Abstract
We investigated the effects of fission yeast replication genes on telomere length maintenance and identified 20 mutant alleles that confer lengthening or shortening of telomeres. The telomere elongation was telomerase dependent in the replication mutants analyzed. Furthermore, the telomerase catalytic subunit, Trt1, and the principal initiation and lagging-strand synthesis DNA polymerase, Polα, were reciprocally coimmunoprecipitated, indicating these proteins physically coexist as a complex in vivo. In a polα mutant that exhibited abnormal telomere lengthening and slightly reduced telomere position effect, the cellular level of the Trt1 protein was significantly lower and the coimmunoprecipitation of Trt1 and Polα was severely compromised compared to those in the wild-type polα cells. Interestingly, ectopic expression of wild-type polα in this polα mutant restored the cellular Trt1 protein to the wild-type level and shortened the telomeres to near-wild-type length. These results suggest that there is a close physical relationship between the replication and telomerase complexes. Thus, mutation of a component of the replication complex can affect the telomeric complex in maintaining both telomere length equilibrium and telomerase protein stability.
The chromosome ends of almost all eukaryotes are capped by telomeres to ensure the integrity of chromosomes and to maintain the overall genome stability. Telomeres consist of simple DNA repeats, which associate with proteins to protect chromosomes from end-to-end fusion, degradation, and inappropriate recombination. Telomeric DNA consists of tandem arrays of short repeats rich in G residues on the strand that runs towards the 3′ end of the chromosome. The telomeric tracts of all organisms are of heterogeneous length, ranging from 28 bases in some ciliates to over 50 kb in mice. However, in a given cell type, the telomeric DNA length is kept within a certain size range. A common structural feature of telomeres from different organisms is a single-stranded G-rich overhang. This G-rich overhang structure and the proteins that bind to it are thought to play an important role in mediating chromosome integrity, since in its absence, chromosome loss and fusion rates are elevated (14, 32, 43, 49, 67, 72, 75, 77).
Telomeres are also essential for the complete replication of eukaryotic chromosomes. DNA synthesis by DNA polymerases has a 5′-to-3′ polarity (38). At the replication fork, the leading-strand synthesis is thought to initiate once and then proceeds continuously. The lagging-strand synthesis is discontinuous throughout and requires repeated initiations by polymerase α (Polα) and primase (9, 12, 73). Due to the polarity of DNA synthesis, the telomeric G-rich strand is synthesized by the leading-strand replication machinery, and the C-rich strand is synthesized by the lagging-strand replication machinery. Removal of the terminal RNA primer by nucleases leaves an 8- to 12-nucleotide gap at the 5′ end of the newly replicated DNA that cannot be refilled by conventional DNA replication, resulting in an end replication dilemma (44, 74). The end replication problem is potentially resolved by telomerase, an unusual reverse transcriptase. The catalytic subunit of telomerase (Trt1) contains an integral RNA molecule with a small template domain that is utilized by telomerase as template to add telomeric repeats onto the 3′ end of the telomere (57). Although telomerase activity is required to maintain the steady-state length of telomeric DNA repeats at the chromosome end, other proteins have been shown to affect telomere length maintenance. In Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast), proteins that bind either directly to telomeric sequences or to telomeric sequence-binding proteins have been shown to positively or negatively influence telomere length (10, 11, 27, 43, 57). In addition, mutations in several checkpoint genes in fission or budding yeast and in Caenorhabditis elegans have been shown to affect telomere length (3, 22, 42, 48, 51, 54, 66).
Mutations of components of budding yeast DNA replication machinery such as POL1 (Polα) or the large subunit of replication factor C (CDC44/RFC1) confer telomere elongation, whereas a rad27 deletion confers destabilized telomere length (1, 15, 60). Moreover, in a POL1 mutant (pol1-17), the telomeric position effect (TPE) is reduced concomitantly with telomere elongation (2). Recent data have shown that fission yeast Polα mediates recruitment of Swi6 to heterochromatin, including that of telomeres to establish silencing (4). By using a novel in vivo assay, it has been shown that telomerase-mediated telomere addition requires the activities of Polα, Polδ, and primase in budding yeast (23). In budding yeast, Cdc13p plays an important role in protecting and maintaining telomeric DNA (31, 56). Budding yeast Cdc13p interacts with the catalytic subunit of DNA Polα (Pol1p) by two-hybrid analysis criteria. Point mutations in either CDC13 or POL1 not only reduce the interaction but also affect telomere length (64). Aphidicolin, an inhibitor of Polα and Polδ, causes abnormal lengthening of the G- and C-strand heterogeneity of telomeres of the ciliate Euplotes (28), and Euplotes telomerase has been shown to physically associate with primase, a component of the lagging-strand machinery (65). Together, these studies suggest that telomere replication by telomerase requires coordination of C- and G-strand syntheses.
Fission yeast is evolutionarily distant from budding yeast. The chromosome organizations of fission yeast, such as centromere structure (18, 19), the replication origin structure (24), and chromosome segregation (5), are more similar to those of mammalian cells than are those of budding yeast. Much less is known about fission yeast telomeres than about their budding yeast counterparts. Fission yeast telomeres consist of about 300 bp of repeat units, which includes a consensus sequence of 5′-TTACAG1-8-3′ on the strand that runs towards the 3′ end (25, 68). To investigate whether telomere length maintenance in fission yeast is similar to that in the evolutionarily distant budding yeast, we have analyzed the telomere length of a large panel of replication mutants. We report here that telomere length is altered in all of the replication mutants analyzed. Using the principal initiation and lagging-strand synthesis DNA polymerase (Polα) as a representative of replication proteins, we show here that Polα and the telomerase catalytic subunit, Trt1, coexist in a complex in vivo. Analysis of the telomere length and the Trt1 protein level in a polα mutant reveals a close physical association between the replication complex and the telomerase complex in S. pombe. Mutations in components of the replication complex, such as Polα, could affect the telomeric complex in maintaining telomere length and telomerase protein stability.
MATERIALS AND METHODS
Fission yeast strains and growth conditions.
All genetic operations were performed as described previously (34). To construct the spp2-9 trt1Δ double mutant strain, the CF248 diploid strain (h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 ade6-M210/ade6-M216 trt1+/trt1::his3+) was sporulated, and the spores were germinated on Edinburgh minimal medium (EMM) (50) lacking histidine to select for trt1::his3+. This strain was crossed with an h− spp2-9 strain (69), which had grown for 100 generations, to generate the spp2-9 trt1Δ mutant. The spp2-9 trt1Δ double mutant was verified by temperature sensitivity and its ability to grow on media lacking uracil and histidine. Tetrads were dissected and germinated on YES (yeast extract plus supplements). The polαts13 strain [pKAN1-C-myc9trt1+ (kanMX Cmyc9trt1+)] (hereafter the polαts13 myc-trt1+ strain) was made by crossing the h+ polαts13 strain (13), which had been grown for 100 generations, and the CF830 (h− leu1-32 ade6-M210 ura4-D18 his3-D1 trt1::his3+ [pKAN1-C-myc9trt1+ (kanMX Cmyc9trt1+)] strain (36). The polαts13 myc-trt1+ mutant strain was verified by Western blotting with anti-myc antibody, temperature sensitivity, and Southern blot analysis of telomere length. The polαts13 ura-tel strain was constructed by crossing the h− polαts13 strain, which had been grown for 100 generations and the FY1872 strain (h90 leu1-32 ade6-210 his3D1 ura4DS/E ade6OTRsph ura4-tel) and was verified by PCR with primers 5′-TGAGGGGATGAAAAATCCCATTG-3′ and 5′-TTCGACAACAGGATTAC-GACCAG-3′ directed against the ura4+gene. The polαts13 strain [pKAN1-Cmyc9trt1+ (kanMX Cmyc9trt1+)] [pART-polα+ (ura4+ polα+)] (hereafter the polαts13 myc-trt1+ + polα+ strain) was constructed by transforming the polαts13 myc-trt1+ strain with pART-polα+, followed by selection for expression of polα+ by shifting the temperature to 36°C to rescue the temperature sensitivity of the polαts13 myc-trt1+ strain. The CF830 (polα+ myc-trt1+), polαts13 myc-trt1+, and polαts13 myc-trt1+ + polα+ strains were grown in yeast extract with G418 (0.1 mg/ml) at 25°C to maintain the episomal plasmid pKAN1-C-myc9trt1+; all other strains were grown in YES at the indicated temperatures.
