Abstract
Cellular processes such as transcription and DNA repair may be regulated through diverse mechanisms, including RNA synthesis, protein synthesis, posttranslational modification and protein degradation. The 26S proteasome, which is responsible for degrading a broad spectrum of proteins, has been shown to interact with several nucleotide excision repair proteins, including xeroderma pigmentosum B protein (XPB), Rad4, and Rad23. Rad4 and Rad23 form a complex that binds preferentially to UV-damaged DNA. The 26S proteasome may regulate repair by degrading DNA repair proteins after repair is completed or, alternatively, the proteasome may act as a molecular chaperone to promote disassembly of the repair complex. In either case, the interaction between the proteasome and nucleotide excision repair depends on proteins like Rad23 that bind ubiquitin-conjugated proteins and the proteasome. While the iteration between Rad4 and Rad23 is well established, it will be interesting to determine what other proteins are regulated in a Rad23-dependent manner.
INTRODUCTION
The regulation of DNA repair is important for cell survival following exposure to DNA-damaging agents. A large array of exogenous and endogenous agents can interact with and cause DNA damage, interfering with essential cellular processes, such as transcription, DNA replication, and cell-cycle progression. Disruption of these processes can lead to cell death. Alternatively, unrepaired or mis-repaired DNA can generate mutations that lead to cellular aging, genetic defects, and carcinogenesis. One major pathway that contributes to the removal of DNA damage is the nucleotide excision repair (NER), whose biochemical mechanism has been characterized extensively. In contrast, the regulation of NER is not well understood. Regulation of NER could be accomplished through changes in RNA transcription, protein translation, protein degradation, or posttranslational modifications.
A burgeoning literature underscores the relevance of the ubiquitin(Ub)/proteasome pathway to many cellular processes. Interactions between the proteins involved in NER and the Ub-mediated protein degradation pathway have been reported for yeast and mammals. The emerging evidence that yeast repair protein Rad4, and its human counterpart XPC, might be targeted for degradation by the 26S proteasome is consistent with a negative role for Ub-mediated proteolysis in NER. In addition, the intact 26S proteasome, or its constituent parts, may serve additional roles in NER, perhaps as molecular chaperones that promote the proper folding of repair proteins or disassembly of protein complexes.
NUCLEOTIDE EXCISION REPAIR
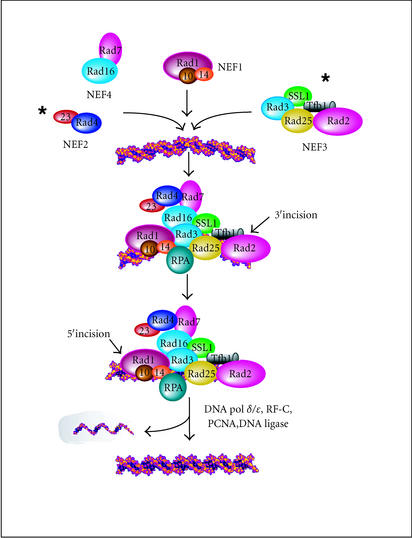
The nucleotide excision repair (NER) pathway in eukaryotes is conserved from yeast to humans. This repair system removes many bulky chemical adducts and UV-induced photoproducts from DNA in a relatively error-free manner. Defects in nucleotide excision repair are associated with increased incidence of cancer. The identification and cloning of genes involved in NER has led to the reconstitution of repair in vitro [1, 2], using approximately 30 purified proteins [3, 4]. Nucleotide excision repair proteins have been purified from yeast cell extracts as functional subassemblies called nucleotide excision repair factors (NEFs). NEF1 contains a damage recognition protein Rad14, and a 5′-endonuclease complex (Rad1/Rad10) that binds preferentially to damaged DNA. NEF2 consists of Rad4 and Rad23, which can also bind preferentially to damaged DNA, and might play a role in recruiting other repair proteins to the sites of DNA lesions. NEF3 contains Rad2, an endonuclease that cleaves on the 3′ side of the DNA lesion, and TFIIH, an RNA polymerase II-associated transcription factor complex. NEF4 consists of Rad7 and Rad16 and, like NEF1 and NEF2, binds preferentially to damaged DNA. As in bacteria, NER in eukaryotic cells is a multistep process that recognizes damaged DNA, generates incisions upstream and downstream from the lesion, and displaces the damaged DNA as part of an oligonucleotide (Figure 1) [2]. The excised oligonucleotide in human extracts is larger than in E coli because the 5′ incision is made farther from the lesion. The gap left after the removal of the lesion-containing oligonucletide is repaired by DNA polymerases δ and ɛ, and ligated to the flanking parental DNA by DNA-ligase. The biochemical details of DNA incision and excision, which have been extensively characterized in yeast, display remarkable mechanistic conservation with the human system [5].
Figure 1.
A model of yeast nucleotide excision repair. The biochemical details of the incision and size of the DNA oligonucleotide that is excised by yeast NER proteins has been determined, and is mechanistically similar to the human system [2, 5]. An asterisk indicates the presence of repair proteins that interact with the proteasome. Mutations in most genes that encode nucleotide excision repair proteins result in severe UV sensitivity. The order of repair complex assembly has been extensively studied, though the biochemical steps that follow DNA excision and gap-filling are unknown. The recycling of RNA polymerase II following exposure to a DNA-damaging agent has been suggested [76]. However, the lack of an in vivo or in vitro assay to measure the recycling, dismantling, and degradation of the repair complex, reveals significant steps in the cellular response to DNA damage that remain to be defined.
NER comprises two subpathways that either target transcribed strands of class II genes, or nontranscribed sequences (that includes the genome overall). These two subpathways share many factors, though some are unique to one subpathway or the other.
GLOBAL GENOMIC REPAIR IN YEAST
The repair of nontranscribing sequences within genes occurs at about the same rate as the repair of overall genomic DNA, and requires the proteins shown in Figure 1, in addition to Rad7 and Rad16 (NEF4), which have been shown to bind UV-damaged DNA [6]. Importantly, Rad16 is a member of the Swi2/Snf2 family of DNA-dependent ATPases that are thought to remodel nucleosomes, and/or displace proteins from chromatin [7]. However, a clear-cut homolog of Rad7 or Rad16 has not been identified in mammals.
