Abstract
Colorectal cancers (CRCs) are characterized by multiple genetic (mutations) and epigenetic (CpG island methylation) alterations, but it is not known whether these evolve independently through stochastic processes. We have recently described a novel pathway termed CpG island methylator phenotype (CIMP) in CRC, which is characterized by the simultaneous methylation of multiple CpG islands, including several known genes, such as p16, hMLH1, and THBS1. We have now studied mutations in K-RAS, p53, DPC4, and TGFβRII in a panel of colorectal tumors with or without CIMP. We find that CIMP defines two groups of tumors with significantly different genetic lesions: frequent K-RAS mutations were found in CIMP+ CRCs (28/41, 68%) compared with CIMP− cases (14/47, 30%, P = 0.0005). By contrast, p53 mutations were found in 24% (10/41) of CIMP+ CRCs vs. 60% (30/46) of CIMP− cases (P = 0.002). Both of these differences were independent of microsatellite instability. These interactions between CIMP, K-RAS mutations, and p53 mutations were preserved in colorectal adenomas, suggesting that they occur early in carcinogenesis. The distinct combinations of epigenetic and genetic alterations in each group suggest that activation of oncogenes and inactivation of tumor suppressor genes is related to the underlying mechanism of generating molecular diversity in cancer, rather than simply accumulate stochastically during cancer development.
Extensive molecular analyses have established that colorectal cancer (CRC) arises as a multistep process involving the accumulation of specific defects in oncogenes and tumor suppressor genes (1). A subset of familial CRCs (hereditary nonpolyposis colorectal cancers) results from germline mutations in mismatch repair genes (1) (2). Microsatellite sequences such as mono- and dinucleotide repeats are frequently altered in these tumors because of the mismatch repair defect (3–5). About 15–20% of nonfamilial CRCs also show microsatellite instability (MSI) characteristic of mismatch repair deficiency, but mutations of mismatch repair genes are not detected in most of these sporadic MSI+ tumors (6, 7). Other genetic defects in CRC include the nearly ubiquitous disruption of APC/β-catenin pathway, frequent activating mutations in the K-RAS oncogene that are thought to arise in large preneoplastic adenomas, and frequent p53 mutations that appear to arise at the adenoma–carcinoma transition (1).
Another potential mechanism underlying CRC progression is epigenetic silencing associated with promoter hypermethylation. Hypermethylation of 5′ CpG islands has been linked to transcriptional repression, and a growing number of genes, including tumor suppressor genes mutated in familial cancer syndromes, have been reported to be inactivated in neoplasms by this epigenetic event (8, 9). Examples of this process in CRCs include inactivation of the cell cycle regulator p16 (10), the growth suppressor ER (11), the angiogenesis inhibitor THBS1 (12), the metastasis suppressor TIMP3 (13), the DNA repair gene MGMT (14), and the mismatch repair gene hMLH1 (15). In fact, microsatellite instability in sporadic CRC appears to be related to epigenetic inactivation of hMLH1 in 70–80% of the cases studied (16, 17).
More recently, a distinct pathway for colorectal tumorigenesis was described, termed CpG island methylator phenotype (CIMP) (18). Tumors affected by this phenotype are characterized by a high degree of concordant CpG island methylation, which affects most of the genes known to be methylated in this tumor type (p16, hMLH1, THBS1). Given that genetic (mutations) and epigenetic (CIMP) changes are common in CRC, both could arise independently of each other, such that CRC might contain an accumulation of genetic/epigenetic changes that occurred randomly and were selected for stochastically during progression. Alternatively, epigenetic changes may mark a distinct group of tumors that have a distinct etiology and molecular profile. To clarify these issues, we examined the mutational status K-RAS, p53, DPC4, and TGFβRII in a series of tumors also characterized for the presence of CIMP. Our results indicate that CRCs develop through distinct epigenetic or genetic pathways that have markedly different molecular profiles.
Materials and Methods
Tumor Samples.
Initially, 41 unselected, sporadic CRCs, paired normal colon tissues, and 64 colorectal adenomas were obtained from patients without a family history of this disease and who were treated at the Johns Hopkins Hospital. Subsequently, 47 additional CRC cases and paired normal colon tissues were also collected. Informed consent for the use of the specimens was obtained from all patients. DNA was extracted by standard methods. The presence or absence of MSI in cancer samples was previously determined (12) according to strict criteria, requiring band shifts at both dinucleotide and mononucleotide tracts. A detailed methylation analysis of the first 41 cancer samples was previously reported (18).
Mutant Allele-Specific Amplification.
