Abstract
Large-scale protein overexpression phenotype screens provide an important complement to the more common gene knockout screens. Here, we have targeted the so far poorly understood Saccharomyces cerevisiae membrane proteome and report growth phenotypes for a strain collection overexpressing ≈600 C-terminally tagged integral membrane proteins grown both under normal and three different stress conditions. Although overexpression of most membrane proteins reduce the growth rate in synthetic defined medium, we identify a large number of proteins that, when overexpressed, confer specific resistance to various stress conditions. Our data suggest that regulation of glycosylphosphatidylinositol anchor biosynthesis and the Na+/K+ homeostasis system constitute major downstream targets of the yeast PKA/RAS pathway and point to a possible connection between the early secretory pathway and the cells’ response to oxidative stress. We also have quantified the expression levels for >550 membrane proteins, facilitating the choice of well expressing proteins for future functional and structural studies.
Keywords: caffeine, paraquat, salt tolerance, yeast
Although a wealth of proteomics data relating to, e.g., protein expression profiles (1), organellar proteomes (2), global protein localization patterns (3, 4), and protein–protein interaction networks (5, 6) has been collected for Saccharomyces cerevisiae, its membrane proteome is underrepresented consistently in such studies to date.
We have recently constructed a clone collection composed of strains overexpressing a total of 617 S. cerevisiae membrane proteins with two or more predicted transmembrane segments (7). Each protein in the collection is genetically fused to a C-terminal topology reporter composed of a hemagglutinin (HA) tag, a part of the Suc2p protein, and the catalytic C-domain of the His4p histidinol dehydrogenase enzyme (8, 9). By analysis of the reporter fusions, the location of the C terminus of each protein relative to the endoplasmic reticulum membrane could be determined, making it possible to produce “constrained” topology models for ≈550 of the proteins in the collection (7).
Because earlier reports using various C-terminal tags to study global protein expression, localization, and complex formation in S. cerevisiae suggest that the stability, localization, and function of most of proteins are not compromised by C-terminal fusions (1, 4, 10), we decided to use the clone collection to examine the effects of overexpression of individual membrane proteins on growth rate under different conditions (basal conditions, salt, oxidative, and caffeine stresses) and to estimate overexpression levels. Protein overexpression screens provide a complementary approach to the widely used gene knockout strategy for large-scale functional annotation (11, 12) and is useful particularly useful for the analysis of essential genes and for gene families that, because of functional redundancy, are unsuitable for targeted deletion studies.
Results and Discussion
Overexpression Levels.
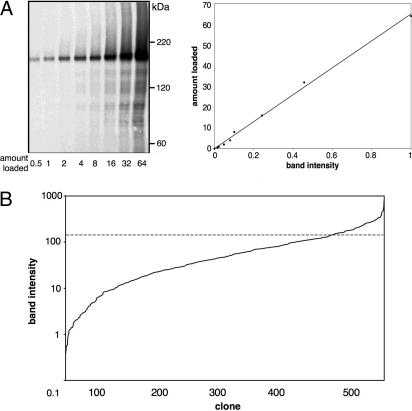
Before the functional studies, we undertook a quantitative Western blot analysis of the entire multicopy plasmid clone collection expressed from the strong TPI1 promoter in the same STY50 strain that was used in the topology mapping study (7, 13). The 122-kDa HA/Suc2/His4C reporter domain helps in reducing protein aggregation and the uneven transfer efficiencies sometimes observed in Western blot analysis of membrane proteins (unpublished data). Pga3p, a 35-kDa protein with two predicted transmembrane segments, was used as an internal standard in the Western blots. Pga3p was produced as a HA/His8/Suc2/His4C fusion protein and purified by using the His8 tag. As shown in Fig. 1A, there is a good linear correlation between the amount of purified protein and the intensity of the Western blot.
Fig. 1.
Quantification of protein expression. (A Left) Increasing amounts (arbitrary units) of purified Pga3p-HA-His-8-Suc2-His4C were loaded onto 6.25% SDS/PAGE and visualized by Western blotting with anti-HA antibody. (A Right) Quantified band intensities versus amount of protein loaded (r2 = 0.997). (B) Relative expression levels for 553 cloned S. cerevisiae proteins. The dashed line indicates an expression level of ≈0.5 mg/liter as estimated from GFP-fusions (see Results and Discussion).