TRANSCRIPTION-COUPLED REPAIR IN YEAST
In addition to the recognition and excision of damaged DNA that occurs throughout the genome, there exists a specific mechanism that recognizes damaged DNA that is present in the transcribed strand of genes that encode mRNA [8, 9, 10, 11]. The preferential repair of the transcribed strand is absolutely dependent upon the transcription by RNA polymerase II [8, 11]. For instance, the removal of cyclobutane pyrimidine dimers (CPDs) from the transcribed strand of RPB2 (which is transcribed by RNA polymerase II), is much more rapid than from the nontranscribed strand. When exponentially growing rpb1-1 cells are shifted to the nonpermissive temperature, they rapidly cease mRNA synthesis, and the repair of the transcribed strand is reduced to that of the nontranscribed strand. Similarly, the transcription defect that is associated with the heat-sensitive kin28ts mutant (that is a component of TFIIK) is accompanied by a defect in transcription-coupled repair [12]. Preferential repair of the transcribed strand of the mammalian DHFR gene is also abolished by the treatment of cells with the RNA polymerase II inhibitor α-amanitin [13, 14]. Collectively, these findings provided compelling evidence that the transcription by RNA pol II contributes to the strand bias in DNA repair. Although it was previously assumed that TCR was confined to genes that are transcribed by RNA polymerase II, recent studies demonstrated that TCR is also present in the transcriptionally active fraction of ribosomal DNA (rDNA), which is transcribed by the RNA polymerase I [15].
TRANSCRIPTION REPAIR-COUPLING FACTORS IN EUKARYOTES
CSB and CSA are the only genes that have been reported to affect transcription-coupled repair in mammalian cells. CSB plays a specific role in resuming RNA synthesis following the UV irradiation [16, 17]. The yeast homolog of the CSB (Rad26) was identified, though its biochemical function is unclear [18, 19]. Rad26 and CSB are members of the Swi2/Snf2 family of DNA-dependent ATPases that are thought to remodel nucleosomes, and/or displace proteins from chromatin [7]. Indeed, CSB has been demonstrated to remodel nucleosomes in vitro [20]. A yeast rad26 disruption mutant displayed similar rate and extent of removal of CPDs from the transcribed and nontranscribed strands of the RPB2 gene [19], revealing a defect in transcription-coupled repair. This defect of the rad26 mutant is consistent with the transcription coupled repair defect of cells that were derived from patients with Cockayne syndrome. However, in contrast to Cockayne cells, rad26 mutants are not UV sensitive, perhaps due to the proficient repair of the overall genomic DNA in yeast [19]. The corresponding lack of UV sensitivity in rad26 mutants could explain why they were not isolated in genetic screens that sought mutants that were sensitive to UV, or defective in overall genomic repair.
The biochemical steps that underlie NER have been carefully elucidated, though the mechanism that enables the repair machinery to distinguish between transcriptionally active and inactive DNA remains enigmatic. While CSA and CSB (RAD26) genes in mammalian and yeast cells clearly play a role in the transcription-coupled repair, their biochemical activities remain to be defined. For instance, it is not known if the CSA or CSB function as transcription-repair coupling factors. There is evidence that CSB can affect transcription elongation in vitro [21], though CSB does not colocalize with RNA pol II in vivo [22]. CSB might deliver CSA to RNA pol II that is stalled at sites of DNA damage [22].
GENETIC EVIDENCE FOR A PROTEOLYTIC ROLE IN DNA REPAIR
Several lines of genetic evidence suggest a role for ubiquitylation and protein degradation in DNA repair. Mutations in the genes encoding Rad6, Ubc13, Mms2, Ufd2, p53, Rad16, Ump1, and Rad23 render cells sensitive to UV irradiation or stressful conditions. Rad6, Ubc13, and Mms2 are required for ubiquitylation of substrate proteins, while Ufd2 and Rad23 are likely to regulate this process [5, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32]. Ump1 is a maturation factor that is required for the assembly of the catalytic subunit of the proteasome [33, 34]. Rad16 is a member of the Swi2/Snf2 family of chromatin remodeling ATPases that contains a zinc finger motif (RING), which is also present in several Ub-specific proteolytic factors [35]. The tumor suppressor protein p53 is targeted for ubiquitylation and degradation by the 26S proteasome [36, 37, 38]. However, DNA damage results in its stabilization, and elevated levels of p53 permit activation of genes that contribute to enhanced survival in response to various environmental stresses. Nucleotide excision repair proteins, Rad4 and XPB, interact with the proteasome, while Rad4 can be copurified with the proteasome. Mutations in the genes encoding Rad4 and XPB cause severe UV sensitivity compared to repair-proficient cells. There is emerging evidence that Rad4, and its human counterpart (XPC), are ubiquitylated and degraded by the 26S proteasome (see section Stabilization of a Repair Protein in a Proteasomal Mutant below). Similarly, it is possible that the ubiquitylation and degradation of other repair proteins could also govern the efficiency of DNA repair.
The expression of several genes that encode components of the ubiquitin/proteasome pathway is induced following DNA damage, consistent with a possible role in DNA repair (see section Many Ubiquitin/Proteasome Genes Are Induced by DNA Damage below). For instance, the expression of both RAD6 and RAD23 is elevated after DNA damage. Additionally, the treatment of cells with methylmethane sulfonate (MMS) resulted in induced expression of several genes in the ubiquitin/proteasome pathway, consistent with the existence of a network that coordinates the regulated expression of ubiquitylation and proteolytic enzymes in response to DNA damage.
TRANSCRIPTION-COUPLED REPAIR AND UBIQUITIN-MEDIATED PROTEIN DEGRADATION
The suggestion that ubiquitylation, and perhaps protein degradation, might play a role in NER was made by Bregman and colleagues [39]. HeLa cells and normal human fibroblasts in culture can ubiquitylate the large subunit of RNA polymerase II (RNA pol II LS) following UV irradiation or treatment with cisplatin [39]. The ubiquitylation of RNA pol II LS was absent in UV-irradiated fibroblasts from Cockayne syndrome patients. In contrast, fibroblasts derived from patients displaying symptoms of xeroderma pigmentosum, another autosomal recessive disorder associated with a DNA repair deficiency, were capable of ubiquitylating RNA pol II LS following UV irradiation [40]. These findings may indicate that the failure to regulate the stability of RNA pol II might underlie the DNA repair-specific defects of Cockayne syndrome.
