Abstract
The candidate tumor-suppressor gene hyaluronidase 2 (HYAL2) encodes a glycosylphosphatidylinositol-anchored cell-surface protein that serves as an entry receptor for jaagsiekte sheep retrovirus, a virus that causes contagious lung cancer in sheep that is morphologically similar to human bronchioloalveolar carcinoma. The viral envelope (Env) protein alone can transform cultured cells, and we hypothesized that Env could bind and sequester the HYAL2 receptor and thus liberate a potential oncogenic factor bound and negatively controlled by HYAL2. Here we show that the HYAL2 receptor protein is associated with the RON receptor tyrosine kinase (also called MST1R or Stk in the mouse), rendering it functionally silent. In human cells expressing a jaagsiekte sheep retrovirus Env transgene, the Env protein physically associates with HYAL2. RON liberated from the association with HYAL2 becomes functionally active and consequently activates the Akt and mitogen-activated protein kinase pathways leading to oncogenic transformation of immortalized human bronchial epithelial cells. We find activated RON in a subset of human bronchioloalveolar carcinoma tumors, suggesting RON involvement in this type of human lung cancer.
The hyaluronidase 2 (HYAL2, also called LUCA2) gene resides in a critical 120-kb region of chromosome 3p21.3 that contains tumor suppressor genes involved in the origin or development of lung and other human malignancies (1). The gene encodes a 473-aa protein that is expressed in all human tissues analyzed including lung (1). HYAL2 is a member of a large family of hyaluronoglucosaminidases (EC 3.2.1.35) but exhibits very low hyaluronidase activity (refs. 2 and 3 and V. Vigdorovich and A.D.M., unpublished results). Recently we showed that HYAL2 is a glycosylphosphatidylinositol-anchored cell-surface protein and serves as a receptor for jaagsiekte sheep retrovirus (JSRV) cell entry (3, 4). In sheep JSRV causes a contagious form of lung cancer that arises from epithelial cells in the lower airway including type II alveolar and bronchiolar epithelial cells (5). In a recent study it was shown that antiserum directed against the JSRV capsid protein crossreacted with 30% of human pulmonary adenocarcinoma samples but not with normal lung tissue or adenocarcinomas from other tissues (6). Collectively these findings support the possibility (7) of a viral etiology for some human lung cancers, particularly the bronchioloalveolar adenocarcinoma (BAC) type, which is morphologically very similar to the sheep tumors (5, 8).
Although there was no known oncogene in JSRV, the JSRV envelope (Env) protein was shown by us (3) and others (9) to transform rodent fibroblast cell lines. The cytoplasmic tail of Env, in particular a YXXM protein motif that is a putative docking site for phosphatidylinositol 3-kinase (PI3-kinase) after tyrosine phosphorylation, is important for fibroblast transformation, which seems to be mediated through the PI3-kinase/Akt pathway (10, 11). Transformation has not been explored yet in epithelial cells, the target for JSRV disease, and the mechanism of transformation might be different compared with that in fibroblasts. We hypothesized an alternative model in which the Env protein could sequester the HYAL2 receptor in the cytoplasm and thus liberate a potential oncogenic factor bound and negatively controlled by HYAL2 in normal bronchial epithelial cells. Here we provide evidence for this model and document a pathway for epithelial cell transformation that involves HYAL2 interactions with the RON receptor tyrosine kinase.
Materials and Methods
Expression Vectors.
Plasmid pHA-Jenv was made by inserting sequences encoding a preprotrypsin signal peptide, a hemagglutinin tag, and JSRV Env amino acids 73–615 into the pCR3.1 expression vector (Invitrogen). Plasmid pJenv-V5 was made by inserting the entire coding sequence of JSRV Env into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen). Both of these plasmids also confer resistance to G418. The pFlag-HYAL2 construct has been described (3). The human wild-type RON cDNA was cloned into the pCI-neo vector (Promega). Kinase-dead RON (RONkd) was obtained by mutagenesis as described (12), and this cDNA and an enhanced GFP (EGFP) cDNA were cloned into the pMSCV-puro vector (CLONTECH) to make pMSCV-puro-RONkd and pMSCV-puro-EGFP, respectively.
Cell Lines.
