Abstract
Site-directed mutagenesis of recombinant Rous sarcoma virus (RSV) integrase (IN) allowed us to gain insights into the protein-protein and protein-DNA interactions involved in reconstituted IN-viral DNA complexes capable of efficient concerted DNA integration (termed full-site). At 4 nM IN, wild-type (wt) RSV IN incorporates ∼30% of the input donor into full-site integration products after 10 min of incubation at 37°C, which is equivalent to isolated retrovirus preintegration complexes for full-site integration activity. DNase I protection analysis demonstrated that wt IN was able to protect the viral DNA ends, mapping ∼20 bp from the end. We had previously mapped the replication capabilities of several RSV IN mutants (A48P and P115S) which appeared to affect viral DNA integration in vivo. Surprisingly, recombinant RSV A48P IN retained wt IN properties even though the virus carrying this mutation had significantly reduced integrated viral DNA in comparison to wt viral DNA in virus-infected cells. Recombinant RSV P115S IN also displayed all of the properties of wt RSV IN. Upon heating of dimeric P115S IN in solution at 57°C, it became apparent that the mutation in the catalytic core of RSV IN exhibited the same thermolabile properties for 3′ OH processing and strand transfer (half-site and full-site integration) activities consistent with the observed temperature-sensitive defect for integration in vivo. The average half-life for inactivation of the three activities were similar, ranging from 1.6 to 1.9 min independent of the IN concentrations in the assay mixtures. Wt IN was stable under the same heat treatment. DNase I protection analysis of several conservative and nonconservative substitutions at W233 (a highly conserved residue of the retrovirus C-terminal domain) suggests that this region is involved in protein-DNA interactions at the viral DNA attachment site. Our data suggest that the use of recombinant RSV IN to investigate efficient full-site integration in vitro with reference to integration in vivo is promising.
Retroviruses have the capability of inserting their DNA genome into the host DNA by the virus-encoded integrase (IN). IN from different retrovirus species possess three similar structural domains even though their amino acid sequences are significantly different (Fig. 1) (3, 13). The N-terminal domain (∼50 residues) contains a zinc binding region (6), promotes IN multimerization (46), and is necessary to catalyze 3′ OH processing and strand transfer. The 3′ OH processing activity is responsible for removal of dinucleotides from the catalytic strands on the blunt-ended viral DNA genome. The central or catalytic core domain (∼180 residues) contains a conserved triad of amino acids comprising the D,D(35)E motif (28) that is involved in coordinating divalent metal ions for enzymatic activities (2, 13). The core is also involved in target binding for strand transfer (1, 21, 27). The C-terminal domain (∼50 residues) binds to the viral DNA at ∼7 to 9 bp from the long terminal repeat (LTR) ends (18, 26, 30), binds to DNA in a nonspecific fashion (3), and also appears to be involved in multimerization of IN (25, 30). The reported subunit structure for purified virion IN from avian myeloblastosis virus (AMV) is a dimer (20, 32), while several retrovirus recombinant INs purified from bacteria are observed as monomers, dimers, and tetramers (12, 13, 14, 25).
FIG. 1.
Structural schematic of RSV IN. (Top) RSV IN (286 amino acids) is divided into three functional domains identified as the N-terminal, catalytic core, and C-terminal regions (13). A hinge region between the core and C-terminal domain is marked as a star (44). (Middle) The highly conserved HH-CC zinc-binding residues and a triad of amino acids comprising the D,D(35)E motif are identified within the box representing IN. The open circles above the box are conserved residues between RSV and HIV-1 IN. The residues marked below the box (D7N, etc.) are residues of RSV IN that have been examined at the genetic level (23, 24, 36, 37), except W233. Residue S282 of RSV IN is phosphorylated. (Bottom) Secondary structural alignments of RSV and HIV-1 residues from amino acids 97 to 146 are shown with permission from Elsevier (5). The blocked sea green-labeled amino acids are identical, and the orange ones are similar. The mutation in RSV IN (P115S) corresponds to the HIV-1 P109S mutation (15, 38).
Models describing potential interactions of IN at the viral DNA ends for concerted integration into the host DNA have been proposed (13, 44). The models are based on several approaches, including complementation of IN mutants (6, 17), crystallographic studies of individual or paired domains of IN (5, 8, 10, 42, 44), cross-linking studies of IN with DNA substrates (18, 19, 22, 26), and protection of viral DNA ends by IN in preintegration complexes (PICs) (4, 7, 9, 33, 43) and in reconstituted nucleoprotein complexes with purified AMV IN with viral DNA substrates (40). The collective theme suggests that IN forms multimers at the viral DNA ends for concerted integration of the two ends into a target, here termed full-site integration.
Attempts to compare the mutant retrovirus IN enzymatic activities in vitro to the observed replication and integration capabilities in vivo of retroviruses carrying the same mutations in the IN gene have been difficult for several reasons. First, recombinant wild-type (wt) and mutant IN have either failed to catalyze full-site integration or have done so in a nonefficient manner, thus offering significant insights into only the 3′ OH processing and half-site integration reactions (3, 16, 35). Half-site integration is defined as insertion of one viral DNA end into a target substrate. Second, even though mutations introduced into IN within the context of the viral genome were defective for integration (class I mutants) (16), others produced pleiotropic effects on the replication of retroviruses, indirectly affecting DNA integration in vivo (class II mutants) (3, 16).
