Abstract
The nemaline myopathies (NMs) are a clinically and genetically heterogeneous group of disorders characterized by nemaline rods and skeletal muscle weakness. Mutations in five sarcomeric thin filament genes have been identified. However, the molecular consequences of these mutations are unknown. Using Affymetrix oligonucleotide microarrays, we have analyzed the expression patterns of >21,000 genes and expressed sequence tags in skeletal muscles of 12 NM patients and 21 controls. Multiple complementary approaches were used for data analysis, including geometric fold analysis, two-tailed unequal variance t test, hierarchical clustering, relevance network, and nearest-neighbor analysis. We report the identification of high satellite cell populations in NM and the significant down-regulation of transcripts for key enzymes of glucose and glycogen metabolism as well as a possible regulator of fatty acid metabolism, UCP3. Interestingly, transcript level changes of multiple genes suggest possible changes in Ca2+ homeostasis. The increased expression of multiple structural proteins was consistent with increased fibrosis. This comprehensive study of downstream molecular consequences of NM gene mutations provides insights in the cellular events leading to the NM phenotype.
Nemaline myopathy (NM) is a slowly progressive or nonprogressive neuromuscular disease, defined by primary proximal muscle weakness associated with a myopathic muscle biopsy, the presence of nemaline bodies in muscle fibers, and the absence of clinical or pathological findings diagnostic of other disorders (1, 2). Although a relatively rare disease, it is the most common of the nondystrophic congenital myopathies, with an estimated incidence of 0.02 per 1,000 live births (3). The unifying genetic and molecular feature of the NMs is that all known disease genes, α-tropomyosinslow (TPM3), β-tropomyosin (TPM2), nebulin (NEB), α-actin (ACTA1), and troponin T1 (TNNT1), encode proteins of the sarcomeric thin filaments. NM is clinically heterogeneous, with severity ranging from neonatal-lethal forms, to congenital or later onset of slowly progressive or nonprogressive mild weakness (1, 2). No strong genotype–phenotype associations have been established to date, and no prognostic markers of disease severity have been identified. At the histological level there may be variation in number, size, and location of nemaline rods, the percentage of fast and slow muscle fibers, and the extent of fibrosis. Although muscles from some patients may contain increased proportions of fast (type 2) fibers, predominance of type 1 fibers is a more common feature of NM, and some patients show exclusively type 1 fibers or poor fiber type differentiation (4). Despite our increasing understanding of the genetic basis for NM, it remains unclear how the thin filament defects affect other aspects of muscle function.
To better understand the molecular pathophysiology of NM skeletal muscles, we used expression profiling to assess transcription levels of up to 21,655 genes by using Affymetrix GeneChips. To identify fundamental molecular changes associated with NM, we studied a panel of 13 skeletal muscle biopsies from 12 patients with a variety of clinical and pathological findings. The resulting data sets define gene expression patterns characteristic of NM skeletal muscle and demonstrate widespread down-regulation of key energy metabolism and muscle differentiation pathways. These patterns are distinct from those of muscles undergoing dystrophic or inflammatory processes (5–8).
Methods
Human Samples.
Thirteen skeletal muscle samples from 12 NM patients were included in the study. The NM cases ranged from severe neonatal lethal, to mild NM and adult cases (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Eleven of the patients had been previously screened for ACTA1 and TPM3 mutations, and one (T80) was positive for an ACTA1 mutation (A.H.B., unpublished data). No TPM3 mutations were found (9). NEB mutations are likely the most common among these patients; however, mutation detection is problematic because the genomic organization of this complex gene is still poorly understood (10). Additional information on the human samples can be found in Supporting Text, which is published as supporting information on the PNAS web site.
RNA Target Preparation and Hybridization.
RNA purification, target preparation, and TaqMan quantitative reverse transcriptase PCR are described in Supporting Text. Only RNA samples with 28S/18S rRNA ratios close to 2 on 1.5% agarose gels, and with absorbance ratios 260 nm/280 nm between 1.9 and 2.1 were processed into targets. Target quality was further controlled for by rejecting data sets if the ratio of expression from representative 3′ and 5′ sequences of the β-actin and the GAPDH genes was >4-fold.
Immunohistochemistry and Immunoblotting.
Antibody sources and dilutions and Western blotting are described in Supporting Text. Indirect immunofluorescence (IF) microscopy was performed on 10-μm-thick cryosections as described (11). Counts of PAX7-positive cells were normalized to 100 myofibers, and a minimum of 1,200 myofibers were counted for each disease class.
Data Analysis.
For each specimen, Affymetrix U95Av2 and U95B datasets were processed by using Affymetrix MAS5.0 to compute signal values for each probe set and resulting datasets for each specimen. Details on the methods of normalization, calculating geometric fold changes, t test analyses, hierarchical clustering, relevance network, and nearest-neighbor analyses can be found in Supporting Text.
Results
NM Muscles Exhibit Unique Patterns of Gene Expression.