BEAS-2B cells were a gift from Curtis C. Harris (National Cancer Institute) and were grown in LHC-8 medium (13). Human embryonic kidney 293 cells (HEK293), lung adenocarcinoma (NCI-H2122), lung alveolar carcinoma (SW-1573), and BAC cell lines (NCI-H358, NCI-H650, NCI-H1650, and NCI-H1781) were from American Type Culture Collection. The RE7 cell line [Madin–Darby canine kidney (MDCK) cells stably expressing human RON] has been described (14). HEK293, lung cancer, and RE7 cell lines were cultured in DMEM containing 10% FBS.
Antibodies.
The antibodies used were anti-RON (C-20) rabbit polyclonal sc-322 and anti-Akt goat polyclonal sc-1618 from Santa Cruz Biotechnology; anti-HYAL2 C-terminal rabbit polyclonal from Research Genetics (Huntsville, AL); anti-mitogen-activated protein kinase (MAPK) monoclonal M12321 from Transduction Laboratories (Lexington, KY); anti-phospho-MAPK monoclonal 9106 and anti-phospho-Akt rabbit polyclonal 9271 from New England Biolabs; anti-Flag M2 monoclonal 05-447 and anti-PY monoclonal (clone 4G10) 05-321 from Upstate Biotechnology (Lake Placid, NY); anti-V5 mouse monoclonal R960-25 from Invitrogen; anti-hemagglutinin rabbit polyclonal antibody 3808-1 from CLONTECH; and rhodamine conjugated goat anti-rabbit secondary antibody 31670 from Pierce. Nonspecific affinity-purified rabbit antibodies were used in preliminary experiments to determine the specificity of all our rabbit affinity-purified antibodies.
BEAS-2B Cell Transfection.
Lipofectamine 2000 (Invitrogen) was used for transfection according to manufacturer protocol. Cells (80% confluent) grown in six-well plates were first overlaid with 2 ml per well of Opti-MEM (Invitrogen) for 20 min and then incubated for 6 h with the DNA–Lipofectamine 2000 complexes (500 μl per well). These were prepared by mixing 5 μg of plasmid DNA, 250 μl of Opti-MEM, and 250 μl of LHC-8 medium and incubated for 20 min at room temperature before transfection. After transfection the mix was removed and replaced by 2 ml of LHC-8. The next day cells were collected from each well and seeded into 10-cm dishes. One day after seeding the cells were treated with selective antibiotics (1 mg/ml G418 or 1 μg/ml puromycin). Under these conditions nontransfected cells died within 5–7 days, whereas stably transfected cells grew as foci in 2–3 weeks, finally replacing the original BEAS-2B cells.
Analysis of Signaling Pathways in Transiently Transfected HEK293 and RE7 Cells by Immunoprecipitation and Western Blotting.
HEK293 and RE7 cells were transiently transfected with pFlag-HYAL2 and/or pJenv-V5 plasmids by using Lipofectamine 2000 according to manufacturer protocol. Forty-eight hours after transfection the cells were starved overnight in medium without serum and lysed in lysis buffer (50 mM Hepes, pH 7.4/150 mM NaCl/10% glycerol/1 mM EDTA/1 mM sodium orthovanadate/10 mM sodium pyrophosphate/100 mM NaF/1% Triton X-100/10 μg/ml leupeptin/10 units/ml aprotinin/1 mM PMSF). Insoluble material was removed by centrifugation. Molecules of interest were immunoprecipitated from the cleared lysates by appropriate antibodies added to lysates together with protein G-agarose. After overnight incubation at 4°C, immunoprecipitates were washed three times with HNTG buffer (50 mM Hepes/150 mM NaCl/0.1% Triton X-100/10% glycerol), 2× SDS/PAGE sample buffer was added, and the samples were boiled for 5 min. Proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes. Protein bands on the membrane were visualized with appropriate antibodies by standard Western blotting procedures. For detection of MAPK and Akt activation, total-cell lysates were used for SDS/PAGE and Western blotting. For some experiments, to visualize the Env–HYAL2 complex formed intracellularly, cells were treated with N-acetyl-Leu-Leu-norleucinal (ALLN; Sigma), an inhibitor of proteasomal enzymes.