Detection of telomeres by Southern blotting.
Genomic DNA was isolated by a glass bead-phenol protocol and analyzed on agarose gels. Agarose gels were stained with ethidium bromide (EtBr) and photographed to ensure that equal amounts of DNA were loaded. A 2.4-kb ApaI fragment of unique S. pombe genomic DNA (22) was also used as a loading control. Detection of telomeres by Southern blotting was carried out as described previously (22).
G-strand overhang assay.
To detect G-strand overhangs, a nondenaturing hybridization assay was used (72). Oligonucleotide probes 5′-GGGTTACAGGTTACAGGGTTAC-3′ (G-specific oligonucleotide) and 5′-GTAACCCTGTAACCTGTAACCC-3′ (C-specific oligonucleotide) were designed based on reference 20. The C- and G-strand-specific probes were end labeled with [γ-32P]ATP (3,000 Ci/mmol; Amersham) by T4 polynucleotide kinase to identical specific activities (9,160 cpm/pmol for the C-strand probe and 9,220 cpm/pmol for the G-strand probe). Labeled probes were purified on MicroSpin G-25 columns (Amersham Pharmacia Biotech). Four microliters of labeled probe (8 nM) was used for each reaction to hybridize to 5 μg of ApaI-restricted genomic DNA (from cells grown for 100 generations). The labeled probes were added to a final volume of 25 μl and then incubated for 12 to 15 h at 50°C. Hybridized samples were separated on a 1.5% agarose gel, stained with EtBr to ensure equal loading, dried, and autoradiographed.
Preparation of protein extracts from S. pombe.
A total of 3 × 108 cells were harvested, washed once in ice-cold stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA [pH 8.0], 1 mM NaN3), and then resuspended in 25 μl of lysis buffer (150 mM HEPES [pH 7.9], 300 mM KCl, 1 mM EDTA, 10% glycerol), with the addition of 1 tablet of Roche Complete Protease Inhibitor Cocktail per 25 ml of buffer, and lysed by vortexing with glass beads. Soluble protein extracts were prepared by centrifugation at 4°C for 10 min, and the protein concentration was determined by the Bradford assay.
Immunoprecipitation and Western blotting.
Five hundred micrograms to 1 mg of total protein was solubilzed in 1 ml of lysis buffer containing 0.2% RNase inhibitor RNasin (Promega) and 0.1% NP-40. Anti-c-myc (9E10) antibody was preadsorbed onto protein G plus/protein A agarose beads (Oncogene Research). Affinity-purified anti-Polα antibody (61) was conjugated with CNBr-activated Sepharose beads according to the manufacturer's instructions. Ten microliters of a 50% slurry of antibody-labeled beads was added to the protein extract, and this mixture was incubated for 2 to 3 h at 4°C with end-over-end rotation. Immunocomplexes were washed four times with lysis buffer and resuspended in 30 μl of sodium dodecyl sulfate (SDS)-sample buffer. Ten microliters was analyzed on SDS-polyacrylamide gel electrophoresis (PAGE) gels (8% polyacrylamide) and then transferred to polyvinylidene difluoride membranes. The membrane was blocked in 5% dry milk powder in phosphate-buffered saline (PBS) and 0.2% Tween 20. Anti-Polα antibody at a 1:4,000 dilution, anti-myc antibody at a 1:500 dilution, and antitubulin antibody at a 1:5,000 dilution were used as the primary antibodies. A rabbit anti-chicken secondary antibody at a 1:1,000 dilution was used for detecting anti-Polα immunoglobulin Y (IgY), and a goat anti-mouse secondary antibody at a 1:3,000 dilution was used for detecting anti-myc. Proteins were detected by the ECL enhanced chemiluminescence system (New England Nuclear). Five milligrams of EtBr per ml was added to the myc-trt1 cell extracts prior to immunoprecipitation.
Cell synchronization.
The myc-trt1+ strain was grown at 25°C to a cell density of 6 × 106 cells per ml before hydroxyurea (HU) was added to a final concentration of 12 mM. The cells were incubated in HU for 4 h and then released into media without HU. Cell samples of 50 ml were taken every 15 min. One milliliter of the cell sample was fixed with ethanol. A total of 3 × 106 fixed cells were stained with propidium iodide for flow cytometry analysis (62). The rest of the cells (49 ml) were used for protein extract preparation.
Telomere position effect.
The polα+ (ura4-tel) (FY1872), polαts13 (ura4-tel), and polαts13 (ura4−) strains were grown to 5 × 106 cells per ml at 25°C. Serial dilutions were applied as spots to PM + adenine-histidine-leucine-uracil (AHLU) (complete), PM + AHL (ura−), and PM + AHLU + 5-fluoroorotic acid (5-FOA) plates and grown at 25°C for 4 days or until colonies appeared.
Ectopic expression of polα+.
Full-length polα+ was expressed from pART-polα+ in the polαts13 myc-trt1+ strain. The myc-trt1+ (CF830), polαts13 myc-trt1, and polαts13 myc-trt1+ + polα+strains were grown at 25°C to 6 × 106 cells per ml, and the cultures were then divided into two halves: one-half was shifted to 36°C to select for cells sustained by pART-polα+, while the other half continued culturing at 25°C. Cell samples were taken at 6 and 15 h after shifting the temperature to 36°C for protein extract and DNA preparation. Western blotting and telomere length analyses were performed as described above.
RESULTS
Telomere length in fission yeast replication mutants is deregulated.
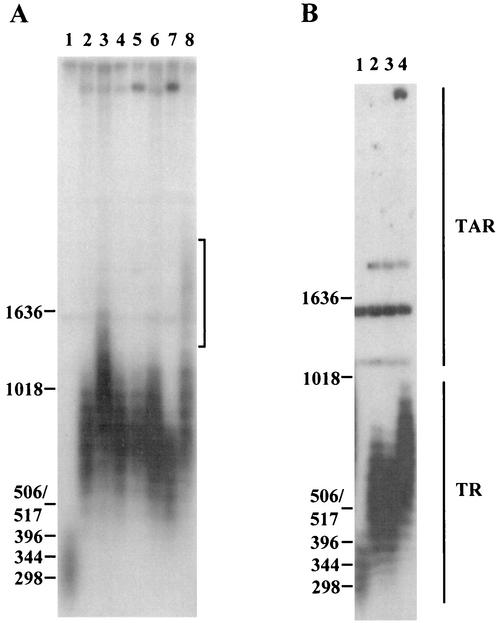
We analyzed the telomere length of 20 mutant alleles of replication genes that are involved in DNA replication. Since there is a typical phenotypic lag of telomere length regulation (22, 45), each mutant strain was grown for ∼100 generations at its respective semipermissive temperature in liquid media. Many of the mutant strains analyzed in the experiments had been propagated for many generations prior to this study, and consequently they had already reached their steady-state telomere lengths before 100 generations of subculture. We chose to routinely analyze strains after 100 generations of subculture and compared them to their respective isogenic parental strains. We first analyzed mutants with mutation of primase and polα that are essential for initiation of Okazaki fragments. All five mutants with mutation of the primase catalytic subunit, spp1 (33), exhibited an average of 400- to 600-bp extensions of telomeres compared to their parental wild-type strains (Fig. 1A, compare lanes 2 to 6 to lane 1). Out of these spp1 mutants, the spp1-9 mutant had the longest telomeres (Fig. 1A, lane 3). The two polα mutants (13) displayed differences in their telomere length after subculture for 100 generations at 25°C. The polαts11 mutant had telomeres about 270 bp longer than those of the wild type (Fig. 1A, lane 7), while the polαts13 mutant had a majority of telomeres 550 bp longer than those of the wild type (Fig. 1A, compare lanes 1 and 8). Longer exposure of the gel revealed that a minor population of the telomeres in the polαts13 strain were ∼1,600 bp longer than wild-type telomeres (appearing as a weak smear in Fig. 1A, lane 8). Of the three spp2 mutants, the mutant containing the gene encoding the primase coupling subunit, spp2-9 (69), exhibited the longest telomeres: ∼300 bp longer than those of the wild type (Fig. 1B, lane 4). These results show that the extent of telomere lengthening in primase and polα mutants is allele dependent.