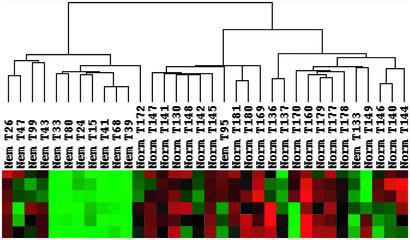
To determine how variations in gene expression correlated with disease status and other experimental variables, such as specimen type or patient age, datasets from 34 NM and normal muscle biopsies (Table 1) were hierarchically clustered by using the cluster and treeview software packages. To facilitate grouping of the specimens based on differences in gene expression, this analysis was restricted to a subset of 2,997 reproducibly expressed transcripts that varied significantly among the datasets from U95Av2 and U95B arrays by first filtering out probe sets with “absent calls” across all samples (n = 8,361) and then removing those with a standard deviation of the signal of <1,500 (n = 13,888). This analysis grouped the 34 specimens into two main branches, one predominantly containing normal muscles and one with all but two of the NM samples (Fig. 1). Interestingly, the five datasets from autopsy specimens from infants with severe NM (T24, T15, T41, T68, and T39) grouped together in the dendrogram, likely reflecting a combination of shared pathology and postmortem and age-related transcriptional patterns. Two pediatric surgical NM specimens from patients with typical NM (T33 and T80) were also closely associated with this group. In contrast, among the normal control specimens, age was not a major determinant of placement on the dendrogram, suggesting that disease-specific processes were distinguishing features of the NM datasets. Autopsy versus surgical origin of specimens may not be discounted as a relevant variable because the five normal specimens obtained at autopsy (T170, T160, T179, T177, T178, and T149) grouped together in the clustering algorithm. As dendrogram patterns may be sensitive to type and number of datasets, the analysis was repeated 10 times after random removal of six datasets (three each NM and normal) to confirm the robustness of this classification. Each of these observations was reproducible in at least 9 of the 10 iterations (not shown).
Figure 1.
NM and normal samples cluster independently by hierarchical clustering. Dendrogram generated by treeview (at top) reveals correlations between data sets for 34 specimens (middle) and identifies separate clusters for NM and normal muscles. The bottom part of the figure depicts signal strengths for a representative gene cluster, in which each row reflects the expression levels of one probe set across all samples. Color indicates relative signal levels, with red indicating the highest and green indicating the lowest expression.
Differentially Expressed Transcripts in NM Versus Normal Samples.
The U95Av2 and U95B arrays used contain 25,115 noncontrol probe sets, representative of ≈11,200 annotated genes and ≈10,455 ESTs as of October 2002. Geometric fold change analysis of all 25,115 probe sets across all NM and normal specimens at a μ − σ cutoff of 0.24 identified 454 reproducibly up-regulated and 312 reproducibly down-regulated probe sets (all fold change values reflect change in NM relative to normal specimens) (Table 2, which is published as supporting information on the PNAS web site). Permutation testing of these datasets revealed a maximal predicted false discovery rate of 0.33 at this cutoff, comparable to previously published analyses (12). The fold values among the 454 overexpressed probe sets ranged between 52 (embryonic myosin heavy chain MYH3) and 1.4 (vesicle amine transport protein 1 VATI). The fold values among the 312 underexpressed probe sets ranged between −1.4 (VE cadherin CDH5) and −16.9 (EST N46863).
Probe sets with altered expression patterns between NM and normal muscles were independently identified by a two-tail unequal variance t test of the signals from all 25,115 probe sets across all 68 datasets. At a threshold of P < 0.001, this analysis identified 406 probe sets with significantly altered signals (128 overexpressed, 278 underexpressed) between NM and normal samples. Their P values ranged from 7 × 10−9 (MYST histone acetyltransferase 1 MYST1) and 0.001 (insulin-stimulated protein kinase 1 ISPK1).
For our analysis, we consider only those probe sets independently identified by both the fold change and t test methods. The final list includes 206 probe sets whose expression we consider reliably changed between NM and normal muscles (136 under-expressed and 70 overexpressed), representative of 193 genes and ESTs (Table 3, which is published as supporting information on the PNAS web site). Of the 206 probe sets, 123 were from U95Av2 and 83 were from U95B arrays. Functional annotation of these 193 genes grouped them into eight major categories, including glucose/glycogen metabolism-related (n = 10), fatty acid metabolism (n = 3), cell cycle control (n = 15), transcription/translation-related (n = 21), ubiquitin-related (n = 7), muscle and nerve (n = 16), Ca2+ homeostasis (n = 9), and signal transduction (n = 13), with 43 miscellaneous “other” and 72 transcripts of unknown function. Because of their multiple functions, certain genes were assigned to more than one group.
Because the hierarchical clustering analysis identified age and autopsy versus surgical origin of specimens as variables that possibly affected overall gene expression patterns, we considered whether signals for any of the 123 reliably changed probe sets from U95Av2 arrays were significantly different between (i) pediatric and adult specimens, and (ii) between those obtained surgically or at autopsy. t test analyses for these variables comparing signals from all 34 specimens revealed that none of these probe sets were significantly different between these groups (P > 0.001).
We also considered whether expression changes of any of the 206 reliably changed probe sets were due to variations in fiber type proportions that characterized the NM muscle specimens (4). Correlation coefficients of signals for each of these versus the percentage of fast fibers across all 34 samples were calculated. Only IMPA2 (r = 0.86), UCP3 (r = 0.86), UBE2D1 (r = 0.83), and PPP1R2 (r = 0.80) had correlation coefficients above 0.8, indicating that the remainder were expressed in a fiber-type-independent manner.