It has been shown that some types of DNA lesions in the transcribed strand can arrest the movement of the transcription complex. In both normal and some xeroderma pigmentosum fibroblasts, the ubiquitylated form of RNA pol II LS was hyperphosphorylated, a form that is associated with the elongating transcription complex [40]. These findings lead the authors to offer a plausible role for ubiquitylation and degradation of the hyperphosphorylated form of RNA Pol II. The recognition of the stalled RNA Pol II by a ubiquitin-protein (E3) ligase could result in ubiquitylation and translocation to the proteasome for degradation. In agreement with this conjecture, Lee et al [41] demonstrated that an RNA pol II elongation complex that is stalled at a site of DNA damage can be ubiquitylated in vitro.
Yeast Rpb1 (RNA pol II LS) is ubiquitylated by the ubiquitin protein (E3) ligase, Rsp5 [42, 43]. However, the ubiquitylation of Rpb1 by Rsp5 is not required for transcription-coupled repair in yeast [44], since a strain expressing a conditional mutant of RSP5 was proficient in transcription-coupled and nucleotide excision repair. These results suggest that the ubiquitylation of Rpb1 could occur after the transcription-coupled repair is achieved, or might even be unlinked to the repair process. However, the lack of a repair defect in rsp5 mutant cells does not preclude a role for proteolysis in transcription-coupled or nucleotide excision repair [45], since there is evidence that the RNA pol II that is stalled at the sites of DNA damage may have several fates. While RNA pol II could be removed from damaged DNA and degraded, it is also conceivable that the Rad26 protects RNA pol II from degradation to enable TCR [46].
We propose that the regulation of NER could be achieved by controlling the abundance of one or more NER proteins, through selective degradation or protein synthesis. In agreement with this conjecture, we note that most NER genes in yeast are constitutively expressed, and only a few are expressed at higher levels following DNA damage. Therefore, an efficient DNA repair would only require the expression or stabilization of one or a few nucleotide excision repair proteins. We presume that once the repair is completed, and the nucleotide excision repair machinery is no longer required, a subset of the NER proteins would be degraded to reduce the NER activity so that the inadvertent incision of DNA structures that are generated during normal cellular transactions are avoided.
THE UBIQUITIN-MEDIATED PROTEIN DEGRADATION PATHWAY
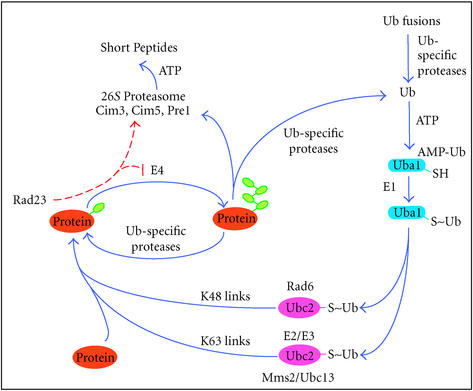
The ubiquitin/proteasome system regulates protein stability and function and is conserved from yeast to humans, similar to the nucleotide excision repair pathway. Cellular processes that are regulated by proteolysis include apoptosis, cell cycle progression, stress responses, development, and transcriptional regulation. Defects in proteasomal function have pleiotropic effects and are implicated in lung cancer, Angelman syndrome, muscle wasting, Parkinson disease, and inflammatory response [47]. However, the ubiquitylation of a protein does not necessarily lead to the destruction of the modified protein. For instance, ubiquitylation can modify protein activity, or promote the targeting of proteins to vesicles during endocytosis [48]. The ubiquitin-mediated protein degradation pathway is depicted in Figure 2. Once the first ubiquitin is attached to a protein, specific ubiquitin chain assembly factors (E4) may promote the formation of multiubiquitin chains, which allow the target protein to be recognized and degraded by the 26S proteasome.
Figure 2.
The yeast ubiquitin-mediated protein degradation pathway (adapted from [49]). The Ubiquitin (Ub) System is a multienzyme process that covalently links Ub to a wide variety of intracellular proteins. Ub is expressed as a fusion with specific ribosomal proteins or as polyubiquitin fusions. Ub-specific proteases generate free Ub that is activated by a Ub-activating (E1) enzyme, in an ATP-dependent reaction. Ub is transferred from E1 to Ub-conjugating (E2), and Ub (E3) ligase enzymes through a series of transesterification reactions, and is finally ligated to lysine residues in the target protein. This last step might require a ubiquitin-protein (E3) ligase. The specificity of the system resides in the E2 and E3 enzymes. Newly identified E4 enzymes may regulate the assembly of multiubiquitin chains on substrates.
THE 26S PROTEASOME
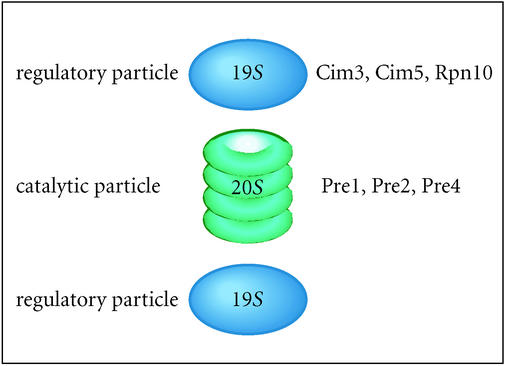
The 26S proteasome consists of two distinct subunits, a 19S regulatory particle and a 20S catalytic core particle [49]. The 20S particle consists of four heptameric rings that form a barrel-shaped protein complex with the catalytic sites confined to the interior surface (Figure 3). Narrow ports at each end inhibit access to the interior. A 19S regulatory subunit spans each end of the 20S particle and restricts access to the catalytic core. Proteins appear to be de-ubiquitinated and unfolded by the 19S regulatory particle, before being funneled into the interior of the 20S catalytic core for degradation. In contrast, ubiquitin is recycled. In yeast and humans, the intact 26S proteasome appears to be the functional form of the proteasome [50, 51], and the association of two 19S subunits with each 20S subunit is detectable by an electron microscopy [50].