Relative expression levels for 553 HA/Suc2/His4C fusion proteins were estimated from the Western blots (Fig. 5, which is published as supporting information on the PNAS web site) and are summarized in Fig. 1B and in Table 1, which is published as supporting information on the PNAS web site. These expression levels correlate well with absolute expression levels measured for a collection of GFP-fused proteins (unpublished data), with a relative expression level of 150 units corresponding roughly to 0.5 mg of protein per liter of culture. Of the 553 proteins, 219 were quantified in an earlier genomewide Western blot-based expression screen where chromosomally TAP-tagged genes expressed from their endogenous promoters were analyzed (1). For these 219 proteins, expression from the strong TPI1 promoter in the high-copy plasmid leads to up to 103-fold enhanced expression levels compared with expression from the chromosomal genes (data not shown).
We could not detect any specific functional classes to be overrepresented among the proteins that displayed low expression (<10 units relative expression level), indicating that expression was not limited by the protein’s biological role. In contrast, among the proteins with the highest degree of expression (>100 units relative expression level), stress response proteins (e.g., Isc1p, Hsp30p, Ist2p, Orm2p, and Orm1p) were found to be significantly overrepresented (P < 0.001; data not shown).
Four hundred seventy-seven of 603 proteins (79%) were detected as a single band of the expected molecular mass, whereas 126 gave rise to two distinct products (Fig. 5). For 54 of the 126 proteins, neither of the two forms was sensitive to Endo H digestion, indicating that the C-terminal orientation is cytosolic in both isoforms (7). For another 56 proteins, the higher molecular mass form was sensitive to Endo H, suggesting that the reporter is located in the endoplasmic reticulum (ER) lumen in only a fraction of these molecules. The banding patterns of the remaining 16 proteins were more complex and could not be interpreted easily (see ref. 7 for a more thorough discussion of the glycosylation-based topology mapping strategy).
Identification of Membrane Proteins That Affect Cell Growth Under Normal Conditions.
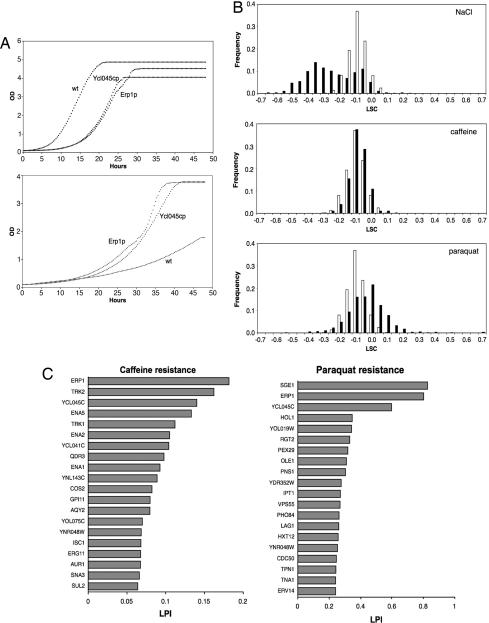
Using the clone collection, we next sought to identify membrane proteins that, when overexpressed, specifically inhibit or promote cell growth under various conditions. To this end, we screened for effects on cell growth caused by the overexpression of individual HA/Suc2/His4C fusions by microcultivation and automated recording of individual growth curves (14) in synthetic defined medium and in three different stress conditions (NaCl, caffeine, and paraquat). Typical growth curves in synthetic defined medium and paraquat for the STY50 strain (transformed with empty vector) and for strains overexpressing Erp1p and Ycl045cp (discussed below) are shown in Fig. 2A. As described in Experimental Procedures, logarithmic strain coefficients (LSCs) were calculated by normalizing the growth rate for each strain to the average growth rate of four reference cultures of the STY50 strain included in each run. For stress conditions, LSC values were normalized further to the LSC values obtained in synthetic defined medium, providing a logarithmic phenotypic index (LPI) that gives a quantitative measure of the gene-by-environment interactions normalized for general growth defects (14).
Fig. 2.
Phenotypic consequences of membrane protein overexpression. (A) Growth curves in synthetic defined medium (Upper) and under oxidative stress (Lower) for the reference strain transformed with empty vector (wt) and strains overexpressing Erp1p and Ycl045cp. (B) Distribution of phenotypes (LSC) during no stress (open bars) and stress (filled bars). Negative LSC values indicate reduced fitness. (C) Phenotypes (LPI values) for the top 20 most stress tolerant (P < 0.001) overexpression strains.