FIG. 1.
Analysis of telomere length in polα and primase mutants. polα and spp1 and spp2 (genes coding for S. pombe primase subunits 1 and 2, respectively) mutants were subcultured for ∼100 generations at their respective semipermissive temperature (polα and spp1 mutants, 25°C; spp2 mutants, 33°C) before harvest. Genomic DNA was restricted with ApaI and separated on 1.2% agarose gels, transferred to Hybond N+ filters, and probed with a 1.9-kb ApaI fragment of pEN42 (55). The positions of DNA size markers and their sizes in base pairs are given on the left. Brackets indicate the weak smear seen in the polαts13 mutant. TR, terminal telomeric repeats; TAR, telomere-associated repeat sequences. (A) Lanes: 1, KG2 (wild type); 2, spp1-4; 3, spp1-9; 4, spp1-14; 5, spp1-19; 6, spp1-21; 7, polαts11; 8, polαts13. (B) Lanes: 1, KG2 (wild type); 2, spp2-7; 3, spp2-8; 4, spp2-9.
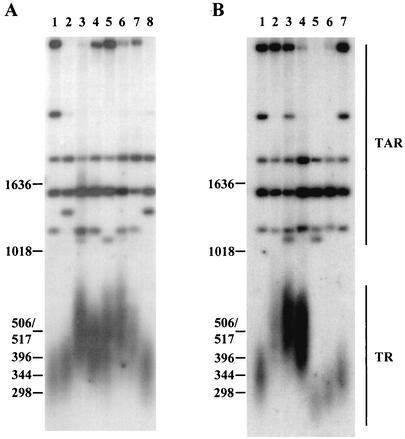
To test whether the aberrant lengthening of telomeres also occurs in replication mutants that are required for elongation of Okazaki fragments and leading strand synthesis, we analyzed the telomere lengths of five polδ mutants (30, 58). All five polδ mutants had on average 100- to 150-bp-longer telomeres than their respective isogenic wild-type strains (Fig. 2A, lanes 3 to 7). Furthermore, mutants with defects in two small subunits of polδ, cdc1 and cdc27 (46), exhibited telomere lengths about 180 bp longer than those of the wild type (Fig. 2B, lanes 2 and 3). Similar to polα and polδ mutants, a mutant with a ligase mutation, cdc17-K42, had telomeres about 160 bp longer than those of the wild-type strain (Fig. 2B, lane 4). Although the exact role of Polɛ in replication is not yet clear, similar to a recent observation in S. cerevisiae (59), a polɛ mutant, the cdc20-M10 strain, exhibited shorter telomeres than the wild-type strain (Fig. 2B, lane 5). A strain with deletion of rad2, encoding a nuclease required for Okazaki fragment maturation (73), also had shorter telomeres than those of the wild-type strain (Fig. 2B, lane 6). Thus, mutations of genes encoding fission yeast replication proteins induce deregulation of telomere length.
FIG. 2.
Analysis of telomere length in replication mutants. Mutants with mutation of polδ subunits (cdc1 and cdc27), ligase (cdc17), polɛ (cdc20), and rad2 were grown at their respective semipermissive temperatures (indicated in parentheses after each mutant) for ∼100 generations. DNA was analyzed as described in the legend to Fig. 1. (A) Lanes: 1, 972 (wild type); 2, SP808 (wild type); 3, polδts1 (25°C); 4, polδts2 (25°C); 5, polδts3 (30°C); 6, cdc6-23 (25°C); 7, cdc6-121 (30°C); 8, SP808. (B) Lanes: 1, 972 (wild type); 2, cdc1-7 (25°C); 3, cdc27-K3 (30°C); 4, cdc17-K42 (30°C); 5, cdc20-M10 (33°C); 6, rad2.d (30°C); 7, 972 (wild type).
Since the two primase subunit spp1-9 and spp2-9 mutants and the polα and polαts13 mutants exhibited substantially longer telomeres than the wild type, we further analyzed whether the telomeric extension in these mutant strains is a G- or C-strand extension as described in Materials and Methods. With C- or G-strand-specific probes of identical radioactive specific activities, the results suggest that the telomere elongation seen in these replication mutants is an extension of the G strand (data not shown).
Mutation at a polα mutant allele, polαts13, affects the silencing at telomere loci.
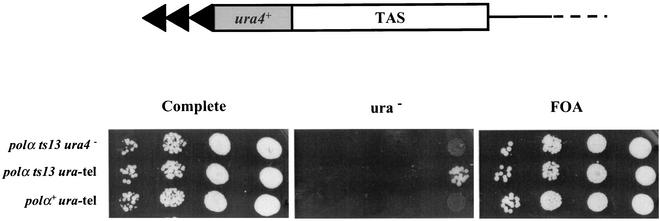
It is known that mutation of genes that causes a telomere length phenotype could also affect the TPE. In S. cerevisiae, the pol1-17 allele causes elongation of telomeres and reduces the TPE (2). We therefore investigated the possible effect of the polαts13, spp1-9, and spp2-9 mutations on silencing at the telomere locus. The polαts13 ura4-tel strain with the ura4+ gene placed at a telomere locus was constructed as described in Materials and Methods. The polαts13 ura4-tel mutant was able to grow on media lacking uracil, indicating a slight reduction of silencing at the telomere locus (Fig. 3). This result corroborates a recent report that showed an interaction between Polα and Swi6, an element required for silencing. In a thermosensitive polα mutant (the swi7-H4 strain), the interaction was impaired and the silencing was defective (4). In contrast, the spp1-9 and spp2-9 mutations do not affect the TPE (data not shown).
FIG. 3.
The telomere position effect is slightly reduced in the polαts13 mutant. Shown is the organization of the telomere region on chromosome II where the ura4+ marker is inserted between the telomere repeats (TR) and the telomere-associated repeat sequences (TAS). Serial dilutions of the polα+ ura4-tel, polαts13 ura4-tel, and polαts13 ura4− mutants were applied as spots on complete, ura− and FOA plates.
The telomere elongation in the polα and primase mutants is telomerase dependent.
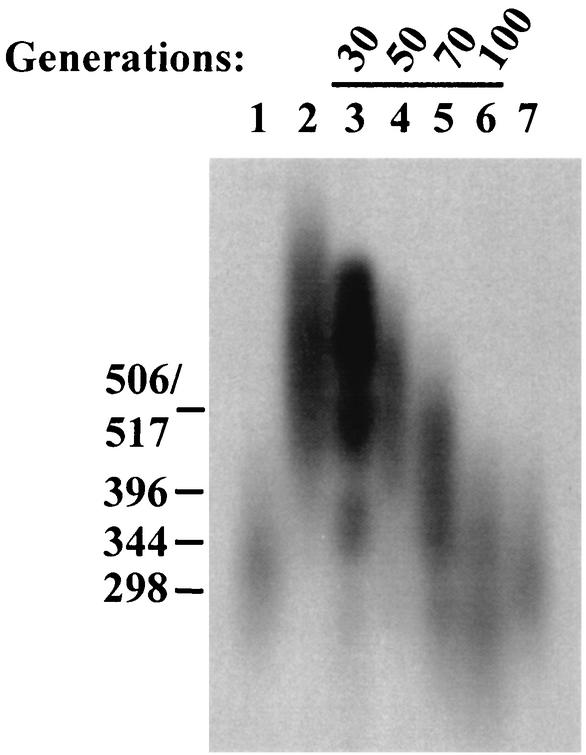
We then investigated whether the telomere length extension occurring in these replication mutants is mediated by telomerase. Attempts were made to generate polαts13 (13) or primase subunit spp1-9 (33) and spp2-9 (69) double mutants in the telomerase catalytic subunit (trt1Δ) deletion background (16, 52, 53) as described in Materials and Methods. However, both random sporulation and crossing followed by tetrad analysis failed to generate spp1-9 trt1Δ or polαts13 trt1Δ double mutants. Mating of the trt1Δ strain with either the spp1-9 or polαts13 mutant strain resulted in one to two spores per tetrad, with none of them being the desired double mutant. Hence, both the spp1-9 and polαts13 strains have a strong synthetic interaction with deletion of trt1+. In contrast, the spp2-9 trt1Δ double mutant was successfully generated. The mutant spp2-9 trt1Δ double mutant was cultured in liquid medium at the semipermissive temperature (33°C) up to 100 generations, and its telomere length was compared to the telomere lengths of the wild type and the spp2-9 single mutant. The telomere length of the spp2-9 single mutant after subculture for 100 generations was about 300 bp longer than that of the wild type (Fig. 4, compare lane 2 to lane 1). After growing for 30, 50, 70, and 100 generations, the spp2-9 trt1Δ mutant exhibited progressively shorter telomere lengths (Fig. 4, lanes 3 through 6). After 100 generations of growth, the telomere length was even shorter than that of the parental wild-type strain (Fig. 4, compare lane 6 to lanes 1 and 7). This result strongly suggests that the telomere lengthening seen in the spp2-9 mutant requires the action of telomerase. By extending the interpretation of our results, the telomere lengthening in the polα and primase mutants may be also mediated by telomerase.
