Abstract
Polycomb group (PcG) proteins play an important role in developmental and epigenetic regulation of gene expression in fruit fly (Drosophila melanogaster) and mammals. Recent evidence has shown that Arabidopsis homologs of PcG proteins are also important for the regulation of plant development. The objective of this study was to characterize the PcG homologs in maize (Zea mays). The 11 cloned PcG proteins from fruit fly and the Enhancer of zeste [E(z)], extra sex combs (esc), and Enhancer of Polycomb [E(Pc)] homologs from Arabidopsis were used as queries to perform TBLASTN searches against the public maize expressed sequence tag database and the Pioneer Hi-Bred database. Maize homologs were found for E(z), esc, and E(Pc), but not for Polycomb, pleiohomeotic, Posterior sex combs, Polycomblike, Additional sex combs, Sex combs on midleg, polyhometoic, or multi sex combs. Transcripts of the three maize Enhancer of zeste-like genes, Mez1, Mez2, and Mez3, were detected in all tissues tested, and the Mez2 transcript is alternatively spliced in a tissue-dependent pattern. Zea mays fertilization independent endosperm1 (ZmFie1) expression was limited to developing embryos and endosperms, whereas ZmFie2 expression was found throughout plant development. The conservation of E(z) and esc homologs across kingdoms indicates that these genes likely play a conserved role in repressing gene expression.
Gene expression patterns in eukaryotes are regulated in response to developmental and environmental cues. Changes in the patterns of gene expression are often the result of specific transcriptional regulators. In many cases, patterns of gene expression must be stably maintained through mitotic cell divisions even though the transcriptional regulator that effected the change in expression is only present transiently. The Polycomb group (PcG) genes of fruit fly (Drosophila melanogaster) stabilize repressed chromatin states during development. Recently, homologs of PcG genes have also been shown to affect developmental gene regulation in other species.
Simon (1995) defined a set of 13 PcG genes in fruit fly based on a common phenotype of homeotic transformation. The homeotic transformations caused by mutations in PcG genes are the result of a failure to maintain transcriptional repression of homeotic genes through development. Biochemical and genetic evidence indicates that the 13 PcG proteins operate in at least two distinct complexes (Franke et al., 1992; Strutt and Paro, 1997; Jones et al., 1998; Sewalt et al., 1998; van Lohuizen et al., 1998; Ng et al., 2000; Tie et al., 2001). One complex includes the PcG proteins E(Z) and ESC, as well as the histone deacetylase RPD3 and the histone-binding p55 proteins (Tie et al., 2001). The second complex includes Polycomb (PC), Posterior sex combs (PSC), Polyhomeotic (PH), Sex combs on Midleg (SCM), and additional uncharacterized proteins (Franke et al., 1992; Shao et al., 1999; Poux et al., 2001). PcG proteins have also been shown to repress expression of introduced (Pal-Bhadra et al., 1997, 1999) and endogenous (Laible et al., 1997) genes in fruit fly. All examples of polycomb-based repression likely operate through formation of a repressive chromatin structure.
Mammalian homologs of all of the cloned PcG proteins, except multisex combs, have been documented (Simon, 1995; Schumacher and Magnuson, 1997; Brock and van Lohuizen, 2001). As in fruit fly, mutations in mammalian PcG genes result in anterior derepression of Hox gene expression and alterations in cellular proliferation patterns (van der Lugt et al., 1994; Alkema et al., 1995; Muller et al., 1995; Akasaka et al., 1996; Core et al., 1997; Gould, 1997). In Caenorhabditis elegans, homologs of three of the 11 cloned PcG proteins, Enhancer of zeste [E(z)], extra sex combs (esc), and Enhancer of Polycomb [E(Pc)], have been reported (Holdeman et al., 1998; Korf et al., 1998; Stankunas et al., 1998). The E(z) and esc homologs (maternal effect sterile-2 [mes-2] and maternal effect sterile-6 [mes-6]) from C. elegans were identified as maternal genes required for germline immortality (Holdeman et al., 1998; Korf et al., 1998). The mes-2 and mes-6 genes are also involved in the silencing of transgenes in germline cells (Kelly and Fire, 1998).
Homologs of E(z) and esc have also been documented in Arabidopsis (Goodrich et al., 1997; Grossniklaus et al., 1998; Ohad et al., 1999). Three E(z)-like genes, curly leaf (clf; Goodrich et al., 1997), Medea (Mea; Grossniklaus et al., 1998), and E(z)-likeA1 (Eza1; Preuss, 1999) and one esc-like gene, fertilization-independent endosperm (fie; Ohad et al., 1999), have been previously described.
Mea (also identified as emb173, fis1, and f644) was identified in a screen for Arabidopsis gametophyte lethal mutations and autonomous endosperm development mutations (Castle et al., 1993; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999). A plant heterozygous for a mea mutation will produce 50% aborted seeds that collapse and do not germinate. It has subsequently been found that Mea is imprinted such that the maternal copy of Mea is expressed endosperm tissues, whereas the paternal copy is not (Kinoshita et al., 1999; Vielle-Calzada et al., 1999). Mea mutants fail to repress endosperm development in the absence of fertilization (Kiyosue et al., 1999). These results indicate that maternal expression of Mea is required to repress endosperm development.
Plants homozygous for clf mutations display curled leaves, altered maturation times, and partial homeotic transformations of floral tissues (Goodrich et al., 1997). Ectopic expression of the homeotic genes Agamous (AG) and Apetela3 (AP3) is also observed in clf homozygotes. In plants with wild-type Clf alleles, AG and AP3 are expressed in floral tissues where Clf mRNA is present. The presence of Clf RNA in cells expressing AG and AP3 indicates that CLF protein is not sufficient to organize suppressive chromatin, but that other targeting factors are also required (Goodrich et al., 1997). Overlapping expression of PcG and homeotic genes also occurs in fruit fly. The coexpression of functional PcG proteins and the genes that they can repress indicates that specific targeting factors are involved in PcG-dependent repression. A third E(z)-like gene, Eza1, is present in the Arabidopsis genome (Preuss, 1999). No phenotype for Eza1 has been reported.
Mutations in the Arabidopsis esc-like gene, fie, produce phenotypes very similar to Mea (Ohad et al., 1996). A female gametophyte with a fie mutant allele will undergo replication of the central cell nucleus and partial endosperm development without a fertilization event (Ohad et al., 1999). This indicates that FIE is involved in repressing endosperm development until fertilization occurs. The similar phenotypes of fie and mea mutants suggests that these two genes may participate together in a silencing complex. This is consistent with the proven direct interaction of E(Z) and ESC in fruit fly (Jones et al., 1998), an interaction also shown to occur between FIE and MEDEA (Luo et al., 2000; Spillane et al., 2000; Yadegari et al., 2000).
The objective of this research was to characterize the PcG genes (defined by Simon, 1995) in maize. Fruit fly and Arabidopsis sequences were used to identify maize sequences homologous to PcG genes. We report the full-length cDNA sequence of three E(z) homologs, two esc homologs, and one E(Pc) homolog, and we describe the likely evolution of these sequences. We also describe expression patterns of these genes in maize.