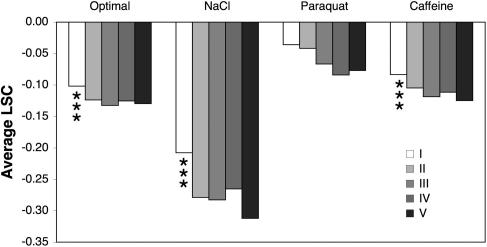
In synthetic defined medium, 529 of 567 (93%) overexpression strains tested had a significantly reduced growth rate (LSC < 0; P < 0.001) as compared to the STY50 reference strain (Fig. 2B, open bars, and Table 1). The general correlation between protein overexpression levels and the severity of the growth phenotype was low (r2 = 0.011; LSC, synthetic defined medium). Except for strains with the most poorly expressed proteins that displayed significantly lower growth defects than the average strain (P < 0.001), no significant deviations from the phenotypic mean were observed for any expression group (Fig. 3). The observed growth phenotypes discussed below therefore cannot be explained as a general overexpression artifact but relate to characteristics of the individual proteins.
Fig. 3.
Correlation between overexpression levels and degree of growth defects. Average growth defect (LSC) of membrane protein overexpression strains were divided into equally sized bins according to level of protein expression (I, <14; II, 14–34; III, 34–70; IV, 70–120; V, >120 units relative expression level). Asterisks indicate significant (P < 0.001) deviation from the mean growth defect of all investigated strains.
Four strains grew significantly better than the reference strain in synthetic defined medium. Two of the four proteins overexpressed in these strains, Rft1p, a flippase required for translocation of Man5GlcNac2-PP-Dol from the cytoplasmic side to the luminal side of the ER membrane (15), and Chs2p, which transfers N-acetylglucosamine to chitin (16), are essential proteins involved in glycosylation processes. The other two, the ferric reductase Fre3p and the iron/copper transporter Pca1p, are involved in maintaining iron/copper homeostasis (17, 18).
In a recent protein overexpression study using an essentially qualitative estimation of growth on solid media, severe growth defects were found for only 15% of the overexpressed S. cerevisiae proteins (11). Among the strains with a severe growth defect, there was a remarkable enrichment of membrane-associated transport proteins, suggesting that growth defects should be unusually frequent after overexpression of membrane proteins. This observation might partly explain the high proportion of growth defects seen here. It should be noted, however, that most of the growth defects we have identified in synthetic defined medium are marginal: 90% of the transformants with a significant growth defect display at the most a 20% decrease in growth rate (LSC > −0.2; Fig. 2B).
Identification of Membrane Proteins Conferring Stress Resistance.
To identify membrane proteins that make cells either more sensitive or more resistant to specific stress conditions, the overexpression strains also were cultivated under salt stress (NaCl), oxidative stress (paraquat), and in the presence of the cAMP phoshodiesterase inhibitor caffeine. Because the STY50 strain carries the dominant hol1-1 mutation (a prerequisite for the topology assay described in ref. 7) that confers hypersensitivity to Na+ by causing a substantially enhanced influx of this ion (19, 20), we first tested whether we could identify proteins that suppress the hol1-1 phenotype in high NaCl. Consistent with the hol1-1 Na+-sensitive phenotype, the distribution of salt stress LSC values was highly skewed with a strong bias toward reduced fitness (Fig. 2B Top, filled bars): 47% of the strains had a specific growth defect in high salt compared with synthetic defined medium (LPI < 0; P < 0.001). Three of the 567 investigated proteins suppressed the hol1-1 phenotype and induced significant tolerance to NaCl (LPI > 0; P < 0.001): the potassium transporter Trk2p, already known from a gene deletion study to be important for Na+ tolerance (21), the iron transporter Arn1p (22), and Ecm27p, a hitherto uncharacterized protein with homology to the Pfam (23) family PF01699 of putative Na+/Ca2+ exchangers.