In order to map the functions associated with the different domains in Rous sarcoma virus (RSV) (Prague A strain) IN for full-site integration, we introduced single amino acid mutations into recombinant IN. RSV IN would be an appropriate candidate for site-directed mutagenesis, since wt recombinant RSV IN has a specific activity for full-site integration similar to that of AMV IN purified from virions (11, 31, 41). The replication capabilities of several RSV IN virus mutants which had appeared to specifically affect viral DNA integration in vivo (Fig. 1) (36, 37) have been mapped; other mutants did not affect integration (23, 24). Here, we correlate in vitro activities of recombinant RSV IN mutants for full-site integration to several RSV PrA IN mutants (A48P and P115S) that possess significant integration defects in vivo. The RSV A48P IN mutation would apparently map in a disordered linker region between the N-terminal and the central catalytic domains, as shown with human immunodeficiency virus type 1 (HIV-1) IN (13, 42). A significant integration defect observed with RSV containing the A48P IN mutation in vivo (37) could not be correlated with a defect for full-site integration or other activities by recombinant A48P IN in vitro, suggesting a possible undiscovered pleiotropic effect for integration with RSV A48P IN in vivo. The RSV P115S IN mutation is located in the turn region between α1 and β4 (Fig. 1) that is in a structurally equivalent region of HIV-1 IN (5) possessing a similar mutation (P109S) (15, 38). Recombinant HIV-1 P109S IN possesses no 3′ OH processing and strand transfer activities, which is consistent with the inability of the HIV-1 virus containing this mutation to integrate its genome into host DNA. However, in this report, recombinant RSV P115S IN possesses the same full-site integration activity as the wt RSV IN. Furthermore, a positive correlation was observed between the temperature-sensitive defect for integration with the RSV P115S IN mutant in vivo (36) and the thermolabile 3′ OH processing, half-site, and full-site integration activities of recombinant P115S IN in vitro. Wt RSV IN is stable under the same heating conditions at 57°C. Substitutions into the highly conserved W233 of recombinant RSV IN suggest that the C-terminal region is involved in multimerization of IN at the LTR terminal attachment (att) sites, as defined by DNase I footprint protection studies (40).
MATERIALS AND METHODS
Linear DNA donors.
A linear 4.5-kbp DNA donor was constructed containing both the avian retrovirus 330-bp wt U3 and wt U5 LTRs (40). An NdeI site was produced at the circle junction of the U3 and U5 ends in the donor plasmid by site-directed mutagenesis. NdeI digestion produces a linear 4.5-kbp donor containing 3′ OH recessed ends analogous to processed viral DNA ends in PIC. In the gain-of-function DNA donor, the fifth and sixth nucleotides were modified to deoxyadenosine at each terminus (40, 41, 47).
Labeling of donors.
The recessed ends on the 4.5-kbp linear donors were 5′ end labeled with [γ-32P]ATP and polynucleotide kinase. The specific activities were ∼2,000 cpm (Cenerkov)/ng of DNA. For DNase I footprint analysis, the 5′ end-labeled 4.5-kbp donors were digested with either NheI or XhoI to produce 3.6-kbp single-end labeled LTR donors (Fig. 2) (40). The fragments were isolated by agarose gel electrophoresis, electroeluted, and concentrated by a Centricon YM-30 filtering device. For 3′ OH processing activity, several of the 3′ OH recessed 4.5-kbp LTR DNA donors were filled in with [32P]dTTP and unlabeled dATP by using Escherichia coli DNA polymerase (Klenow fragment) at 7°C, which results in the labeling of the penultimate T nucleotide (39).
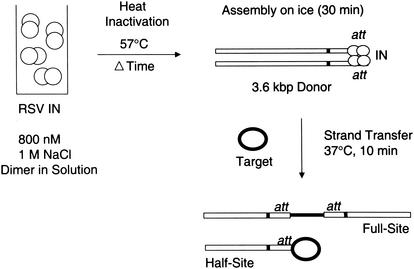
FIG. 2.
Strategy used for heat inactivation of RSV IN in solution. (Left) RSV IN (paired open circles) was diluted to 800 nM in a 1 M NaCl buffer prior to heating at 57°C. (Right) At various times after heating, IN was placed in assembly buffer with 32P-labeled donor DNA. IN was allowed to assemble onto the att sites of a 3.6-kbp LTR donor (or 4.5-kbp donor) prior to the addition of target (darkened circles) and strand transfer. The full- and half-site products obtained with the 3.6-kbp LTR donors were resolved by agarose gel electrophoresis.
Full-site integration and 3′ OH processing assays.
Assembly of macromolecular complexes capable of producing higher quantities of full-site integration products than half-site integration products at low IN concentrations (<20 nM) was described previously (40). Briefly, IN was assembled with either 3.6- or 4.5-kbp donors (10 ng) in the presence of 330 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 8% polyethylene glycol (6,000 Da), 20 mM HEPES (pH 7.5) on ice for 30 min as indicated. This assembly time, to apparent equilibrium, is sufficient for maximum production of stable complexes capable of either half-site or full-site integration (40) (data not shown). The standard volume was 20 μl or multiples thereof. The concentrations of wt or mutant RSV IN for assembly with donors were varied as indicated. After assembly, the integration reactions were initiated by the addition of supercoiled pGEM-3 (50 ng) as target (2.8 kbp) and immediately incubated at 37°C for 10 min. The integration reactions were stopped by addition of sodium dodecyl sulfate and proteinase K followed by phenol extraction. The reaction products were subjected to agarose gel electrophoresis, and the amounts of donor incorporated into the target were determined with a phosphorimager apparatus (40). For 3′ OH processing activities, the assay buffer described above for strand transfer was used unless indicated otherwise. The release of labeled dinucleotides from the filled-in 4.5-kbp donors by IN was measured as acid-soluble counts (39).
DNase I footprinting.