Figure 3.
The structure of the 26S proteasome. Two 19S regulatory complexes straddle the openings to the 20S particle, and control access to catalytic sites within the 20S complex. Subunits in the base of the 19S regulatory particle are believed to perform a gating function that regulates the entry of unfolded proteolytic substrates into the catalytic chamber of the core particle [50]. Heat-sensitive conditional mutants in the two distinct particles of the proteasome have greatly assisted the functional characterization of the proteasome.
The 19S regulatory particle can bind multiubiquitin chains and consists of at least 17 proteins. The translocation of unfolded proteolytic substrates into the 20S catalytic core [49] is highly processive, as degradation intermediates are rarely detected. The six homologous AAA ATPases in the 19S particle are thought to unfold substrate proteins in an ATP-dependent manner, and are essential for proteasome function. Conditional mutations in these ATPases, including cim3-1 (Sug1/Rpt6), sug2-1 (Sug2/Rpt4), and cim5-1 (Rpt1/Yta3) have been isolated, and their characterization has suggested that the ATPase subunits can discriminate among cellular substates, and at least one has been shown to directly recognize multiubiquitin chains. The subcellular distribution of the proteasome is a question of considerable interest, and recent reports have not clarified this issue. There is a compelling evidence that the proteasome in yeast is located primarily at the junction of the nuclear envelope and the endoplasmic reticulum [52, 53]. However, there is uncertainty as to whether the intact 26S proteasome contributes to all proteasomal functions, or if the 19S regulatory subunits can function independently of the 20S catalytic core [51].
The expression of a number of genes involved in NER is induced following the DNA damage, probably to hasten the removal of DNA adducts. The induced proteins are presumably degraded to restore basal levels of expression following the completion of repair. Although it is not known if nucleotide excision repair is regulated by the protein degradation, there exist several tantalizing links between these pathways. Weeda et al [54] demonstrated that the XPB subunit of human transcription initiation/repair factor IIH (TFIIH) interacts with hSUG1, the human homolog of the 19S regulatory subunit Cim3. XPB is the human counterpart of Rad25. Microinjection of cDNA encoding mouse SUG1 into the fibroblast nuclei led to a dramatic decrease in transcription, though there was no evidence that XPB was ubiquitiylated and degraded. Additionally, the human homologue of Cim5 (MSS1), interacts with basal transcription factors TBP, TFIIB, TFIIH, and TFIIF [55]. Weeda et al [54] speculated that the proteasome might activate proteins by processing inactive precursors. In support of this idea, it has been shown that the activation of the transcription factors NFκB and a sterol-induced regulator in yeast require proteolytic activiation of precursor proteins. Another link to proteolysis was described by Schauber et al [29] who found that Rad23 and Rad4 could be copurified with the 26S proteasome, though it was not clear if Rad4 was degraded. Rad23 can interact with other proteins involved in nucleotide excision repair [5, 32] as well as the 19S proteasome regulatory particle [28, 29]. Schauber et al [29] proposed that repair protein complex could be disassembled or degraded upon completion of repair, through a specific interaction between Rad23 and the proteasome. These studies lead the authors to suggest that Rad23 might escort proteins to the 26S proteasome for destruction. Alternatively, it is possible that the proteolytic activities of Rad23 are unrelated to an independent role in promoting the assembly of the nucleotide excision repair complex [56]. Both scenarios are consistent with the DNA repair defect of rad23Δ mutants [57].
MANY UBIQUITIN/PROTEASOME GENES ARE INDUCED BY DNA DAMAGE
Jelinsky and colleagues [58] used gene chip technology to examine the transcription profile of the S cerevisiae genome in response to DNA damage. Exponentially growing cultures were exposed to DNA-damaging agents and biotin-labeled cRNA was made and hybridized to an oligonucleotide array. Remarkably, almost a third of the genes showed altered expression after exposure to the DNA-damaging agent. Similarly, treatment of cells with methylmethane sulfonate (MMS) resulted in changes in the expression of ∼25% of yeast genes. Some sets of genes were expressed soon after treatment, while other genes were expressed either late or transiently. Furthermore, the transcriptional response to DNA damage was cell cycle dependent. One of the genes whose expression is increased following DNA damage is NPR1. Npr1 protein is a putative regulator of Rsp5 (a ubiquitin protein ligase), Doa4 (a ubiquitin-specific protease), UBC13 (a subunit of a bipartite ubiquitin-conjugating-enzyme), and subunits of the 26S proteasome. Sequence analysis of the promoters of many genes encoding proteolytic factors revealed putative DNA binding sites for transcription factors. Interestingly, a significant number of promoters in genes involved in Ub-mediated proteolysis contain a conserved nonameric sequence, the proteasome-associated control element (PACE), that could coordinate transcriptional activation in response to DNA damage. There is evidence that the transcription factor Rpn4 is required for stimulating transcription from PACE-containing promoters [59]. Thus, there may exist a regulatory network that coordinates the expression of ubiquitylation and proteolytic enzymes in response to DNA damage.
IN VITRO NUCLEOTIDE EXCISION REPAIR AND THE PROTEASOME
Russell et al [28] found that the addition of antibodies against the proteasome subunit Sug1 (Cim5) to a reconstituted NER reaction, inhibited repair. Furthermore, extracts prepared from yeast strains that contained mutant Sug1 or Sug2 were moderately defective in accomplishing nucleotide excision repair. Because a similar decrease in NER was not observed in yeast strains that expressed defective 20S subunits, it was proposed that NER does not require the proteolytic activity of the proteasome. In agreement with this result, the proteasome inhibitor lactacystin did not reduce the capacity of yeast extracts to carry out nucleotide excision repair in vitro. Furthermore, immunoprecipitation studies suggested that the 19S regulatory complex might function as an independent entity, distinct from the intact 26S proteasome that contains the 20S core particle. Additional support for this idea comes from identification of subunits of the 19S regulatory particle in the RNA pol II transcription complex [60], and the recent demonstration that the base of the 19S regulatory particle is recruited to promoters by Gal4 in vitro [61]. Collectively, these studies suggest that the link between NER and the proteasome might not involve proteolysis.