K-RAS mutations were determined by mutant allele-specific amplification, performed as described (19). In brief, 50 ng of genomic DNA was amplified by mutant allele-specific PCR primers, which exclusively amplify the mutated alleles of codon 12 and codon 13 of the K-RAS gene. PCR products were electrophoresed on 2% agarose gels and were visualized by ethidium bromide staining.
Single-Stranded Conformational Polymorphisms (SSCP) and Sequencing.
PCR was performed as described previously by using primers that amplify exons 2–11 of p53 (20) as well as exons 3, 5, 6, and 7 of TGFβRII (21). The primers used to amplify DPC4 exons 8–11 were designed based on the genomic DNA sequence (22), and their sequences are available at www.med.jhu.edu/methylation/primers.html. For SSCP, 4 μl of PCR products were mixed with 4 μl of loading buffer (95% formamide/20 mM EDTA/0.05% xylene cyanol/0.05% bromophenol blue), were denatured at 90°C for 3 min, were cooled on ice for 2 min, and were electrophoresed in nondenaturing polyacrylamide gels at a controlled temperature by using the Dcode Mutation Detection System (Bio-Rad). The presence of abnormally migrating bands was confirmed by three different conditions (with 5% glycerol at 20°C, with 5% glycerol at 10°C, and without glycerol at 4°C). The shifted bands were excised from gels and were reamplified by using the same set of primers. The PCR products were then purified by the PCR Purification System (Promega), and mutations were determined by direct sequencing using an Automated Sequencer (Applied Biosystems). In addition, direct sequencing of exon 5 to exon 8 of p53 was performed in five cases that showed loss of heterozygosity (LOH) by SSCP but no detected band shifts.
Methylated CpG Island Amplification.
Methylation of p16, MINT 1, 2, 12, 17, 27, and 31 in colorectal adenomas was determined by methylated CpG island amplification exactly as described (23). These loci were selected based on the fact that methylation was not detected in normal tissues using methylated CpG island amplification and Southern blot analysis. One hundred nanograms of methylated CpG island amplification PCR products were blotted onto nylon membranes and were hybridized by using 32P-labeled probes. Thirty nanograms of a DNA fragment from p16 exon 1 (10) and MINT clones (23) were labeled by random priming and were used as probes. Each sample was blotted in duplicate. Each filter included mixtures of a positive control and a negative control. The filters were exposed to a phosphor screen for 24–72 hr and were developed by using a PhosphorImager (Molecular Dynamics). Tumors that showed methylation of three or more loci (of six) were defined as CIMP+. All others were defined as CIMP−. A detailed protocol for methylated CpG island amplification is available at www.med.jhu.edu/methylation.
Methylation-Specific PCR (MSP) and Bisulfite-PCR.
The methylation status of p16, hMLH1, MINT2, and MINT31 was also determined by MSP (p16) or bisulfite-PCR (all others). In brief, 2 μg of genomic DNA was treated with Na-bisulfite for 16 hr. After purification, 2 μl of aliquot was used as template for PCR reactions. Methylation of p16 was determined by MSP as described (24). Methylation of hMLH1, MINT2, and MINT31 was determined by bisulfite-PCR followed by restriction digestion as described (25). In brief, 20–40 μl of the amplified products were digested with restriction enzymes that distinguish methylated from unmethylated sequences, were electrophoresed on 3% agarose or 5% acrylamide gels, and were visualized by ethidium bromide staining. Primer sequences, condition for PCR, and restriction enzymes used are available at www.med.jhu.edu/methylation/primers.html. For p16, MINT2, and MINT31, the results obtained by methylated CpG island amplification were 97% concordant with the results obtained by MSP (p16) or bisulfite-PCR (MINT2 and MINT31). Both MCA and bisulfite/PCR provide semiquantitative results. The loci selected for analysis are unmethylated (<1%) in normal tissues. Therefore, any tumor showing ≥5% methylation was considered positive. Because the tumors were not microdissected and contain a variable amount of contaminating normal tissues, no attempt was made to distinguish levels of methylation beyond positive/negative.
Results
K-RAS Mutations in CRCs With or Without CIMP.
The recently described hypermethylator phenotype in CRC termed CIMP affects about half of all cases and includes ≈70% of sporadic MSI+ cases through methylation of hMLH1 (18). CIMP was originally defined by using a panel of six newly cloned, differentially methylated CpG islands and required that the tumors methylated three or more of these loci. This threshold of three loci was based on the distribution of methylation: most cases (38/41 or 93%) had methylation of either zero or one loci, or at least four loci. Thus, changing the threshold to two loci or four loci would not appreciably change the CIMP classification. Moreover, all three cases that had only three MINT loci methylated also methylated p16, suggesting a true hypermethylator phenotype in these cases.
