Abstract
The Gene Expression Profile Analysis Suite (GEPAS) has been running for more than four years. During this time it has evolved to keep pace with the new interests and trends in the still changing world of microarray data analysis. GEPAS has been designed to provide an intuitive although powerful web-based interface that offers diverse analysis options from the early step of preprocessing (normalization of Affymetrix and two-colour microarray experiments and other preprocessing options), to the final step of the functional annotation of the experiment (using Gene Ontology, pathways, PubMed abstracts etc.), and include different possibilities for clustering, gene selection, class prediction and array-comparative genomic hybridization management. GEPAS is extensively used by researchers of many countries and its records indicate an average usage rate of 400 experiments per day. The web-based pipeline for microarray gene expression data, GEPAS, is available at http://www.gepas.org.
INTRODUCTION
It is quite common that the introduction of a new technology is accompanied by claims and promises which on many occasions cannot be fulfilled. This hype is then followed by a wave of disappointment against the technology. Fortunately, as it is reaching a certain degree of maturity, DNA microarray technologies do not seem to have followed this fate. During an initial period, DNA microarray publications were dealing with issues such as reproducibility and sensitivity. Many classical microarray papers dating from the late nineties were mere proof-of-principle experiments (1,2), in which only cluster analysis was applied. Later, sensitivity became a main concern as a natural reaction against quite liberal interpretations of microarray experiments made by some researchers, such as the fold criteria to select differentially expressed genes. It was soon obvious that genome-scale experiments should be carefully analysed because many apparent associations happened merely by chance (3). In this context, different methods for the adjustment of P-values, which are considered standard today, started to be extensively used (4,5). More recently the use of microarrays as predictors of clinical outcomes (6), despite not being free of criticisms (7), fuelled the use of the methodology because of its practical implications. There are still some concerns with the cross-platform coherence of results but it seems clear that intra-platform reproducibility is high (8) and, despite the fact that gene-by-gene results are not always the same, the biological themes emerging from the different platforms are increasingly consistent (9). That points to the importance of the interpretation of experiments in terms of their biological implications instead of a mere comparison of lists of genes (10,11).
Keeping a pace with the trends mentioned above, Gene Expression Profile Analysis Suite (GEPAS) has been growing during the last 4 years. In the first release it was more oriented towards clustering and data preprocessing (12). Successive releases showed a package more oriented towards gene selection, class prediction and the functional annotation of experiments (13,14). The version presented here include several new modules, some of which are new while other ones constitute already available tools completely rewritten including new functionalities. GEPAS is not a simple web server, but it constitutes one of the largest resources for integrated microarray data available over the web. It has been working for more than four years having by the end of year 2005 an average of 400 experiments analysed per day summing up over all of their modules. GEPAS is used by researches worldwide as can be seen in the usage map, where all the sessions are mapped to its geographic location (http://bioinfo.cipf.es/access_map/map.html). It also offers on-line tutorials that can be used in courses. In the new version (3.0) we present new modules for the normalization of Affymetrix experiments, for differential gene expression, for the evaluation of cluster quality and another module for array-comparative genomic hybridization (Array-CGH) data management. Also, another conceptual novelty is the connection of GEPAS to the PupaSuite tools (15–17), which offers the possibility of analysing polymorphisms at the light of the results of the gene expression analysis.