Double end-labeled 4.5-kbp DNA donors were digested with either NheI or XhoI (Fig. 2) to isolate 3.6-kbp single end-labeled U3 or U5 donors, respectively, on agarose gels for DNase I footprinting and strand transfer (40). For DNase I footprint analysis, RSV IN and the single end-labeled donors were assembled on ice or at 14°C, as indicated. An aliquot was removed from the mixture for measuring integration activity just prior to the addition of DNase I (final concentration, 750 ng/ml). The nucleoprotein complexes in the assembly mixture were further incubated with DNase I for 90 s at 14°C. The DNase I reactions were stopped by the addition of phenol, and the DNA products were subjected to denaturing 10% polyacrylamide gel electrophoresis. The dried gels were analyzed by a phosphorimager and exposure to X-ray films.
Site-directed mutagenesis, expression, and purification of RSV IN.
Standard oligonucleotide site-directed mutagenesis of RSV PrA IN was performed by several methods described by different manufacturers. The entire IN gene in each mutant DNA clone was sequenced to verify the DNA constructs. The RSV IN residues that were mutated are A48P, P115S, W233F, W233E, and W233A, with the last residue specifying the mutation. Wt and mutant RSV IN were expressed in E. coli BL21(DE3) pLysS cells (Novagen Corporation) (32). The cultures were induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of ∼1.6 and allowed to grow for an additional 3.5 h. Generally, 400-ml cultures yielding ∼5 g (wet weight) of cells were sufficient for purification. Cells were harvested by centrifugation and stored at −70°C prior to use.
The lysis, sonication, and washing steps with a 0.1 M NaCl buffer followed by a 1 M NaCl extraction step of the pelleted IN-DNA complex, as well as the column chromatography buffers, were similar to those previously described for larger-scale purification of wt RSV IN (32). Briefly, 25 ml of lysis buffer was added to 5 g of cells on ice with gentle shaking for 30 min. After a light sonication, the homogenized materials were subjected to high-speed centrifugation, and the pellets were suspended with 18 ml of a 0.1 M NaCl buffer. After a series of low-salt (0.1 M NaCl) washes and centrifugation steps, IN was extracted from the pellet with 3.5 ml of a 1 M NaCl buffer per gram (wet weight) of bacteria. The high-salt extract contained the majority of IN at a reasonable purity of ∼60 to 70%. The extracts were applied to a Pharmacia SP-Sepharose HiTrap column (5 ml) for further purification and removal of nucleic acids (32). The major nucleic acid peak preceded the elution of IN at 0.85 M NaCl. The protein peak (OD280) representing the majority of IN was loaded onto a second SP-Sepharose column, which usually resulted in preparations with >90% purity. Heparin affinity HP HiTrap columns (5 ml) were used to purify IN to near homogeneity. RSV IN eluted from heparin-Sepharose columns at 0.75 M NaCl. The standard chromatography buffers used for both columns were identical to that previously described (50 mM HEPES-NaOH [pH 7.5], 1 mM dithiothreitol, 1 mM EDTA, and 10 mM MgSO4) (32) except that 10% glycerol was present in the heparin-Sepharose procedure. Fractions were divided into aliquots and frozen at −70°C for storage. There were no apparent differences in the column elution profiles between wt and the different RSV mutant INs. Protein concentrations were determined by the absorbance at 280 nm, where one OD unit corresponds to a concentration of 1.87 mg/ml (32). Concentrations were calculated as IN dimers (64 μg/ml is equal to 1,000 nM).
Heat inactivation analysis of wt and mutant RSV IN.
RSV wt and mutant (P115S) IN preparations were diluted to a final concentration of 800 nM by using the above-described heparin-Sepharose column buffer containing 1 M NaCl on ice. The diluted IN mixtures (20 μl) were then subjected to heat inactivation at 57°C (Fig. 2) for various times, as indicated. Prior to heating, an aliquot (2 μl) of diluted IN was transferred to a tube containing 198 μl of full-site integration reaction buffer with donor stored on ice. IN was allowed to assemble onto the donor for 30 min. After assembly, aliquots were then taken both for strand transfer analysis at 37°C for 10 min and for DNase I footprint analysis at 14°C for 90 s. After continued heating of diluted IN at 57°C, the above procedures were applied to each aliquot taken at specific time intervals. For heat inactivation studies that measured strand transfer activities only, the gain-of-function double-ended LTR donor was used. For combined strand transfer and DNase I footprinting studies, gain-of-function single-end LTR donors (either U3 or U5 ends) were used. IN was assembled at 4, 6, 8, 10, and 12 nM in the assembly tube with donor present. The filled-in gain-of-function double-ended LTR donor was used for the heat inactivation studies analyzing the 3′ OH processing activity of IN.
Calculations for determining the t1/2.
To define the half-life (t1/2) of the heat-labile P115S IN, we used the exponential decay equation N = Noe−λt to find the decay constant λ of each independent full-site and half-site integration reaction. No is defined as the number of molecules at the beginning, when the IN activity is at its maximum, generating the highest amount of full- and half-site products. N is the number of molecules at a given time t (in our case, the amount of full-site or half-site product generated at a given time). Once the decay constant is found, we inserted that value in the half-life equation t1/2 = (ln 2)/λ as the λ. To confirm this calculation, another method to obtain the half-life of P115S IN was to plot the number of molecules versus time on a semilog graph. We took the common log of the number of molecules at time t (in our case, the percentage of full- or half-site products) and plotted it on the log side of the graph versus time (t). The times required for 50% of the P115S IN to be inactivated, or the t1/2, were determined. Both methods produced similar results.
RESULTS
Molecular insights into avian retrovirus full-site integration by site-directed mutagenesis of IN.
Several recombinant RSV INs containing mutations that had previously appeared to specifically affect integration in vivo were purified and studied (Fig. 1) (36, 37). In addition to measurement of strand transfer (full- and half-site) (Fig. 2) and 3′ OH processing activities, examination of whether these mutations (A48P and P115S) and others at the C terminus of IN affected the ability of recombinant IN to assembly onto and to protect ∼20 bp at the viral LTR termini encompassing the att site was conducted (40).