In contrast to the highly skewed distribution of salt phenotypes, the LSC distributions of caffeine and paraquat phenotypes more closely resembled the distribution seen in synthetic defined medium (Fig. 2B Middle and Bottom, filled bars). Caffeine is an inhibitor of cAMP phosphodiesterases in the PKA/RAS signal transduction pathway and induces abnormally high PKA activity via enhanced intracellular levels of cAMP. Hence, overexpression of proteins that are negatively regulated by the PKA pathway may be expected to mitigate the effects of high PKA activity and, hence, confer caffeine tolerance, in line with a recent study where relevant downstream targets of Pho85p were identified by systematic gene overexpression in a pho85Δ strain (11). We found overexpression of 70 membrane proteins to confer significantly increased tolerance to caffeine (LPI > 0; P < 0.001). Among the top 20 hits were two major functional classes (Fig. 2C Left): proteins involved in the biosynthesis of glycosylphosphatidylinositol (GPI) anchors and its sphingolipid precursors, including Gpi1p (24), Isc1p (25), and Aur1p (26), and regulators of intracellular Na+/K+ homeostasis. Notably, both components of the main K+ transport system (Trk1p and Trk2p) (27) the three major membrane transporters mediating Na+ export (Ena1p, Ena2p, and Ena5p) (28, 29), and the previously identified Na+ tolerance protein Sna3p (30) scored among the most caffeine-tolerant overexpression strains. The Ena1p-induced caffeine tolerance may be related to the fact that PKA activity influences the nucleocytosolic localization of the basic leucine zipper repressor Sko1p (31) that, in turn, represses ENA1 expression by binding to the cyclicAMP response-like element in the ENA1 promoter (32).
Overexpression of proteins involved in GPI anchor biosynthesis and the Na+/K+ homeostasis system thus mitigates the toxic effects of caffeine, suggesting that regulation of these processes constitute major downstream targets of the yeast PKA/RAS pathway. In agreement with these results, other genes in GPI anchor biosynthesis, including GPI1 (33), and components of the saline response (34) earlier have been linked to the activity of the PKA pathway.
Paraquat induces oxidative stress by redox-cycling, shunting electrons from the electron transport chain into oxygen, forming superoxide radicals (35). Consequently, perturbations of the oxidative stress protection systems by removal of reactive oxygen species scavengers or proteins regulating the homeostasis of electron acceptors, such as iron and copper, lead to hypersensitivity to paraquat (J.W. and A.B., unpublished data). It therefore may be expected that overexpression of such proteins should confer increased tolerance to superoxide anions generated by paraquat. We identified 29 membrane proteins that confer significant tolerance to paraquat (LPI > 0; P < 0.001) when overexpressed (Fig. 2C Right). These proteins represent a wide diversity of functions, such as Rgt2p and Hxt12p involved in glucose sensing and transport (36, 37), the lipid biosynthesis proteins Ole1p and Ipt1p (38, 39), and the endosomal, lipid-translocation-associated protein Cdc50p (40). In particular, overexpression of the multidrug transporter Sge1p (41), the COPII vesicle transport protein Erp1p (42), and the functionally unclassified protein Ycl045cp induces hypertolerance to paraquat.
Functional Inferences from Hierarchical Clustering of Overexpression Phenotypes.
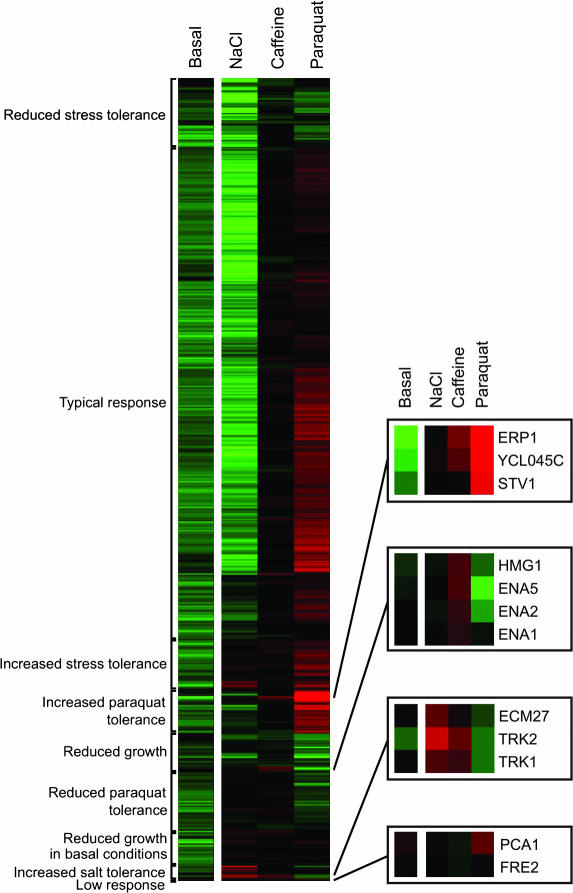
To complement the statistical analysis, similarities in overexpression phenotypes of 515 membrane proteins were analyzed by hierarchical clustering (ref. 43; Fig. 4). The 515 membrane proteins segregated into nine major clusters (correlation coefficient >0.5). We find several subclusters of functionally related proteins suggesting that the growth data captures relevant correlations between different proteins. Notably, the two potassium transporters Trk1p and Trk2p form a distinct subcluster that also includes the poorly characterized protein Ecm27p (cf. above). An equally tight cluster is formed by the dominant sodium exporters Ena1p, Ena2p, and Ena5p. The distinct profile of the sodium exporters are shared by Hmg1p, one of the two 3-hydroxymethylglutaryl coenzymes in the mevalonate synthesis pathway, suggesting a function in ion homeostasis. In contrast to overexpression of Trk1p or Trk2p, overexpression of Ena1, Ena2, or Ena5 does not confer increased tolerance to NaCl in the hol1-1 background but improves growth in caffeine. The phenotypically most distinct cluster is formed by Pca1p and Fre2p, proteins involved in iron/copper import and reduction.