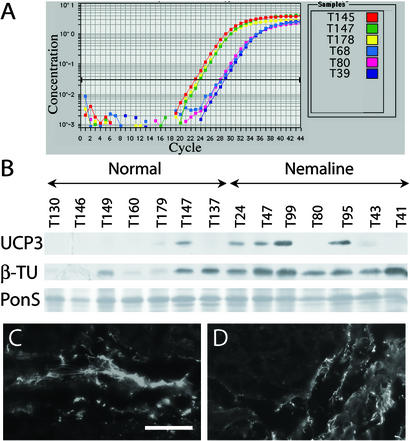
To independently confirm the validity of the microarray data on relative mRNA expression levels, we used TaqMan real-time quantitative reverse transcriptase PCR to analyze six genes from different functional categories across a subset of samples. Microarray fold change values for these genes were recalculated for this subgroup of samples. In each case, the fold changes were similar in direction and magnitude, although the GeneChip data sometimes showed a more modest change between NM and normal specimens (Table 4, which is published as supporting information on the PNAS web site, and Fig. 2A).
Figure 2.
Confirmatory studies of UCP3 and β-tubulin, at the transcript and protein levels. (A) UCP3 gene expression levels by quantitative reverse transcriptase PCR, for three representative normal (T145, T147, and T178) and NM (T68, T80, and T39) samples. UCP3 mRNA concentration is higher in normal than NM samples, and therefore peaks at an earlier cycle. (B) Immunoblotting reveals higher UCP3 protein levels in NM than in normal samples (Top). Specimens are indicated above. Ponceau S staining (PonS, Bottom) was used to ensure even loading of all protein samples. β-Tubulin also appears increased by immunoblotting in NM samples (β-TU, Middle). IF for β-tubulin in representative samples T146 (normal) (C) and T41 (NM) (D) reveals localization to connective tissue and not muscle fibers. (Bar = 50 μm.)
Relevance Network and Nearest-Neighbor Analysis of Significantly Changed Probe Sets in U95Av2 and U95B GeneChips.
The 206 reliably changed probe sets identified by the fold change and t test analysis represent transcripts that, on average, are altered between NM and normal muscles. However, as the NM specimens are heterogeneous with respect to genotype and pathological findings, we do not expect that all these transcripts are necessarily altered in the same way in all specimens. To identify potential subgroups of coordinately regulated transcripts represented by these 206 probe sets, we analyzed their signals by using relevance network analysis across normal and NM samples separately. At a correlation coefficient cutoff >0.8, 17 networks of 2–10 probe sets and one with 36 probe sets (network 17) were identified within the 13 NM datasets (RelNet 1, which is published as supporting information on the PNAS web site). Similar analysis of the 21 normal datasets revealed 19 networks; however, the largest contained only 8 probe sets (range 2–8) (RelNet 2, which is published as supporting information on the PNAS web site). Of the 36 probe sets in NM network 17, 23 represented known genes, including 5 involved in glucose or glycogen metabolism, indicating that these members of this pathway were coordinately down-regulated in the NM muscles (Fig. 5, which is published as supporting information on the PNAS web site). A number of probe sets for ESTs or hypothetical proteins of unknown function are also associated with this network, suggesting possible functional relationships among some of these.
To identify additional transcripts that were coordinately regulated with any of the 206 reliably changed probe sets, we conducted a nearest-neighbor analysis between signals for each of these and the 25,115 other probe sets across all 34 data sets, using a correlation coefficient cutoff of 0.6 and reporting no more than the 50 nearest neighbors meeting these criteria (Nearest Neighbors available at www.tch-genomics.org/beggslab/). The resulting tables provide a rich source of information about genes and ESTs coregulated with each of the transcripts altered in NM muscle.
Correlation of Transcript Levels with Protein Levels and Localization in Normal and NM Muscles.
Alterations in transcript levels do not necessarily correlate with changes in protein levels. Furthermore, as skeletal muscle is a complex organ with multiple cell types, altered protein levels need to be correlated with particular cell populations. To address these issues, we analyzed several proteins representative of different physiologic processes for which the GeneChip analysis suggested alterations in NM muscle.
Fold change analysis of the GeneChip data identified up-regulation (1.7- to 3.2-fold) in NM muscles of transcripts for α- and β-tubulin subunits (TUBB and TUBA3) as well as S100A4, which is involved in tubulin polymerization (13) (Table 3). Immunoblotting confirmed that these were paralleled by a similar increase in β-tubulin protein (Fig. 2B). IF studies revealed that β-tubulin was predominantly localized to perimysial connective tissue (Fig. 2 C and D), suggesting that differences observed at the transcriptional level are a molecular correlate of the mildly increased connective tissue often seen in NM muscle (4).
This increased fibrosis may be driven by the 9.2-fold over-expression of TGFB2, which is known to stimulate the synthesis of extracellular matrix (14). Although both immunoblotting and IF analyses failed to demonstrate an appreciable increase in TGFβ2 (transforming growth factor β2) protein level in NM muscle (not shown), it may be that increased TGFβ2 production is not detectable because of loss of this secreted peptide from the processed tissues, as other soluble proteins, such as GFP, are undetectable by immunohistochemistry if muscle tissue is not previously fixed with paraformaldehyde or other cross-linking agents (15).