RESULTS
The 11 cloned (two other PcG genes have not been cloned) fruit fly proteins defined as the PcG by Simon (1995) were used as queries to identify homologs in Arabidopsis and maize (Table I). The top TBLASTN score from searches of the complete Arabidopsis genome sequence is shown for each PcG protein. The existence of E(z) and esc homologs in Arabidopsis has been previously documented (Goodrich et al., 1997; Grossniklaus et al., 1998; Ohad et al., 1999). The BLAST scores of searches with E(Z) and ESC proteins were much higher than the scores for searches with any of the other PcG proteins. In addition, two sequences with significant similarity to E(Pc) were detected. The two Arabidopsis proteins with significant similarity to E(PC) (AAG10815 and AAC17077) are much shorter than the fruit fly E(Pc), which is 2,033 amino acids. Further analysis of these sequences indicates that they contain the domains found in E(Pc) homologs from fruit fly, mouse, C. elegans, and Saccharomyces cerevisiae.
Table I.
PcG homologs in the Arabidopsis genome
| PcG Proteins | Best Score in Arabidopsis | Likely Arabidopsis Homologs |
|---|---|---|
| esc (extra sex combs) | AF129516 (4e-73) | Fie |
| E(z) (Enhancer of zeste) | AF100163 (7e-69) | Clf, Eza1, and Medea |
| E(pc) enhancer of polycomb | AC011808 (2e-20) | AAG10815 and AAC17077 |
| pho (pleiohomeotic) | AL391716 (4e-13) | 0 |
| mxc (multi sex combs) | T00677 (2e-10) | 0 |
| Pc (Polycomb) | AB006706 (5e-07) | 0 |
| psc (posterior sex combs) | AB026655 (2e-06) | 0 |
| asx (additional sex combs) | AC007651 (0.18) | 0 |
| pcl (polycomblike) | AL132966 (0.019) | 0 |
| ph (polyhomeotic) | AC006592 (0.23) | 0 |
| scm (sex combs on midleg) | AJ292982 (0.82) | 0 |
| super sex combs (sxc) | Not cloned in fruit fly | N.A. |
| sex combs extra (sce) | Not cloned in fruit fly | N.A. |
A search for Polycomb-group homologs in Arabidopsis. The protein sequences of the fruit fly PcG proteins were used to perform TBLASTN searches of the Arabidopsis genomic sequence. The fruit fly sequences used were as follows: esc, S58672; E(z), AAC46462; E(Pc), AAF58641; pho, AAF59378; mxc, AAF27644; Pc, CAA39229; psc, CAA41965; asx, CAA04568; pcl, AAA64457; ph, CAA45211 and S23632; scm, AAB57632. The accession no. for the Arabidopsis sequence with the best BLAST score for each search is shown in the second column (the BLASTP score in parentheses). The number of Arabidopsis homologs for each PcG protein is indicated in the last column. The esc and E(z) homologs have been previously documented (Goodrich et al., 1997; Grossniklaus et al., 1998; Ohad et al., 1999). The accession nos. for the predicted protein sequences of the two E(Pc) homologs found in the Arabidopsis genome are shown in parentheses.
The top two BLAST hits for all Polycomb proteins were analyzed based on length, organization of the gene, and similarity of conserved domains. Based on BLAST scores, gene organization, and conservation in critical domains, we determined that only homologs of E(z), esc, and E(Pc) exist in Arabidopsis. Relatively strong BLAST hits to pleiohomeotic (pho), mxc, Pc, and psc were found, but subsequent analysis determined that they were not PcG homologs. The Arabidopsis proteins most similar to pho showed homology within a zinc finger domain, but not to any other regions of pho. Therefore, these zinc-finger proteins were determined to not be orthologs of pho. The proteins most similar to mxc contained an RNA binding motif, but were different in length and composition throughout the rest of the protein and were clearly not orthologs of mxc. The search for Pc homologs identified chromodomain-containing proteins that were members of other types of gene families. The lack of homology between Pc and these proteins outside the chromodomain clearly indicated that no Pc homologs exist in Arabidopsis. Psc candidates were found to have homology in a RING finger domain, but no homology outside this domain. Therefore, these candidates were determined to be RING finger proteins, but not Psc homologs.
The 11 cloned PcG proteins from fruit fly and the E(z), esc, and E(Pc) homologs from Arabidopsis were used as queries to perform TBLASTN searches against the public maize expressed sequence tag (EST) database and the Pioneer Hi-Bred database. Maize homologs were found for E(z), esc, and E(Pc), but not for Pc, ph, Psc, Polycomblike (Pcl), Additional Sex combs (Asx), Scm, pho, or mxc. The full-length sequences of the maize E(z), esc, and E(Pc) genes were then obtained and characterized.
Maize Has Three E(z) Homologs
The Mez1 cDNA is 3,180 bp in length and produces a predicted protein of 933 amino acids (Fig. 1). The Mez2 cDNA is 3,025 bp in length and encodes a putative protein of 893 amino acids. The Mez3 cDNA is 3,149 bp in length and encodes a putative 896-amino acid protein. Mez1 maps to the short arm of chromosome 6 (bin 6.01–6.02) and Mez2 maps to the short arm of chromosome 9 (bin 9.04; data not shown). Mez3 has not been mapped.
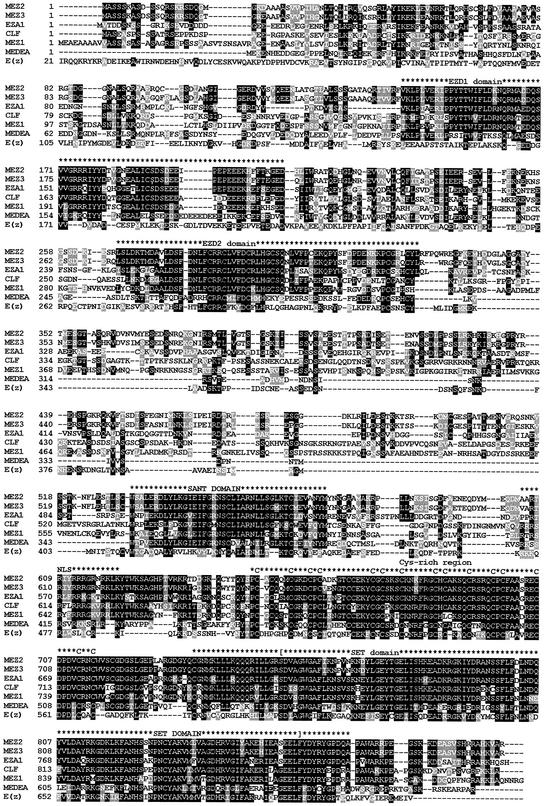
Figure 1.
Alignment of plant and animal E(z)-like sequences identifies two new conserved domains, EZD1 and EZD2. The sequences of the maize E(z)-like proteins, MEZ1, MEZ2, and MEZ3, were aligned with the Arabidopsis E(z)-like proteins, CLF (AAC23781), MEDEA (AAC39446), and EZA1 (T01127), and the fruit fly E(Z) sequence using ClustalW. The alignment was then colored using Boxshade (http://www.ch.embnet.org/software/BOX_form.html) to shade conserved residues in black and similar amino acids in gray. The position of the EZD1, EZD2, SANT Cys-rich, and SET domains are indicated above the alignments. A putative nuclear localization signal (NLS) is also indicated. The brackets identify the region for the phylogenetic analysis shown in Figure 2.