GENERAL OVERVIEW
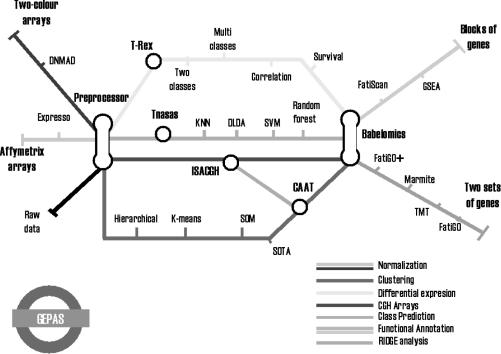
GEPAS aims to tackle the most common problems in microarray data analysis in a simple but rigorous way. Thus, after an essential step of normalization, there are different ‘workflows’, or sequences of steps, that can be followed, depending on the aim of the experiment: class discovery, differential gene expression, class prediction or genomic copy number estimation, just to cite the most common objectives of microarray experiments. Class discovery, either in genes or in experiments, is achieved by using clustering methods. GEPAS includes some commonly used clustering methods such as hierarchical clustering (18), SOTA (19,20), SOM (21), K-means (22) and SOM-Tree (23). The evaluation of cluster quality, a scarcely addressed issue, has been implemented here in the Cluster Accuracy Analysis Tool (CAAT) module (see below). Differential gene expression implies finding genes with significant differences in expression between two or more classes, related to a continuous experimental factor (e.g. the concentration of a metabolite) or to survival data. A new, more complete module for differential gene expression is presented in this new version of GEPAS (see below). The module Tnasas for class prediction implements different classifiers, such as diagonal linear discriminant analysis (DLDA) (24), nearest neighbour (NN) (25), support vector machines (SVM) (26), random forest (27) and shrunken centroids (PAM) (28) of known efficiency as class predictors using microarray data (24). Cross-validation error is calculated in a way to avoid the well-known selection bias problem (29,30). See Tnasas help (http://tnasas.bioinfo.cipf.es/cgi-bin/docs/tnasashelp) for a more detailed description of the methods and error estimation strategy. Array-CGH (31) can be analysed through the module ISACGH that allows predicting copy number, relating these values to gene expression and performing functional annotation through the babelomics (11) suite. Finally, functional annotation is carried out with the babelomics suite which can be used either as an independent suite or as an integrated part of the GEPAS. Figure 1 illustrates, following the metaphor of a subway line, the interconnections of the different tools in the GEPAS environment.
Figure 1.
Map of GEPAS functionalities as a subway line. Data (Affimetrix, two-colour or raw) are introduced from the left side and pass through the preprocessor. Then different types of analyses can be performed: gene selection (T-rex) in different situations (two or more classes, correlation or survival; see text for details) or class discovery (Tnasas) are two types of supervised analyses. Array-CGH data can be analysed through the red line ISACGH. Unsupervised analysis can also be performed using different methods. CAAT allows to map co-expressed genes on their chromosomal coordinates allowing the study of RIDGES (54). All the tools end up in Babelomics (11), that allows for two different types of analysis: comparison of two sets of genes of analysis or blocks of functionally related genes.
NORMALIZATION AND PREPROCESSING
GEPAS now implements normalization facilities for both two-colours and Affymetrix arrays. DNMAD (32) module performs normalization in two-colour arrays using print-tip loess (33) with a number of different options. DNMAD can input Genepix (Axon instruments) GPR files. The module expresso normalizes Affymetrix CEL files using standard Bioconductor (34) tools; in particular the package affy (35). Besides its friendly web interface we provide the user with the speed and above all the physical memory available in our server.
More information can be found in the corresponding tutorial web pages (http://bioinfo.cipf.es/docus/courses/on-line.html).
In addition, the preprocessor (36) module performs some preprocessing of the data (log-transformations, standardizations, imputation of missing values and so on).
CLUSTERING AND CLUSTER QUALITY ESTIMATION
Despite the fact that clustering is one of the most popular—albeit often improperly used (30)—methodologies in the analysis of microarray data there are very few alternatives for the estimation of the quality of the results found. We have included a module, CAAT, which provides many options for the visualization and intuitive manipulation of hierarchical and non-hierarchical clustering results. Many visualization modes, browsing options and cluster extraction possibilities are currently available. Moreover, CAAT provides some descriptive measures about each partition (average profiles, standard deviation profiles, inter and intra-cluster distances) as well as a global estimation of cluster quality by the silhouette method (37), which performs well, in noisy situations, such as microarray analysis (38). CAAT submits data to other tools such as the Babelomics (11) functional annotation suite or to ISACGH (Figure 1).