For quantitative measurements, our standard time for strand transfer was 10 min at 37°C. The production of full-site integration products with wt RSV IN at 4, 6, 8, and 10 nM was linear at each concentration for approximately 20 min (data not shown). After 40 min of incubation at 37°C, the maximum amount of donor incorporated into full-site products at these concentrations of IN was approximately 50%. The efficiency of the full-site integration reactions catalyzed by these assembled RSV IN nucleoprotein complexes is equivalent to those observed with purified cytoplasmic PIC from virus-infected cells (7, 9, 43).
A significant defect for integration in vivo does not correlate with a defect for full-site integration in vitro.
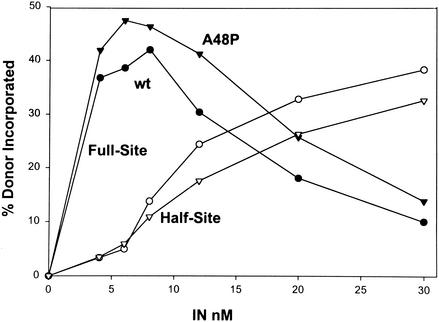
Earlier mutagenesis studies with RSV PrA IN suggested that modifying residue A48 to P resulted in a significant defect for integration in vivo (36). Viral DNA synthesis and viral polymerase (pol) proteins appeared to be normal upon infection by the virus possessing the A48P mutation. Virus replication displayed a delayed growth phenotype, and integration into the host chromosome was decreased ∼80% in comparison to integrated wt viral DNA. We introduced this mutation (A48P) into the recombinant RSV IN clone. Under standard assay conditions for half- and full-site integration, RSV A48P IN activity was very similar to wt RSV IN activity (Fig. 3). The 3′ OH processing activity of A48P IN was also similar to wt IN activity (data not shown). The results show that even though this mutation affected viral DNA integration in vivo, the mutation did not affect the enzymatic activities of recombinant mutant IN in vitro. This observation suggests that the A48P IN mutation produced an unknown pleiotropic effect in the RSV replication cycle that indirectly affects integration in vivo and would be a class II mutant (16).
FIG. 3.
Stand transfer analysis of wt and mutant A48P IN. wt RSV (circles) and A48P (triangles) IN at the indicated concentrations were assayed for strand transfer activity as described in Materials and Methods. Solid and open symbols indicate full-site and half-site products, respectively. The percentage of donor incorporated into each product was determined by using a phosphorimager.
A temperature-sensitive RSV IN mutant for replication in vivo correlates with IN thermolabile integration activities in vitro.
The introduction of a mutation into RSV IN (P115S) produced a virus that was temperature-sensitive for replication with a specific defect for integration in vivo (37). To determine whether the mutation introduced into recombinant RSV IN produced a thermolabile protein, a series of preliminary heat-inactivation experiments with IN in solution were performed to determine the optimum temperature for subsequent heat inactivation experiments with both wt and mutant RSV IN (data not shown). The strategies for investigating the thermolabile properties of RSV P115S and wt IN as control are illustrated in Fig. 2. Previous studies have demonstrated that recombinant RSV wt IN is a dimer in solution at 800 nM (32). Sedimentation analysis of P115S IN (initial concentrations of IN layered onto the gradients were between 1,000 and 3,000 nM) on 5 to 15% glycerol gradients containing the 1 M NaCl dilution buffer used in the heat inactivation experiments showed that it also sediments as a dimer (data not shown).
Both wt and P115S IN were diluted in the heat inactivation buffer to 800 nM prior to heating at 57°C for various lengths of time. At the indicated times, aliquots were removed and placed into reaction mixtures containing DNA donor substrate on ice. IN was allowed to assemble onto the donor to investigate whether the P115S mutation affected the assembly properties observed with wt IN as well as its strand transfer activities. The ability of wt RSV IN to assemble onto the donor att site and to perform strand transfer activities was stable upon heating of IN in solution at 57°C for 15 min (Fig. 4A and 4B, left) while P115S IN was thermolabile during the same time (Fig. 4A and 4B, right).
FIG. 4.
Heat inactivation of RSV wt and P115S IN. (A) Purified wt (left) and P115S (right) were diluted to 800 nM prior to heat inactivation at 57°C. At the indicated times, aliquots of IN were taken from the heated samples and added to assay mixtures containing gain-of-function double-ended LTR donor on ice. After 30 min, the assembled IN-DNA complexes were assayed for strand transfer at 37°C, and the products were analyzed by agarose gel electrophoresis. For both IN, the final concentrations in the assay mixtures were 8 nM. The half- and full-site products as well as the input donor are indicated on the left of each gel. Lanes Neg and 0 represent input donor sample without IN and analysis of IN without heating, respectively. (B) (Left) The percentage of DNA products from wt IN shown in panel A (circles) were determined with a phosphorimager along with results for another independent experiment for wt IN (triangles). (Right) The percentage of donor incorporated for both DNA products (circles) with P115S IN shown in panel A. The inset defines a semilog plot of the data for determining the t1/2 (see Materials and Methods).
Determining the inactivation rates for half- and full-site integration.
The strategy of heating IN in solution at 57°C prior to strand transfer analysis allowed us to assess the quantity of active IN dimers remaining after various times of heat inactivation (Fig. 2). After assembly of IN onto recessed LTR donor substrates, the time required to inactivate 50% (t1/2) of IN molecules necessary for each strand transfer activity was determined. The quantity of each strand transfer product produced is determined by the initial concentration of IN in the reaction mixture, i.e., at high IN concentrations, the production of half-site products is favored over that of full-site products (Fig. 3) (40).