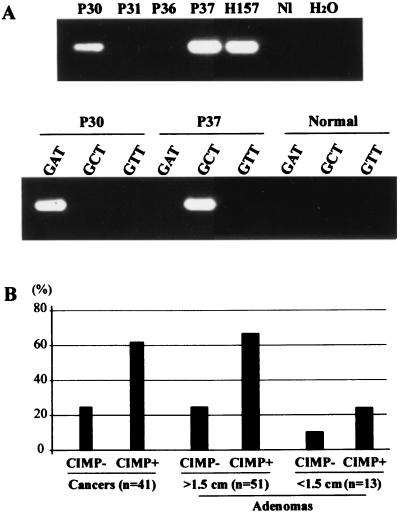
To study the interaction between CIMP and known genetic events in CRC, we began by determining the K-RAS mutational status of a panel of 41 unselected cancers previously typed for the presence of CIMP and MSI. Of 41 cases, 21 (51%) were CIMP+ and averaged 4.6/6 loci methylated whereas the 20 CIMP− cases averaged 0.3/6 loci methylated. Nine cases (22%) were MSI+, of which five were also CIMP+. Screening for K-RAS mutations at codons 12 and 13 was performed by mutant allele-specific amplification (19) (Examples in Fig. 1A, summarized in Fig. 1B and Table 1; details are shown in Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). K-RAS mutations were found in 13/21 (62%) CIMP+ tumors and 5/20 (25%) CIMP− tumors, and the difference between the two groups was statistically significant (P = 0.028 by Fisher's exact test). This difference was preserved when MSI+ cases were excluded [13/16 (81%) K-RAS mutations in CIMP+ cases vs. 4/16 (25%) in CIMP− cases, P = 0.004].
Figure 1.
Mutational analysis of K-RAS in colorectal tumors. Mutations were detected by mutant allele-specific amplification. (A). PCR reactions were first performed by using a primer mixture to detect six different mutations of K-RAS codon 12 (Upper). A second PCR reaction was then performed by using specific primers to detect the exact mutation in each case (Lower). For example, p30 was found to have a mutation by using the primer mix (Upper), and the mutation was determined to be a GGT to GAT change by using specific primers (Lower). P30, P31, P36, and P37, colorectal adenomas; H157, a human lung cancer cell line that has a mutation in codon 12 of K-RAS. “Normal” refers to normal colon (a negative control). (B) Frequencies of K-RAS mutations in colorectal tumors with or without CIMP. The frequencies of K-RAS mutations in each population were expressed as percentages. The number of tumors examined is shown at the bottom. The adenomas were divided into <1.5 cm and ≥1.5 cm.
Table 1.
Summary of epigenotype/genotype interactions in colorectal tumors
| Group | Epigenotype | MINT methylation* | p16 methylation | K-RAS mutations | p53 mutations | TGFβRII mutations | MSI |
|---|---|---|---|---|---|---|---|
| Cancer, first series, n = 41 | CIMP+, n = 21, 51%; | 4.7 | 62%† | 62%§ | 24%‡ | 33% | 24% |
| CIMP−, n = 20, 49% | 0.3 | 0% | 25% | 68% | 21% | 21% | |
| Cancer, second series, n = 47 | CIMP+, n = 20, 42%; | 4.0 | 40%‡ | 75%‡ | 25%§ | ND | ND |
| CIMP−, n = 27, 58% | 0.3 | 0% | 33% | 63% | ND | ND | |
| Adenomas, n = 45 | CIMP+, n = 22; | 4.5 | 55%† | 68%‡ | 5% | ND | ND |
| CIMP−, n = 23 | 0.4 | 0% | 13% | 22% | ND | ND | |
| All cases, n = 133 | CIMP+, n = 63; | 4.4 | 53%† | 68%† | 18%† | ND | ND |
| CIMP−, n = 70 | 0.3 | 0% | 24% | 51% | ND | ND |
ND, not determined.
Average of six.
P < 0.0001.
P < 0.01.
P < 0.05 (compared to CIMP− cases).
p53 Mutations in CRCs With or Without CIMP.