There is more detailed information in the CAAT documentation (http://bioinfo.cipf.es/docus/courses/on-line.html).
DIFFERENTIAL GENE EXPRESSION
This version of GEPAS includes new methods for differential gene expression analysis under different conditions. The old module pomelo has been replaced by the new module T-rex (Tools for RElevant gene seleXion) which is much faster and offers new tests for different situations. T-rex distinguishes among four conceptually different testing cases:
- Finding genes differentially expressed between two discrete classes (e.g. case/control and so on). A number of authors (39,40) have found that the classical t-statistic, which was widely used in early work on the analysis of differential expression, can be highly unreliable for microarray data. Problems arise mainly as a consequence of statistical issues relating to the SD term in the denominator of the t-statistic. For example, many non-differentially expressed genes may by chance have small observed SDs, which may cause these genes to be erroneously selected. GEPAS now also implements different new tests:
- The t-test, which is still available.
- An empirical Bayes methodology that allows fitting hierarchical mixture models to identify differentially expressed genes (41). One of the advantages of this methodology is that it fits a global model taking into account all genes in the dataset.
- A novel test for the analysis of microarray data by combining inference for differential expression and variability (CLEAR-test) (J. Valls, M. Grau, X. Sole, P. Hernandez, D. Montaner, J. Dopazo, M. A. Peinado, G. Capella, M. A. G. Pujana and V. Moreno, manuscript submitted). Most tests evaluate differential expression by using estimated variability, but no inference is made in terms of the variability itself. CLEAR-test evaluates both whether genes show large fold changes and whether their variability is high.
- A data-adaptive approach to the analysis of differential expression, in which an effective test statistic is learned directly from microarray data. This approach has been shown to ameliorate many of the problems associated with both the t-statistic and simple moderated statistics like SAM (42), and to produce good results under a range of conditions (43).
Finding genes differentially expressed between more than two classes (e.g. different types of cancers and so on) Together with the classical ANOVA methodology we make available the same CLEAR test mentioned above (41). While the mathematical treatment of this kind of data is similar to that of two classes, in our tools, we separate the case when more than two classes are available because of its different conceptual implications.
Finding genes whose expression is correlated to a continuous variable (e.g. the level of a metabolite). Regression analysis of gene expression on any numerical independent variable has been implemented. C routines have been compiled for the particular architecture of our computers in order to achieve the maximal speed. Estimates of Pearson's and Spearman's correlation coefficients as well as P-values for testing the null hypothesis of no correlation can also be obtained with T-rex.
Finding genes whose expression is related to survival times. GEPAS uses C routines to estimate a Cox proportional hazards regression model (44). Right censored data are allowed as well as replicates in the survival times. Censoring variables should be provided by the researcher together with survival times that may be replicated.
When appropriate, P-values adjusted for multiple testing are provided. Three methodologies are implemented. One of them controls the FWER (family-wise error rate) (45) while the others control the FDR (false discovery rate) (46). Our implementations make use of the p.adjust function in the stats R package and the qvalues package (47) from Bioconductor.
FUNCTIONAL ANNOTATION
Functional annotation of the experiments gives clues to the researcher for the interpretation of the experiment. There are a number of tools that make use of gene functional annotations to try to understand the global changes in gene expression in microarray experiments (48), but probably one of the most complete packages in this respect is the Babelomics suite (11,49). This suite of programs for functional annotation of genome-scale experiments has undergone a deep modification described in detail elsewhere (49). In brief, Babelomics can now compare two groups of genes and test simultaneously for the significant over-abundance of diverse biological themes such as GO terms, KEGG pathways, Interpro motifs, Swissprot keywords, Transfac® motifs, CisRed motifs, relative abundance in tissues and bioentities extracted from PubMed, with the proper multiple testing adjustment. This is carried out by the FatiGO+ module, the evolution of the FatiGO program (50). Additionally there are two modules designed to search for functionally related blocks of genes that are co-ordinately over- or under-expressed using both the FatiScan (51) or the GSEA (52) algorithms.