Results and Discussion
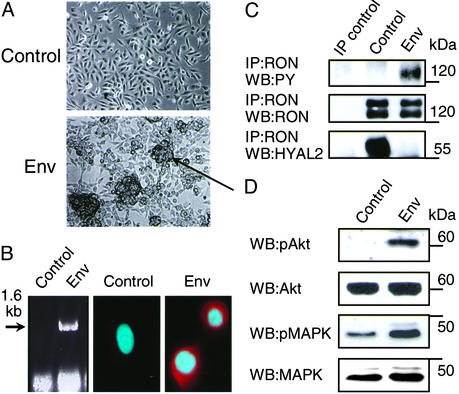
The BEAS-2B cell line was established by infection of normal human bronchial epithelial cells with an adenovirus-12 simian virus 40 (SV40) hybrid virus (13). The cells retain epithelial features, grow indefinitely in culture, express the SV40 large T, show minimal karyotypic changes, retain the ability to undergo squamous differentiation, and are not able to form tumors in nude mice (13, 15). Interestingly, it was shown recently that expression of the SV40 early region was an essential first event in the multistep transformation of primary normal human lung epithelial cells (16, 17). We transfected the BEAS-2B cells with a JSRV Env expression plasmid. Six weeks later the stably transfected cells had grown into numerous piled-up foci (Fig. 1A). These were expanded and analyzed further. First, we showed by RT-PCR and immunofluorescence microscopy that the Env transgene was expressed in these cells (Fig. 1B). Second, using immunoprecipitation and Western blotting with specific antibodies we showed that in BEAS-2B control cells the HYAL2 protein was associated with an inactive/dormant form of the receptor tyrosine kinase RON (MST1R, also known as Stk in the mouse) (Fig. 1C). In Env-transformed cells RON was not associated with HYAL2 anymore and became activated probably through autophosphorylation (Fig. 1C) indicating that the glycosylphosphatidylinositol-anchored HYAL2 protein negatively regulates RON receptor signaling. The implied ability of HYAL2 to bind hyaluronan present in the extracellular matrix may strengthen its association with RON further and thereby block RON signaling pathways. Fig. 1D shows that activation of RON in Env-transformed cells is associated with constitutive activation of Akt (19) and MAPKs (20). The natural ligand for RON is macrophage-stimulating protein (MSP); however, in this case RON activation was ligand-independent, because we could not detect MSP in culture medium exposed to BEAS cells or BEAS cells transfected with the JSRV Env expression plasmid (<0.1 nM), whereas MSP could be clearly detected in medium to which purified MSP was added at 0.1 and 1 nM concentrations (see Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site, www.pnas.org). Consequently, activation of Akt and MAPK pathways would drive proliferation and promote cell survival, both necessary for malignant transformation and maintenance of the transformed phenotype (21). Both Akt and MAPK have been shown to be activated directly by kinase-active RON involving their upstream targets in each of the respective pathways (22, 23).
Figure 1.
Effects of JSRV Env expression in BEAS-2B cells. BEAS-2B cells were transfected with pHA-Jenv (Env) or the empty pCR3.1 expression vector (Control) and grown in the presence of G418 (see Materials and Methods). (A) Cells were photographed 6 weeks after transfection. The arrow points to a transformed focus. (B) RT-PCR was used to detect the presence of Env mRNA in transfected cells (Left), and immunofluorescence staining was used to detect the expression and subcellular localization of JSRV Env protein (Center and Right). For staining, cells grown on coverslips were fixed, permeabilized, and processed as described (18). Cells were stained with anti-hemagglutinin antibody then secondary goat anti-rabbit antibody conjugated with rhodamine and were counterstained with 4′,6-diamidino-2-phenylindole. (C) RON receptor was immunoprecipitated (IP) from cell lysates by anti-RON antibodies. RON tyrosine phosphorylation was detected by Western blotting (WB) with anti-PY antibodies (Top). RON was detected by using anti-RON antibodies (Middle). The two bands represent mature RON (lower band) and its intracellular precursor (upper band). HYAL2 was detected with anti-HYAL2 antibodies (Bottom). IP control, immunoprecipitations of BEAS-2B control cell lysates with nonspecific rabbit IgG antibodies in place of RON antibodies followed by Western blotting as indicated. (D) Activation of Akt and MAPK in cell lysates was determined by Western blotting with anti-phospho-Akt and anti-phospho-MAPK antibodies. The amount of Akt and MAPK (the two bands represent p42 and p44 ERK1/ERK2) was determined by reprobing of the membrane with anti-Akt and anti-MAPK antibodies. The positions of molecular mass markers are indicated at right.