The MEZ2 and MEZ3 putative proteins are 89% identical and the nucleotide sequences of the genes are 92% identical. Based on the high degree of sequence homology, Mez2 and Mez3 are likely the genome duplicates resulting from the paleotetraploid origin of maize. It is common to find two closely related sequences in the maize genome due its evolutionary history (Gaut and Doebley, 1997). The amino acid sequences of MEZ1 and MEZ2 are 42% identical and 56% similar to each other over their entire lengths. The nucleotide sequences of Mez1 and Mez2 are 52% identical.
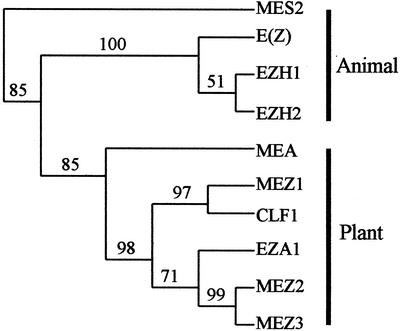
Arabidopsis contains at least three E(z)-like genes, and mutations in two of these genes, Mea and Clf, show distinct phenotypes. We attempted to determine which of the Arabidopsis E(z)-like genes that Mez1, Mez2, and Mez3 were most closely related to. The SET domain sequences of all E(z)-like proteins (indicated in Fig. 1) were aligned using ClustalW. This alignment was then processed using PHYLIP, and a parsimonious tree was constructed (Fig. 2). The tree shows grouping of the Arabidopsis clf and the maize Mez1. When the full-length protein sequences were used for the alignments, the same tree was produced. This suggests that Mez1 is a clf-like gene in maize, whereas Mez2 and Mez3 are likely to be Eza1 homologs. The low degree of nucleotide similarity between Mez1 and Mez2/3 corroborates the notion that these genes may have distinct evolutionary origins. The orthology of monocot E(z) homologs with Eza1 and Clf1, rather than Medea is supported by overall sequence similarity as well as by the fact that Medea has approximately 300 fewer amino acids between the EZD2 and SANT domains, which are present in the monocot proteins (Fig. 3). No gene more similar to Medea than to Eza1 or Clf1 was detected in any monocot EST or genomic sequence.
Figure 2.
Relationships of maize and Arabidopsis E(z)-homologs. The SET domains (indicated in Fig. 1) from plant and animal homologs of E(z) were aligned using ClustalW and were analyzed using PHYLIP parsimonious methods to generate a parsimonious tree. The bootstrap values are indicated at nodes in the tree. The sequences used are MES-2 (AAC27124), EZH1 (AAC50778), EZH2 (AAC51520), E(Z) (AAC46462), MEZ1 (AF443596), MEZ2 (AF443597), MEZ3 (AF443598), CLF1 (AAC23781), EZA1 (T01127), and MEDEA (AAC39446). The phylogenetic analysis reveals that MEZ1 is most orthologous to CLF1, whereas MEZ2 and MEZ3 are orthologs of EZA1.
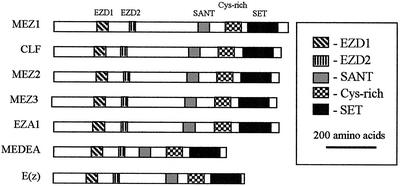
Figure 3.
Schematic diagram of E(z) homologous proteins. The relative positions of the EZD1, EZD2, SANT, Cys-rich, and SET domains found in MEZ1, MEZ2, CLF1, MEDEA, EZA1, and E(Z) are indicated by the shaded boxes. Identities of each protein are shown at the right, and the structures are oriented with the amino terminal on the left and the carboxy terminal on the right. The legend at the right indicates the shading pattern for each domain and the scale of the drawings.
Alignment of E(z) Homologs Identifies Five Conserved Domains
Alignments of plant and animal E(z) homologs were used to identify conserved domains (Fig. 1). We first searched for previously annotated domains in the MEZ protein sequences using the SMART program (Schultz et al., 2000). A Cys-rich region and a SET domain (Fig. 3) near the C terminus of the proteins are conserved among all E(z) homologs. The Cys-rich region has 15 invariant Cys residues with a conserved spacing pattern in all E(z) homologs. The spacing of the Cys residues in all E(z) homologs is unique and is different from other Cys-rich zinc finger domains involved in DNA binding. The SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain found at the C-terminal end of the protein is also highly conserved among all E(z) homologs. In addition to the SET domain, a SANT (SWI3, ADA2, N-CoR, and TFIIIB′′ DNA-binding domains) domain was identified by SMART in the plant and animal proteins (Fig. 3). SANT domains are often involved in nonspecific DNA binding (Aasland et al., 1996).
In addition to the domains identified by SMART, two additional regions of conservation are present in the plant and animal E(z)-like proteins. These domains were analyzed by BLASTP and are not found in any other sequences outside of E(z) homologs. Therefore, we have named these two domains Enhancer of zeste domain1 (EZD1) and Enhancer of zeste domain2 (EZD2) because they represent conserved domains specific to the E(z) family. EZD1 is a highly conserved acidic region of 70 amino acids in the N-terminal region (Figs. 1 and 3), originally noted by Grossniklaus et al. (1998) as an acidic domain. The EZD1 domain contains a significant proportion of charged residues (34%–39%), with seven more acidic residues than basic residues. The function of this domain is not known. EZD1 is highly conserved between MEZ1, MEZ2, MEZ3, CLF, and EZA1. Although the primary sequence is less conserved in MEA and animal E(z)-like proteins, a similar distribution of charged residues exists. EZD2 is a small, highly conserved region of 44 amino acids near amino acid 250 of the plant and animal E(z)-like proteins. This corresponds to the C5 region noted by Goodrich et al. (1997). The EZD2 domain contains five highly conserved Cys residues and is composed primarily of polar or charged residues.
Maize Contains Two ESC/FIE Homologs
Two homologs of fruit fly esc were isolated from maize, ZmFie1 (Zea mays fertilization independent endosperm 1) and ZmFie2 (Zea mays fertilization independent endosperm 2). The ZmFIE1 and ZmFIE2 proteins are 76% identical and 85% similar over their entire lengths (Fig. 4). The nucleotide sequences of ZmFie1 and ZmFie2 are 83% identical to one another. ZmFie1 maps to chromosome 4 (bin 4.05), and ZmFie2 maps to chromosome 10 (bin 10.03; data not shown). It is likely that ZmFie1 and ZmFie2 are genome duplicates that arose from the ancient polyploidization of maize. The map position of these genes is consistent with ancient polyploidy as the origin of the duplication (Gaut, 2001).
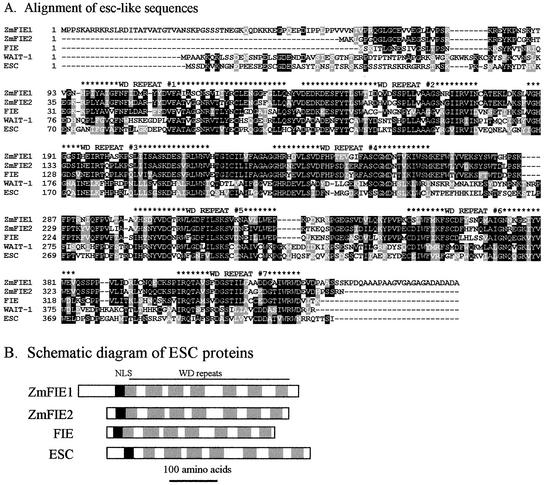
Figure 4.