Despite its general scope (Babelomics is not restricted to microarrays but applicable to any type of large-scale experiment), and the possibility of being used alone as an independent resource, the Babelomics suite has been fully integrated into GEPAS. Modules of gene selection (T-rex) or class prediction (tnasas) can submit the genes selected as relevant to the FatiGO+ module for testing against the rest of genes. Likewhise, the modules for clustering (hierarchical, k-means, SOM, SOTA) through their cluster' viewers or through CAAT, can submit the genes within the selected cluster to be tested against the rest of genes. Similar operation can be performed from within ISACGH, with the genes contained in the selected chromosomal region. Moreover, arrangements of genes can be sent from T-rex to the FatiScan to test blocks of functionally related genes tha are co-ordinately over- or under-expressed. Sets of arrays can also be submitted to GSEA with the same purpose.
ARRAY-CGH
Genetic aberrations, which are the molecular basis of many diseases, have classically been studied through CGH. The introduction of microarray-based CGH methods (array-CGH) has revolutionized this methodology in terms of resolution and throughput (31,53) but, at the same time, has generated a need for new algorithms and software for dealing with this type of data. We have included in GEPAS a new module, ISACGH, which completely replaces the old viewer InSilicoCGH (14). ISACGH includes two new and efficient methods for accurate estimation of genomic copy number from array-CGH hybridization data, integrated into a web-based system that allows, for the first time, the combined study of gene expression and genomic copy number. Several visualization options offer a convenient representation of the results. Moreover, the link to the Babelomics (11,49) tools allows, for the first time in a tool of this type, the production of functional annotations (using different relevant biological information such as gene ontology, pathways, etc.) for the detected chromosomal regions of interest (amplified or deleted). We use the DAS technology (Distributed Annotation System; see http://www.biodas.org/), that allows a remote mapping of information (our predictions) from a server (our server) to a client (Ensembl), to represent the ISACGH predictions and data onto the Ensembl chromosomal coordinates.
ISACGH generically maps data onto their chromosomal coordinates. So, beyond to map genomic hybridisations any other data can be mapped. Thus CAAT can send to ISACGH groups of co-expressing genes, which might be useful for defining regions of increased gene expression, also known as RIDGES (54).
Polymorphisms affecting gene expression
Although the study of regulatory polymorphisms is not new, there has been a recent revival of interest in them mainly because of the availability of high-throughput data and methodologies that allows their characterisation (55). The corresponding GEPAS modules (CAAT, tnasas and T-rex) have a unique feature in this regard: the possibility of connecting the genes found to be regulated in a microarray experiment to possible regulatory SNPs in such genes. In particular, clustering and gene selection methods can be connected to the PupaSuite (15–17).
DISCUSSION
GEPAS is a long-term project that aims to provide the scientific community with an advanced set of tools for microarray data analysis, without renouncing to an easy and intuitive use. It has been running uninterruptedly for more than four years and has grown to include more tools as new algorithms were introduced in the microarray data analysis arena (12–14). The GEPAS team has intended to deliver a coherent set of state-of-the-art and widely established algorithms, running away from building a simple collection of as-much-as-possible tools. Actually, any new tool included is the response to a new or emerging requirement requested by our users. As the Functional Genomics node of the Spanish Institute of Bioinformatics (INB; http://www.inab.org) and being part of the Spanish Network of Cancer Centers (RTICCC; http://www.rticcc.org) we have a direct contact with researchers from which we get much of the feedback necessary to build up a useful tool. GEPAS, integrated with the Babelomics suite (11,49), provides the tools for performing the most common analyses of microarray data. Moreover, it has been conceived as a workflow that helps the user to carry out a series of consecutive steps of analysis with simple mouse clicks. GEPAS has been designed to take full advantage of the properties of the web: connectivity, cross-platform functionality and remote usage. Its modular architecture allows easy implementation of new tools and facilitates the connectivity of GEPAS from and to other web-based tools.