Without heating, the specific activities of RSV P115S IN (Fig. 4A and 4B, right [zero time]) for both full- and half-site integration were similar to those observed for wt RSV IN (Fig. 4A and 4B [zero time]). With heating of IN in solution, the decay rate for half-site integration activity of P115S IN was calculated to have an average t1/2 of 1.9 min for six independent experiments at several different IN concentrations (Fig. 4 and 5), as summarized in Table 1. For determining the t1/2 values (Fig. 4B, see the inset semilog plot), half-site activity was taken as 100% at zero time. Inactivation of P115S IN in solution is immediate, as is evident by the decrease in half-site integration activities with time. Wt RSV IN activities (half- and full-site) were essentially stable upon identical heating and assay conditions (Fig. 4A and B, left) (data not shown).
FIG. 5.
Heat inactivation of RSV P115S IN in solution. (A) In three independent analyses, P115S IN (800 nM) was subjected to heat inactivation for various times at 57°C prior to being assayed for strand transfer activities at 8 nM. The corresponding solid (full-site) and open (half-site) symbols represent individual paired experiments. (B) As described for panel A, P115S IN was subjected to heat inactivation and assayed at 6 nM (triangles) and 12 nM (circles); solid and open symbols represent full- and half-site products, respectively. The inset depicts an analysis at 4 nM IN. See Table 1 for a summary of the data.
TABLE 1.
Heat inactivation of thermolabile RSV IN (P115S)
| IN concn (nM) | Experiment no.a |
t1/2 (min)b
|
|
|---|---|---|---|
| Full-site | Half-site | ||
| 6 | 1 | 1.6 | 1.8 |
| 8 | 1 | 1.2 | 2.1 |
| 2 | 1.7 | 1.6 | |
| 3 | 2.3 | 2.5 | |
| 4 | 1.7 | 1.8 | |
| 12 | 1 | 1.5 | 1.8 |
| Averagec | 1.7 | 1.9 | |
Experiment number for the indicated specific concentration of IN.
IN was heated inactivated at 57°C in solution, and the time (min) to decrease production of each product by 50% was determined by the formula t1/2 = (ln2)/λ where λ is derived from N = N0e−λt. See Materials and Methods.
Average of results of the six experiments.
With heating of IN in solution, there were two observed phases of full-site integration activity with P115S IN at 8 nM, presumably because IN is in excess at this concentration in the reaction mixture prior to heating (see Fig. 3). First, there was always a slight increase in full-site integration activity prior to a plateau and stable phase for activity (Fig. 4B and 5A). The decay phase results in a decrease of full-site integration activity that is calculated to have an average t1/2 of 1.7 min, similar to that of half-site integration (Fig. 4B) (Table 1). To calculate the t1/2 for full-site integration, 100% activity was assigned as the highest value at or just prior to the decay phase (Fig. 4B, see inset) (Table 1). These above results suggest that RSV P115S IN is a class I mutant (16).
As shown in Fig. 5A and Table 1, similar inactivation rates of IN in solution for both the half- and full-site integration activities were obtained with separate independent experiments at 8 nM with P115S IN. In each of these experiments, wt RSV IN was heat inactivated and assayed at the same concentrations as mutant IN. In contrast to mutant P115S IN, wt RSV IN remained heat stable in the 15 min for full-site integration, as shown in Fig. 4 (data not shown). Similar t1/2 values were also obtained for both strand transfer activities with P115S IN at either 6 or 12 nM in the reaction mixtures (Fig. 5B) (Table 1), with wt RSV IN being stable at these concentrations also. The results showed that similar inactivation rates were obtained for both half- and full-site reactions at various IN concentrations in the assembly mixtures.
Calculations for defining the number of IN subunits at the viral DNA ends.
With P115S IN at 4 nM prior to heating, the initial amounts of products for full- and half-site integration activities were ∼30 and ∼8%, respectively (Fig. 5B, inset). As previously shown (Fig. 3), excess IN in the reaction mixture results in a subsequent decrease in full-site activity relative to half-site activity (40). With P115S IN at 12 nM in the reaction mixtures, both the half- and full-site integration activities prior to heating were similar, at ∼20% each (Fig. 5B). The apparent reason for an increase in full-site integration activity at 12 nM upon heating of IN in solution was the proportional decrease in the number of active IN molecules in solution. A smaller increase in full-site activity was also observed upon heating with the reactions at 8 nM (Fig. 4B and 5A) but was nearly negligible at either 6 (Fig. 5B) or 4 nM (Fig. 5B, inset). This decrease in active IN molecules upon heating is tabulated in Table 2 as a function of time and protein concentrations in the reaction mixture. Using the last time point for maximum full-site integration activity at all of the IN concentrations studied (Fig. 4 and 5) and the determined t1/2 value of 1.7 min (Table 1), the percentages of active IN molecules in solution at a specific time were determined in Table 2. The calculated number of remaining active IN dimers per donor end was in the range from 5 to 9. This calculation excludes the binding of IN at donor molecules for half-site integration as well as the potential nonspecific binding of IN internally on the donor. The results suggest that the actual number of subunits at each donor end required for full-site integration is unknown.
TABLE 2.
Calculation defining the minimal number of RSV P115S IN dimers required for maximum full-site integration activitya
| Initial IN concn (nM) | Time of maximum activity prior to inactivation (min) | % Active IN leftb | Calculated IN left (nM) | IN dimers per viral endc |
|---|---|---|---|---|
| 4 | 1 | 67 | 2.7 | 7.2 |
| 6 | 3 | 29 | 1.8 | 4.8 |
| 8d | 3 | 29 | 2.4 | 6.4 |
| 12 | 3 | 29 | 3.5 | 9.3 |
The maximum full-site integration activity was determined from Fig. 4 and 5. It was defined as the last time point showing maximum full-site activity. The average t1/2 for full-site integration was 1.7 min, as defined in Table 1.