Fig. 4.
Hierarchical clustering of overexpression phenotypes recorded in synthetic defined medium (basal conditions; LSC values) and different stress conditions (NaCl, caffeine, and paraquat; LPI values). Green indicates a reduced growth rate, and red indicates an increased growth rate relative to the reference STY50 strain.
As a final example, overexpression of Erp1p, Ycl045cp, and Stv1p dramatically reduces the growth rate in synthetic defined medium, and this effect is completely reversed when cells are grown in the presence of paraquat. Stv1p is a subunit of a V type ATPase of the Golgi apparatus and endosomes (44), and Erp1p is a COPII vesicle transport protein. Ycl045cp has the same predicted topology as Erp1p (they are both class I membrane proteins with a large luminal domain and a short C-terminal cytoplasmic tail), and we suggest that Ycl045cp is also part of the ER-Golgi transport machinery. Consistent with this proposal, a recent genomewide epistatic miniarray analysis (45) has identified genetic interactions between YCL045C and GET1 (involved in Golgi-ER trafficking), PMR1 (Ca2+ transport into the Golgi), and SSH1 (protein translocation into the ER).
Conclusions
In an effort to functionally characterize the S. cerevisiae membrane proteome, we have recorded growth rate effects induced by overexpression of nearly 600 membrane proteins in liquid synthetic defined medium and under three different stress conditions (NaCl, paraquat, and caffeine). We also have estimated overexpression levels and checked whether each protein is produced as a single or multiple molecular species.
By Western blotting, we estimate that 15% of the membrane proteins are overexpressed to >0.5 mg/liter without explicit attempts to optimize production levels and only a slightly lower value than found in a recent study (12), where a PGAL promoter and a His6/HA/ZZ tag were used. As for E. coli (46), we find no strong correlation between expression levels and mRNA or protein characteristics such as codon usage, protein length, or number of transmembrane helices (data not shown), and the expression data thus will facilitate the choice of well expressing proteins, e.g., for structural studies.
A small subset of the membrane proteins migrate on SDS/PAGE as two distinct bands. Of 126 such proteins, 54 were insensitive to Endo H digestion of the C-terminal HA/Suc2/His4C reporter domain, suggesting that the C terminus of both forms is in the cytosol (7) and, hence, that the two forms either have different N-termini or have undergone some partial posttranslational modification not related to N-linked glycosylation. For 56 proteins, the two forms have opposite C-terminal orientations as assessed by Endo H digestion of the topology reporter. In the latter group, we expect to find dual topology proteins (47) or proteins that are dually targeted to different compartments such as the ER and mitochondria.