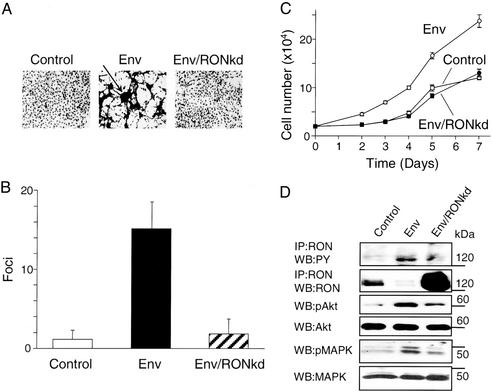
To characterize further the properties of the Env-transformed BEAS-2 cells, we first examined their growth potential in culture as compared with control cells transfected with an empty vector. The transformed cells formed 15 times more foci (Fig. 2 A and B) and grew much faster and to a higher density than nontransformed cells (Fig. 2C). We next examined whether Env-induced transformation mediated by activated RON could be reversed by ectopic expression of RONkd (12) or HYAL2. Overexpression of RONkd blocked production of transformed foci by JSRV Env (Fig. 2 A and B) and blocked Env-induced increases in cell-growth rate and maximum density (Fig. 2C). Ectopically overexpressed RONkd abrogated the activation of Akt and MAPK pathways (Fig. 2D) and reversed the growth properties of the transformed cells (Fig. 2 A–C), indicating that RON activation is essential for both transformation and maintenance of the transformed phenotype. HYAL2 overexpression also blocked transformation by Env (≈0.4 foci versus ≈20 foci per cm2 after 4 days of growth). Our results show that HYAL2 negatively regulates the function of the RON receptor tyrosine kinase in bronchial epithelial cells.
Figure 2.
Reversion of Env-induced transformation of BEAS-2B cells by stable expression of RONkd. BEAS-2B cells were transfected with equal amounts of the following plasmids: Control, pCR3.1 and pMSCV-puro-EGFP; Env, pHA-Jenv and pMSCV-puro-EGFP; Env/RONkd, pHA-Jenv and pMSCV-puro-RONkd. Transfected cells were cultured in the presence of G418 and puromycin to select for the expression of both plasmids. (A) Cells photographed 6 weeks after transfection. The arrow indicates a focus of Env-transformed cells. (B) Control, Env, and Env/RONkd cells were fixed and stained with Giemsa dye, and foci were counted by using a microscope. Bars represent the number (mean ± SE) of foci counted in 25 fields (×16 magnification) in three independent experiments. (C) Cells were seeded at 2 × 104 cells per well in duplicate wells of 24-well plates. Cell numbers were measured at the indicated time points by using a 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) cell-growth determination kit (Sigma). The number of cells was determined from an MTT calibration curve generated for each cell line. Each experimental point represents the mean ± SE of three independent experiments. (D) RON, Akt, MAPK, and their tyrosine-phosphorylated forms were detected as described for Fig. 1. The intensity of the RON bands is noticeably less in the Env-transformed cells as compared with Fig. 1C because of rapid turnover and recycling of activated RON during continuous culture of transformed cells. The dense RON band in Env/RONkd cells represents overexpressed RONkd. The positions of molecular mass markers are indicated at right. IP, immunoprecipitated; WB, Western blot.
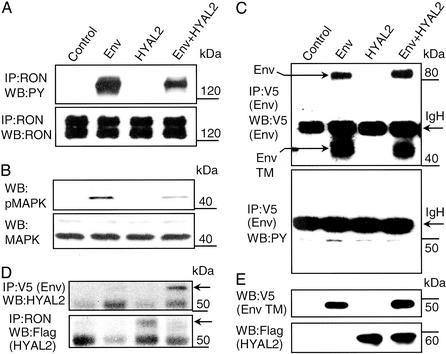
BEAS cells are difficult to transfect, so for further studies we used highly transfectable MDCK (which do not express RON or MSP) and HEK293 cell lines to allow facile detection of protein interactions and modifications such as tyrosine phosphorylation. We first used a variant of MDCK cells (RE7) stably expressing high levels of human RON (14) to study the effects of Env and/or HYAL2 expression. JSRV Env induced ligand-independent RON and MAPK activation, both of which were reduced by coexpression of HYAL2 (Fig. 3 A and B). In this system we did not observe tyrosine phosphorylation of the Env protein (Fig. 3C). Also, in these cells overexpression of Env disrupted RON–HYAL2 association by competitively binding HYAL2 (Fig. 3D). HYAL2 was expressed at similar levels in the presence or absence of Env, and Env expression was not affected by HYAL2 coexpression in these cells (Fig. 3E).