The user of GEPAS ranges from the experimentalist with not much experience in bioinformatics and no deep statistical skills, interested only in data analysis, to the bioinformatician that invokes some of the tools remotely for different purposes.
GEPAS is running in a high-end cluster (with 20 dedicated AMD Opteron CPUs at 2.4 GHz) with a large amount of RAM (6 GB). This allows to use tools (e.g. normalization tools are highly RAM-consuming) that usually are beyond the capabilities of the hardware available to many end users.
In addition, there is a teaching programme related to GEPAS (see http://bioinfo.cipf.es/docus/courses/courses.html) with on-line tutorials that can be freely used (http://bioinfo.cipf.es/docus/courses/on-line.html).
Although other alternatives are available for microarray data analysis, there is no other similar resource over the web with the number of possibilities offered by GEPAS.
Acknowledgments
We are deeply acknowledged to our external advisors Sandrine Dudoit, Yves Moreau and John Quackenbush for their comments and support. This work is supported by grants from Fundació La Caixa, Fundación BBVA, MEC BIO2005-01078 and NRC Canada-SEPOCT Spain. The Functional Genomics node (INB) is supported by Genoma España. Funding to pay the Open Access publication charges for this article was provided by Genoma España.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou C.M., Jeffrey S.S., van de Rijn M., Rees C.A., Eisen M.B., Ross D.T., Pergamenschikov A., Williams C.F., Zhu S.X., Lee J.C., et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl Acad. Sci. USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge H., Walhout A.J., Vidal M. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini Y., Yekutieli D. The control of false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 5.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 8.Moreau Y., Aerts S., De Moor B., De Strooper B., Dabrowski M. Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Bammler T., Beyer R.P., Bhattacharya S., Boorman G.A., Boyles A., Bradford B.U., Bumgarner R.E., Bushel P.R., Chaturvedi K., Choi D., et al. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 10.Al-Shahrour F., Dopazo J. In: Data analysis and visualization in genomics and proteomics. Azuaje F., Dopazo J., editors. West Sussex, UK: Wiley; 2005. pp. 99–112. [Google Scholar]
- 11.Al-Shahrour F., Minguez P., Vaquerizas J.M., Conde L., Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero J., Al-Shahrour F., Diaz-Uriarte R., Mateos A., Vaquerizas J.M., Santoyo J., Dopazo J. GEPAS: A web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero J., Vaquerizas J.M., Al-Shahrour F., Conde L., Mateos A., Diaz-Uriarte J.S., Dopazo J. New challenges in gene expression data analysis and the extended GEPAS. Nucleic Acids Res. 2004;32:W485–W491. doi: 10.1093/nar/gkh421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaquerizas J.M., Conde L., Yankilevich P., Cabezon A., Minguez P., Diaz-Uriarte R., Al-Shahrour F., Herrero J., Dopazo J. GEPAS, an experiment-oriented pipeline for the analysis of microarray gene expression data. Nucleic Acids Res. 2005;33:W616–W620. doi: 10.1093/nar/gki500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conde L., Vaquerizas J., Dopazo H., Arbiza L., Reumers J., Rousseau F., Schymkowitz J., Dopazo J. PupaSuite: finding functional SNPs for large-scale genotyping purposes. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl071. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conde L., Vaquerizas J.M., Ferrer-Costa C., de la Cruz X., Orozco M., Dopazo J. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res. 2005;33:W501–W505. doi: 10.1093/nar/gki476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conde L., Vaquerizas J.M., Santoyo J., Al-Shahrour F., Ruiz-Llorente S., Robledo M., Dopazo J. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sneath P., Sokal R. Numerical Taxonomy. San Francisco: W.H. Freeman; 1973. [Google Scholar]
- 19.Dopazo J., Carazo J.M. Phylogenetic reconstruction using an unsupervised growing neural network that adopts the topology of a phylogenetic tree. J. Mol. Evol. 1997;44:226–233. doi: 10.1007/pl00006139. [DOI] [PubMed] [Google Scholar]
