shapiro E, Pellicer A, Sun TT, Wu XR. p53 deficiency provokes urothelial proliferation and synergizes with activated Ha-ras in promoting urothelial tumorigenesis. Oncogene. 2004;23:687–696. doi: 10.1038/sj.onc.1207169. [DOI] [PubMed] [Google Scholar]
- 16.Zhang ZT, Pak J, Huang HY, Shapiro E, Sun TT, Pellicer A, Wu XR. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene. 2001;20:1973–1980. doi: 10.1038/sj.onc.1204315. [DOI] [PubMed] [Google Scholar]
- 17.??el-Marjou A, Delouvee A, Thiery JP, Radvanyi F. Involvement of epidermal growth factor receptor in chemically induced mouse bladder tumour progression. Carcinogenesis. 2000;21:2211–2218. doi: 10.1093/carcin/21.12.2211. [DOI] [PubMed] [Google Scholar]
- 18.Yao R, Wang Y, Lubet RA, You M. Differentially expressed genes associated with mouse lung tumor progression. Oncogene. 2002;21:5814–5821. doi: 10.1038/sj.onc.1205422. [DOI] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 21.Kroft SH, Oyasu R. Urinary bladder cancer: mechanisms of development and progression. Lab Invest. 1994;71:158–174. [PubMed] [Google Scholar]
- 22.Theodorescu D, Cornil I, Fernandez BJ, Kerbel RS. Overexpression of normal and mutated forms of HRAS induces orthotopic bladder invasion in a human transitional cell carcinoma. Proc Natl Acad Sci USA. 1990;87:9047–9051. doi: 10.1073/pnas.87.22.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vageli D, Kiaris H, Delakas D, Anezinis P, Cranidis A, Spandidos DA. Transcriptional activation of H-ras, K-ras and N-ras protooncogenes in human bladder tumors. Cancer Lett. 1996;107:241–247. doi: 10.1016/0304-3835(96)04372-8. [DOI] [PubMed] [Google Scholar]
- 24.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 25.Theodorescu D, Cornil I, Sheehan C, Man MS, Kerbel RS. Ha-ras induction of the invasive phenotype results in up-regulation of epidermal growth factor receptors and altered responsiveness to epidermal growth factor in human papillary transitional cell carcinoma cells. Cancer Res. 1991;51:4486–4491. [PubMed] [Google Scholar]
- 26.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y. Dbl family guanine nucleotide exchange factors. Trends Biochem Sci. 2001;26:724–732. doi: 10.1016/s0968-0004(01)01973-9. [DOI] [PubMed] [Google Scholar]
- 28.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 29.Seraj MJ, Harding MA, Gildea JJ, Welch DR, Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- 30.Gildea JJ, Seraj MJ, Oxford G, Harding MA, Hampton GM, Moskaluk CA, Frierson HF, Conaway MR, Theodorescu D. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- 31.Joneson T, Bar-Sagi D. Suppression of Ras-induced apoptosis by the Rac GTPase. Mol Cell Biol. 1999;19:5892–5901. doi: 10.1128/mcb.19.9.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 33.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 34.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 35.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 36.Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;424:1009–1013. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- 37.Paggi MG, Baldi A, Bonetto F, Giordano A. Retinoblastoma protein family in cell cycle and cancer: a review. J Cell Biochem. 1996;62:418–430. doi: 10.1002/(SICI)1097-4644(199609)62:3%3C418::AID-JCB12%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 38.Paggi MG, Giordano A. Who is the boss in the retinoblastoma family? The point of view of Rb2/p130, the little brother. Cancer Res. 2001;61:4651–4654. [PubMed] [Google Scholar]
- 39.Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 40.Nicolas E, Ait-Si-Ali S, Trouche D. The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein. Nucleic Acids Res. 2001;29:3131–3136. doi: 10.1093/nar/29.15.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 42.Chen GC, Guan LS, Yu JH, Li GC, Choi Kim HR, Wang ZY. Rb-associated protein 46 (RbAp46) inhibits transcriptional transactivation mediated by BRCA1. Biochem Biophys Res Commun. 2001;284:507–514. doi: 10.1006/bbrc.2001.5003. [DOI] [PubMed] [Google Scholar]
- 43.Guan LS, Li GC, Chen CC, Liu LQ, Wang ZY. Rb-associated protein 46 (RbAp46) suppresses the tumorigenicity of adenovirustransformed human embryonic kidney 293 cells. Int J Cancer. 2001;93:333–338. doi: 10.1002/ijc.1338. [DOI] [PubMed] [Google Scholar]
- 44.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 45.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 46.Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA. 2003;100:8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 48.Yan S, Krebs S, Leister KJ, Wenner CE. Perturbation of EGF-activated MEK1 and PKB signal pathways by TGF-beta1 correlates with perturbation of EGF-induced cyclin D1 and DNA synthesis by TGF beta1 in C3H 10T1/2 cells. J Cell Physiol. 2000;185:107–116. doi: 10.1002/1097-4652(200010)185:1<107::AID-JCP10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 50.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]