Figure 3.
Effects of Env and HYAL2 expression on RON activation in RE7 epithelial cells. RE7 cells were transiently transfected with an empty vector (Control), pJenv-V5 (Env), pFlag-HYAL2 (HYAL2), or both pJenv-V5 and pFlag-HYAL2 (Env+HYAL2). Two days later cells were incubated overnight in DMEM without serum in the presence of the proteasomal inhibitor ALLN (see Materials and Methods). After incubation the cells were lysed and analyzed by immunoprecipitation (IP) and Western blotting (WB). (A and B) RON, MAPK, and their tyrosine-phosphorylated forms were detected as described for Fig. 1. (C) Env was immunoprecipitated from transiently transfected RE7 cells with anti-V5 antibody. Expression was detected by Western blotting with anti-V5 antibody (Upper). The upper arrow on the left shows the position of the full-length Env protein. The lower arrow on the left points on the transmembrane domain (TM) of the Env protein generated by cleavage of the Env protein with endoplasmic proteases. Tyrosine phosphorylation of the Env protein was probed with anti-PY antibody (Lower). The arrows on the right show the positions of Ig heavy chains (IgH). (D) HYAL2 was detected in Env precipitates with anti-HYAL2 antibodies and in RON immunoprecipitates with anti-Flag antibody. Arrows on the right show the positions of HYAL2. (E) Expression of Env was detected with anti-V5 antibody and HYAL2 was detected with anti-Flag antibody in RE7 total-cell lysates. The positions of molecular mass markers are indicated at the right in all panels.
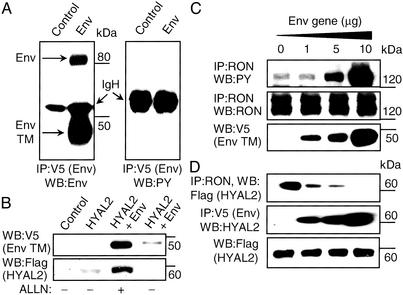
We next analyzed these interactions in HEK293 cells, which express very low levels of endogenous RON and HYAL2 proteins. In this system we showed that (i) the Env protein was processed properly but not tyrosine-phosphorylated (Fig. 4A), (ii) RON tyrosine phosphorylation was directly dependent on the level of Env expression (Fig. 4C), and (iii) Env competed with RON for binding to HYAL2 (Fig. 4D). Note that in these experiments (Fig. 4 C and D) and those described above with MDCK cells (Fig. 3), the cells were treated with the proteasomal inhibitor ALLN to inhibit protein degradation. Without such treatment, HYAL2 is degraded to undetectable levels in the presence of Env (Fig. 4B). Interactions in transiently transfected MDCK and HEK293 cells involving Env, HYAL2, and RON and the impact on RON activation are consistent with the proposed mechanism of Env-induced transformation of immortalized bronchial epithelial cells.
Figure 4.
Effects of Env expression in HEK293 cells. (A) HEK293 cells were transiently transfected with an empty vector (Control) or pJenv-V5 (Env) plasmid. Env protein was detected as described for Fig. 3. The upper arrow on the left indicates the position of the full-length Env protein, and the lower arrow indicates the position of the transmembrane domain of the Env protein. Tyrosine phosphorylation of the Env protein was probed with anti-PY antibody (Right). The positions of Ig heavy chain (IgH) proteins are shown. IP, immunoprecipitated; WB, Western blot. (B) HEK293 cells were transiently transfected with an empty vector (control), HYAL2, or HYAL2 and Env expression vectors. Forty-eight hours after transfection the cells were treated with or without ALLN proteasome inhibitor overnight, and total-cell lysates were prepared and analyzed as indicated. (C) HEK293 cells were transiently transfected with 5 μg of pCIneo-RON wild type, 5 μg of pFlag-HYAL2, and increasing amounts of pJenv-V5 plasmid. Two days later cells were incubated overnight in DMEM without serum in the presence of the proteasomal inhibitor ALLN (see Materials and Methods). After incubation the cells were lysed and analyzed. RON and tyrosine-phosphorylated RON was detected as described for Fig. 1. Env protein was detected in total-cell lysates with anti-V5 antibodies. Only the Env transmembrane bands are shown, but the full-length Env showed parallel changes in abundance. (D) HEK293 cells were transiently transfected as described for B. The presence of HYAL2 in RON immunoprecipitates was detected by anti-Flag antibody (Top). The presence of HYAL2 in Env immunoprecipitates was detected by anti-HYAL2 antibodies (Middle). HYAL2 was detected in total-cell lysates with anti-Flag antibody (Bottom). The positions of molecular mass markers are indicated at the right in all panels.