Using high-throughput growth studies in synthetic defined medium, we find that overexpression of most of the membrane protein–HA/Suc2/His4C fusions moderately reduces growth rates, whereas overexpression of four proteins involved in glycosylation (Rft1p and Chs2p) and iron/copper homeostasis (Fre3p and Pca1p) significantly increase growth rate. Rft1p is an essential ER protein involved in the assembly of the dolichol-linked oligosaccharide donor Dol-PP-GlcNAc2Man8Glc3 for N-linked glycosylation (15). This protein is proposed to be a limiting component in transferring Man5GlcNac2-PP-Dol precursor from the cytoplasmic side to the luminal side of the ER membrane, and the available evidence indicates that N-glycosylation efficiency increases when Rft1p is overexpressed (15, 48). Constitutive overexpression of Rft1p thus might facilitate N-linked glycosylation and the production of glycoproteins. Chs2p is an essential protein (49) responsible for synthesis of chitin, a major fungal cell wall polysaccharide important for septum formation during budding of the daughter cell (16). Interestingly, this protein is found to be localized at the bud neck at the end of mitosis and then undergoes rapid degradation (50). Possibly, the constitutive overexpression of Chs2p might alter bud formation and lead to an increased rate of cytokinesis.
Under NaCl stress, half of the strains tested exhibit significant growth defects compared with synthetic defined medium, most likely caused by the hol1-1 mutation present in the STY50 strain, which dramatically increases the influx of sodium ions (19). Three proteins induce significant tolerance to NaCl in the hol1-1 background: the potassium transporter Trk2p, the iron transporter Arn1p, and the previously uncharacterized putative Na+/Ca2+ exchanger Ecm27p. In particular, the increased NaCl tolerance conferred by overexpression of Trk2p makes physiological sense because it is mainly the balance between K+ and Na+ that determines the physiological response to salt, and externally added potassium can suppress the detrimental effects from sodium (51, 52).
More than 70 proteins induce significant resistance to caffeine, an inhibitor of cAMP phosphodiesterases in the PKA/RAS pathway, when overexpressed. Many of the top hits are involved either in the biosynthesis of GPI anchors or in K+ and/or Na+ transport, suggesting that control of the GPI anchor and Na+/K+ homeostasis systems is a major function of the PKA/RAS pathway.
Finally, some 30 proteins representing a wide variety of functions induce significant resistance to the oxidative stress agent paraquat. Three proteins stand out as particularly interesting in this context: Stv1p, Erp1p, and Ycl045cp. Overexpression of these proteins leads to a drastic reduction in growth rate in synthetic defined medium but strongly increases growth rate compared with the reference strain in oxidative stress conditions. Stv1p is a component of a Golgi- or endosome-localized V-type ATPase, and Erp1p is an ER protein involved in the formation of COPII vesicles. It thus seems likely that Ycl045cp also is involved in ER-Golgi trafficking, pointing to a possible connection between the early secretory pathway and the cells’ response to oxidative stress.
Experimental Procedures
Yeast Genetic Manipulations.
The construction of the expression library in plasmid pJK90 is described in ref. 7. To construct the pJK91 plasmid carrying a His8 tag between the HA tag and the start of SUC2 in pJK90 (9), a His8 fragment was amplified from an unpublished plasmid (a kind gift from N. Meindl-Beinker, Stockholm University). Yeast homologous recombination was carried out in strain STY50 (MATa, his4-401, leu2-3, -112, trp1-1, ura3-52, hol1-1, SUC2::LEU2) (13) with XhoI-linearized pJK90 (9) and the His8 PCR product. Transformants were selected on -Ura plates, plasmid was isolated, and the correct sequence was confirmed by DNA sequencing. PGA3 was subcloned into pJK91 by PCR amplification followed by homologous recombination with SmaI-linearized pJK91 in STY50.
Quantification of Protein Expression.
For quantification of protein expression by Western blot analysis, Pga3p was selected as a standard and cloned as a HA-HIS8-SUC2-HIS4C fusion (see the previous section). This construct was transformed into strain STY50 (13), and the fusion protein was purified as follows. A 250-ml cell culture in -Ura medium was grown at 30°C to OD600 = 2, and the membrane fraction was prepared as described in ref. 53. The membrane pellet was resuspended in 2 ml of buffer (20 mM phosphate, pH 7.4/0.5 M NaCl/1% Triton X-100/10 mM imidazole), the sample was centrifuged at 13,000 × g at 4°C for 5 min, and the pellet was discarded. Further purification of His-tagged protein was done by using the HisTrap HP kit (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s protocol. For calibration, a standard curve was prepared by loading different amounts of purified protein on a 6.25% SDS gel, followed by Western blotting with an anti-HA antibody and quantification in a Fuji LAS-1000Plus charge-coupled device camera light box by using Image Gauge v. 3.4 software. To estimate the relative expression levels of the cloned proteins, a standard amount of purified Pga3p fusion protein was included together with the cell lysates prepared from a 1-ml culture of the yeast transformants on each gel. Western blots were quantified, and relative expression levels were calculated by normalizing against the intensity of the purified Pga3p.