The formula used to calculate the % active IN left (N) as a function of time is N = N0e−λt.
Calculated number of IN dimers per viral DNA end at maximum full-site integration activity at the chosen time of 1 or 3 min. For illustration, at 3 nM IN, the calculated number of IN dimers per end is 8 (4.5-kbp double-ended LTR substrate).
Average of four experiments.
Heat inactivation of RSV P115S IN results in loss of 3′ OH processing activity and binding to the viral att site for full-site integration.
To further define the defect with RSV P115S IN, we performed a 3′ OH processing reaction. The 3′ OH recessed gain-of-function 4.5-kbp donor ends were converted into blunt-end donors by a fill-in reaction (39). As shown in Fig. 6A, we determined that the specific activities for the 3′ OH processing activities of wt and P115S IN at various concentrations were the same. Employing a similar procedure used for strand transfer heat inactivation studies (Fig. 2), we demonstrated that the t1/2 for 3′ OH processing of RSV P115S IN was 1.6 min (average of results of three independent experiments), while wt IN activity was essentially heat stable under the same conditions (Fig. 6B). The inactivation curve for 3′ OH processing by P115S IN at 20 nM is similar to the curve observed for strand transfer at 12 nM (Fig. 5B); i.e., the enzyme is in excess prior to heating with the subsequent loss of active IN molecules after heating.
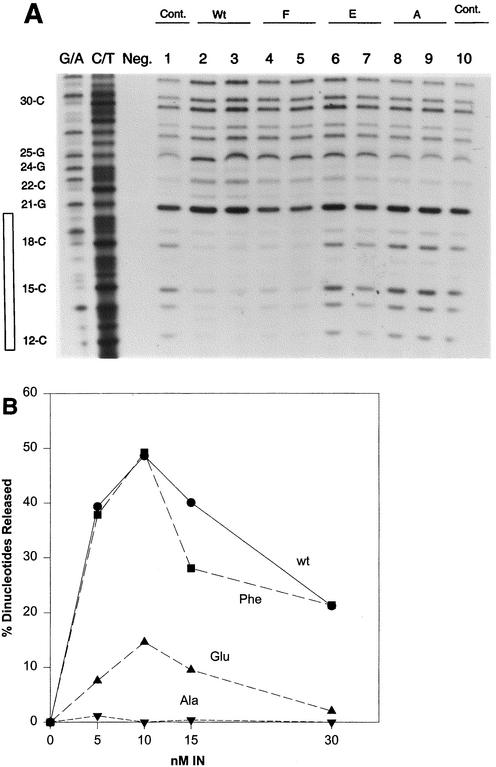
FIG. 6.
Heat inactivation of wt RSV and P115S IN in solution for 3′ OH processing activity. (A) Filled-in gain-of-function double end-labeled 4.5-kbp LTR donor was assembled with either wt or P115S at various IN concentrations under strand transfer conditions, except that the NaCl concentration was 300 mM. After assembly for 30 min on ice, the assay mixtures were placed at 37°C for 15 min. Acid-soluble counts were determined (y axis) as the percentage of dinucleotides released from the input DNA substrate. Negligible acid-soluble counts were observed with a filled-in non-LTR DNA substrate (data not shown). (B) wt and P115S IN were diluted to 2,000 nM in buffer and heated at 57°C for the indicated times. Aliquots were taken from the heated samples and allowed to assemble on the blunt-ended substrate for 30 min prior to 3′ OH processing for 15 min at 37°C. The final IN concentration in the assay mixtures was 20 nM. The percentage of dinucleotides released is indicated on the left.
We investigated whether the physical binding of wt and P115S IN to the att site was disrupted upon heating. A similar protocol for heat inactivation of IN was developed for our DNase I protection studies, except that the 3.6-kbp single-ended gain-of-function U3 donor was used as substrate (Fig. 2). DNase I protection studies using wt RSV IN showed that IN was able to protect the viral donor ends which mapped ∼20 bp from the viral DNA end with efficient and stable full-site integration activity (Fig. 7). In contrast, although P115S IN at 8 nM possessed initial efficient full-site integration activity as well as the ∼20-bp DNase I protection pattern, subsequent heating at 57°C simultaneously inactivated both the full-site integration activity and the DNase I protection (data not shown).
FIG. 7.
DNase I footprint protection analysis with purified RSV wt and W233F IN. Titration of wt and W233F IN (indicated above the lanes) for strand transfer activities using the gain-of-function single-ended U5 LTR donor under standard conditions was performed. The percentages of input donor incorporated into the full- and half-site products were determined with a phosphorimager (see text for data). Separate aliquots of each strand transfer reaction at different IN concentrations were subjected to DNase I protection analysis. Lanes 1 to 4, wt IN; lanes 7 to 10, W233F IN; lanes Neg., DNA without DNase I treatment; lanes G/A and G/T, G/A and G/T chemical markers. The concentrations in the lanes are as follows: lanes 1 to 5, wt IN at 10, 6, 4, 2, and 0 (Cont) nM, respectively; lanes 6 to 10, 0 (Cont), 2, 4, 6, and 10 nM W233F IN, respectively.
In summary, prior to heating, the specific activities of P115S IN for 3′ OH processing and half- and full-site strand transfer activities were similar to those of wt IN. Heating of P115S IN in solution at 57°C disrupts all of these above properties, suggesting that a common function is altered for these activities.
Is the C-terminal domain of IN important for protein-DNA interactions at the viral att site for full-site integration?