IN VIVO NUCLEOTIDE EXCISION REPAIR IN PROTEASOME MUTANTS
Conditional mutations in the 19S regulatory subunit of the 26S proteasome result in increased nucleotide excision repair in vivo [45], in contrast to the in vitro results with protein extracts. Repair of both the transcribed and nontranscribed strands of an RNA polymerase II-transcribed gene was increased in the absence of proteasome function, suggesting that proteolysis played a negative role in NER. In agreement with this conjecture, the over-expression of Rad4-hemagglutinin (Rad4-HA) led to increased repair of the nontranscribed strand of a reporter gene (see [45]).
STABILIZATION OF A REPAIR PROTEIN IN A PROTEASOMAL MUTANT
Pulse-chase studies revealed that Rad4-HA levels were rapidly reduced in a wild-type strain [62]. However, treatment with the UV-mimetic 4-nitro-1-quinoline (4-NQO) resulted in transient stabilization of Rad4-HA. Similarly, Rad4-HA was stabilized in yeast proteasome mutants, suggesting that the Ub/proteasome pathway mediated its degradation. To determine if Rad4-HA was ubiquitylated, we examined its levels in cim5-1 at the nonpermissive temperature. Incubation with antiubiquitin antibodies revealed that immunoprecipitated Rad4-HA was multiubiquitylated in vivo. Over-expression of Rad23, a partner of Rad4, inhibited the multiubiquitylation of Rad4-HA, consistent with a previously described role for Rad23 in transiently stabilizing proteolytic substrates. Thus, Rad4-HA is likely to be ubiquitylated and degraded in a Rad23-dependent manner.
Similar results have been reported for mouse cells lacking both homologues of Rad23 (mHR23A and mHR23B) [63]. Homozygous loss of either mHR23A or mHR23B results in viable knockout mice, though a double mHR23A/B knockout mouse was inviable. Although the double mutant mouse was not viable, cell lines were established from the embryos and characterized for DNA repair-specific defects. In the absence of both mHR23A and B, mXPC (the mouse homolog of Rad4) was undetectable. Significantly, treating the double knockout cells with a proteasome inhibitor resulted in detectable XPC, consistent with the yeast results. These results suggest that mHR23A and B can interact with XPC and prevent its multiubiquitylated and degradation.
Cells from xeroderma pigmentosum patients of complementation group E have a defect in the repair of nontranscribed sequences. Two proteins that contribute to this defect are p48 and p125, which form a heterodimeric complex called UV-DDB that recognizes UV-damaged DNA. Intriguingly, the expression of p48 is regulated by the tumor suppressor p53, whose levels are also altered by DNA damage [64]. The p48 subunit binds to a specific E3 ubiquitin-ligase complex CUL-4A [65, 66, 67], and is subsequently ubiquitylated and degraded.
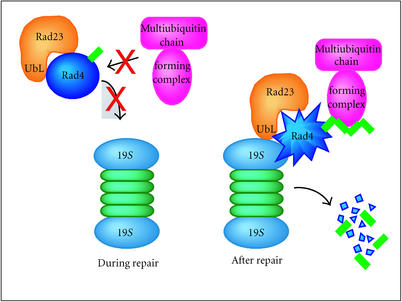
Collectively, these findings lead us to suggest that the proteins required for the nucleotide excision repair, or regulation of NER could be constitutively degraded by the 26S proteasome (Figure 4). However, in the absence of proteasome function, the repair proteins are expected to accumulate and increase the repair capacity of the cell. We hypothesize that following the completion of DNA repair, the repair proteins whose levels were induced are degraded by the proteasome. This mechanism of regulation could prevent improper incision of DNA structures that arise naturally during cellular processes such as transcription, replication, and recombination. This model also predicts that a failure to properly control the levels of specific repair proteins might cause deleterious effects, such as genomic instability.
Figure 4.
A model for regulation of repair by protein degradation. The ubiquitylation and degradation of specific repair proteins is inhibited in the presence of DNA damage. However, upon completion of repair, factors such as Rad23 might translocate repair proteins to the proteasome to promote degradation. Rad4 is a potential proteolytic target for Rad23, though there may exist other repair proteins that are degraded by the proteasome following the completion of DNA repair. It is also conceivable that negative regulators of NER, which suppress the activity of this repair pathway, are degraded following exposure to DNA-damaging agents.
THE ROLE OF RAD23 IN PROTEOLYSIS AND REPAIR
Recent observations provide support for the hypothesis that Rad23 proteins have proteolytic functions. Watkins et al [56] initially noticed that the amino-terminal domain of Rad23 (UbL) bore a striking resemblance to the sequence of ubiquitin (Ub), and intriguingly, Ub could functionally replace UbL [56]. At the time, it was proposed that Rad23 might play a role in ubiquitin/proteasome-mediated protein degradation since the only known function for Ub was its well documented effects in proteolysis. Recent studies have shown that Rad23 can bind the proteasome through its UbL domain, and can inhibit the assembly of substrate linked multi-Ub chains in a reconstituted system [68]. Additionally, a conserved motif called the ubiquitin-associated (UBA) domain has been reported to bind Ub, multi-Ub chains, and ubiquitinated cellular proteins [69, 70]. The UBA motif is present in a diverse array of regulators of signal transduction, DNA repair and proteolysis, and it seems likely that the interaction with ubiquitinated proteins provides these pathways with a previously unknown link to the proteolytic system. Other binding partners of the UBA domain have also been described, and their interaction with Rad23 can affect its DNA repair properties. Other genetic and biochemical studies are also consistent with a proteolytic role for Rad23. For instance, the loss of Rad23 in yeast cells caused stabilization of specific model substrates that was compounded by the simultaneous loss of Rpn10, a proteasome associated multi-Ub chain binding protein.