FIG. 4.
Telomere shortening in an spp2-9 trt1::his3+ double mutant is telomerase dependent. The spp2-9 mutant cells (grown for ∼100 generations at the semipermissive temperature [33°C) were crossed with a haploid trt1::his3+ strain as described in Materials and Methods. An spp2-9 trt1::his3+ colony was grown in liquid medium until the total number of progeny reached 5 × 108 cells. Assuming cell death to be insignificant, this is equivalent to ∼30 cell divisions since the meiotic segregation. Numbers in parentheses in lanes 3 through 6 correspond to the number of cell divisions after the meiotic segregation. Lanes: 1, 972 strain (wild type); 2, spp2-9 (100 generations); 3, spp2-9 trt1::his3+, 30 generations; 4, 50 generations; 5, 70 generations; 6, 100 generations; 7, 972 strain (wild type).
Polα and Trt1 physically interact in vivo.
Studies of S. cerevisiae and Euplotes have suggested a coordination of replication of the two strands at the telomere (23, 28, 47, 65). Our results indicate that Trt1 is responsible for the aberrant telomere extension in the polα and primase mutants (Fig. 4). Furthermore, polαts13 and trt1Δ strains showed a strong synthetic phenotype. These data bring to mind a physical interaction between Polα and telomerase. To test this possibility, coimmunoprecipitation experiments were performed with a myc-tagged trt1+ strain ectopically expressed at a moderate level from its own promoter in a trt1Δ background (CF830) (36), and a wild-type nontagged trt1 strain as a negative control. An ∼130-kDa myc-tagged Trt1 protein, but no protein of the size of Polα, was detected when cell extracts prepared from the myc-trt1+ strain were probed with the anti-myc monoclonal antibody 9E10 (Fig. 5A, lane 1). When the same cell extract was probed with polyclonal anti-Polα antibody (61), three protein species of Polα were detected—a major band of 180 kDa and minor amounts of 165 and 155 kDa, which are degraded forms of Polα (61): however, no protein band of the same size as myc-tagged Trt1 was detected (Fig. 5A, lane 2). These results indicate that the anti-myc and anti-Polα antibodies do not cross-react with Polα and myc-Trt1, respectively.
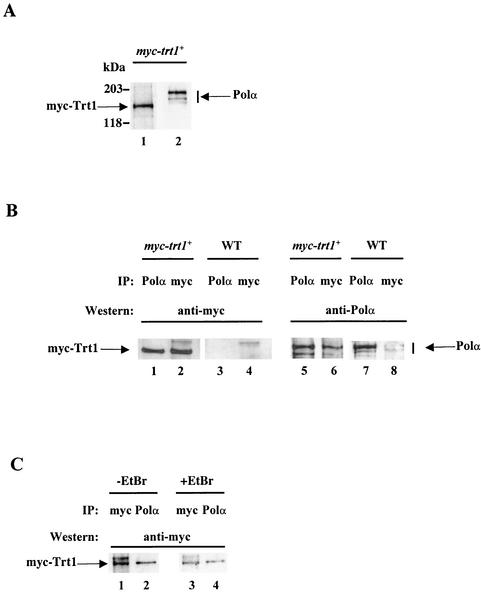
FIG. 5.
Coimmunoprecipitation of myc-Trt1 and Polα. (A) Antibodies against Polα and c-myc do not cross-react. Protein extracts from the myc-trt1+ strain CF830 were Western blotted with anti-c-myc antibody (lane 1) and anti-Polα antibody (lane 2). (B) Coimmunoprecipitation of myc-Trt1 and Polα. Protein extracts from the myc-trt1+ strain and the nontagged wild-type strain (WT) were immunoprecipitated with anti-Polα antibody or anti-c-myc antibody, followed by transfer to membranes. Membranes were probed with either anti-c-myc antibody (lanes 1 through 4) or anti-Polα antibody (lanes 5 through 8). (C) Coimmunoprecipitation of Trt1 and Polα is not due to association with nucleic acid. EtBr (+EtBr) was added to the myc-trt1+ extract at 5 mg/ml prior to immunoprecipitation (lanes 3 and 4). EtBr-treated extracts and nontreated extracts (−EtBr) were immunoprecipitated with anti-Polα antibody or anti-c-myc antibody. The membrane was probed with anti-c-myc antibody.
When immunoprecipitating Polα with anti-Polα from myc-trt1+ cell extracts and probing with anti-myc antibodies, a myc-Trt1 protein was detected (Fig. 5B, lane 1). As a control, myc-Trt1 was detected from the same cell extracts when immunoprecipitated and probed with anti-myc antibodies (Fig. 5B, lane 2). No myc-Trt1 was detected in immunoprecipitates made from the nontagged wild-type strain (Fig. 5B, lane 3 and 4). Reciprocally, Polα was detected in the anti-myc immunoprecipitates from the myc-trt1+ strain when probed with anti-Polα antibody (Fig. 5B, lane 6). To verify that this was indeed Polα, the anti-Polα immunoprecipitate from the same cell extract was probed with anti-Polα antibody. It exhibited proteins of the size of Polα (Fig. 5B, lane 5). Anti-Polα-antibodies detected Polα in anti-Polα immunoprecipitates from lysates of the nontagged wild-type trt1 strain (Fig. 5B, lane 7), whereas no Polα could be detected in the anti-myc immunoprecipitates from the same cell extract (Fig. 5B, lane 8). Thus, Trt1 and Polα can be reciprocally coimmunoprecipitated by these antibodies.
To ascertain that the coimmunoprecipitation of Trt1 and Polα was not caused by a random nonspecific coassociation with DNA or RNA fragments in the cell extract, EtBr was added to the myc-trt1+ cell extract prior to immunoprecipitation. myc-Trt1 was detected in the anti-Polα-immunoprecipitates from cell extracts with or without EtBr added (Fig. 5C, lanes 2 and 4). Thus, the coimmunoprecipitation of Trt1 and Polα was not due to nonspecific random association of these proteins with nucleic acids in the cell extract. Together, these results strongly suggest that telomerase and DNA Polα physically interact in vivo.
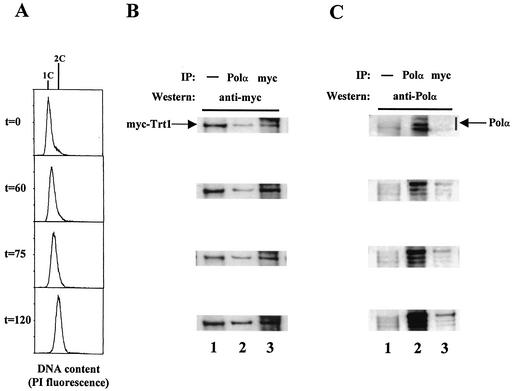
We then investigated whether the association of Trt1 and Polα is cell cycle dependent. The myc-trt1+ strain was arrested in early S phase with HU at 25°C for 4 h and then released into fresh medium to continue growing at 25°C. After release from the HU arrest, the cells took approximately 60 min to recover and progress through S phase. Cell samples were removed at 0, 60, 75, and 120 min after release from the HU block for flow cytometry analysis (Fig. 6A) and for preparation of protein extracts for coimmunoprecipitation experiments (Fig. 6B and C). Cells arrested in HU for 4 h at 25°C displayed a 1C DNA profile (Fig. 6A, t = 0 min). The cells gradually shifted towards a greater-than-1C fluorescence-activated cell sorter (FACS) profile, and after 120 min, cells had a 2C DNA profile, indicating that cells had completed S phase and entered the G2 phase (t = 120 min). Since S. pombe has a very abbreviated G1 phase, no G1-phase cells are observed in the 120-min cell sample. Throughout S phase and in G2 phase, myc-Trt1 was detectable in the anti-Polα immunoprecipitates (Fig. 6B, lane 2) and the anti-myc immunoprecipitates (Fig. 6B, lane 3). Polα was effectively detected by the anti-Polα antibody in the anti-Polα immunoprecipitate (Fig. 6C, lane 2). Furthermore, Polα coimmunoprecipitated with myc-Trt1 as cells progressed through S phase and entered the G2 phase, although the level was low at t = 0 min (Fig. 6C, lane 3, t = 0, 60, 75, and 120 min). These experiments indicate that Trt1 and Polα interact throughout S and G2 phases of the cell cycle.