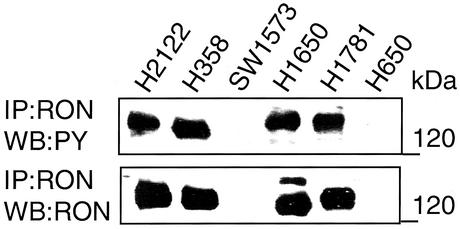
We tested the prediction that RON activation could be involved in the development of human lung cancer. Indeed, we detected RON activation, as measured by RON tyrosine phosphorylation, in three of four human BAC cell lines (Fig. 5, lanes 2 and 4–6) and in one lung adenocarcinoma (Fig. 5, lane 1), indicating a possible role of RON in BAC pathogenesis. It should be emphasized that mutational analysis of RON in these cell lines and other lung tumor samples did not reveal any mutations (data not shown) (see Supporting Text). Recently, overexpression of RON in distal lung epithelial cells in transgenic mice has been show to induce peripheral adenocarcinomas (24), further supporting a role for RON in lung cancer. Future studies would be necessary to elucidate the mechanisms of HYAL2 inactivation in BAC and other common lung adenocarcinomas. These studies need to focus on understanding the regulation of cellular RON by HYAL2 as well as the molecular mechanism(s) by which HYAL2 impacts RON silencing and its possible interaction with hyaluronan in the extracellular matrix.
Figure 5.
RON tyrosine phosphorylation in human BAC cell lines. RON and tyrosine-phosphorylated RON were detected as described for Fig. 1. Two milligrams of total protein was analyzed in each lane. The positions of molecular mass markers are indicated at the right. IP, immunoprecipitated; WB, Western blot.
The mechanism of JSRV Env transformation of epithelial cells presented here (summarized in Fig. 6) is quite different from that proposed for 208F rat and NIH 3T3 mouse fibroblasts, where the cytoplasmic tail of the Env protein is thought to interact with PI3-kinase to stimulate the PI3-kinase/Akt pathway leading to transformation (10, 11). Indeed, mouse Hyal2 seems to play no role in transformation of NIH 3T3 mouse cells, because the Hyal2 protein from mouse NIH 3T3 cells functions very poorly as a receptor for JSRV and binds JSRV Env very weakly if at all, and overexpression of the mouse Hyal2 does not suppress transformation by JSRV Env, as expected if Hyal2 functions as a tumor suppressor (27). Furthermore, NIH 3T3 cells do not express Stk, the mouse ortholog of RON, thus the model for transformation we propose for epithelial cells cannot be active in these mouse fibroblasts. It has been reported that a tyrosine residue at position 590 (Y590) of the intracellular portion of the JSRV Env is essential for transformation of NIH 3T3 cells by Env, implying phosphorylation of Y590 and subsequent activation of Akt by PI3-kinase bound to Y590 (5, 10). In contrast, we have not found phosphorylation of Y590 in MDCK (Fig. 4C) or HEK293 (Fig. 4A) cells, again suggesting that the mechanism of Env transformation is different in fibroblasts and epithelial cells.
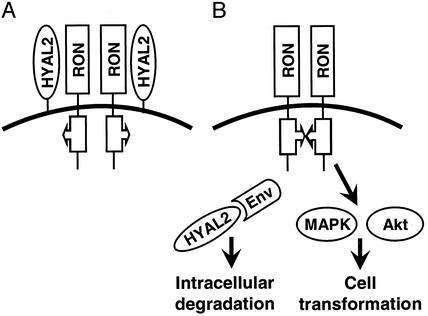
Figure 6.
Model of Env-mediated transformation of human bronchial epithelial cells. (A) BEAS-2B cells express RON receptor tyrosine kinase (Fig. 1). RON is expressed on the cell surface as an inactive dimer (25). Association of HYAL2 with RON prevents RON activation. (B) JSRV interacts with HYAL2 via the Env protein. This interaction leads to viral entry and subsequent sequestration of HYAL2 by expressed Env and intracellular degradation of HYAL2-Env complex by a proteasomal-dependent mechanism (Figs. 3D and 4 C and D). Removal of HYAL2 from the RON complex may cause conformational changes in the RON kinase domain triggering RON catalytic activity. RON constitutive activation induces activation of oncogenic pathways involving MAPK and Akt causing cell transformation. Human and animal BAC tumors are commonly composed of three types of cells, namely Clara, mucin-producing, and alveolar type II cells, none of which express RON in normal lung (26). However a common progenitor cell for these cell types, which probably gives rise to BAC, may reside in the bronchiolar compartment and may express RON (26). The heterogeneity of the differentiation phenotype within BAC lesions has been well established.