Maize contains two esc homologs. A, The protein sequences of the two maize esc homologs, ZmFIE1 (AY061964), ZmFIE2 (AY061965), Arabidopsis FIE (AAD23584), fruit fly ESC (AAF53124), and the human WAIT-1 (AAC68675) were aligned using ClustalW. The identities and amino acid position for each sequence is shown on the left. The alignment was shaded with Boxshade such that the conserved residues are colored black and the similar residues are gray. The location of the WD40 repeats is indicated above the alignment. B, The protein sequences of ZmFIE1, ZmFIE2, FIE, ESC, and WAIT-1 are shown as schematic diagrams with the location of the WD40 repeats and a putative NLS indicated. The position of a putative NLS is indicated by a black box, and the gray boxes indicate the positions of the WD repeats. The scale of the drawings is indicated.
An alignment of esc-like proteins is shown in Figure 4A. All previously cloned esc-like proteins contain seven WD-40 repeats (Ng et al., 1997; Korf et al., 1998; Ohad et al., 1999). ZmFIE1 and ZmFIE2 also contain seven WD-40 repeats, which are indicated in Figure 4. The WD-40 repeats are more highly conserved between the maize ZmFIE proteins and the fruit fly ESC and Arabidopsis FIE than other regions of the proteins. A putative nuclear localization signal is also found near the N terminus of the plant esc-like proteins. The ZmFIE1 protein contains a unique 58-amino acid extension at the N-terminal end and a unique 22-amino acid extension at the 3′ end relative to ZmFIE2.
Plants Contain Homologs of E(Pc)
Two predicted proteins with significant similarity to E(Pc) were detected in the Arabidopsis genome. These proteins were named AtEPL1 (AAG10815) and AtEPL2 (AAC17077). AtEpl1 is located on chromosome 1 at 25cM (BAC F19K19) and is represented by a single EST (AV559019) from green silique tissue. AtEpl2 is also located on chromosome 1 at 117 cM (yeast artificial chromosome YUP8H12R) and is not represented by any Arabidopsis ESTs. The Arabidopsis proteins are 67% identical and 79% similar to each other.
We searched for ESTs from maize with significant similarity to the Arabidopsis Epc genes. A single EST (BE511872) with significant similarity to the AtEpl1 and AtEpl2 genes was found. The maize E(Pc) homolog, ZmEpl101, is 52% identical and 70% similar to AtEpl2. In addition to the ZmEpl101 EST from maize, E(pc) homologous ESTs were also found in soybean (Glycine max), tomato (Lycopersicon esculentum), potato (Solanum tuberosum), wheat (Triticum aestivum), and barley (Hordeum vulgare). This indicates that E(Pc) homologs are present and expressed in a variety of plant species.
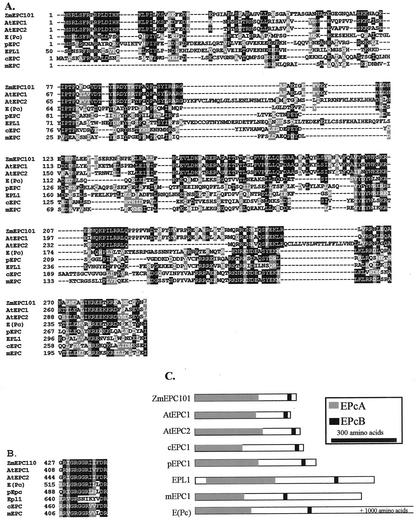
E(Pc) homologs have previously been identified in mammals, yeast, and C. elegans (Stankunas et al., 1998). Alignments of the known E(Pc) homologs with the plant EPL proteins revealed significant conservation of the previously characterized EPcA domain (Stankunas et al., 1998; Fig. 5A). A portion of the EPcB domain is strongly conserved in all eukaryotes with the exception of the yeast Epl1 gene (Fig. 5B). The EPcC domain identified in fruit fly and mammals (Stankunas et al., 1998) is not observed in yeast, C. elegans, or higher plants.
Figure 5.
Plants contain sequences similar to E(Pc). A, The protein sequences the EPcA domain of ZmEPL101 (AF443599), Arabidopsis EPL1 (AtEPL1–AC011808), Arabidopsis EPL2 (AtEPL2–YUP8H12R), C. elegans EPC1 (cEPC1–CAC35840), Schizosaccharomyces pombe EPC1 (pEPC1–T41631), S. cerevisiae EPL1 (NP 116629), M. musculus EPC1 (mEPC1–AAC64272), and fruit fly E(PC) (AAF58641) were aligned using ClustalW. The identity and position within the protein for each sequence is shown at the left. This alignment was shaded using Boxshade to show conserved residues in black and similar residues in gray. B, The portion of the EpcB domain that is conserved in plants and animals was aligned using ClustalW. The protein and amino acid position in the protein are indicated on the left. C, The structures of the EPL proteins are represented by drawings. The size and location of the EpcA and EpcB domains are indicated by the shaded boxes. The identity of each structure is shown on the left and the structures are orientated with the amino terminus on the left. Only the N-terminal 1,023 amino acids of the fruit fly E(PC) are represented. The legend on the right indicates the shading used for each domain and the scale of the drawings.
Mez Genes in Maize Are Expressed throughout Plant Development, and Mez2 Shows a Tissue-Specific Alternative Splicing Pattern
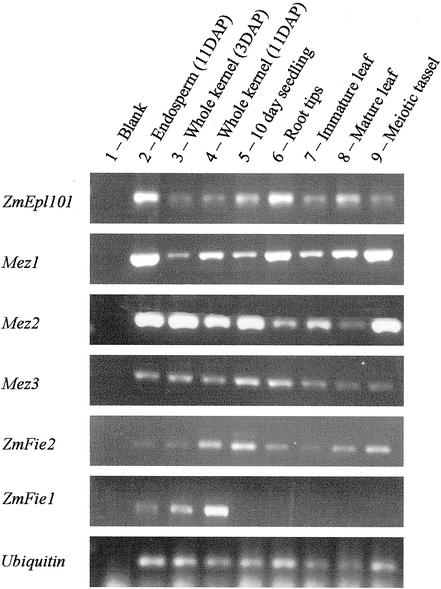
The expression level of the maize PcG homologs is low and difficult to detect by northern-blot analysis. In addition, it is not possible to generate specific probes for the pairs of duplicate genes, ZmFie1/ ZmFie2 and Mez2/Mez3, due to the high degree of nucleotide identity between these two sequences. Therefore, we used reverse transcriptase (RT)-PCR analysis to determine the expression pattern of the PcG genes of maize (Fig. 6). ZmEpl101, Mez1, Mez2, and Mez3 transcripts were successfully amplified in all tissues tested. In most cases, there was some variation in the amount of product amplified from different tissues; this may reflect slight differences in the amount of cDNA used in the amplification reaction and not the exact expression level.
Figure 6.
Expression pattern of the maize PcG genes. RT-PCR was performed on RNA isolated from various maize tissues to determine the expression patterns of the maize PcG genes. The sequence amplified is indicated on the left, and the source of the RNA for each lane is indicated above the pictures (1, blank; 2, endosperm (11 DAP); 3, whole kernel (3 DAP); 4, whole kernel (11 DAP); 5, 10-d-old seedling; 6, root tips; 7, immature leaf; 8, mature leaf; and 9, meiotic tassel).