UCP3 was one of the most clearly down-regulated transcripts, with three GeneChip probe sets reporting 5.1- to 12.6-fold reductions in NM muscles. This result was confirmed by the TaqMan analysis, which supported an even greater fold change. Remarkably, however, immunoblotting repeatedly revealed an apparent increase in UCP3 protein levels in a series of NM biopsies (Fig. 2B), suggesting that UCP3 transcription may be negatively regulated in the presence of increased levels of protein.
PAX7-Positive Satellite Cell Numbers Are Increased in NM Muscles.
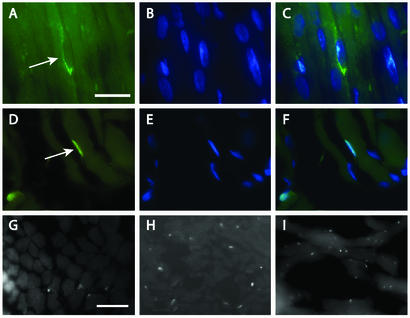
IF staining for neural cell adhesion molecule 1 (NCAM1), whose transcript was up-regulated 3 fold in NM muscle (Table 3), revealed increased expression in NM muscles, predominantly in the sarcolemma of many myofibers as well as in mononucleate cells closely apposed to the myofibers (Fig. 3 A–C). The GeneChip analysis also identified a slight (1.6-fold) increase of CDK4 transcription in NM muscles. Although immunoblotting for cyclin-dependent kinase 4 (CDK4) failed to show any detectable differences between NM and normal muscles, IF studies revealed increased numbers of CDK4-positive nuclei in NM muscles (data not shown). As with NCAM1, these nuclei also appeared to be closely associated with, but outside, the myofibers. Both NCAM1 and CDK4 are known to be expressed in proliferating myoblasts and satellite cells (16, 17). To test whether some or all of these NCAM1/CDK4-positive nuclei might represent satellite cells, we stained similar sections for PAX7, a satellite cell-specific marker for skeletal muscle (18). Anti-PAX7 antisera appeared to stain a subset of the NCAM1-positive cells and yielded a pattern consistent with that seen for CDK4 (Fig. 3 D–I). Remarkably, anti-PAX7 antisera stained 1.7 ± 2 nuclei per 100 myofibers in three normal muscle specimens (n = 1,568 fibers counted) and 17.6 ± 6 nuclei per 100 myofibers in six NM muscles (n = 1,246 fibers counted), confirming a 10-fold increase of these cells in NM muscles (P < 0.0008).
Figure 3.
Identification of satellite cells in NM muscle. Indirect IF for NCAM1 (A–C) or PAX7 (D–F) is shown in green. 4′,6-Diamidino-2-phenylindole (DAPI)-counterstained nuclei (B, C, E, and F) are shown in blue. Putative satellite cells (arrows) are located peripheral to the myofibers and are surrounded by NCAM1-positive plasma membrane. (G–I) Lower-power views of satellite cells stained for PAX7 (appearing as small white dots) in normal biopsy, T137 (G), and two NM biopsies, T41 (H) and T43 (I). [Bars = 20 μm (A–F) and 50 μm (G–I).]
Discussion
Overview.
The aim of this study was to identify physiological alterations and molecular pathways perturbed in NM skeletal muscles. All patients with NM share certain pathological and clinical features that enable the diagnosis of NM. We chose to study muscle specimens from patients of all ages, disease states, and genders, to minimize the bias introduced by each of these factors, and to focus on those transcriptional changes most closely associated with the broader NM phenotype. Geometric fold change analysis of 25,115 probe sets on U95Av2 and U95B arrays produced a list of 766 probe sets with signals altered between NM and normal muscles. An independent t test analysis of the same data, using a threshold of P < 0.001, generated a list of 406 probe sets with significantly altered sample means. The intersecting list contains 206 probe sets and likely represents a highly conservative estimate of transcripts altered in NM skeletal muscles.
Although uncontrollable experimental variables and the statistical nature of our analysis suggest that not all 206 probe sets identified in Table 3 represent reproducibly altered transcripts, several lines of evidence support the assumption that many are reliable indicators of pathophysiologic alterations in NM muscles. First, in a number of cases (e.g., UCP3, NFATC3, CYP2J2, etc., Table 3), analysis of the GeneChip data reproducibly identified multiple probe sets for a given gene as significantly up- or down-regulated. An additional 14 transcripts with multiple probe sets were also reproducibly identified by the fold change analysis alone (Table 2). Second, for all six genes studied, the quantitative reverse transcriptase PCR analysis confirmed the GeneChip data, similar to our previous experience for 12 transcripts altered in Duchenne muscular dystrophy muscles (8). Third, coordinate changes in a number of different functionally related members of the same physiological pathways (see below) provide circumstantial evidence that these represent significant differences between NM and normal muscles. Finally, the observation that UCP3 transcript levels correlated inversely with protein levels is an important reminder that it is not possible to automatically equate changes in gene expression with protein levels. Further detailed studies of each potentially perturbed pathway or process will be required to confirm and expand on our findings at the transcriptional level.
Satellite Cells and Cell Cycle Control in NM Muscles.