In contrast to most acutely transforming retroviruses that express oncogenes derived from cellular genes, JSRV, its close relative enzootic nasal tumor virus (ENTV), avian hemangioma virus (AHV) (28), and spleen focus-forming virus (SFFV) are members of a very small group of acutely transforming retroviruses that express oncogenic Env proteins. Similar to JSRV, the cell-entry receptor for ENTV is HYAL2, and it is likely that the mechanism of transformation by ENTV is similar to that of JSRV (4, 11). The AHV Env can induce transformation and/or apoptosis, but the mechanism is unknown. SFFV is replication-defective and encodes a truncated Env protein that does not mediate virus entry. It is interesting that a naturally occurring truncated form of Stk, the mouse ortholog of RON, is required for SFFV oncogenesis in mice (29). This short form of Stk lacks the ligand-binding domain present in Stk but retains the transmembrane and cytoplasmic tyrosine kinase domains of Stk. Transformation is mediated through a complex of the erythropoietin receptor, the SFFV Env, and this short form of Stk (30, 31).
Our investigation of the molecular interactions leading to transformation of immortalized human bronchial epithelial cells by the Env protein of JSRV revealed a tumor-suppressor function for HYAL2 (summarized in Fig. 6). An important challenge for future investigations will be to determine whether JSRV or a similar human virus might be involved in the natural history of BAC and whether a subset of these tumors might be associated with other perhaps epigenetic mechanisms of HYAL2 inactivation. We speculate that treatment of BAC patients with tyrosine kinase-type inhibitors might be beneficial in view of possible RON involvement in BAC carcinogenesis as suggested by this work and the recently published observations that even brief oncogene inactivation can produce sustained tumor regression (32).
Supplementary Material
Acknowledgments
We thank Curtis Harris for providing the BEAS-2B cells and Carl Barrett, Curtis Harris, Ed Leonard, George Klein, Harvey Pass, Alan Rabson, Al Knudson, and Bert Zbar for critically reading the manuscript. This work was supported by National Cancer Institute Contracts NO1-CO-56000 and NO1-CO-12400 and National Institutes of Health Grants DK47754 and HL54881.
Abbreviations
- HYAL2
hyaluronidase 2
- JSRV
jaagsiekte sheep retrovirus
- BAC
human bronchioloalveolar carcinoma
- Env
envelope
- PI3-kinase
phosphatidylinositol 3-kinase
- kd
kinase-dead
- EGFP
enhanced GFP
- HEK
human embryonic kidney
- MDCK
Madin–Darby canine kidney
- MAPK
mitogen-activated protein kinase
- ALLN
N-acetyl-Leu-Leu-norleucinal
- MSP
macrophage-stimulating protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lerman M I, Minna J D the International Lung Cancer Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 2.Lepperdinger G, Strobl B, Kreil G. J Biol Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 3.Rai S K, Duh F M, Vigdorovich V, Danilkovitch-Miagkova A, Lerman M I, Miller A D. Proc Natl Acad Sci USA. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirks C, Duh F M, Rai S K, Lerman M I, Miller A D. J Virol. 2002;76:2141–2149. doi: 10.1128/jvi.76.5.2141-2149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmarini M, Fan H. J Natl Cancer Inst. 2001;93:1603–1614. doi: 10.1093/jnci/93.21.1603. [DOI] [PubMed] [Google Scholar]
- 6.De las Heras M, Barsky S H, Hasleton P, Wagner M, Larson E, Egan J, Ortin A, Gimenez-Mas J A, Palmarini M, Sharp J M. Eur Respir J. 2000;15:330–332. doi: 10.1034/j.1399-3003.2000.16b23.x. [DOI] [PubMed] [Google Scholar]
- 7.Angeloni D, Lerman M I. Sourcebook on Asbestos Diseases. Vol. 23. Philadelphia: LexisNexis; 2001. pp. 169–209. [Google Scholar]