The C-terminal region of IN has been implicated to be essential for binding to nucleotides located 7 to 9 bp from the viral DNA ends (18, 19, 30) as well as to be important for multimerization (25, 30). Residue W233 in RSV IN is highly conserved among retroviruses, including W235 in the structurally similar C-terminal region (residues 220 to 270) in HIV-1 IN (8, 16, 29, 44). This conserved residue in HIV-1 IN has been suggested to be involved in multimerization of IN and/or protein-DNA interactions, as shown by MuA transposase-mediated PCR footprinting with purified HIV-1 PIC derived from cells whose infecting virions contained either wt or mutant W235E IN (9). We produced a series of recombinant RSV PrA IN mutants (W233F, W233E, and W233A) that paralleled the same series of HIV-1 W235 IN mutants studied both in vivo and in vitro (9, 16). Each RSV W233 mutant was investigated for full-site strand transfer, 3′ OH processing, and the physical association of IN at the viral att site by DNase footprinting (40).
The conservative structural mutation of W233F in RSV IN resulted in wt activities for all three functional assays. Titration of wt and W233F IN between 2 and 10 nM IN produced similar DNase protection patterns mapping ∼20 bp from the viral ends (Fig. 7). In simultaneously performed assays, full- and half-site integration activities for wt and W233F at 4 nM (Fig. 7, lanes 3 and 8) were ∼15 and ∼4%, respectively (data not shown). The assembled nucleoprotein complexes with either wt or W233F IN at 8 nM were stable for at least 240 min at 14°C, as shown by DNase protection analysis (data not shown) and integration activity (∼23 and ∼11% full- and half-site integration activities for each IN, respectively). Similar stabilities were observed with reconstituted AMV IN-viral DNA (3.6 kbp) complexes (40). Both RSV wt and W233F IN possess similar 3′ OH processing activities at 140 mM (Fig. 8B) or at 300 mM NaCl (data not shown). The in vitro data obtained with RSV W233F IN is consistent with the observation that the W235F mutation in HIV-1 IN does not affect virus replication (16).
FIG. 8.
DNase I footprint protection and 3′ OH processing analyses of wt, W233F, W233E, and W233A IN. (A) The assay conditions were as described for Fig. 7. IN bearing the amino acid indicated above pairs of lanes was examined at 5 and 10 nM (left and right lanes of each pair, respectively). Lanes G/A and C/T, G/A and G/T chemicals markers; lane Neg., DNA without DNase I treatment; lane 1, no IN (Cont.), lanes 2 and 3, wt IN; lanes 4 and 5, 6 and 7, 8 and 9, W233F, W233E, and W233A IN, respectively; lane 10 (Cont.), repeat of lane 1. (B) The same proteins examined for panel A were analyzed for 3′ OH processing activities as described for Fig. 6, except that the assay was at 140 mM NaCl without PEG present. The assays were performed at 37°C for 10 min. Shown are results for wt IN (wt), W233F IN (Phe), W233E IN (Glu), and W233A IN (Ala). The concentrations used with each IN are indicated at the bottom.
In contrast to the conservative amino acid substitution at W233, the substitution of either E or A into W233 produced significantly more defective proteins for strand transfer (Table 3), DNase protection (Fig. 8A, lanes 6 to 10), or 3′ OH processing (Fig. 8B). The 3′ OH processing activity assay (Fig. 8B) was performed at 140 mM NaCl (39) because W233E is inhibited ∼50% in the presence of 300 mM NaCl relative to 140 mM (data not shown). Decreasing the NaCl concentration to 140 mM in the strand transfer assay for W233E increased its activity by ∼30% compared to that at 330 mM NaCl, as shown in Table 3. However, essentially all of the products were half-site integration reactions (data not shown) at the lower salt concentrations, as shown previously for wt RSV or AMV IN dependency on higher NaCl concentrations for full-site integration (32, 39). Additional protein titration experiments with either W233E or W233A up to 25 nM IN did not result in any significant increases in strand transfer activities or specific DNase protection at the viral termini (Fig. 8) (data not shown).
TABLE 3.
Comparison of full and half-site integration activities for wt RSV IN and RSV IN with mutations at position W233a
| IN variant | Concn (nM) | Full-site activity | Half-site activity |
|---|---|---|---|
| wt | 5 | 16.7 | 2.7 |
| 10 | 11.0 | 5.4 | |
| W233F | 5 | 16.0 | 5.3 |
| 10 | 9.8 | 6.5 | |
| W233E | 5 | 1.9 | 0.2 |
| 10 | 1.8 | 0.2 | |
| W233A | 5 | 0 | 0 |
| 10 | 0 | 0 |
The values for activity represent the percentage of donor incorporated into target DNA for full and half-site integration products produced under standard reaction conditions. The DNase I footprints associated with these activities are shown in Fig. 8A.
The results suggest that the conservative aromatic substitution of F at W233 in RSV IN results in wt full-site integration activity and multimerization functions necessary for protection of ∼20 bp at the viral att site. Substitution of either E or A in position RSV W233 was detrimental for these functions, similar to what was observed with HIV-1 IN in the context of the PIC (9, 16), suggesting that this residue plays a critical role in protein-protein and protein-DNA interactions that occur at the viral att site.
DISCUSSION
The reconstitution of recombinant wt RSV IN-LTR donor complexes capable of efficient full-site integration activity has permitted us to begin examining the potential roles of individual amino acids and domains in full-site integration. The enzymatic activities of recombinant IN containing the A48P mutation possess wt IN activities even though the virus carrying this mutation was defective for integration in vivo (36). The P115S mutation in IN produced a protein that possesses thermolabile properties for 3′ OH processing and strand transfer activities consistent with the observation that this same mutation in RSV produces a temperature-sensitive defect for integration in vivo (37). The results of conservative and nonconservative mutations introduced into W233 located in the C-terminal region of IN suggest that this domain is important for protein-DNA interactions at the viral att site.