We next studied the mutational status of p53 in this same panel of cancers. To detect mutations of p53, the entire coding region of the gene was screened by SSCP, and all mutations were confirmed by excision of the shifted bands and direct sequencing (Fig. 2A). Overall, p53 mutations were found in 18/40 (45%) cases. Four additional cases had intragenic p53 LOH (determined by using polymorphisms in exon 2, 3, or 4 of the gene) but no detectable mutations by SSCP. p53 mutations were found in 5/21 (24%) CIMP+ cancers and 13/19 (68%) CIMP− tumors, and this difference was statistically significant (P = 0.01 by Fisher's exact test). Furthermore, three of four cases with intragenic p53 LOH but no detectable mutations belonged to the CIMP− group. Thus, overall, 6/21 (29%) CIMP+ cases showed p53 mutations and/or LOH whereas 16/19 (84%) CIMP− cases showed such alterations (P = 0.0005 by Fisher's exact test). These differences between CIMP+ and CIMP− cases were independent of the MSI status.
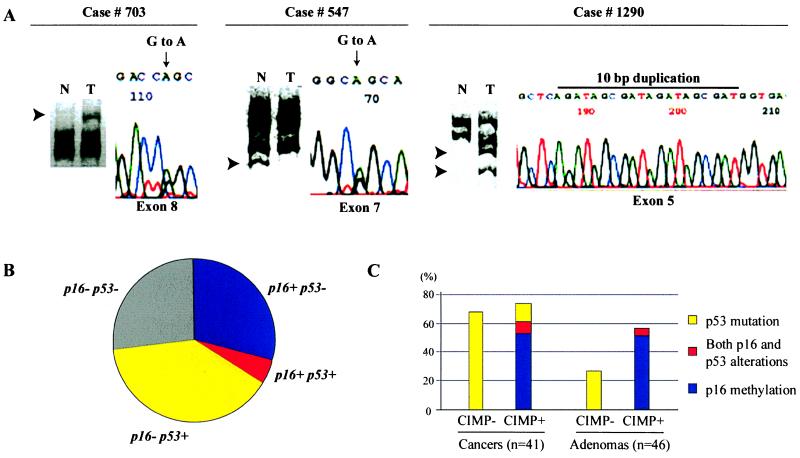
Figure 2.
Mutations of p53 and methylation of p16 in colorectal tumors. (A) Detection of p53 mutations by SSCP and direct sequencing. Aberrantly migrating SSCP bands (reflecting base pair changes) are indicated by arrows (left part of each panel). Shifted bands were excised from gels and were reamplified by using the same set of primers, and direct sequencing was performed by using automated sequencers (right part of each panel). Tumor 703 has a band shift in exon 8 caused by a mutation in codon 282 (CGG to CAG). Tumor 547 has a band shift in exon 7 caused by a mutation in codon 245 (GGC to AGC). Tumor 1290 has a band shift in exon 5 caused by a 10-bp duplication that creates a premature stop codon. N, normal tissue; T, colorectal cancer. (B) Frequencies of alterations of p53 and p16 in CRCs. Forty-one CRCs are divided into four categories based on the presence of alterations of p53 (p53+) and/or p16 (p16+). Alterations of p16 and p53 were inversely correlated (P = 0.0007, Fisher's Exact test). (C) Summary of alterations of p53 and p16 in colorectal tumors with or without CIMP. The frequencies of alterations in p53 and p16 in cancers and adenomas are expressed as percentages. n, number of tumors analyzed.
p53 and p16 alterations have been implicated as alternative mechanisms of overcoming senescence in various cells (26). We therefore sought to determine whether there was a specific interaction between p16 methylation and p53 mutations in CRC (Fig. 2B). In this series, we found a strong inverse correlation between these two events: 11/40 (28%) cases had p16 methylation only, 16/40 (40%) cases had p53 mutation only, 2/40 (5%) cases had both events, and 11 (28%) cases had neither p16 nor p53 mutations (P = 0.016 by Fisher's exact test). This inverse correlation was maintained when we considered CIMP+ cases alone and appeared to be independent of the MSI status. Interestingly, the two cases that had both p16 methylation and p53 mutation had very low levels of methylation (<10% by Southern blot analysis), indicating perhaps that only a fraction of the cells were affected. In addition, we note that 12/18 (67%) cases with K-RAS mutations had concomitant inactivation of either p53 or p16.
Mutations of Other Genes.
To determine whether the distinct differences in the genotypes of CIMP+ and CIMP− cancers is applicable to other genetic alterations, we used SSCP to examine the mutational status of TGFβRII and DPC4 in these cancers (summarized in Table 1; details are shown in Table 2). TGFβRII has a poly(A) tract in exon 3 that is frequently mutated in MSI+ cancers (21, 27). As expected, we found exon 3 TGFβRII mutations in 7/8 (88%) MSI+ cancers and 0/32 (0%) MSI− cancers. In MSI− cancers, however, we found four mutations in the kinase domain of TGFβRII. Three of the four mutations were in CIMP+ cases. We found only one DPC4 mutation, which was in a CIMP− MSI− tumor that had a K-RAS mutation but no p53 inactivation.