Satellite cells are mononucleate myogenic precursors capable of differentiating into myoblasts. They are located inside the basal lamina, and they participate in postnatal myofiber formation and regeneration (18). Satellite cell numbers have been shown to increase after experimental myonecrosis in mice (19) and are increased in younger patients with Duchenne muscular dystrophy and neurogenic atrophy, but not in other muscular dystrophies or inflammatory myopathies (20). Although PAX7 expression, used as a marker for satellite cells, was not detectably altered in the GeneChip data, we identified a 10-fold increase in satellite cells through the observation of increased NCAM1 (+3-fold) and CDK4 (+1.6-fold) transcription, both of which are expressed in myoblasts and their precursors. IF studies showed that NCAM1 was present on many myofibers and both NCAM1 and CDK4 were found in increased numbers of mononucleate cells in NM muscles. As with PAX7, gene expression levels of additional satellite muscle cell markers, MyoD1, Myf5, desmin, GLUT1, GLUT4, BCL2, MET, α-7 integrin, and M-cadherin, did not show significant changes in NM samples, probably because of a dilution effect, as satellite cells make up only a few percent of total myonuclei (19).
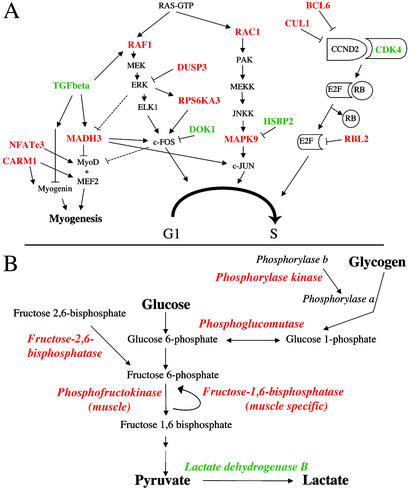
Overall, the cyclin D2/CDK4 pathway that promotes progression from G1 to S phase and suppresses skeletal muscle differentiation (17) appears to be up-regulated in NM muscles, as CDK4 was overexpressed 1.6-fold and three inhibitors of this pathway CUL1, RBL2, and BCL6, were underexpressed 1.6- to 2.4-fold (Fig. 4A, Table 3). Furthermore, FIBP, a gene believed to promote mitogenesis and normally highly expressed in skeletal muscle (21), is overexpressed 1.8-fold. Although satellite cells may account for some portion of these alterations, additional cell populations are also likely involved. Because fibroblast proliferation and increased fibrosis are not significant features of NM muscle, and similar changes were not seen in Duchenne muscular dystrophy muscles, for which fibrosis is a prominent feature (8), we suggest that this possible up-regulation of the CDK4 pathway may reflect both increased satellite cell and myoblast proliferation.
Figure 4.
Highlights of key changes in cell cycle, differentiation, and metabolism-related transcripts. (A) Representative steps from pathways involved in cell cycle regulation and myogenesis. Significantly underexpressed (in red) and overexpressed (in green) genes were identified by using fold and t test analysis (Table 3). The CCND2/CDK4 pathway seems to be overall promoted, whereas the c-Fos and c-Jun pathways appear inhibited at the transcriptional level. Myogenesis-related genes are underexpressed. (B) Overall underexpression of key enzymes from the glucose and glycogen metabolic pathways (Table 3).
Because NM muscle is not characterized by pathological evidence for regeneration, our finding of a 10-fold increase in satellite cell numbers was unexpected. Satellite cell numbers were equally increased in all NM biopsies examined, regardless of the presence or absence of developmental or neonatal isoforms of MHC (Table 1), suggesting that they were not actively participating in a regenerative process. This may be related to the observation that other measures of muscle differentiation appear overall inhibited. This hypothesis is suggested by the observation that TGFB2, a potent inhibitor of skeletal muscle differentiation (22), was up-regulated 9.2-fold, whereas CARM1, MADH3, and NFATC3, all inhibitors of the myogenin/MyoD pathways, were underexpressed by 1.9- to 4.6-fold (Fig. 4A) (23–25). Finally, alterations in several modulators of the c-Fos and c-Jun pathways (Fig. 4A) suggest a reduction in levels of these two proliferation-promoting transcription factors, again consistent with the idea that extensive regeneration-associated myoblast proliferation does not occur despite up-regulation of the cyclinD2/CDK4 pathway.
Evidence for Altered Energy Metabolism in NM Muscle.
Several genes directly or indirectly involved in the glycolytic pathway had significantly altered expression in NM muscles (Fig. 4B, Table 3). 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (PFKFB1), fructose-1,6-bisphosphatase (FBP2), phosphoglucomutase (PGM1), and phosphofructokinase (PFKM), all highly expressed in skeletal muscle and directly involved in glycolysis and/or gluconeogenesis, were underexpressed in NM specimens. Furthermore, GeneChip signals for all these transcripts were highly correlated with each other as seen by relevance network analysis (correlation coefficient cutoff ≥80% for PFKFB1, FBP2, and PGM1) of both NM and normal samples (Fig. 5), as well as nearest-neighbor analysis (r > 0.75 between all four transcripts), implying coordinate regulation of transcription for multiple key enzymes in this pathway. Transcription of the cardiac/slow (type 1 fiber) isoform of lactate dehydrogenase (LDHB) was up-regulated 2.6-fold, but a probe-set signal for LDHA, the predominant skeletal muscle fast isoform, was unchanged. The net effect on lactate dehydrogenase activity, which catalyzes conversion of pyruvate to lactate, is unclear.