FIG. 6.
myc-Trt1 and Polα interact in the S and G2 phases. The myc-trt1+ strain was incubated with HU for 4 h at 25°C, and the cells were then released from the HU block. (A) Flow cytometry analysis. Cell samples were removed every 15 min for FACS analysis and protein extract preparation. After 4 h of HU arrest, immediately before release, a cell sample was taken (t = 0). The cells at t = 0 have a 1C content. After 60 min (t = 60), the peak has started to move towards 2C content. After 75 min (t = 75), the cells have a 1.5C content. Finally, after 120 min, the cells have a 2C content corresponding to G2-phase cells. (B) myc-Trt1 in Polα immunoprecipitates. Cell extracts were prepared from samples released from the HU block at the times indicated and subjected to Western blotting with anti-c-myc antibody (lane 1), immunoprecipitated with anti-Polα or anti-c-myc antibody, and probed with anti-c-myc antibody (lanes 2 and 3). (C) Polα in anti-myc immunoprecipitates. The study was performed as described for panel B, but the membrane was probed with anti-Polα antibody.
The interaction between Polα and Trt1 is compromised in a polα mutant exhibiting long telomeres.
To test the status of interaction between Trt1 and Polα in a polα mutant (the polαts13 strain) that exhibited abnormally long telomeres (Fig. 1A, lane 8), we constructed a strain containing the polαts13 allele and myc-trt1+ and analyzed the interaction of these two proteins. As expected, myc-Trt1 was detected in anti-myc immunoprecipitates (Fig. 7A, lane 1) and also coimmunoprecipitated with Polα (Fig. 7A, lane 2), and Polα was detected in anti-Polα immunoprecipitates from cell lysates of the myc-trt1+ (polα+) strain (Fig. 7A, lane 3). In contrast, although myc-Trt1 was immunoprecipitated by identical amounts of anti-myc antibody from myc-trt1+ (polα+) (Fig. 7A, lane 1) and polαts13 myc-trt1+ cell extracts (Fig. 7B, lane 1), no detectable myc-Trt1 was found in the anti-Polα immunoprecipitates from the polαts13 myc-trt1+ cell extract (Fig. 7B, lane 2). This is not due to the failure of anti-Polα antibody to immunoprecipitate the mutant Polα protein, because mutant Polα protein was readily detectable in the anti-Polα immunoprecipitates from polαts13 myc-trt1+ cell extract (Fig. 7B, lane 3).
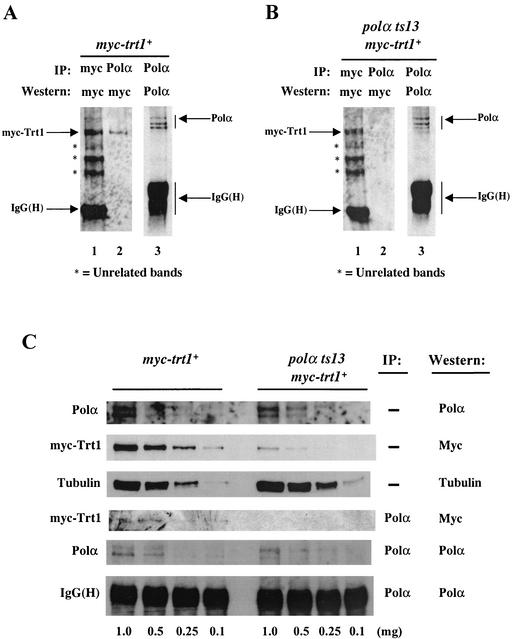
FIG. 7.
The interaction between myc-Trt1 and Polα is compromised in a polαts13 background. The myc-trt1+ (polα+) strain and the polαts13 myc-trt1+ mutant strain were grown at 25°C. Cells were harvested, and protein extracts were prepared. Asterisks indicate three unrelated bands, which were used as loading controls. (A) Coimmunoprecipitation of myc-Trt1 from myc-trt1+ (polα+) cells. Extracts from the myc-trt1+ cells were immunoprecipitated (IP) with anti-c-myc antibody (lane 1) or anti-Polα antibody (lanes 2 and 3) and probed with anti-c-myc antibody. (B) Coimmunoprecipitation of myc-Trt1 from polαts13 myc-trt1+ cells. Extracts from polαts13 myc-trt1+ cells were immunoprecipitated with anti-c-myc antibody (lane 1) or anti-Polα antibody (lanes 2 and 3) and probed with anti-c-myc antibody. (C) Comparison of the myc-Trt1 and Polα protein levels and their efficiency at coimmunoprecipitation in myc-trt1+ (polα+) and polαts13 myc-trt1+ cells. Cell extracts were diluted to 1, 0.5, 0.25, and 0.1 mg of total protein extract of myc-trt1+ (polα+) and polαts13 myc-trt1+ cells. One percent of each diluted extract was loaded onto SDS gels followed by Western blotting with anti-Polα antibody to determine the level of Polα proteins or anti-myc antibody to determine the level of myc-Trt1 protein. Polα proteins were immunoprecipitated with anti-Polα antibody from the same diluted extracts as in the three top lanes: myc-trt1+ (polα+) and polαts13 myc-trt1+. Ten microliters of anti-Polα immunoprecipitates was loaded onto SDS gel followed by Western blotting with anti-Polα antibody to determine the levels of Polα protein and with anti-myc antibody for coimmunoprecipitation of Polα and myc-Trt1.
To investigate why the coimmunoprecipitation of myc-Trt1 was compromised in the polαts13 myc-trt1+ mutant, we tested whether protein levels of Polα or myc-Trt1 were altered in the polαts13 myc-trt1+ mutant. Cell extracts of 0.1 to 1 mg of protein from the myc-trt1+ (polα+) and polαts13 myc-trt1+ strains were Western blotted to determine the levels of the Polα and myc-Trt1 proteins (Fig. 7C, top two panels). Comparable levels of Polα protein were present in both myc-trt1+ (polα+) cells and the polαts13 myc-trt1+ mutant (Fig. 7C, first panel). Surprisingly, the level of myc-Trt1 protein was severely reduced in the polαts13 myc-trt1+ mutant (Fig. 7C, second panel). The level of myc-Trt1 in polαts13 myc-trt1+ mutant cell extract was about 10% of the myc-Trt1 level in the myc-trt1+ (polα+) cell extract: compare the myc-Trt1 protein level in 0.1 mg of myc-trt1+ (polα+) cell extract with the myc-Trt1 protein level in 1 mg of polαts13 myc-trt1+ cell extract. We then analyzed the interaction of myc-Trt1 and Polα in these cell extracts. With comparable levels of Polα protein immunoprecipitated from myc-trt1+ (polα+) and polαts13 myc-trt1+ cells, myc-Trt1 was detectable in the anti-Polα immunoprecipitates from myc-trt1+ (polα+) cell extracts, but not in polαts13 myc-trt1+ extracts. Furthermore, with comparable levels of myc-Trt1 presented in 0.1 mg of myc-trt1+ (polα+) extract and in 1 mg of polαts13 myc-trt1+ cell extract, myc-Trt1 protein was again detectable only in the anti-Polα-immunoprecipitates from 0.1 mg of myc-trt1+ (polα+) cell extract, but not from 1 mg of polαts13 myc-trt1+ cell extract (Fig. 7C, panels 4 and 5). Moreover, although 1 mg of polαts13 myc-trt1+ cell extract had a much higher level of Polα protein than 0.1 mg of myc-trt1+ (polα+) cell extract (Fig. 7C, panel 1), myc-Trt1 was only detectable in the anti-Polα-immunoprecipitates from myc-trt1+ (polα+) cell extract and not in those from polαts13 myc-trt1+ extract. These results suggest that the polαts13 mutation not only causes an abnormal telomere lengthening, but also severely reduces the cellular Trt1 protein level and compromises the ability of Trt1 and Polα to physically coexist as a complex in vivo.