K-RAS and p53 Mutations in a Separate Group of CRCs.
To determine the reproducibility of the interactions between CIMP, K-RAS, and p53 changes, we studied a separate group of unselected 47 CRCs for both epigenetic and genetic changes. Of the 47 cases, 20 (43%) were CIMP+ based on previously determined criteria (18) and averaged 4.0/6 loci methylated whereas the 27 CIMP− cases averaged 0.3/6 loci methylated. Nine cases had p16 methylation and four had hMLH1 methylation. All of these were CIMP+. As in the first series, there were no significant differences in age, gender, stage, or location in the colon between CIMP+ and CIMP− cases. Combining the two series, CIMP+ tumors tended to be more frequent among proximal tumors (57% of proximal tumors were CIMP+ vs. 35% of distal tumors), and patients with CIMP+ tumors tended to be older (mean age 68 years old vs. 63 years old for CIMP− tumors). These trends, however, were not statistically significant.
The results of K-RAS and p53 mutations in this group are summarized in Table 1, and details are shown in Table 3, published as supplemental data. K-RAS mutations were present in 15/20 (75%) CIMP+ cases vs. 9/27 (33%) CIMP− cases (P = 0.008). This difference in the rate of K-RAS mutations is essentially identical to that observed in the first series. p53 mutations were present in 5/20 (25%) CIMP+ cases vs. 17/27 (63%) CIMP− cases. This difference is largely similar to what was seen in the first series and was statistically significant (P = 0.02). Overall (combining the two cancer series), 28/41 (68%) CIMP+ cases had K-RAS mutations compared with 14/47 (30%) CIMP− cases (P = 0.0005). By contrast, p53 mutations were found in 24% (10/41) of CIMP+ CRCs vs. 65% (30/46) of CIMP− cases (P = 0.0002). These differences in K-RAS and p53 mutation rates were independent of tumor location in the colon.
Epigenetic and Genetic Interactions in Colorectal Adenomas.
To determine whether the epigenotype/genotype interactions we observed in CRC are the result of early events in cancer progression, we studied 64 colorectal adenomas of various sizes for the presence of CIMP, p16 methylation, K-RAS mutations, and p53 mutations by using methods similar to those described above. The methylation status of the same six CpG islands used to define the CIMP phenotype in cancers was determined by methylated CpG island amplification and was confirmed by bisulfite-PCR for two loci. Among the 64 colorectal adenomas, 38 (59%) had methylation of three or more of these loci and were defined as CIMP+ (Fig. 3). CIMP+ adenomas averaged 4.0/6 methylation events per tumor, which was identical to CIMP+ cancers. Similarly, CIMP− adenomas averaged 0.3/6 methylation events per tumor, which was the same rate seen in CIMP− carcinomas. There was a significant difference in the rate of CIMP positivity cases between small (<1.5 cm) and large (>1.5 cm) adenomas (23% vs. 73% respectively, P = 0.004 by Fisher's exact test). Interestingly, six cases (9%) showed partial methylation (5–10%) at multiple loci, suggesting that only a portion of the tumor cells were affected by CIMP. These cases were considered CIMP+ for all subsequent analyses. The CIMP phenotype in adenomas was independent of age, gender, or location (proximal vs. distal).
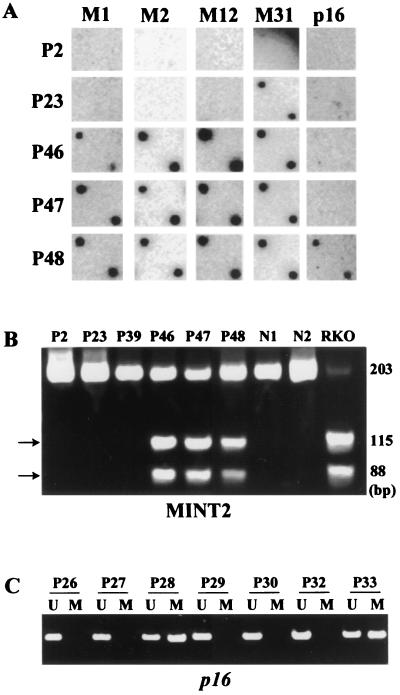
Figure 3.