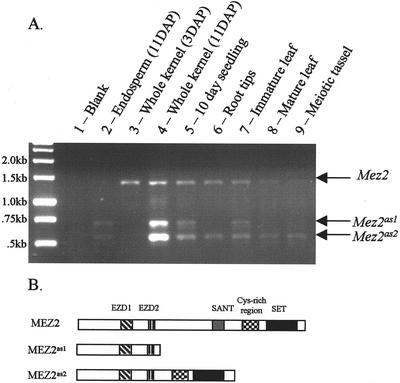
Multiple transcripts were observed when the entire coding sequence of Mez2 was PCR amplified (data not shown). Three transcripts were characterized from Mez2, including the full-length transcript and two alternatively spliced products (Fig. 7A). The alternative splice products are named Mez2 alternative splice 1 (Mez2as1), which is 2,385 bp in length and Mez2 alternative splice 2 (Mez2as2), which is 2,234 bp in length. Relative to Mez2, Mez2as1 lacks a 659-bp fragment and Mez2as2 is missing a fragment of 810 bp. The missing fragment in Mez2as1 corresponds to basepair 1,017 to 1,675 of the full-length transcript and causes a frameshift resulting in the production of a truncated protein of 341 amino acids (Fig. 7B). The deletion in the Mez2as2 corresponds to basepair 1,017 to 1,826 of the full-length Mez2 transcript and results in a 624-amino acid protein that is missing the SANT domain (Fig. 7B).
Figure 7.
Alternative splicing of Mez2. A, PCR amplification of Mez2 transcripts from various maize tissues. The expected size of the amplified band is 2.2 kb. The presence of multiple bands indicates alternative splicing of the Mez2 transcript. Different tissues displayed different patterns of splicing for Mez2. The three bands amplified from ear tissue (indicated by the arrows at the right of the picture) were sequenced and are designated at Mez2, Mez2as1, and Mez2as2. The products amplified from embryo tissue were also cloned and sequenced and were all found to correspond to Mez2as2. B, Schematic diagrams of the alternative splicing event and the altered proteins. The shading pattern is identical to that used in Figure 3. Mez2as1 introduces a frameshift and will produce a truncated protein that lacks the SANT, NLS, Cys-rich, and SET domains. Mez2as2 produces an in-frame deletion that removes only the SANT domain.
We tested cDNA from eight different maize tissues to determine whether the splicing of Mez2 transcripts is tissue specific (Fig. 7A). Amplification from whole kernel (11 d after pollination [DAP]), 10-d whole seedling, and immature leaf cDNA revealed the presence of the three transcripts observed previously. Low levels of Mez2as1 and Mez2as2 transcripts were detected in endosperm cDNA (11 DAP), whereas no band corresponding to the full-length transcript was observed. We cannot rule out the presence of a low level of the full-length transcript in this tissue. Mez2as1 and Mez2as2 transcripts were not observed in whole kernel tissues isolated 3 DAP. In root tips, mature leaf, and meiotic tassel, the Mez2as1 was not observed. The presence of the same alternative splice products has been noticed in multiple genotypes (B73, Mo17, and W22) and in tissue taken from different sources (data not shown). Although it is possible that the alternative splice pattern is influenced by environmental stimuli, we have not noticed differences in the splicing patterns taken from the same tissues (data not shown).
ZmFie2 Is Expressed throughout Development, Whereas ZmFie1 Expression Is Limited to the Early Embryo and Endosperm
The presence of ZmFie1 and ZmFie2 transcripts in various plant tissues was tested by RT-PCR (Fig. 6). ZmFie2 transcripts were detected in all tissues tested. This is similar to the expression pattern of the Mez genes. ZmFie1 transcripts were only found in kernels after pollination, but not in any other plant tissues tested. The ZmFie1 transcripts were detected in embryo (data not shown) and endosperm tissue.
DISCUSSION
PcG proteins play an important role in the maintenance of gene silencing in animals. In this study, we searched the complete Arabidopsis genome sequence and maize EST databases for homologs of PcG proteins. This exhaustive search identified homologs for only three of the 11 cloned fruit fly PcG proteins, E(z), esc, and E(Pc). Biochemical studies have found that the PcG proteins of animals are organized into at least two complexes, the E(z)/esc complex (Jones et al., 1998; Sewalt et al., 1998) and the PRC1 complex, which includes Pc, Ph, Scm, and Psc (Shao et al., 1999). The role of the E(PC) protein is not well characterized, and current evidence suggests that it operates independently of the E(Z)/ESC complex. Genetic evidence from fruit fly suggests that the functions of all PcG proteins are necessary for repression of homeotic genes (Simon, 1995). The genetic data include similar phenotypes of single mutants as well as studies of double mutants. The observations that E(z) and esc homologs function to repress gene expression in organisms that lack other PcG proteins,such as plants and C. elegans (Goodrich et al., 1997; Holdeman et al., 1998; Kelly and Fire, 1998; Korf et al., 1998), indicates that the E(z)/esc complex is capable of repression of transcription in the absence of the PRC1 complex.
In our searches, we failed to find homologs of any proteins in the PRC1 complex. The fact that the E(Z)/ESC complex is capable of repressing transcription in the absence of PRC1 raises the question of what, if anything, is fulfilling the function of the PRC1 complex in plants and C. elegans. No catalytic activities have been defined for any of the fruit fly proteins found in the PRC1 complex. In vitro, the PRC1 complex of fruit fly interferes with SWI/SNF-dependent chromatin remodeling activities (Shao et al., 1999). If the primary role of the PRC1 complex is inhibition of SWI/SNF-dependent chromatin remodeling, then plants may contain a different group of proteins, targeted by E(Z)/ESC, that interfere with SWI/SNF activity. In an alternate manner, the E(Z)/ESC complex may be capable of silencing transcription independent of PRC1 function. The role of the PRC1 complex in fruit fly and mammals may be to enhance or stabilize this silencing, whereas in plants and C. elegans this supplementary function is absent.
The E(Z) and ESC Proteins Can Provide Two Mechanisms for Repression
We have found multiple homologs of E(z) and esc in maize, and previous studies have documented similar homologs in Arabidopsis (Goodrich et al., 1997; Grossniklaus et al., 1998; Ohad et al., 1999; Preuss, 1999). Our sequence analysis of the MEZ1, MEZ2, and MEZ3 proteins revealed the presence of multiple domains that are conserved between plant and animal E(Z)-like proteins. The most conserved domain is the SET domain, which is found near the C terminus of all E(Z)-like proteins. The SET domain of the Su(var)3-9-like proteins and mammalian G9a protein has been demonstrated to act as a histone methyltransferase (Rea et al., 2000; Lachner et al., 2001; Nakayama et al., 2001; Tachibana et al., 2001). These proteins contain Cys-rich regions on both sides of the SET domain. The presence of both Cys-rich regions was proposed to be required for targeting histone methyltransferase activity to histone H3 (Rea et al., 2000). In this same study, other proteins such as EZH2 and HRX, which contain only one Cys-rich region, did not possess detectable histone methyltransferase activity. The maize E(Z)-like proteins like fruit fly and mammalian E(Z) proteins only contain a Cys-rich region on the N-terminal side of the SET domain. Due to the high conservation of the SET domain, it is likely that the plant E(Z)-like proteins are protein methyltransferases. Further research will determine the if E(Z)-like proteins are capable of histone methylation in vivo or if they methylate other proteins.