Ectopic expression of polα+ restores myc-Trt1 level and shortens telomeres in the polαts13 mutant.
As shown above in Fig. 7C, the myc-Trt1 protein level in the polαts13 myc-trt1+ mutant strain was significantly lower than that in the myc-trt1 (polα+) strain. It is possible that the mutant Polα in the polαts13 strain might affect the stability of Trt1 in the Polα-Trt1-containing protein complex, causing Trt1 to become unstable. To test this hypothesis, we transformed pART-polα+ into the polαts13 myc-trt1+ strain (hereafter the polαts13 myc-trt1+ + polα+ strain) to express the wild-type polα gene. The myc-trt1+(polα+), polαts13 myc-trt1+, and polαts13 myc-trt1+ + polα+ strains were cultured at 25°C to early log phase and then shifted to 36°C for 6 h to select for cells sustained by the ectopic expression of polα+. At 25°C, when there is no selection for pART-polα+, most cells have probably lost the plasmid, and the myc-Trt1 level is comparable to that of polαts13 myc-trt1+ strain (Fig. 8A, lane 2 and 3). The polαts13 myc-trt1+ strain without the ectopic expression of polα+ is viable at 36°C for 6 h, but then starts to gradually lose viability, whereas the polαts13 myc-trt1+ strain transformed with pART-polα+ is viable. After 6 h at 36°C, the myc-Trt1 level is increased in the polαts13 myc-trt1+ + polα+ strain compared to that in the polαts13 myc-trt1+ strain (Fig. 8A, compare lanes 5 and 6). After 15 h at 36°C, the level of myc-Trt1 in the polαts13 myc-trt1+ + polα+ strain is comparable to that of the myc-trt1+ (polα+) strain expressing wild-type Polα (Fig. 8B, compare lanes 1 and 2), with equal amounts of protein extracts analyzed, as shown by the tubulin levels used as the control. These experiments indicate that ectopic expression of polα+ is able to restore the myc-Trt1 level in the polαts13 myc-trt1+ strain, suggesting that Polα affects the physical status of Trt1.
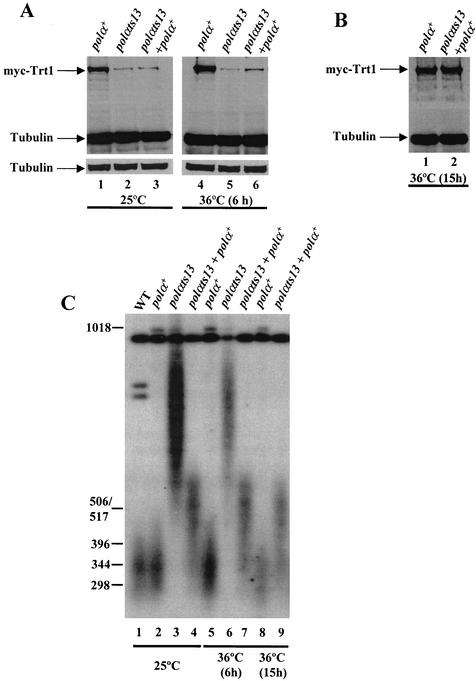
FIG. 8.
Ectopic expression of polα+ in the polαts13 mutant can restore the myc-Trt1 protein and telomere length to near-wild-type level. (A) Ectopic expression of polα+ increases the cellular myc-Trt1 protein in polαts13 cells. Prior to the experiment, the polαs13 myc-trt1+ + polα+ (polαts13 + polα+) transformants had been grown at 36°C to select for expression of polα+. The myc-trt1+ (polα+), polαts13 myc-trt1+ (polαts13), and polαs13 myc-trt1+ + polα+ (polαts13 + polα+) strains were grown at 25°C to a cell density of 6 × 106 cells per ml (lanes 1 through 3) and then shifted to 36°C and continued to culture for 6 h (lanes 4 through 6). (B) Ectopic expression of polα+ restores myc-Trt1 to the wild-type level. The myc-trt1+ (polα+) and polαs13 myc-trt1+ + polα+ (polαts13 + polα+) cells were grown at 36°C for 15 h, and cell extracts were analyzed as described for panel A. (C) Ectopic expression of polα+ reduces telomere length in the polαts13 mutant. DNA was isolated from the same cell extracts as in panels A and B for analysis of the telomere lengths by Southern blotting. Lanes: 1, KG2 (wild type); 2, myc-trt1+ (polα+) (25°C); 3, polαts13 myc-trt1+ (polαts13) (25°C); 4, polαs13 myc-trt1+ + polα+ (polαts13 + polα+) (first selected at 36°C and then shifted to 25°C); 5, myc-trt1+ (polα+) (36°C, 6 h); 6, polαts13 myc-trt1+ (polαts13) (36°C, 6 h); 7, polαs13 myc-trt1+ + polα+ (polαts13 + polα+) (36°C, 6 h); 8, myc-trt1+ (polα+) (36°C, 15 h); 9, polαs13 myc-trt1+ + polα+ (polαts13 + polα+) (36°C, 15 h).
To test whether ectopic expression of polα+ also affects the telomere length in the polαts13 myc-trt1+ strain, genomic DNA was prepared from the same strains shown in Fig. 8A and B, and the telomere lengths were analyzed by Southern blotting. Interestingly, ectopic expression of a functional polα+ gene rapidly restored the telomere length to near-wild-type length in the polαts13 myctrt1+ mutant (Fig. 8C). These results indicate that Polα plays a significant role in maintaining the Trt1 protein level and also the telomere length of cells, supporting the finding of a close physical association between Polα and Trt1 in vivo.
DISCUSSION
In this study, we analyzed telomeres of a large panel of fission yeast replication mutants and found telomere length abnormalities in all mutants analyzed (Fig. 1 and 2). Analysis of one of the replication mutants, the primase spp2-9 mutant, shows that the abnormal telomere lengthening is telomerase dependent (Fig. 4), inferring that this may also be the case for other replication mutants. Importantly, we show that Polα associates with Trt1 in a complex in vivo and that the status of Polα influences telomere length as well as the cellular level of the Trt1 protein (Fig. 5, 7, and 8). Below, we discuss these findings and their possible effects on the cell biology.
Do replication proteins of fission yeast and budding yeast affect telomere length differently?
In this study, we analyzed 20 replication mutants for their telomere length (Fig. 1 and 2). Among these mutants analyzed, mutation of ligase (cdc17-K42) induces abnormal telomere lengthening (Fig. 2B, lane 4). In contrast to the fission yeast ligase mutant (cdc17-K42), an S. cerevisiae mutant with a ligase mutation (cdc9) has telomeres of wild-type length (2). In addition, deletion of fission yeast rad2 (S. pombe homolog of Fen1/RAD27), which is essential for maturation of Okazaki fragments (9, 73), induces telomere shortening (Fig. 2B, lane 6). In budding yeast, deletion of RAD27 (the homolog of S. pombe rad2+) causes destabilization of telomeres but not a shortening of telomeres (60). The differences seen in telomere length regulation of ligase and of rad2+/RAD27 mutants between the two yeasts might simply be due to allele specificity. On the other hand, they might reflect a difference in the effect of replication proteins on telomere length regulation between the two yeasts.
In budding yeast, telomerase-mediated telomere addition requires Polα, primase and Polδ, but not Polɛ (23). Although the exact role of Polɛ in replication is not yet clear (9, 12, 73), mutation of polɛ (cdc20-M10) in both fission yeast and budding yeast, similar to fission yeast rad2, induced shortening of telomeres (Fig. 2B, lane 5) (59). This suggests that in fission yeast, polɛ+ and rad2+ regulate telomere length through a different mechanism from that of the polα+, polδ+, and ligase genes. Since Polα, primase, and Polδ are all thought to be involved in lagging-strand synthesis, a different kind of telomere length regulation seen in the polɛ mutant may reflect a unique and distinct role of Polɛ in replication.