Several lines of evidence have predicted a role for the ubiquitin/proteasome system in DNA repair. Perhaps the first DNA repair protein to be directly associated with protein degradation is Rad6/Ubc2, which encodes a ubiquitin-conjugating enzyme. The RAD6/UBC2 gene had been extensively studied and was shown to play a role in providing resistance to various types of DNA damage, meiotic recombination and sporulation, and induced mutagenesis, consistent with a role in the postreplication bypass repair. Rad6/Ubc2 is also required for retrotransposition and proper growth, and its catalytic activity as an E2 enzyme is required for all its known functions. However, the specific targets of Rad6/Ubc2, which are related to its DNA repair specific functions, are still unknown. Similarly, two other proteins that play a role in postreplication repair are Mms2 and Ubc13, which comprise a bipartite ubiquitin-conjugating enzyme. Intriguingly, Mms2/Ubc13 assemble a unique type of multi-Ub chain that is mediated by linkages involving lysine-63 (K63) in Ub. The K63-linked multi-Ub chains are among the most abundant ubiquitinated proteins in yeast cells, and these species are not detected in cells that express the K63R Ub mutant. A subset of these bands was present at higher levels following exposure of yeast cells to UV light. Spence et al [30] reported that yeast mutants that were unable to assemble K63 multi-Ub chains were highly sensitive to DNA damage, since they were sensitive to MMS and UV light, and had reduced levels of induced-mutagenesis, consistent with the defects associated with mutations in RAD6/UBC2. Surprisingly, however, the K63R mutation was able to partially suppress the defects of a rad6/ubc2 null mutant. The RAD6 epistasis pathway includes two additional members that encode potential ubiquitin protein (E3) ligases. One of these is Rad18, a single-stranded DNA-dependent ATPase that forms a high affinity complex with Rad6/Ubc2, and can potently stimulate the ubiquitin-conjugating activity of Rad6/Ubc2 on a test substrate. Additionally, Rad5 is an ATPase/helicase that displays similarity to the SNF2/SWI2 family of chromatin remodeling factors. Both Rad18 and Rad5 are putative RING-type E3 proteins that, in concert with Rad6/Ubc2, function in error-free repair. Evidence for the existence of a large complex containing both putative E3 factors (Rad18, Rad5), as well as both E2 enzymes (Rad6/Ubc2, Mms2/Ubc13), suggests that distinct types of multi-Ub chains could be assembled on DNA repair specific targets. Rad6/Ubc2 has been shown to assemble K48 chains, which are recognized by the 26S proteasome to promote degradation of substrates. In contrast, the K63 chains that are assembled by Ubc13/Mms2 do not appear susceptible to degradation, raising the possibility that K63 multi-Ub chains might compete with K48 chains to modulate the in vivo stability of DNA repair proteins, possibly in response to DNA damage. A clear precedence for this type of regulation is evident by the competition between a ubiquitin-like protein (SUMO-1) and Ub for ligation to physiological substrates. For instance, attachment of SUMO-1 results in the stabilization of IκBα, while conjugation to Ub results in its degradation by the proteasome. Similar findings have been reported for Mdm2, an E3 protein that regulates the stability of p53 in response to DNA damage.
Rad23 and its counterparts in humans (hHR23A and hHR23B) interact with many other proteins, including 3-methyladenine-DNA glycosylase (MPG), Png1, ataxin-3, and ubiquitin. The interaction with MPG is of particular interest because the hHR23/MPG complex binds alkylated DNA with high affinity. The C-terminal 68 amino acids of hHR23B that interact with MPG differ from the residues that bind XPC. In yeast, a protein involved in deglycosylation of misfolded proteins, Png1, also interacts with the C-terminus of Rad23. Interestingly, Png1 is a peptide-N-glycanase (PNGase) that shares a common transglutaminase fold with Rad4, suggesting that these proteins may have evolved from a common ancestoral PNGases [71]. Interestingly, the interaction between Png1 and the C-terminal UBA domain of Rad23 prevents the interaction with Rad4, though Rad4 binds a distinct region in Rad23. Because Png1 and Rad4 compete for interaction with Rad23, it is conceivable that the repair capacity of the cell is influenced by the availability of Rad23. In agreement with this idea, it was found that over-expression of Png1 prevented Rad23/Rad4 association, and was accompanied by severe sensitivity to UV light. A plausible interpretation of these results is that Rad4 is constitutively degraded in the absence of damaged DNA, and it is specifically stabilized in a Rad23/Rad4 in the presence of damaged DNA. This complex (NEF2) might then be competent for promoting the assembly of other repair proteins at the sites of lesions.
THE 26S PROTEASOME NEGATIVELY REGULATES DNA REPAIR
We speculate that DNA damage results in increased levels of specific repair proteins. However, the successful completion of DNA repair may be followed by the controlled degradation of repair proteins by the proteasome. The maintenance of low levels of specific repair factors could serve to carefully regulate the activity of the repair proteins, to prevent improper activities on DNA structures that occur naturally during DNA replication, recombination, and transcription. As described above in Stabilization of a Repair protein in a Proteasomal Mutant, Rad4 appears to be a substrate of the 26S proteasome, and its stability is likely to be controlled by Rad23. In agreement with this conjecture, we overexpressed Rad4 in wild-type cells and observed increased repair in both strands of the RPB2 gene. These findings are consistent with the notion that specific regulatory proteins are degraded in the absence of DNA damage, but stabilized in the presence of lesions to promote repair. Rad4 may be representative of this class of proteins, and its elevated levels following DNA damage could increase transcription-coupled and genomic nucleotide excision repair.