Underexpression of several key enzymes for glycogen metabolism and pathway regulators in NM samples indicate a broad disruption of glycogen metabolism and further support the overall picture of a reduction of glycolytic pathway enzyme levels (Fig. 4B). By nearest-neighbor analysis, expression of RPS6KA3 was strongly correlated with PHKB and PPP1R2 (r = 0.75 and 0.74, respectively), and members of both the glucose and glycogen metabolism pathways were present in a single large relevance network from NM, but not normal, GeneChip datasets (Fig. 5), indicating that coordinate down-regulation of these transcripts was a feature of NM but not normal muscles. Increased intramuscular glycogen stores are a known feature of some NM muscle biopsies (4). Although quantitative analysis of muscle glycogen content in two NM and two normal muscles yielded results within the normal range (D.S., unpublished data), the transcriptional alterations discussed above do suggest a reduced reliance on glycolytic energy production above and beyond that expected from a shift from fast to slow fiber types in many of the specimens. It is interesting to speculate that these changes may be the molecular basis for glycogen deposits in some NM muscles.
Of note, a number of glycolytic enzymes, including phosphofructokinase (PFK) and lactate dehydrogenase, bind actin, an interaction affected by the presence of tropomyosin and troponin (26). Genes for all three of these proteins encode components of the sarcomeric thin filaments and may be mutated in NM. It is interesting to consider whether the sarcomeric disruption seen in NM could affect the activation of metabolic enzymes, and consequently their levels of transcription.
Data from several GeneChip probe sets provide evidence for energy production by increased use of fatty acid oxidation in NM muscle. The products of the PPARD and RXRA genes form heterodimers that act to block acyl-CoA oxidase (27). PPARD also negatively modulates PPARA and PPARG activity, both encoding proteins that positively regulate fatty acid metabolism in skeletal muscle (28, 29). Therefore down-regulation of PPARD and RXRA, 2.7- and 2.4-fold, respectively, is predicted to promote fatty acid oxidation in NM skeletal muscles.
Of the known reliably changed genes, the largest transcriptional down-regulation in NM muscles was seen for uncoupling protein 3 (UCP3) (range 5.1- to 12.6-fold for three probe sets) yet, remarkably, by immunoblotting, UCP3 protein levels were increased in a number of NM muscles compared with normal samples (Fig. 2 A and B). This observation suggests the existence of a negative feedback loop where increased protein levels down-regulate UCP3 gene expression, consistent with previous observations of an inverse correlation between mRNA and protein levels. A number of factors influence UCP3 levels, but in normal muscles, UCP3 protein is most abundant in human fast type 2B fibers (30), whereas mRNA expression is directly correlated with percentage of slow type 1 fibers (31). Our data suggest that UCP3 regulation is reversed in NM muscles, as mRNA levels were reduced in all NM muscles but were proportional to slow type 1 fiber numbers (three probe sets: UCP3 34421_g_at r = 0.72, UCP3 34420_at r = 0.84, and UCP3S 34422_r_at r = 0.37). We saw no clear correlations between fiber type proportions and UCP3 protein levels in NM muscles (r = 0.49 for type 1 fibers). UCP3 is thought to be involved in the regulation of mitochondrial fatty acid transport and glucose metabolism in skeletal muscle, as well as possibly protection against excessive production of reactive oxygen species and regulation of whole-body energy metabolism (32). As UCP3 may enhance fatty acid oxidation, overall increases at the protein level are consistent with a shift from glycolytic to oxidative pathways of energy production.
Alterations in Ca2+ Regulation and Related Pathways.
Several genes involved in Ca2+ homeostasis had significantly altered expression patterns. ATP2A1, ANX7, CASQ2, DMPK, and HSBP2 are involved in Ca2+ translocation from the cytosol to the sarcoplasmic reticulum lumen and in Ca2+ homeostasis in muscle contraction (33–35). Their aberrant expression patterns suggest an overall increase of cytosolic Ca2+ (Table 3).
Increase in cytosolic Ca2+ could also be induced through a second significantly changed pathway involving arachidonic acid. Differential expression of FADS1, PAFAH1B1, and CYP2J2 supports the increase of arachidonic acid and its eicosanoid products. Arachidonic acid and/or its metabolites have been implicated in the regulation of intracellular Ca2+ homeostasis (36).
Elevated cytosolic Ca2+ has been shown to lead to insulin resistance (37). A number of our findings are suggestive of a mild form of insulin resistance in NM, with several of the genes underexpressed in our study (namely PPP1R2, RPS6KA3, RAS, RAF, MEK, MAPK, PEA15, and LPIN1) being, directly or indirectly, associated with insulin metabolism (38–41). Hexokinase and insulin receptor substrate (IRS1), two more proteins with potentially significant roles in insulin resistance, were down-regulated by 4.6- and 1.8-fold, respectively (but P > 0.001) (42). One of the primary steps in insulin signaling involves activation of phosphatidylinositol 3-kinase, followed by rapid rise in phosphorylated phosphatidylinositols. The significantly down-regulated IMPA2 is involved in resynthesis of phosphatides, and is also known to have a negative correlation with Ca2+ levels. The strong negative correlation of IMPA2 with insulin-like growth factor 2 (IGF2) (r = −0.73), CASQ2 (r = −0.76) and DMPK (r = −0.69) supports the link between Ca2+, insulin, and inositol signaling, across all 34 muscle specimens (web supplement Nearest Neighbors).