How might the replication complex influence the telomere homeostasis?
Studies of telomeres in the ciliate Euplotes (28, 63, 65) and budding yeast (1, 2, 15, 23, 27, 47, 64) have led to the proposal that G-strand extension and C-strand synthesis require a coordinated regulation. Since C-strand synthesis requires the lagging-strand replication proteins, it is reasonable to assume that synthesis of the two telomeric strands also requires a tight coordination of the lagging-strand replication complex and the telomerase-containing telomeric complex (27). Studies of budding yeast have shown that Pol1p (Polα) interacts with Cdc13p by two-hybrid analysis; mutations in an N-terminal region of Pol1p abolish the interaction and result in longer telomeres (64). Cdc13p also interacts with Est1p (64), which also associates with the telomerase RNA (26, 40, 56). This interaction chain implies that Cdc13p might recruit both telomerase and Polα to the telomeres in budding yeast; hence Cdc13p may be a key linchpin in telomere homeostasis.
In this study, we have shown that the Trt1 and Polα proteins coexist in a complex in vivo (Fig. 5), and the physical and/or functional status of Polα significantly affects the telomerase protein stability and telomere length maintenance (Fig. 7 and 8). It is not yet known whether the two proteins directly interact or whether other proteins mediate the interaction. One S. pombe protein, Taz1, has been identified as a telomere binding protein (21, 29). Taz1 is an ortholog of human telomere repeat binding factors (TRFs). Taz1 is neither a structural nor a functional homolog of Cdc13p (20, 39). Another telomere end binding protein, Pot1, was recently identified in fission yeast and humans (10, 11). It is not yet clear whether Pot1 is an ortholog of Cdc13 and whether Pot1 is a linchpin of the interaction between the lagging strand replication complex component Polα and Trt1. Thus, the coordination of G- and C-strand syntheses in S. pombe may be mediated by a yet-to-be-identified protein. It is also possible that the coordination of the synthesis of G and C strands in fission yeast is by a direct interaction between the Polα-containing lagging-strand replication complex and the telomerase-containing telomeric complex.
The polαts13 mutant at 25°C has a growth rate similar to that of the wild type; however, polαts13 cells exhibit a slight cdc phenotype with a normal nuclear morphology (13), indicating that 25°C is the semipermissive temperature of this mutant. Moreover, at 25°C, the polαts13 mutant exhibits an elevated mutation rate (41); hence, at this temperature, Polα is semidysfunctional. Here we showed that at 25°C, the polαts13 mutant allele induced an aberrant lengthening of telomeres (Fig. 1), a mild reduction of TPE (Fig. 3), a severely compromised interaction with Trt1 (Fig. 7B), and a significant decrease in the cellular Trt1 protein level (Fig. 7B and C). It is possible that a semidysfunctional Polα adversely affects the coordination between the lagging strand replication complex and the telomerase complex. The impaired physical coordination between these two protein complexes would make Trt1 prone to degradation, resulting in a decrease in the cellular Trt1 level, and a mild reduction in silencing at the telomere loci due to a change of the heterochromatin structure at the telomere. In support of this hypothesis, ectopic expression of wild-type polα at the restrictive temperature can progressively restore the cellular level of Trt1 protein and reduce telomere length to near-wild-type length (Fig. 8). However, the recovery rates of Trt1 protein level and the telomere length induced by ectopic expression of polα+ in the polαts13 are not perfectly correlated. This could be due to a delayed telomere effect. Furthermore, the telomere length also seemed to have reached a new steady-state level, almost as short as the wild-type telomere length (Fig. 8C). This telomere-shortening phenomenon was already seen at the first time point after establishment of the polα+ transformants (Fig. 8C, lane 4) and is most likely due to the preselection of polα+ transformants at 36°C. The fact that telomeres do not quite revert to wild-type length could also be due to an altered extension of heterochromatin in the region near the telomere during the previous phase when the telomeres were abnormally long. If this were the case, it would require many generations to restore the chromatin near telomeres to the original state. Nonetheless, these results suggest that the Polα protein plays a significant role in maintaining telomeric complex stability and telomere homeostasis.
There are two paradoxical findings in our studies. First, Polα and Trt1 interact in early S phase (Fig. 6). At this phase of the cell cycle, no telomere replication presumably takes place. Budding yeast telomere addition at the de novo end occurs in M phase, although telomerase activity can be measured in vitro in both G1- and M-phase cells (23). The finding that Polα and Trt1 coimmunoprecipitate at low levels throughout S phase suggests that a fraction of Polα may constitutively associate with telomerase during the entire S phase. This also implies that another essential factor or factors required for the telomerase-mediated telomere addition may be associating with telomerase in a cell cycle-dependent manner (23).
The second paradox is how the polαts13 myc-trt1+ mutant, with a significantly reduced level of Trt1 protein, can exhibit lengthened telomeres. A likely explanation is that cells might have excess amounts of potentially active telomerase, which is usually tightly regulated by the coordination between the telomerase complex, the replication complex, and/or perhaps other proteins. In the polαts13 mutant, a dysfunctional Polα might perturb the tight coordination of these proteins in regulating the telomerase-mediated telomere addition. Furthermore, a perturbed telomeric protein complex in the polαts13 mutant may allow the chromosome ends to be more accessible to extension by low levels of residual telomerase in an aberrant manner (27). Thus, it is possible that the abnormal telomere lengthening observed in the polαts13 strain is a consequence of perturbation of the organization of protein complexes at the telomere end.
Mutations of replication proteins not only contribute to telomere dysfunction but also promote genomic instability.
Telomere dysfunction due to the absence of functional telomerase has been shown to increase mutation rate, impair DNA repair, enhance ionizing radiation sensitivity, and induce genomic instability in tumorigenesis (7, 17, 35, 70, 76). We have previously reported that specific mutations of S. pombe polα, polδ, primase (spp1 and spp2), and ligase (cdc17-K42) confer a mutator phenotype characterized by deletion of sequences flanked by short direct repeats and small sequence alterations (41). Deletion of the fission yeast rad2+ gene, similar to deletion of the homologous gene in budding yeast, RAD27 (71), induces duplication of sequences flanked by short direct repeats (41). Comparison of the panel of replication mutant alleles that confer a mutator phenotype to those that exhibit telomere length alterations shows that all of the replication mutants (especially those implicated in lagging-strand synthesis) that confer a mutator phenotype also exhibit telomere length deregulation (41) (Fig. 1 and 2). However, some replication mutant alleles, such as those in the two polδ mutants (polδts2 and cdc6-23), and a polɛ mutant (cdc20-M10), exhibit deregulation of telomere length, but do not display a mutator phenotype (41). These results suggest that the replication genes contribute to telomeric complex stability as well as genomic mutation avoidance in an allele-specific manner. The underlying mechanisms in these mutants causing a mutator phenotype may also differ from those causing telomere deregulation. Studies of budding yeast est1Δ cells have suggested that telomerase contributes to inhibition of chromosome instability (35). Studies of mammalian cells and rodent models without telomerase RNA have shown that telomerase dysfunction in certain genetic contexts can facilitate cancer development by compromising chromosome integrity (6-8, 17, 37, 76). In this study, we have shown that mutations of replication proteins can affect the homeostasis of telomeres. Using Polα as a paradigm, we demonstrate that mutation of Polα has a significant effect on telomere length maintenance and telomerase protein levels. It is not yet known to what extent the observed mutator phenotypes induced by replication mutants are indirectly contributed by a telomere homeostasis abnormality. Nonetheless, the results of this study and our previous studies (41) suggest that mutations of replication genes, especially those whose products are involved in the lagging-strand synthesis, could have a major impact on the overall genomic stability.
Acknowledgments
We thank members of the Wang laboratory for helpful discussion and Thomas R. Cech for providing us the CF830 and CF248 strains and Robin Allshire for providing the FY1872 strain. We especially thank Toru Nakamura, Peter Baumann, and Steven Artandi for valuable advice during the course of the study.