Methylation analysis of multiple CpG islands in colorectal adenomas. (A) Methylation of multiple CpG islands detected by methylated CpG island amplification. Methylated CpG island amplification products from colon adenomas (P2–P48) were blotted on a nylon membrane and were hybridized with one of the MINT clones (indicated on the top), as well as a p16 probe. M2, MINT2; M1, MINT1; M12, MINT12; and M31, MINT31. Tumors P46, P47, and P48 showed methylation of multiple loci, indicating the presence of CIMP. Tumor P23 showed methylation of MINT31 only. (B) Methylation analysis of MINT2 by bisulfite-PCR. PCR products were digested with BstUI, which cleaves CGCG sequences that are retained after bisulfite treatment only when both cytosines are methylated. Methylated alleles are shown by arrows. Tumors P46, P47, and P48 are methylated at this locus. No methylated alleles were detected in PCR products from normal colon. RKO is a colon cancer cell line used as a positive control. (C) Methylation analysis of p16 in colorectal adenomas by MSP. U, unmethylated allele-specific primers; M, methylated allele-specific primers. Tumor P28 and P33 showed methylated alleles.
We next determined the rates of K-RAS mutation, p53 mutation, and p16 methylation in this group of adenomas (summarized in Table 1; details are shown in Table 4, published as supplemental data). Just like in carcinomas, K-RAS mutations were more frequent in CIMP+ cases (23/38, 61%) than in CIMP− cases (5/26, 19%, P = 0.002 by Fisher's exact test). In both CIMP+ and CIMP− tumors, K-RAS mutations were more frequent in larger adenomas (Fig. 1B). The frequency of K-RAS mutations in large CIMP+ and CIMP− adenomas was identical to that found in CIMP+ and CIMP− carcinomas, respectively.
Mutations of p53 were determined for 23 CIMP+ adenomas and 23 CIMP− adenomas (Fig. 2C). Just like in carcinomas, p53 mutations were more frequently detected in CIMP− adenomas (5/23, 22%) than in CIMP+ adenomas (2/23, 9%), although this difference did not reach statistical significance. Among CIMP− cases, p53 mutations were more frequent in cancers than in adenomas (65% vs. 22%, P = 0.0009 by Fisher's exact test).
As previously seen in carcinomas, methylation of p16 was exclusively detected in the CIMP+ group, being seen in 16/38 (42%) CIMP+ adenomas and 0/26 (0%) CIMP− adenomas. Among CIMP+ cases, methylation of p16 appeared to be slightly lower in adenomas than in carcinomas (42% vs. 51%, P = NS). The inverse relationship between p16 methylation and p53 mutations was preserved in adenomas. Finally, methylation of hMLH1 was detected in 0 of 64 colorectal adenomas (data not shown), which is consistent with previous results showing a low frequency of MSI in this preneoplastic lesion (28, 29).
Mutational Spectra of K-RAS and p53 in CIMP+ vs. CIMP− Tumors.
We next analyzed the specific types of mutations observed in the entire group of tumors. Overall, 70 tumors had K-RAS mutations, and 51 of these (73%) were CIMP+. For K-RAS, the relative number of G to A, G to T, and G to C changes was identical for the two CIMP groups (55, 34, and 11% in CIMP+ cases vs. 56, 28, and 17% in CIMP− cases). Interestingly, all 10 cases with codon 13 mutations were CIMP+, which was marginally different than the distribution of codon 12 mutations (41/60 CIMP+, P = 0.05). For p53, 47 tumors overall had confirmed mutations, of which 68% were CIMP−. In this limited series, the spectrum of mutations and codon involvement was similar between the two CIMP groups.
Discussion
In this report, we have examined the interactions between epigenetic and genetic alterations in sporadic colorectal tumors. By characterizing the mutational status of four major genes (K-RAS, p53, DPC4, and TGFβRII) that are known to be altered in CRCs in a panel of tumors also typed for hypermethylation and MSI status, we observed several findings: (i) K-RAS mutations are significantly higher in CIMP+ cancers compared with CIMP− cases; (ii) p53 mutations are significantly higher in CIMP− cases and are inversely correlated with methylation of p16; and (iii) the CpG island methylator phenotype, p16 methylation, and p53 mutation are all relatively early events in the progression of CRC because they occur at a significant frequency in preneoplastic adenomas. The overall frequency of each genetic and epigenetic alteration we observed here is broadly similar to those reported previously (3, 29), suggesting that our results are not related to a failure to detect mutations in a group of cases. Our results, which have been observed in two independently studied groups of cancers and one group of adenomas, demonstrate that there are clear differences between the genotypes of CIMP+ and CIMP− tumors, suggesting important interactions between genetic and epigenetic changes in this tumor type.