Alignments of the two ZmFIE proteins with Arabidopsis FIE, fruit fly ESC, and mammalian WAIT-1 revealed several conserved features. The maize ZmFIE1 and ZmFIE2 proteins contain seven WD repeats, the same as found in all other characterized ESC-like proteins (Ng et al., 1997; Ohad et al., 1999). The spacing of the WD repeats found in ZmFIE1 and ZmFIE2 is also consistent with that observed in other ESC-like proteins. This suggests that the ZmFIE proteins found in plants are likely to form a β-propeller structure similar to that predicted for fruit fly ESC (Ng et al., 1997). The sequence conservation of ZmFIE proteins with the fruit fly and mammalian proteins makes it reasonable to propose that these proteins are involved in similar protein-protein interactions. The ESC-like proteins have been documented to physically interact with two other proteins. A direct interaction between E(Z) and ESC homologs has been demonstrated in fruit fly, mice, C. elegans, and Arabidopsis (Jones et al., 1998; Sewalt et al., 1998; Luo et al., 2000; Spillane et al., 2000; Yadegari et al., 2000; Xu et al., 2001). The ESC-like proteins in fruit fly and mouse directly interact with Rpd3 histone deacetylase proteins (van der Vlag and Otte, 1999; Tie et al., 2001). These interactions provides a mechanism for ESC-like proteins to link the catalytic activities of protein methylation by E(Z)-like proteins with histone deacetylation by Rpd3-like proteins.
The E(Z)/ESC complex could perform two distinct functions in repressing gene expression. First, the E(Z)/ESC contains a histone deacetylase protein, which can mediate a repressive chromatin structure. Histone acetylation states are relatively unstable and require constant presence of a histone deacetylase to be maintained (Jenuwein, 2001). Second, the SET domain of E(z) is predicted to be involved in protein methylation. Unlike acetylation, methylation often tends to be more stable (Jenuwein, 2001). Although the role of histone acetylation in regulating chromatin states is defined (Cheung et al., 2000), the role of E(z)-mediated protein methylation is less apparent. Protein methylation may be involved in potentiating interactions with other proteins by creating specific binding sites. For example, the SET domain protein Su(var)3-9 methylates Lys 9 of histone H3 (Rea et al., 2000). Heterochromatin protein HP1 will bind specifically to methylated histone H3, but not to unmodified histone H3 (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001). In animals, the PcG proteins include a SET domain protein, E(z), and a chromodomain protein, Polycomb. Homologs of the Polycomb gene are not present in plants, but other chromodomain containing genes, such as chromomethylases, are present. Therefore, the E(Z)/ESC complex provides a combination of a reduction in histone acetylation coupled with targeted protein methylation that likely results in a stable repressive chromatin state.
Plants Contain Proteins Similar to E(Pc)
We have identified homologs of a third PcG protein, E(Pc), from Arabidopsis and maize. The Arabidopsis Epl) sequences that we found are located at two unlinked locations on chromosome 1. The maize ZmEpl101 gene is homologous to the Arabidopsis genes throughout the coding sequence. Unlike E(z) and esc, the size and organization of E(Pc) homologs differs between species. The fruit fly E(PC) protein is 2,033 amino acids. The homologs of E(Pc) from other species lack the C-terminal 1,400 amino acids present in the fruit fly protein and average 560 amino acids in length (Stankunas et al., 1998). All of the E(Pc) homologs contain two conserved domains, EpcA and EpcB. The EpcA domain encompasses approximately the first 200 amino acids of all E(Pc) homologs, whereas the EpcB domain is 13 amino acids. Stankunas et al. (1998) identified the EPcC domain as a region of conservation between the fruit fly and mammalian E(PC) protein sequences that is not present in C. elegans or S. cerevisiae homologs. Our alignments show that like the C. elegans or S. cerevisiae homologs, the plant E(Pc) homologs do not contain the EPcC domain identified in fruit fly and mammalian sequences (Stankunas et al., 1998).
To date, E(PC) has not been found in complexes associated with any other PcG proteins. Unlike the other PcG group genes, mutations in E(Pc) do not display homeotic transformations alone (Sato et al., 1983). However, mutations in E(Pc) enhance the homeotic transformations observed when other PcG genes are mutated (Sato et al., 1984). This suggests that E(Pc) plays a role distinct from other PcG proteins. In addition, a homolog of E(Pc), Epl1, has been found in S. cerevisiae, a species that does not contain homologs of any other PcG proteins (Stankunas et al., 1998). The presence of an E(Pc) homolog in a species that lacks all other PcG proteins suggests that E(Pc) homologs may have a basic chromatin function independent of PcG proteins, but that is required for PcG silencing. In support of the idea that E(Pc) plays a broader role in chromatin regulation, the human E(Pc) homolog has been shown to repress and activate transcription (Shimono et al., 2000). In a similar manner, another study found that the yeast E(Pc) homolog Epl1 was present in the NuA4 histone acetyltransferase complex involved in transcriptional activation (Galarneau et al., 2000). The plant E(Pc) homologs Epl1, Epl2, and ZmEpl101 are likely to be proteins involved in chromatin-based regulation of gene expression, but may operate independently of E(z) and esc homologs.
Expression of Multiple E(Z) and ESC Proteins May Allow Specialization of Function in Plants
Plants contain multiple E(z) and esc homologs. Arabidopsis contains three homologs of E(z): Clf, Eza1, and Mea. We have documented the presence of three E(z) homologs in maize. The first gene, Mez1, does not have a closely related nucleotide sequence in the maize genome and is most likely to be the maize ortholog of Clf based on our phylogenetic analysis. The other two E(z) homologs, Mez2 and Mez3, are highly related to each other and are predicted to be the orthologs of Eza1 based on sequence phylogeny. No Mea homologs are observed in the public monocot EST databases, or rice (Oryza sativa) genomic sequences (searches done February 8, 2002). This could simply reflect the fact that Mea is not highly expressed and is therefore not represented as an EST, or it could indicate that monocots do not have a Mea homolog. If there is no Mea present in monocots, another E(z) protein is likely performing a function in monocots analogous to Mea's function in dicots. This could also reflect differences in the regulation of development in monocots and dicots.
In addition to having three distinct genes encoding E(z) homologs, maize also displays alternative splicing of at least one E(z) gene, Mez2. The alternative splicing pattern of Mez2 could provide distinct functions arising from the same gene in different tissues and cells. We successfully amplified transcripts corresponding to multiple splice products from many tissues. However, the alternative splicing pattern was distinct in some tissues, indicating a developmental control of the splice pattern of Mez2. The presence of multiple transcripts in some tissues may be due to simultaneous presence of these transcripts in all cells of that tissue or may be due to cell type-specific expression of certain transcripts of Mez2.