This study was supported by grants from National Cancer Institute of the National Institutes of Health to T.S.W and grants from the Swedish Cancer Fund (2163-B00-11XAC) and the Swedish Research Council (K2002-31X-14197-01A) to P.S. M.D. was a predoctoral fellow supported by the Sweden-America Foundation, the Swedish Institute, and Lennander's Foundation.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams Martin, A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, S., and J. Hodgkin. 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403:159-164. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed, S., S. Saini, S. Arora, and J. Singh. 2001. Chromodomain protein Swi6-mediated role of DNA polymerase α in establishment of silencing in fission yeast. J. Biol. Chem. 276:47814-47821. [DOI] [PubMed] [Google Scholar]
- 5.Allshire, R. C. 1995. Elements of chromosome structure and function in fission yeast. Semin. Cell Biol. 6:55-64. [DOI] [PubMed] [Google Scholar]
- 6.Artandi, S. E. 2002. Telomere shortening and cell fates in mouse models of neoplasia. Trends Mol. Med. 8:44-47. [DOI] [PubMed] [Google Scholar]
- 7.Artandi, S. E., S. Chang, S. L. Lee, S. Alson, G. J. Gottlieb, L. Chin, and R. A. DePinho. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406:641-645. [DOI] [PubMed] [Google Scholar]
- 8.Artandi, S. E., and R. A. DePinho. 2000. Mice without telomerase: what can they teach us about human cancer? Nat. Med. 6:852-855. [DOI] [PubMed] [Google Scholar]
- 9.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 10.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 11.Baumann, P., E. Podell, and T. R. Cech. 2002. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 13.Bhaumik, D., and T. S.-F. Wang. 1998. Mutational effect of fission yeast Polα on cell cycle events. Mol. Biol. Cell 9:2107-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackburn, E. H. 1991. Structure and function of telomeres. Nature 350:569-573. [DOI] [PubMed] [Google Scholar]
- 15.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 16.Cech, T. R., T. M. Nakamura, and J. Lingner. 1997. Telomerase is a true reverse transcriptase. A review. Biochemistry (Moscow) 62:1202-1205. [PubMed] [Google Scholar]
- 17.Chin, L., S. E. Artandi, Q. Shen, A. Tam, S. L. Lee, G. J. Gottlieb, C. W. Greider, and R. A. DePinho. 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97:527-538. [DOI] [PubMed] [Google Scholar]
- 18.Clarke, L. 1990. Centromeres of budding and fission yeasts. Trends Genet. 6:150-154. [DOI] [PubMed] [Google Scholar]
- 19.Clarke, L., M. Baum, L. G. Marschall, V. K. Ngan, and N. C. Steiner. 1993. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harbor Symp. Quant. Biol. 58:687-695. [DOI] [PubMed] [Google Scholar]
- 20.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 21.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828-831. [DOI] [PubMed] [Google Scholar]
- 22.Dahlen, M., T. Olsson, G. Kanter-Smoler, A. Ramne, and P. Sunnerhagen. 1998. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol. Biol. Cell 9:611-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerase α and δ. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 24.Dubey, D. D., S. M. Kim, I. T. Todorov, and J. A. Huberman. 1996. Large, complex modular structure of a fission yeast DNA replication origin. Curr. Biol. 6:467-473. [DOI] [PubMed] [Google Scholar]
- 25.Duffy, M., and A. Chambers. 1996. DNA-protein interactions at the telomeric repeats of Schizosaccharomyces pombe. Nucleic Acids Res. 24:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans, S. K., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 28.Fan, X., and C. M. Price. 1997. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell 8:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusion. Mol. Cell 7:55-63. [DOI] [PubMed] [Google Scholar]
- 30.Francesconi, S., H. Park, and T. S.-F. Wang. 1993. Fission yeast with DNA polymerase δ temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 21:3821-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths, D. J. F., V. F. Liu, P. Nurse, and T. S.-F. Wang. 2001. Role of fission yeast primase catalytic subunit in the replication checkpoint. Mol. Biol. Cell 12:115-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. C. King (ed.), Handbook of genetics 1, vol. I. Plenum Press, New York, N.Y.
- 35.Hackett, J. A., D. M. Feldser, and C. W. Greider. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106:275-286. [DOI] [PubMed] [Google Scholar]
- 36.Haering, C. H., T. M. Nakamura, P. Baumann, and T. R. Cech. 2000. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 97:6367-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan, D. 2000. Benefits of bad telomeres. Nature 406:573-574. [DOI] [PubMed] [Google Scholar]
- 38.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 39.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471-483. [DOI] [PubMed] [Google Scholar]
- 40.Lin, J. J., and V. A. Zakian. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93:13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, V. F., D. Bhaumik, and T. S.-F. Wang. 1999. Mutator phenotype induced by aberrant replication. Mol. Cell. Biol. 19:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longhese, M. P., V. Paciotti, H. Neecke, and C. Lucchini. 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics 155:1577-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundblad, V. 2000. DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res. 451:227-240. [DOI] [PubMed] [Google Scholar]
- 44.Lundblad, V. 1997. The end replication problem: more than one solution. Nat. Med. 3:1198-1199. [DOI] [PubMed] [Google Scholar]
- 45.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacNeill, S. A., S. Moreno, N. Reynolds, P. Nurse, and P. A. Fantes. 1996. The fission yeast cdc1 protein, a homologue of the small subunit of DNA polymerase δ, binds to Pol3 and cdc27. EMBO J. 15:4613-4628. [PMC free article] [PubMed] [Google Scholar]
- 47.Marcand, S., V. Brevet, C. Mann, and E. Gilson. 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10:487-490. [DOI] [PubMed] [Google Scholar]
- 48.Matsuura, A., T. Naito, and F. Ishikawa. 1999. Genetic control of telomere integrity in Schizosaccharomyces pombe: rad3(+) and tel1(+) are parts of two regulatory networks independent of the downstream protein kinases chk1(+) and cds1(+). Genetics 152:1501-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 50.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 51.Naito, T., A. Matsuura, and F. Ishikawa. 1998. Circular chromosome formation in a fission yeast mutant defective on ATM homologues. Nat. Genet. 2:203-206. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomere. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nimmo, E. R., G. Cranston, and R. C. Allshire. 1994. Telomere-associated chromosome breakage in fission yeast in variegated expression of adjacent genes. EMBO J. 13:3801-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 57.Nugent, C. I., and V. Lunblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 58.Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167-178. [DOI] [PubMed] [Google Scholar]
- 59.Ohya, T., Y. Kawasaki, S. Hiraga, S. Kanbara, K. Nakajo, N. Nakashima, A. Suzuki, and A. Sugino. 2002. The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 277:28099-28108. [DOI] [PubMed] [Google Scholar]
- 60.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park, H., S. Francesconi, and T. S.-F. Wang. 1993. Cell cycle expression of two replicative DNA polymerases α and δ from Schizosaccharomyces pombe. Mol. Biol. Cell 4:145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 63.Price, C. 1999. Telomeres and telomerase: broad effects on cell growth. Curr. Opin. Genet. Dev. 9:218-224. [DOI] [PubMed] [Google Scholar]
- 64.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and telomerase-associated Est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 65.Ray, S., Z. Karamysheva, L. Wang, D. E. Shippen, and C. M. Price. 2002. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol. Cell. Biol. 22:5859-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandell, L. L., D. E. Gottschling, and V. A. Zakian. 1994. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl. Acad. Sci. USA 91:12061-12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugawara, N. 1989. Ph.D. thesis. Harvard University, Cambridge, Mass.
- 69.Tan, S., and T. S.-F. Wang. 2000. Analysis of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol. Cell. Biol. 20:7853-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiagalingam, S., S. Laken, J. K. Willson, S. D. Markowitz, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2001. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. USA 98:2698-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 72.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 73.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 74.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 75.Wellinger, R. J., and D. Sen. 1997. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33:735-749. [DOI] [PubMed] [Google Scholar]
- 76.Wong, K. K., S. Chang, S. R. Weiler, S. Ganesan, J. Chaudhuri, C. Zhu, S. E. Artandi, K. L. Rudolph, G. J. Gottlieb, L. Chin, F. W. Alt, and R. A. DePinho. 2000. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 26:85-88. [DOI] [PubMed] [Google Scholar]
- 77.Zakian, V. A. 1995. ATM-related genes: what do they tell us about functions of the human gene? Cell 82:685-687. [DOI] [PubMed] [Google Scholar]