The cause of the high frequency of K-RAS mutations in CIMP+ cancers remains unclear. One possible explanation is that CIMP+ tumors inactivate DNA repair gene(s) (other than mismatch repair) and that this results in a higher frequency of mutation. O6-MGMT may be a candidate for such a gene because its inactivation could increase the rate of G to A mutations, a common mechanism of K-RAS activation (30). However, many K-RAS mutations in CIMP+ tumors are non-G to A, suggesting that another gene could be involved in the process. Given the high rate of TGFβRII mutations in MSI+ cases (7/8, 88%) and the high rate of K-RAS mutations in CIMP+ MSI− cases (44/49, 90%), one attractive hypothesis is that both of these genetic events inactivate a common critical pathway for CRC progression. Indeed, many CRCs are TGFβ unresponsive despite having no TGFβRII mutation (31), and experimental evidence suggests that K-RAS mutations can largely overcome the growth-inhibiting effects of TGFβ treatment (32). If this hypothesis is correct, the low rate of K-RAS mutations in CIMP− cases could be related to (unidentified) alterations in one or several other genes in the K-RAS/TGFβ pathway. Interestingly, 24 of 42 CRCs (57%) that have K-RAS mutations also have p16 or p53 alterations. It was previously shown that, although oncogenic K-RAS efficiently transforms immortalized rodent cell lines, it fails to transform primary cells because altered K-RAS signaling can induce senescence, mediated in part by up-regulation of p53 and p16 (33, 34). Thus, it was predicted that K-RAS mutations would occur concurrently with alterations in senescence-related genes (35), and our results are consistent with these observations.
One of the interesting results of this study is the inverse correlation between p16 and p53 inactivation. About 65% of the CRCs we examined showed either p16 or p53 inactivation, but only 5% showed both events simultaneously. These results indicate, as suggested previously (35), that colorectal tumor cells have an absolute requirement for inactivation of either p16 or p53 (in addition to other pathways). Furthermore, at least in primary tumors, cancer cells that have both p16 and p53 alterations do not appear to have a growth advantage compared with tumors that have only one of these alterations. Interestingly, both of these molecular changes start early in CRC pathogenesis because 42% of CIMP+ adenomas showed p16 methylation, and 22% of CIMP− adenomas showed mutations of p53. A possible explanation for these results comes from the apparent requirement for either p16 or p53 inactivation (but not both) for overcoming cellular senescence, as documented in various models (33, 36, 37). However, p16 and p53 also play roles distinct from this senescence-response, and the simultaneous inactivation of both genes may be important for late steps of tumorigenesis. Indeed, studies in pancreatic xenografts (38) and bladder cancer cell lines (39) demonstrated frequent coexistence of p16 and p53 alterations.
By studying both epigenetic and genetic defects in CRC, we identified at least two major groups of tumors. CIMP+ tumors have a high degree of CpG island methylation and a high frequency of classical genetic changes such as MSI, K-RAS, and TGFβRII mutations. CIMP− tumors, on the other hand, are characterized primarily by p53 mutations, with relatively few additional “classical” genetic changes. We propose that this latter group may then evolve along a distinct pathway characterized by chromosomal instability (40), including a significant degree of gene amplification and deletion. This division of CRC into two major molecular pathways appears early, being identifiable in adenomas. Further studies are needed to verify this predicted difference in chromosomal changes between the two groups. Furthermore, a strikingly important issue is the identification of molecular changes that are responsible for the malignant transformation of preneoplastic polyps. Indeed, when divided into CIMP+ and CIMP− tumors, the rates of p16, p53, and K-RAS alterations are fairly high in preneoplastic adenomas, suggesting that other events mediate this critical transformation step.
It is now well recognized that, during cancer progression, inactivation of one of the components of multiple critical pathways (senescence, apoptosis, cell cycle regulation, etc.) is required in virtually all of the tumors, regardless of how each component is activated/inactivated (mutation, deletion, and methylation). However, our results show that the ultimate molecular profile of each cancer is very different, dependent on the major pathway selected by the cancer for generating molecular diversity (genetic or epigenetic). Therefore, genotyping of one or two known oncogenes and tumor suppressor genes is not enough for the molecular classification of CRCs. We propose that determining the precise genetic and/or epigenetic alterations of each cancer may be important to determining treatment strategies and predicting prognosis. Indeed, the subset of tumors that show a high degree of methylation may be appropriate for treatment with methylation inhibitors (41). On the other hand, tumors that develop through different pathways (e.g., chromosomal instability) may be more sensitive to drugs such as DNA damaging agents. Obviously, more studies are necessary to clarify the causes and the clinical implications of genetic and epigenetic alterations in colorectal tumorigenesis.