ADDITIONAL RESPONSES TO DNA DAMAGE INVOLVING THE PROTEASOME
Not all cellular responses to DNA damage are directly related to the process of repair, since other consequences involving cell cycle arrest (checkpoint) and recovery, also involve proteolysis. Indeed, Rad23 is known to participate in a G2/M-phase transition during the cell cycle, and has been shown to control the in vivo levels of Pds1, an important regulator of cell cycle progression and DNA damage response. Furthermore, the expression of many proteins involved in transcriptional regulation is induced, and proteins are activated or stabilized in response to DNA damage. For instance, the levels of the transcription factor Gcn4, in response to UV light and amino acid deprivation, are controlled by proteolysis and by posttranscriptional mechanisms. Gcn4 is a bZip protein of the AP-1 family that includes AP-1 and c-Jun. The protein kinase Gcn2 is essential for controlling Gcn4 protein levels in response to amino acid starvation. Gcn4 is rapidly turned over in cells growing in nutrient rich media. Gcn4 is ubiquitylated by the ubiquitin-conjugating enzymes Rad6 and Cdc34/SCFCdc34 [72, 73], and degraded by the 26S proteasome. As expected, the degradation of Gcn4 is reduced in rad6Δ and cdc34ts mutants, and in a cim5-1 proteasome mutant [72, 73]. Exposing cells to UV radiation or shifting glucose-conditioned cells to glucose-deficient medium results in the stabilization of Gcn4, and increased transcription from Gcn4-dependent genes [74, 75]. These results point to proteolytic effects that are manifested by DNA damage-inducing conditions, that may be quite unrelated to the enzymology of DNA repair itself.
CONCLUSIONS
Ubiquitin has been shown to participate in a variety of biochemical activities in addition to proteolysis. The ligation of mono-Ub to histone H2A and H2B has been known for a long time, though the significance of this modification is unknown. Recent studies have shown that the mono-ubiquitylation of numerous cell surface receptors promotes internalization and accurate localization to the vacuole or lysosome for degradation. While it was long presumed that a substrate-linked multiubiquitin chain was the singular feature that distinguished proteolytic from nonproteolytic substrates, it is now quite clear that the nature of the Ub∷Ub linkage within the chain defines the fate of the targeted protein. These findings make clear that ubiquitin conjugation could have diverse biochemical effects. Consequently, the effect of the Ub/proteasome pathway in nucleotide excision repair will have to be assessed on a case-by-case basis, since it could promote the degradation of some repair factors, while altering the activity of others in a nonproteolytic manner. If Rad23-like proteins are deficient or absent in a cell, then nondegraded substrate proteins whose activity is normally regulated by mono- and di-ubiquitylation may get multiubiquitylated and subsequently degraded by the 26S proteasome. Even in the case of Rad23, for which proteolytic effects have been clearly ascribed, a nonproteolytic role involving Ub and the proteasome (or its subunits) in protein folding and/or disassembly of protein complexes has to be considered.
Acknowledgments
ACKNOWLEDGMENTS
The authors thank their colleagues, Lori Lommel and Mike Hampsey, for helpful comments and conversations. This work was supported by a grant from the New Jersey State Commission on Cancer Research (02-1083-CCR-SO) (KSS) and the National Institutes of Health (1R01 CA83875) (KM).
References
- Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270(22):12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- Huang J C, Svoboda D L, Reardon J T, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89(8):3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Biggerstaff M, Shivji M K, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80(6):859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270(6):2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- Guzder S N, Bailly V, Sung P, Prakash L, Prakash S. Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J Biol Chem. 1995;270(15):8385–8388. doi: 10.1074/jbc.270.15.8385. [DOI] [PubMed] [Google Scholar]
- Guzder S N, Sung P, Prakash L, Prakash S. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J Biol Chem. 1998;273(11):6292–6296. doi: 10.1074/jbc.273.11.6292. [DOI] [PubMed] [Google Scholar]
- Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23(14):2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon S A, Lawrence D A. Strand-selective repair of DNA damage in the yeast GAL7 gene requires RNA polymerase II. J Biol Chem. 1992;267(32):23175–23182. [PubMed] [Google Scholar]
- Mellon I, Hanawalt P C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Bedoyan J, Gupta R, Thoma F, Smerdon M J. Transcription, nucleosome stability, and DNA repair in a yeast minichromosome. J Biol Chem. 1992;267(9):5996–6005. [PubMed] [Google Scholar]
- Sweder K S, Hanawalt P C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci USA. 1992;89(22):10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Tasseron-de Jong J G, Verhage R A, Brouwer J. Defective Kin28, a subunit of yeast TFIIH, impairs transcription-coupled but not global genome nucleotide excision repair. Mutat Res. 1998;409(3):181–188. doi: 10.1016/s0921-8777(98)00060-3. [DOI] [PubMed] [Google Scholar]
- Carreau M, Hunting D. Transcription-dependent and independent DNA excision repair pathways in human cells. Mutat Res. 1992;274(1):57–64. doi: 10.1016/0921-8777(92)90043-3. [DOI] [PubMed] [Google Scholar]
- Christians F C, Hanawalt P C. Inhibition of transcription and strand-specific DNA repair by α-amanitin in Chinese hamster ovary cells. Mutat Res. 1992;274(2):93–101. doi: 10.1016/0921-8777(92)90056-9. [DOI] [PubMed] [Google Scholar]
- Conconi A, Bespalov V A, Smerdon M J. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc Natl Acad Sci USA. 2002;99(2):649–654. doi: 10.1073/pnas.022373099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, Odijk H, de Wit J, et al. Molecular cloning of the human DNA excision repair gene ERCC-6. Mol Cell Biol. 1990;10(11):5806–5813. doi: 10.1128/mcb.10.11.5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers J H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71(6):939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- Huang M E, Chuat J C, Galibert F. A possible yeast homolog of human active-gene-repairing helicase ERCC6+ . Biochem Biophys Res Commun. 1994;201(1):310–317. doi: 10.1006/bbrc.1994.1703. [DOI] [PubMed] [Google Scholar]
- van Gool A J, Verhage R, Swagemakers S M, et al. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6 . EMBO J. 1994;13(22):5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Van den Boom V, Schnitzler G, et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20(20):7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94(21):11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers J H, Tanaka K. Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci USA. 2002;99(1):201–206. doi: 10.1073/pnas.012473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R J, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci USA. 1991;88(16):7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J Biol Chem. 1999;274(39):28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Hofmann R M, Pickart C M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96(5):645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis A L, Howley P M. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274(26):18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Madura K, Prakash S, Prakash L. Expression of the Saccharomyces cerevisiae DNA repair gene RAD6 that encodes a ubiquitin conjugating enzyme, increases in response to DNA damage and in meiosis but remains constant during the mitotic cell cycle. Nucleic Acids Res. 1990;18(4):771–778. doi: 10.1093/nar/18.