The putative proteins produced by the alternatively spliced transcripts of Mez2 are likely to provide different functions. The MEZ2as2 protein is similar in size and domain composition to MEA (with the exception of the SANT domain). The MEZ2as2 form predominates in embryonic tissues, which is the location of Mea function in Arabidopsis. It is possible that through alternative splicing, Mez2 provides Mea and Eza1 functions in maize. The MEZ2as1 protein, which is present in kernel, seedling, and immature leaf tissue, lacks the C-terminal two-thirds of the protein, including the SANT, Cys-rich, and SET domains. The truncated protein produced from Mez2as1 transcripts might negatively regulate function of the E(z)/esc complexes by binding ESC proteins in a nonfunctional complex. The production of the MEZ2as2 protein, which is missing a 270-amino acid region that includes the SANT DNA-binding domain, but still possesses a SET domain, may alter the localization or activities of the complex. The alternative splicing of Mez2 may allow for this gene to provide distinct functions in different tissues.
In this study, we have also documented that maize contains multiple esc homologs, ZmFie1 and ZmFie2. In the other organisms in which the PcG proteins have been studied (fruit fly, humans, mice, C. elegans, and Arabidopsis), only one esc homolog has been found. Based on the chromosomal locations and high degree of nucleotide identity, it is likely that the duplication of these genes is due to the allotetraploid history of maize (Helentjaris, 1995; Gaut and Doebley, 1997). This duplication event may have allowed specialization of the function of ZmFie1 and ZmFie2 in expression pattern or function. We have found that ZmFie1 is only expressed in kernel tissues, whereas ZmFie2 is expressed in all tissues tested. The sequence of ZmFie1 is also different from esc homologs, containing N- and C-terminal extensions. ZmFIE1 may have evolved kernel-specific functions that are important in the development of the maize endosperm or embryo.
The E(Z) and ESC proteins function as a complex. The domain of the E(Z) and MEA proteins that interacts with ESC and FIE has been mapped to the N-terminal region of the protein (Jones et al., 1998; Luo et al., 2000; Spillane et al., 2000; Yadegari et al., 2000). All three of the MEZ proteins from maize as well as the two putative proteins produced by alterative splicing of Mez2 contain the N-terminal region of the protein and therefore are predicted to interact with the maize ZmFIE proteins. Evidence from Arabidopsis indicates that MEA and EZA1 interact with FIE (Luo et al., 2000). Assuming that the three full-length MEZ proteins and the two alternative splice isoforms can interact equally with both ZmFIE proteins, up to 10 distinct E(z)/esc complexes could be formed. The multiplicity of complexes could allow for a number of specialized roles in regulating gene expression. Furthermore, the expression and splicing pattern of Mez and ZmFie genes is tissue specific, which would regulate the exact set of E(Z)/ESC complexes present in a given tissue.
Epigenetic regulation of gene expression involving homologs of E(z) and esc is conserved across diverse species. These proteins offer a pathway for epigenetic regulation separate from DNA methylation. The main role of PcG proteins in plants and in animals may be to maintain the gene expression patterns determined by developmental decisions. This type of repression must be reset at meiosis each generation. In contrast, repression mediated by DNA methylation provides a meiotically heritable mechanism for gene silencing. There may be instances where PcG-mediated and DNA methylation-mediated silencing overlap, such as the regulation of AG and AP3 (Finnegan et al., 1996; Goodrich et al., 1997). However, we believe PcG-mediated silencing is primarily a source of epigenetic memory during development, whereas DNA methylation plays a central role in heritable gene silencing for genome protection.
MATERIALS AND METHODS
Cloning of Mez Genes
Fruit fly (Drosophila melanogaster) E(z) (AAC46462) was used in a TBLASTN search of the Pioneer Hi-Bred EST database. Three contigs with significant similarity to E(z) were discovered and named Mez1, Mez2, and Mez3. Other SET domain-containing proteins were also identified, but were not included in this research because they had greater similarity to proteins other than to E(z). RACE was performed to obtain full-length cDNA sequence of Mez1 and Mez2. Full-length Mez3 was obtained from an EST clone that contained the entire coding region. RACE reactions were performed using the Marathon cDNA kit (CLONTECH, Palo Alto, CA) on cDNA produced from 1-week-old Mo17 seedlings. Advantage2 polymerase (CLONTECH) was used in the RACE reactions. The primers used in the RACE reactions were Mez1F1 (5′-GGGTGTGGTGATGGTACATTGG-3′), Mez1R2 (5′-CAGCTTGTCACCCATTCTGTATGCG-3′, Mez2R3 (5′-TGCCTCGTCCTTCTTTGATCCTTCG-3′), and Mez2F3 (5′-CTCACAAGGAAGCAGACAAACGCGG-3′). RACE products were gel purified and cloned into pGEM-T Easy (Promega, Madison, WI). All sequencing was performed using BigDye terminator cycle sequencing on an ABI sequencer (PerkinElmer Applied Biosystems, Foster City, CA). Sequencing reactions were done in a 10 μL volume with 200 to 400 ng of DNA and 10 pg of primer. The cycling conditions used were 95oC for 2 min, 70 cycles at 95o for 15 s, 55oC for 20 s, 60oC for 4 min, followed by 72oC for 7 min.
Cloning of ZmFie Genes
Arabidopsis FIE (AF129516) was used in a TBLASTN search of the Pioneer Hi-Bred maize (Zea mays) EST database. Two contigs with significant similarity were found and were named ZmFie1 and ZmFie2. Both contigs contained at least one full-length EST. The sequence of the ZmFie genes was obtained by sequencing the full-length clones as described above.
Cloning of ZmEpl101
The Arabidopsis genome sequence was searched using fruit fly E(Pc) as a query. Two sequences were identified in this search, AtEpl1 and AtEpl2. The AtEPL1 (AC011808) protein sequence was then used to perform a TBLASTN search of the public maize EST database (http://www.zmdb.iastate.edu/). One EST (BE511872) with significant similarity was found. We obtained the remaining sequence for the ZmEpl101 gene by RACE-PCR. The primers used were Mepc1R1 (5′-GTCCGGAGAAGAGGATTCCATCGATC-3′) and Mepc1R2 (5′-CCTCGTCCAATCCTACCTCGACACC-3′).
Phylogenetic Analysis
The SET domains from all E(z)-like proteins were aligned using ClustalW (the exact region used is indicated in Fig. 1). This alignment was then submitted to the PHYLIP server at http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html. The protpars feature was used, with bootstrapping performed before analysis. One hundred replicates were examined to determine bootstrap values. The consensus tree was then displayed with bootstrap values.