This down-regulation of calcium-regulated signaling molecules might reflect a negative-feedback loop where the chronic increased cytoplasmic Ca2+ levels lead to a compensatory reduction in transcription of corresponding genes, similarly to observations in muscular dystrophies (5, 43). Together with the known mutations in sarcomeric thin filament proteins, these changes in Ca2+ homeostasis may have direct consequences for sarcomere structure and muscle contraction. By interfering with insulin and inositol signaling these changes could also affect a number of other pathways, including the cell cycle and energy metabolism (44).
Finally, it is interesting to note that several proteins involved in the ubiquitination pathway of protein degradation are down-regulated, suggesting alterations in protein turnover, perhaps related to the presence of abnormal thin filament proteins found in nemaline rods.
Conclusion
Gene expression profiling of NM muscles has identified transcriptional changes related to a number of pathways not previously known to be affected in this disorder. Increased numbers of satellite cells, in the context of several other changes in expression of cell cycle and transcription factors, suggest an abortive regenerative response in NM muscles. Our data also suggest that Ca2+ level changes may contribute to clinical weakness. We hypothesize that these modifications are linked with down-stream aberrations in cell cycle regulation and reduction in glucose energy metabolism. Whereas transcripts for multiple mitochondrial genes involved in energy metabolism were down-regulated in Duchenne muscular dystrophy muscle (5), we observed the greatest changes in transcripts for glycolytic energy production. This transcriptome-wide analysis of NM provides insights into the molecular pathways linking genotype to phenotype, and it may provide a starting point in the search for therapeutic approaches.
Supplementary Material
Acknowledgments
We thank Mei Han for outstanding technical assistance, Ana Morales for ascertainment of patients and specimens, Emanuela Gussoni for many helpful discussions and assistance concerning satellite cells, and Emanuela Gussoni and Kinga Tomczak for critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 AR44345 (to A.H.B.) and P01 NS40828 (to A.H.B., L.M.K., and I.S.K.), and by the Muscular Dystrophy Association and the Joshua Frase Foundation (A.H.B. and L.M.K.). L.M.K. is an investigator of the Howard Hughes Medical Institute. The PAX7 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa Department of Biological Sciences.
Abbreviations
- CDK4
cyclin-dependent kinase 4
- IF
immunofluorescence
- NCAM
neural cell adhesion molecule
- NM
nemaline myopathy
References
- 1.North K N, Laing N G, Wallgren-Pettersson C. J Med Genet. 1997;34:705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanoudou D, Beggs A H. Trends Mol Med. 2001;7:362–368. doi: 10.1016/s1471-4914(01)02089-5. [DOI] [PubMed] [Google Scholar]
- 3.Wallgren-Pettersson C. Commentationes Physico-Mathematicae. 1990;111:1–102. [Google Scholar]
- 4.Ryan M M, Ilkovski B, Strickland C D, Schnell C, Sanoudou D, Midgett C, Houston R, Muirhead D, Dennett X, Shield L K, et al. Neurology. 2003;60:665–673. doi: 10.1212/01.wnl.0000046585.81304.bc. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y W, Zhao P, Borup R, Hoffman E P. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter J D, Khanna S, Kaminski H J, Rao J S, Merriam A P, Richmonds C R, Leahy P, Li J, Guo W, Andrade F H. Hum Mol Genet. 2002;11:263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg S A, Sanoudou D, Haslett J N, Kohane I S, Kunkel L M, Beggs A H, Amato A A. Neurology. 2002;59:1170–1182. doi: 10.1212/wnl.59.8.1170. [DOI] [PubMed] [Google Scholar]
- 8.Haslett J N, Sanoudou D, Kho A T, Bennett R R, Greenberg S A, Kohane I S, Beggs A H, Kunkel L M. Proc Natl Acad Sci USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wattanasirichaigoon D, Swoboda K J, Takada F, Tong H Q, Lip V, Iannaccone S T, Wallgren-Pettersson C, Laing N G, Beggs A H. Neurology. 2002;59:613–617. doi: 10.1212/wnl.59.4.613. [DOI] [PubMed] [Google Scholar]
- 10.Pelin K, Hilpela P, Donner K, Sewry C, Akkari P A, Wilton S D, Wattanasirichaigoon D, Bang M L, Centner T, Hanefeld F, et al. Proc Natl Acad Sci USA. 1999;96:2305–2310. doi: 10.1073/pnas.96.5.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.North K N, Beggs A H. Neuromuscul Disord. 1996;6:229–235. doi: 10.1016/0960-8966(96)00361-6. [DOI] [PubMed] [Google Scholar]
- 12.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakshmi M S, Parker C, Sherbet G V. Anticancer Res. 1993;13:299–303. [PubMed] [Google Scholar]