The majority of the proteins (>84%, 462 of 553) were detected as a single band, 78 of 553 (14%) proteins gave rise to two distinct products of different molecular mass, and 13 of 553 (2%) produced three products of different molecular mass (Fig. 5). In the latter cases, if two or three bands were of approximately equal intensity, they were merged in the quantification; if not, only the major band was quantified.
Growth Assay.
Overexpression strains in the STY50 background were inoculated in 350 μl of synthetic defined medium (unstressed condition) as described in ref. 14, in synthetic defined medium containing 0.7 M NaCl, 650 μg/ml caffeine, or 300 μg/ml paraquat. Two consecutive precultures were run, each for 48 h. For experimental runs, strains were cultivated for 47 h with duplicates on separate 100-well plates (14). The rate of growth was calculated as reported in ref. 14. In each growth plate, four replicates of the reference STY50 strain transformed with empty plasmid were included and used for normalization in the calculation of LSC defined as:
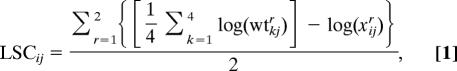 |
where wtkj is the doubling time of the kth measurement of the reference strain in environment j, xij is the doubling time of strain i in environment j, and r indicates the run.
The LSC value describes the growth of the mutant in relation to the growth of the reference strain (14). For the statistical test of the null hypothesis that LSC in unstressed conditions equals 0, a threshold of 3.3 mean SEs (corresponding to P < 0.001) was applied.
For analysis of specific gene-by-environment interactions, LSC in stress conditions was normalized to LSC in no-stress conditions, forming an LPI (LPI = LSCstress − LSCnostress) (14). Statistical tests of the null hypothesis that LPI equals 0 were done by using a threshold of three mean SEs. In order not to reject the null hypothesis only because of a single extreme value, a two-tailed, two-sample, Student’s t test also was applied (the combined statistical tests corresponded to P < 0.001). Growth data also were analyzed by hierarchical clustering of synthetics defined medium (LSC) and stress phenotypes (LPI). Clustering was performed by using Cluster 3.0 (43), applying an uncentered similarity metric and average linkage clustering.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council, The Marianne and Marcus Wallenberg Foundation, and the Swedish Foundation for Strategic Research and by the Swedish Cancer Foundation (G.v.H.) and the Swedish Knowledge Foundation (K.M.).
Abbreviations
- ER
endoplasmic reticulum
- GPI
glycosylphosphatidylinositol
- HA
hemagglutinin
- LPI
logarithmic phenotypic index
- LSC
logarithmic strain coefficient
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O’Shea E. K., Weissman J. S. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 2.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schonfisch B., Perschil I., Chacinska A., Guiard B., et al. Proc. Natl. Acad. Sci. USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., et al. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 5.von Mering C., Krause R., Snel B., Cornell M., Oliver S. G., Fields S., Bork P. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 6.Miller J. P., Lo R. S., Ben-Hur A., Desmarais C., Stagljar I., Noble W. S., Fields S. Proc. Natl. Acad. Sci. USA. 2005;102:12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H., Melén K., #x00D6;sterberg M., von Heijne G. Proc. Natl. Acad. Sci. USA. 2006;103:11142–11147. doi: 10.1073/pnas.0604075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deak R., Wolf D. J. Biol. Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 9.Kim H., Melén K., von Heijne G. J. Biol. Chem. 2003;278:10208–10213. doi: 10.1074/jbc.M300163200. [DOI] [PubMed] [Google Scholar]
- 10.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 11.Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., et al. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strahl-Bolsinger S., Scheinost A. J. Biol. Chem. 1999;274:9068–9075. doi: 10.1074/jbc.274.13.9068. [DOI] [PubMed] [Google Scholar]
- 14.Warringer J., Blomberg A. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 15.Helenius J., Ng D. T., Marolda C. L., Walter P., Valvano M. A., Aebi M. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- 16.Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Duran A., Cabib E. J. Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun C. W., Bauler M., Moore R. E., Klebba P. E., Philpott C. C. J. Biol. Chem. 2001;276:10218–10223. doi: 10.1074/jbc.M010065200. [DOI] [PubMed] [Google Scholar]