- 8.Palmarini M, Fan H, Sharp J M. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 9.Maeda N, Palmarini M, Murgia C, Fan H. Proc Natl Acad Sci USA. 2001;98:4449–4459. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmarini M, Maeda N, Murgia C, De-Fraja C, Hofacre A, Fan H. J Virol. 2001;75:11002–11009. doi: 10.1128/JVI.75.22.11002-11009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti A, Murgia C, Liu S-L, Mura M, Cousens C, Sharp M, Miller A D, Palmarini M. J Virol. 2002;76:5387–5394. doi: 10.1128/JVI.76.11.5387-5394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danilkovitch-Miagkova A, Angeloni D, Skeel A, Donley S, Lerman M I, Leonard E J. J Biol Chem. 2000;275:14783–14786. doi: 10.1074/jbc.C000028200. [DOI] [PubMed] [Google Scholar]
- 13.Reddel R R, Ke Y, Gerwin B I, McMenamin M G, Lechner J F, Su R T, Brash D E, Park J B, Rhim J S, Harris C C. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 14.Wang M H, Ronsin C, Gesnel M C, Coupey L, Skeel A, Leonard E J, Breathnach R. Science. 1994;266:117–119. doi: 10.1126/science.7939629. [DOI] [PubMed] [Google Scholar]
- 15.Ke Y, Reddel R R, Gerwin B I, Miyashita M, McMenamin M, Lechner J F, Harris C C. Differentiation (Berlin) 1988;38:60–66. doi: 10.1111/j.1432-0436.1988.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 16.Lundberg A S, Randell S H, Stewart S A, Elenbaas B, Hartwell K A, Brooks M W, Fleming M D, Olsen J C, Miller S W, Weinberg R A, Hahn W C. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 17.Hahn W C, Dessain S K, Brooks M W, King J E, Elenbaas B, Sabatini D M, DeCaprio J A, Weinberg R A. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins S. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R F, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 2002. , Unit 14.6. [Google Scholar]
- 19.Chan T O, Tsichlis P N. Sci. STKE. 2001. http://stke.sciencemag.org/cgi/content/full/sigtrans , http://stke.sciencemag.org/cgi/content/full/sigtrans;2001/66/pe1. ;2001/66/pe1. [DOI] [PubMed] [Google Scholar]
- 20.Gutkind J S. Sci. STKE. 2002. http://stke.sciencemag.org/cgi/content/full/sigtrans , http://stke.sciencemag.org/cgi/content/full/sigtrans;2000/40/re1. ;2000/40/re1. [Google Scholar]
- 21.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 22.Danilkovitch A, Donley S, Skeel A, Leonard E J. Mol Cell Biol. 2000;20:2218–2227. doi: 10.1128/mcb.20.6.2218-2227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanhaesebroeck B, Alessi D R. Biochem J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y-Q, Zhou Y-Q, Fisher J H, Wang M-H. Oncogene. 2002;21:6382–6386. doi: 10.1038/sj.onc.1205783. [DOI] [PubMed] [Google Scholar]
- 25.Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro M, Gallo K A, Godowski P J, Comoglio P M. EMBO J. 1994;13:3524–3532. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto O, Iwama A, Amitani R, Takehara T, Yamaguchi N, Yamamoto T, Masuyama K, Yamanaka T, Ando M, Suda T. J Clin Invest. 1997;99:701–709. doi: 10.1172/JCI119214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S-L, Duh F-M, Lerman M I, Miller A D. J Virol. 2003;77:2850–2858. doi: 10.1128/JVI.77.5.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alian A, Sela-Donenfeld D, Panet A, Eldor A. Virol. 2000;276:161–168. doi: 10.1006/viro.2000.0550. [DOI] [PubMed] [Google Scholar]
- 29.Persons D A, Paulson R F, Loyd M R, Herley M T, Bodner S M, Bernstein A, Correll P H, Ney P A. Nat Genet. 1999;23:159–165. doi: 10.1038/13787. [DOI] [PubMed] [Google Scholar]
- 30.Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. J Virol. 2001;75:7893–7903. doi: 10.1128/JVI.75.17.7893-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkelstein L D, Ney P A, Liu Q P, Paulson R F, Correll P H. Oncogene. 2002;21:3562–3570. doi: 10.1038/sj.onc.1205442. [DOI] [PubMed] [Google Scholar]
- 32.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg C D, Bishop J M, Felsher D W. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.