- 20.Herrero J., Valencia A., Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- 21.Kohonen T. Self-organizing maps. Berlin: Springer-Verlag; 1997. [Google Scholar]
- 22.Hartigan J., Wong M. A k-means clustering algorithm. Appl. Stat. 1979;28:100–108. [Google Scholar]
- 23.Herrero J., Dopazo J. Combining hierarchical clustering and self-organizing maps for exploratory analysis of gene expression patterns. J. Proteome Res. 2002;1:467–470. doi: 10.1021/pr025521v. [DOI] [PubMed] [Google Scholar]
- 24.Dudoit S., Fridlyand J., Speed T. Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 2002;97:77–87. [Google Scholar]
- 25.Ripley B. Pattern recognition and neural networks. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- 26.Vapnik V. Statistical Learning Theory. NY: Wiley; 1998. [DOI] [PubMed] [Google Scholar]
- 27.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 28.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambroise C., McLachlan G.J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl Acad. Sci. USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R., Radmacher M.D., Dobbin K., McShane L.M. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J. Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 31.Mantripragada K.K., Buckley P.G., de Stahl T.D., Dumanski J.P. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Vaquerizas J.M., Dopazo J., Diaz-Uriarte R. DNMAD: web-based diagnosis and normalization for microarray data. Bioinformatics. 2004;20:3656–3658. doi: 10.1093/bioinformatics/bth401. [DOI] [PubMed] [Google Scholar]
- 33.Smyth G., Yang Y., Speed T. In: Functional Genomics: Methods and Protocols. Brownstein M., Khodursky A., editors. Vol. 224. Totowa, NJ: Humana Press; 2003. pp. 111–136. [Google Scholar]
- 34.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 36.Herrero J., Diaz-Uriarte R., Dopazo J. Gene expression data preprocessing. Bioinformatics. 2003;19:655–656. doi: 10.1093/bioinformatics/btg040. [DOI] [PubMed] [Google Scholar]
- 37.Rousseeuw P. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 38.Azuaje F. A cluster validity framework for genome expression data. Bioinformatics. 2002;18:319–320. doi: 10.1093/bioinformatics/18.2.319. [DOI] [PubMed] [Google Scholar]
- 39.Baldi P., Long A.D. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 40.Cui X., Churchill G.A. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kendziorski C.M., Newton M.A., Lan H., Gould M.N. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 2003;22:3899–3914. doi: 10.1002/sim.1548. [DOI] [PubMed] [Google Scholar]
- 42.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee S., Roberts S.J., van der Laan M.J. Data-adaptive test statistics for microarray data. Bioinformatics. 2005;21:ii108–ii114. doi: 10.1093/bioinformatics/bti1119. [DOI] [PubMed] [Google Scholar]
- 44.Klein J.P., Moeschberger M.L. Survival Analysis: Techniques for Censored and Truncated Data. New York: Springer; 2003. [Google Scholar]
- 45.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- 46.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R Stat. Soc. [Ser B] 1995;57:289–300. [Google Scholar]
- 47.Storey J., Taylor J., Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J. R Stat. Soc. [Ser B] 2004;66:187–205. [Google Scholar]
- 48.Khatri P., Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Shahrour F., Minguez P., Tarraga J., Montaner D., Alloza E., Vaquerizas J.M., Conde L., Blaschke C., Vera J., Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl172. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 51.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. Discovering molecular functions significantly related to phenotypes by combining gene expression data and biological information. Bioinformatics. 2005;21:2988–2993. doi: 10.1093/bioinformatics/bti457. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albertson D.G., Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12:R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- 54.Caron H., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 55.Wang D.G., Fan J.B., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]