- 18.Rad M. R., Kirchrath L., Hollenberg C. P. Yeast. 1994;10:1217–1225. doi: 10.1002/yea.320100910. [DOI] [PubMed] [Google Scholar]
- 19.Gaber R. F., Kielland-Brandt M. C., Fink G. R. Mol. Cell. Biol. 1990;10:643–652. doi: 10.1128/mcb.10.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright M. B., Howell E. A., Gaber R. F. J. Bacteriol. 1996;178:7197–7205. doi: 10.1128/jb.178.24.7197-7205.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warringer J., Ericson E., Fernandez L., Nerman O., Blomberg A. Proc. Natl. Acad. Sci. USA. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heymann P., Ernst J. F., Winkelmann G. FEMS Microbiol. Lett. 2000;186:221–227. doi: 10.1111/j.1574-6968.2000.tb09108.x. [DOI] [PubMed] [Google Scholar]
- 23.Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., et al. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leidich S. D., Orlean P. J. Biol. Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- 25.Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y. A. J. Biol. Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 26.Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. J. Biol. Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 27.Ko C. H., Gaber R. F. Mol. Cell. Biol. 1991;11:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garciadeblas B., Rubio F., Quintero F. J., Banuelos M. A., Haro R., Rodriguez-Navarro A. Mol. Gen. Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 29.Wieland J., Nitsche A. M., Strayle J., Steiner H., Rudolph H. K. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nylander M., Heino P., Helenius E., Palva E. T., Ronne H., Welin B. V. Plant Mol. Biol. 2001;45:341–352. doi: 10.1023/a:1006451914231. [DOI] [PubMed] [Google Scholar]
- 31.Pascual-Ahuir A., Posas F., Serrano R., Proft M. J. Biol. Chem. 2001;276:37373–37378. doi: 10.1074/jbc.M105755200. [DOI] [PubMed] [Google Scholar]
- 32.Proft M., Serrano R. Mol. Cell. Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobering A. K., Watanabe R., Romeo M. J., Yan B. C., Specht C. A., Orlean P., Riezman H., Levin D. E. Cell. 2004;117:637–648. doi: 10.1016/j.cell.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Park J. I., Collinson E. J., Grant C. M., Dawes I. W. J. Biol. Chem. 2005;280:2529–2535. doi: 10.1074/jbc.M407900200. [DOI] [PubMed] [Google Scholar]
- 35.Hauptmann N., Cadenas E. Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 36.Ozcan S., Dover J., Johnston M. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruckeberg A. L. Arch. Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 38.Stukey J. E., McDonough V. M., Martin C. E. J. Biol. Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 39.Dickson R. C., Nagiec E. E., Wells G. B., Nagiec M. M., Lester R. L. J. Biol. Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- 40.Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrenhofer-Murray A. E., Seitz M. U., Sengstag C. Yeast. 1998;14:49–65. doi: 10.1002/(SICI)1097-0061(19980115)14:1<49::AID-YEA199>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 42.Marzioch M., Henthorn D. C., Herrmann J. M., Wilson R., Thomas D. Y., Bergeron J. J., Solari R. C., Rowley A. Mol. Biol. Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manolson M. F., Wu B., Proteau D., Taillon B. E., Roberts B. T., Hoyt M. A., Jones E. W. J. Biol. Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- 45.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., et al. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 46.Daley D. O., Rapp M., Granseth E., Melén K., Drew D., von Heijne G. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 47.Rapp M., Seppälä S., Granseth E., von Heijne G. Nat. Struct. Mol. Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- 48.Helenius J., Aebi M. Semin. Cell Dev. Biol. 2002;13:171–178. doi: 10.1016/s1084-9521(02)00045-9. [DOI] [PubMed] [Google Scholar]
- 49.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 50.Chuang J. S., Schekman R. W. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camacho M., Ramos J., Rodriguéz-Navarro A. Curr. Microbiol. 1981;6:295–299. [Google Scholar]
- 52.Gomez M. J., Luyten K., Ramos J. FEMS Microbiol. Lett. 1996;135:157–160. doi: 10.1111/j.1574-6968.1996.tb07982.x. [DOI] [PubMed] [Google Scholar]
- 53.Kim H., von Heijne G., Nilsson I. J. Biol. Chem. 2005;280:20261–20267. doi: 10.1074/jbc.M412213200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.