Supplementary Material
Acknowledgments
We thank Dr. Stanley Hamilton for providing the colon tumor samples and for helpful discussions. This work was supported by grants from the National Institute of Health (Colon Cancer Spore Grant CA62924) and the American Cancer Society (Grant RPG9909801MGO). M.T. is a postdoctoral fellow from Japan Society for Promotion of Science. N.A. is supported by National Institutes of Health Training Grant 1-T32-DK07713. J.-P.I. is a Kimmel Foundation Scholar.
Abbreviations
- CRC
colorectal cancer
- MSI
microsatellite instability
- CIMP
CpG island methylator phenotype
- SSCP
single-stranded conformational polymorphisms
- LOH
loss of heterozygosity
- MSP
methylation-specific PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Peltomaki P, de la Chapelle A. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 3.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 4.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 5.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, Nicolaides N C, Markowitz S, Willson J K, Parsons R E, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 7.Moslein G, Tester D J, Lindor N M, Honchel R, Cunningham J M, French A J, Halling K C, Schwab M, Goretzki P, Thibodeau S N. Hum Mol Genet. 1996;5:1245–1252. doi: 10.1093/hmg/5.9.1245. [DOI] [PubMed] [Google Scholar]
- 8.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P J. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 9.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 10.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 11.Issa J P, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 12.Ahuja N, Mohan A L, Li Q, Stolker J M, Herman J G, Hamilton S R, Baylin S B, Issa J P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 13.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M, Hamilton S R, Burger P C, Baylin S B, Herman J G. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 15.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 16.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J P J, Markowitz S, Willson J K V, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham J M, Christensen E R, Tester D J, Kim C Y, Roche P C, Burgart L J, Thibodeau S N. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 18.Toyota M, Ahuja N, Ohe-Toyota M, Herman J G, Baylin S B, Issa J P J. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichii S, Takeda S, Horii A, Nakatsuru S, Miyoshi Y, Emi M, Fujiwara Y, Koyama K, Furuyama J, Utsunomiya J. Oncogene. 1993;8:2399–2405. [PubMed] [Google Scholar]
- 20.Rhei E, Bogomolniy F, Federici M G, Maresco D L, Offit K, Robson M E, Saigo P E, Boyd J. Cancer Res. 1998;58:3193–3196. [PubMed] [Google Scholar]
- 21.Lu S L, Zhang W C, Akiyama Y, Nomizu T, Yuasa Y. Cancer Res. 1996;56:4595–4598. [PubMed] [Google Scholar]
- 22.Moskaluk C A, Hruban R H, Schutte M, Lietman A S, Smyrk T, Fusaro L, Fusaro R, Lynch J, Yeo C J, Jackson C E, et al. Diagn Mol Pathol. 1997;6:85–90. doi: 10.1097/00019606-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Toyota M, Ho C, Ahuja N, Jair K-W, Ohe-Toyota M, Baylin S B, Issa J P J. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 24.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Z, Laird P W. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeager T R, DeVries S, Jarrard D F, Kao C, Nakada S Y, Moon T D, Bruskewitz R, Stadler W M, Meisner L F, Gilchrist K W, et al. Genes Dev. 1998;12:163–174. doi: 10.1101/gad.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 28.Samowitz W S, Slattery M L. Gastroenterology. 1997;112:1515–1519. doi: 10.1016/s0016-5085(97)70032-5. [DOI] [PubMed] [Google Scholar]
- 29.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et al. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 30.Zaidi N H, Pretlow T P, O'Riordan M A, Dumenco L L, Allay E, Gerson S L. Carcinogenesis. 1995;16:451–456. doi: 10.1093/carcin/16.3.451. [DOI] [PubMed] [Google Scholar]
- 31.Grady W M, Myeroff L L, Swinler S E, Rajput A, Thiagalingam S, Lutterbaugh J D, Neumann A, Brattain M G, Chang J, Kim S J, et al. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 32.Kretzschmar M, Doody J, Timokhina I, Massague J. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 34.Lin A W, Barradas M, Stone J C, van Aelst L, Serrano M, Lowe S W. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberg R A. Cell. 1997;88:573–575. doi: 10.1016/s0092-8674(00)81897-8. [DOI] [PubMed] [Google Scholar]
- 36.Sugrue M M, Shin D Y, Lee S W, Aaronson S A. Proc Natl Acad Sci USA. 1997;94:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozenblum E, Schutte M, Goggins M, Hahn S A, Panzer S, Zahurak M, Goodman S N, Sohn T A, Hruban R H, Yeo C J, Kern S E. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 39.Markl I D, Jones P A. Cancer Res. 1998;58:5348–5353. [PubMed] [Google Scholar]
- 40.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 41.Bender C M, Pao M M, Jones P A. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.