RT-PCR Analysis
RT-PCR was used to assess expression patterns due to the relatively low expression of the maize PcG homologs, and to the fact that most of the genes were duplicated. Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA) from 10 tissues from the inbred B73 (endosperm [11 DAP], whole kernel [3 DAP], whole kernel [11 DAP], 10 d seedling [whole plant included], root tips, immature leaf [leaves 3–5], mature leaf [fully expanded leaf 10], and meiotic tassel). One microgram of total RNA was used to make cDNA with the SMART cDNA synthesis kit according to manufacturer's instructions (CLONTECH). PCR reactions were performed in a 25-μL total volume containing approximately 0.5 ng of cDNA, 5 pm each primer, 1 unit of Taq polymerase (Promega), 2.5 μL of 10× reaction buffer, 2 μL of 25 mm MgCl2, and 0.3 μL of 25 mm dNTPs. Primers used for the RT-PCR reactions were Ubi1F1 (5′-TAAGCTGCCGATGTGCCTGCGTCG-3′) and Ubi1R1 (5′-CTGAAAGACAGCACATAATGAGCACAGGC-3′) for Ubiquitin; Mepc1F1 (5′-CCGGCCATGGCGAAGCTG-3′) and Mepc1R2 (5′-CCTCGTCCAATCCTACCTCGACACC-3′) for ZmEpl101; Mez1F1 (5′-GGGTGTGGTGATGGTACATTGG-3′) and Mez1R1 (5′-CGGGACCTAACTCTACGGATGG-3′) for Mez1; Mez2F8 (5′-CCCCTGTTTTGCAGCCAGTCGTGA-3′) and Mez2R8 (5′-GGTGAGAGAAGGATGCCTCGTCC-3′) for Mez2; Mez3F3 (5′-AGTATGTGTTGGATGCTTATCGCAAGG-3′) and Mez3R2 (5′-GGTTGTCAGTTTGTCACCTTCCGACC-3′) for Mez3; Mesc1–5 (5′-TTTGCAAGTTGTGGCATGGA-3′) and Mesc1R2 (5′-CCCAACTTTCAACATTCGAAGCATTC-3′) for ZmFie2; and FLMesc2F1 (5′-CAACATCTGGCACAGCATGC-3′) and Mesc2R3 (5′-GTTGCCTATTGCCATCTGGTTGGAG-3′) for ZmFie1. Conditions of the PCR were as follows: 94oC for 2 min, 35 cycles at 94oC for 30 s, 63oC for 30 s, and 72oC for 2 min, followed by 72oC for 7 min. Amplified products were separated in a 1% (w/v) agarose Tris borate-EDTA gel and were visualized by ethidium bromide staining. The primers chosen flanked introns and are expected to produce different sized transcripts from genomic DNA and cDNA. The ZmEpl101 primers amplify a 450-bp cDNA product and an approximately 1,200-bp genomic product. The Mez1 primers would amplify a 717-bp cDNA product and a 1,235-bp genomic product, the Mez2 primers would amplify a 556-bp cDNA product and a 1,083-bp genomic product, the Mez3 primers would amplify a 509-bp cDNA product and a 1,211-bp genomic product, the ZmFie1 primers would amplify 1,135-bp cDNA product and a 4,008-bp genomic product, and the ZmFie2 primers would amplify 711-bp cDNA product and a 1,748-bp genomic product.
Analysis of Mez2 Alternative Splicing
Alternative splicing of Mez2 was indicated by the presence of multiple bands when using primers that amplified the entire coding sequence. The primers used were Mez2F6 (5′-CGCAGCTGATACGGCAAGTCCAATCG-3′) and Mez2R2 (5′-GTATCATCCGGAGCGACTCTTCAGC-3′). These primers are expected to produce a 2,594-bp cDNA product and a >9-kb genomic fragment. To characterize the alternative splice products, PCR was performed using 10 ng of B73 cDNA isolated from ear tissue and Amplitaq Gold DNA polymerase (PerkinElmer Applied Biosystems). The cycling conditions were 95oC for 5 min, 35 cycles at 95oC for 30 s, 65oC for 30 s, and 72oC for 3.5 min, followed by 72oC for 7 min. The amplified products were separated by electrophoresis in a 1% (w/v) low melting point agarose Tris borate-EDTA gel and were observed by ethidium bromide staining. Three alternative splice products were consistently observed and each band was excised and sequenced. The tissue-specific distribution of the different splice products was assayed using RT-PCR on the same tissues used for RT-PCR analysis. The primers used for these reactions were Mez2F10 (5′-CCATGTGAGAAGCAACCCTACAGC-3′) and Mez2R10 (5′-CCCAACCTGCAACATCAGATCTTCC-3′). The reaction conditions and cycling times were the same as those used for the RT-PCR reactions.
Footnotes
This research was supported by the U.S. Department of Agriculture (National Needs Fellowship no. 98–38420–5832) and by the National Science Foundation (grant no. 9975930).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010742.
LITERATURE CITED
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- Alkema MJ, van der Lugt NM, Bobeldijk RC, Berns A, van Lohuizen M. Transformation of axial skeleton due to overexpression of bmi-1in transgenic mice. Nature. 1995;374:724–727. doi: 10.1038/374724a0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Brock HW, van Lohuizen M. The Polycomb group: no longer an exclusive club? Curr Opin Genet Dev. 2001;11:175–181. doi: 10.1016/s0959-437x(00)00176-3. [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- Core N, Bel S, Gaunt SJ, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thalianaresults in abnormal plant development. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Gen Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by Medea, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Helentjaris T. Atlas of duplicated sequences. Maize Newslett. 1995;69:67–81. [Google Scholar]
- Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a DrosophilaPolycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z)proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsisendosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB et al. Control of fertilization-independent endosperm development by the Medea polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Fan Y, Strome S. The Polycomb group in Caenorhabditis elegansand maternal control of germline development. Development. 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiaetelomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsisseeds. Proc Natl Acad Sci USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Gaunt S, Lawrence PA. Function of the Polycombprotein is conserved in mice and flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Ng J, Hart CM, Morgan K, Simon JA. A DrosophilaESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J, Li R, Morgan K, Simon J. Evolutionary conservation and predicted structure of the Drosophilaextra sex combs repressor protein. Mol Cell Biol. 1997;17:6663–6672. doi: 10.1128/mcb.17.11.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu Y-C, Williams C, Repetti P, Fischer RL. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: Gene silencing of alcohol dehydrogenase by white-Adhtransgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression of nonhomologous transgenes in Drosophilainvolves mutually related endogenous sequences. Cell. 1999;99:35–46. doi: 10.1016/s0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- Preuss D. Chromatin silencing and Arabidopsisdevelopment: a role for polycomb proteins. Plant Cell. 1999;11:765–768. doi: 10.1105/tpc.11.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sato T, Hayes PH, Denell RE. Homeosis in Drosophila: maternal affect of the Enhancer of Polycomb locus and its interaction with Polycomband related loci. Dev Genet. 1984;4:185–198. [Google Scholar]
- Sato T, Russell MA, Denell RE. Homeosis in Drosophila: a new enhancer of Polycomband related homeotic mutations. Genetics. 1983;105:357–370. doi: 10.1093/genetics/105.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- Sewalt RG, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DP, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Murakami H, Hasegawa Y, Takahashi M. RET finger protein is a transcriptional repressor and interacts with enhancer of polycombthat has dual transcriptional functions. J Biol Chem. 2000;275:39411–39419. doi: 10.1074/jbc.M006585200. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophiladevelopment. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J. Interaction of the Arabidopsispolycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol. 2000;10:1535–1538. doi: 10.1016/s0960-9822(00)00839-3. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Berger J, Ruse C, Sinclair DA, Randazzo F, Brock HW. The enhancer of polycomb gene of Drosophilaencodes a chromatin protein conserved in yeast and mammals. Development. 1998;125:4055–4066. doi: 10.1242/dev.125.20.4055. [DOI] [PubMed] [Google Scholar]
- Strutt H, Paro R. The polycomb group protein complex of Drosophila melanogasterhas different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The DrosophilaPolycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Fong Y, Strome S. The Caenorhabditis elegansmaternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc Natl Acad Sci USA. 2001;98:5061–5066. doi: 10.1073/pnas.081016198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB et al. Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell. 2000;12:2367–2382. doi: 10.1105/tpc.12.12.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]