- 14.Border W A, Ruoslahti E. J Clin Invest. 1992;90:1–7. doi: 10.1172/JCI115821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaBarge M A, Blau H M. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin T J, Fazeli M S, Doherty P, Walsh F S. J Cell Biochem. 1996;61:502–513. doi: 10.1002/(sici)1097-4644(19960616)61:4<502::aid-jcb3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.Lazaro J B, Bailey P J, Lassar A B. Genes Dev. 2002;16:1792–1805. doi: 10.1101/gad.U-9988R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seale P, Sabourin L A, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki M A. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 19.Reimann J, Irintchev A, Wernig A. Neuromuscul Disord. 2000;10:276–282. doi: 10.1016/s0960-8966(99)00118-2. [DOI] [PubMed] [Google Scholar]
- 20.Maier F, Bornemann A. Muscle Nerve. 1999;22:578–583. doi: 10.1002/(sici)1097-4598(199905)22:5<578::aid-mus5>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 21.Kolpakova E, Wiedlocha A, Stenmark H, Klingenberg O, Falnes P O, Olsnes S. Biochem J. 1998;336:213–222. doi: 10.1042/bj3360213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pena T L, Chen S H, Konieczny S F, Rane S G. J Biol Chem. 2000;275:13677–13682. doi: 10.1074/jbc.275.18.13677. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J M, Zhao X, Wei Q, Paterson B M. EMBO J. 1999;18:6983–6993. doi: 10.1093/emboj/18.24.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S L, Loffler K A, Chen D, Stallcup M R, Muscat G E. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 25.Delling U, Tureckova J, Lim H W, De Windt L J, Rotwein P, Molkentin J D. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke F M, Masters C J. Biochim Biophys Acta. 1975;381:37–46. doi: 10.1016/0304-4165(75)90187-7. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Hon M, Evans R M. Proc Natl Acad Sci USA. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapsys N M, Kriketos A D, Lim-Fraser M, Poynten A M, Lowy A, Furler S M, Chisholm D J, Cooney G J. J Clin Endocrinol Metab. 2000;85:4293–4297. doi: 10.1210/jcem.85.11.6973. [DOI] [PubMed] [Google Scholar]
- 29.Muoio D M, Way J M, Tanner C J, Winegar D A, Kliewer S A, Houmard J A, Kraus W E, Dohm G L. Diabetes. 2002;51:901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- 30.Hesselink M K, Keizer H A, Borghouts L B, Schaart G, Kornips C F, Slieker L J, Sloop K W, Saris W H, Schrauwen P. FASEB J. 2001;15:1071–1073. [PubMed] [Google Scholar]
- 31.Calsbeek D J, Thompson T L, Dahl J A, Stob N R, Brozinick J T, Jr, Hill J O, Hickey M S. Am J Physiol. 2002;283:E631–E637. doi: 10.1152/ajpendo.00449.2001. [DOI] [PubMed] [Google Scholar]
- 32.Schrauwen P, Hesselink M. J Exp Biol. 2002;205:2275–2285. doi: 10.1242/jeb.205.15.2275. [DOI] [PubMed] [Google Scholar]
- 33.Maloney J A, Wheeler-Clark E S. J Hypertens. 1996;14:65–74. [PubMed] [Google Scholar]
- 34.Herr C, Smyth N, Ullrich S, Yun F, Sasse P, Hescheler J, Fleischmann B, Lasek K, Brixius K, Schwinger R H, et al. Mol Cell Biol. 2001;21:4119–4128. doi: 10.1128/MCB.21.13.4119-4128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benders A A, Groenen P J, Oerlemans F T, Veerkamp J H, Wieringa B. J Clin Invest. 1997;100:1440–1447. doi: 10.1172/JCI119664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Striggow F, Ehrlich B E. Biochem Biophys Res Commun. 1997;237:413–418. doi: 10.1006/bbrc.1997.7152. [DOI] [PubMed] [Google Scholar]
- 37.Zemel M B. Mol Cell Biochem. 1998;188:129–136. [PubMed] [Google Scholar]
- 38.Sturgill T W, Ray L B, Erikson E, Maller J L. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 39.Dent P, Lavoinne A, Nakielny S, Caudwell F B, Watt P, Cohen P. Nature. 1990;348:302–308. doi: 10.1038/348302a0. [DOI] [PubMed] [Google Scholar]
- 40.Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti C G, Andreozzi F, Cafieri A, Tecce M F, Formisano P, Beguinot L, Beguinot F. EMBO J. 1998;17:3858–3866. doi: 10.1093/emboj/17.14.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huffman T A, Mothe-Satney I, Lawrence J C., Jr Proc Natl Acad Sci USA. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen K F, Shulman G I. Am J Cardiol. 2002;90:11G–18G. doi: 10.1016/s0002-9149(02)02554-7. [DOI] [PubMed] [Google Scholar]
- 43.Bakay M, Zhao P, Chen J, Hoffman E P. Neuromuscul Disord. 2002;12, Suppl. 1:S125–S141. doi: 10.1016/s0960-8966(02)00093-7. [DOI] [PubMed] [Google Scholar]
- 44.Bevan P. J Cell Sci. 2001;114:1429–1430. doi: 10.1242/jcs.114.8.1429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.