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S J, Reed S H, Huang W, Friedberg E C, Johnston S A. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3(6):687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391(6668):715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas A L, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spek P J, Smit E M, Beverloo H B, et al. Chromosomal localization of three repair genes: the xeroderma pigmentosum group C gene and two human homologs of yeast RAD23. Genomics. 1994;23(3):651–658. doi: 10.1006/geno.1994.1554. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wei S, Reed S H, et al. The RAD7, RAD16, and RAD23 genes of Saccharomyces cerevisiae: requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol Cell Biol. 1997;17(2):635–643. doi: 10.1128/mcb.17.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P C, Hockendorff J, Johnson E S, Varshavsky A, Dohmen R J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92(4):489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- Mieczkowski P, Dajewski W, Podlaska A, Skoneczna A, Ciesla Z, Sledziewska-Gojska E. Expression of UMP1 is inducible by DNA damage and required for resistance of S. cerevisiae cells to UV light. Curr Genet. 2000;38(2):53–59. doi: 10.1007/s002940000136. [DOI] [PubMed] [Google Scholar]
- Ulrich H D, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19(13):3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J M, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272(44):28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- Ford J M, Baron E L, Hanawalt P C. Human fibroblasts expressing the human papillomavirus E6 gene are deficient in global genomic nucleotide excision repair and sensitive to ultraviolet irradiation. Cancer Res. 1998;58(4):599–603. [PubMed] [Google Scholar]
- Wagenknecht B, Hermisson M, Eitel K, Weller M. Proteasome inhibitors induce p53/p21-independent apoptosis in human glioma cells. Cell Physiol Biochem. 1999;9(3):117–125. doi: 10.1159/000016308. [DOI] [PubMed] [Google Scholar]
- Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93(21):11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273(9):5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- Lee K B, Wang D, Lippard S J, Sharp P A. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc Natl Acad Sci USA. 2002;99(7):4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94(8):3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudenon S L, Huacani M R, Wang G, McDonnell D P, Huibregtse J M. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(10):6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel L, Bucheli M E, Sweder K S. Transcription-coupled repair in yeast is independent from ubiquitylation of RNA pol II: implications for Cockayne's syndrome. Proc Natl Acad Sci USA. 2000;97(16):9088–9092. doi: 10.1073/pnas.150130197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel L, Chen L, Madura K, Sweder K. The 26S proteasome negatively regulates the level of overall genomic nucleotide excision repair. Nucleic Acids Res. 2000;28(24):4839–4845. doi: 10.1093/nar/28.24.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudstra E C, Gilbert C, Fellows J, et al. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415(6874):929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- Vu P K, Sakamoto K M. Ubiquitin-mediated proteolysis and human disease. Mol Genet Metab. 2000;71(1-2):261–266. doi: 10.1006/mgme.2000.3058. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106(5):527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22(10):383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Hendil K B, Hartmann-Petersen R, Tanaka K. 26S proteasomes function as stable entities. J Mol Biol. 2002;315(4):627–636. doi: 10.1006/jmbi.2001.5285. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel P M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17(21):6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel P M. GFP-labelling of 26S proteasomes in living yeast: insight into proteasomal functions at the nuclear envelope/rough ER. Mol Biol Rep. 1999;26(1-2):131–135. doi: 10.1023/a:1006973803960. [DOI] [PubMed] [Google Scholar]
- Weeda G, Rossignol M, Fraser R A, et al. The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res. 1997;25(12):2274–2283. doi: 10.1093/nar/25.12.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi S, Shimbara N, Tamura T. Tissue and cell distribution of a mammalian proteasomal ATPase, MSS1, and its complex formation with the basal transcription factors. Biochem Biophys Res Commun. 2000;279(2):568–573. doi: 10.1006/bbrc.2000.3969. [DOI] [PubMed] [Google Scholar]
- Watkins J F, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13(12):7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J P, Smerdon M J. Rad23 is required for transcription-coupled repair and efficient overrall repair in Saccharomyces cerevisiae . Mol Cell Biol. 1996;16(5):2361–2368. doi: 10.1128/mcb.16.5.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky S A, Estep P, Church G M, Samson L D. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20(21):8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450(1-2):27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Swaffield J C, Bromberg J F, Johnston S A. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature. 1992;357(6380):698–700. doi: 10.1038/357698a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston S A. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296(5567):548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Lommel L, Ortolan T, Chen L, Madura K, Sweder K. Protcolysis of a nucleotide excision repair protein by the 26S proteasome. Curr Genet. 2002;2002 doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- Ng J M.Y. Erasmus University; Rotterdamm, The Netherlands: 2001. Mammalian RAD23 homologs: multifunctional proteins in DNA repair and development [Ph.D. thesis] [Google Scholar]
- Hwang B J, Ford J M, Hanawalt P C, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96(2):424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem. 1999;274(50):35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol. 2001;21(20):6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem. 2001;276(51):48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- Ortolan T G, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2(9):601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- Bertolaet B L, Clarke D J, Wolff M, et al. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8(5):417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- Wilkinson C R, Seeger M, Hartmann-Petersen R, et al. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3(10):939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin E V, Aravind L. Peptide-N-glycanases and DNA repair proteins, Xp-C/Rad4, are, respectively, active and inactivated enzymes sharing a common transglutaminase fold. Hum Mol Genet. 2001;10(16):1627–1630. doi: 10.1093/hmg/10.16.1627. [DOI] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13(24):6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimoun A, Holtzman T, Weissman Z, et al. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell. 2000;11(3):915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach I, Licht R, Frohnmeyer H, Engelberg D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J Biol Chem. 2001;276(20):16944–16951. doi: 10.1074/jbc.M100383200. [DOI] [PubMed] [Google Scholar]
- Stitzel M L, Durso R, Reese J C. The proteasome regulates the UV-induced activation of the AP-1-like transcription factor Gcn4. Genes Dev. 2001;15(2):128–133. doi: 10.1101/gad.863801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx D A, Mason R, van Hoffen A, et al. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci USA. 2000;97(19):10503–10508. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]