Surprisingly, recombinant A48P IN possesses wt IN activities (Fig. 3), because this same mutation, when introduced into the context of the RSV genome, produced an apparent severe defect at the integration level without affecting either the processing or levels of pol and viral structural proteins and the quantity of viral DNA synthesized (36). We will need to reinvestigate what other possible defects in the replication cycle are produced by the A48P mutation. As previously observed with other retrovirus systems (3, 16), it is clear that mutations introduced into IN must be examined at multiple levels to have a better understanding of how these mutations affect the replication cycle of the virus.
The thermolabile properties of RSV P115S IN are consistent with the temperature-sensitive replication properties of the virus containing this same mutation in IN (37). To our knowledge, this is the first thermolabile IN identified that specifically affected viral DNA integration at the nonpermissive temperature in vivo. Without heating, RSV P115S IN at 4 nM in the reaction mixture produced the maximum amount of full-site integration products (∼30% of donor incorporated into full-site products in 10 min) under our assay conditions (Fig. 5B, see inset). As shown previously (40), the production of half-site products in relationship to full-site products is significantly affected by the initial concentration of IN in the reaction mixtures (Fig. 3). Without heating, P115S IN between 6 nM and 12 nM was in excess for producing the maximum number of complexes capable of full-site integration events (∼30%) (Fig. 4 and 5). Upon heating, a stable phase of full-site integration activity was observed that was consistent with IN being in excess for this reaction. During the stable phase, the ratio of full-site products to half-site products produced also increased with time of heating, supporting the idea that the initial concentration of IN dictates what products are produced (Fig. 3), i.e., that at low protein concentrations (Fig. 5B, inset), full-site integration is favored over half-site integration (40). In the decay phase for full-site integration, the inactivation rate was similar to that observed for half-site integration (Table 1) as well as that for the 3′ OH processing reaction. Since the inactivation rates were similar, the number of subunits required per viral att site for enzymatic activities may also be similar. Assuming that a dimer is required for 3′ OH processing and half-site integration, the results would suggest that the synapsed complexes required for full-site integration may be a dimer of dimers, or a tetramer (13, 19, 42, 44). In addition, the results suggest that the defect associated with P115S IN serves a common function needed for all three enzymatic activities.
The minimal number of IN dimers per donor end to achieve the ∼30% donor incorporation into full-site products was calculated and was defined to be in the range between 5 and 9 (Table 2). This somewhat high number can be due to IN binding DNA nonspecifically; IN also has the capacity to form extended multimers on viral DNA substrates at higher protein concentrations (3, 40). The results suggest that it is not possible by this experimental approach to accurately measure the number of IN dimers per donor end that is necessary and sufficient for full-site integration.
The results show that prior to heating, the mutation in RSV P115S IN does not affect the ability of IN to bind onto the att site for either 3′ OH processing or strand transfer, nor does it interfere with the ability of IN to interact with the target substrate for strand transfer or producing a DNase I protection pattern at the viral att site. The RSV P115S IN mutation maps near a region on HIV-1 IN (residue S119 in α-helix 2) (Fig. 1, bottom) that was shown to be involved in target site selection (1, 21, 27). The results also suggest that without heating, the substitution of S at P115 of RSV IN does not disrupt near secondary structural features, including D121 within the active site (Fig. 1). In contrast, the P109S mutation in HIV-1 IN (Fig. 1) renders the recombinant protein inactive in vitro, and the same mutation in the context of the virus produces an integration-defective class I mutant in vivo (15, 16, 38). The structural defect(s) that are produced upon heating of RSV P115S IN in solution, causing loss of enzymatic activities, are unknown.
The C-terminal domain of IN appears to have multiple functions at the in vitro levels for various enzymatic activities, including binding to the viral DNA at ∼7 to 9 bp from the viral ends (18, 19, 26), involvement with the core domain to produce tetramers in solution (25), and promotion of oligomerization of IN (30). Structure-based sequence alignment of the C-terminal domains of RSV and HIV-1 IN confirmed their similarities (44). Mutational analysis of highly conserved residue W235 to F in HIV-1 IN permitted wt virus replication (16). W235E prevented protection of the viral DNA ends by IN in isolated PIC, and these PIC were not capable of full-site integration activity (class I mutant) (9, 16). W235A prevented virus replication (16). Mutagenesis of recombinant RSV IN at the analogous W233 produced somewhat parallel results. Conversion of W233 to the conservative F residue with RSV IN did not alter either its ability to produce the ∼20-bp DNase I footprint (Fig. 7 and 8A, lanes 2 to 5) or its 3′ OH processing activity (Fig. 8B). Interestingly, the W233E mutation abolished the ability of IN to produce the ∼20-bp DNase I footprint (Fig. 8A, lanes 6 and 7), inhibited its ability to promote the 3′ OH processing reaction (Fig. 8B), and even more severely inhibited its strand transfer activities (Table 3). The W233A mutation in RSV IN rendered it inactive (Fig. 8) (Table 3). In summary, the results suggest that W233 in RSV IN plays a critical role in functions necessary for multimerization of IN, as demonstrated by the DNase I footprint analysis and strand transfer data, but is not necessarily as critical for 3′ OH processing. Besides the potential multimerization effect, the W233E mutation in RSV IN may have also affected its ability to bind to the 3′ OH recessed ends in a productive manner, as suggested by results obtained with HIV-1 PIC containing the same parallel mutation in IN (9). Multiple effects are surely possible, because the C-terminal domain has both cis and trans capabilities for strand transfer (13). With RSV IN, modification of S262 to P in the C-terminal domain prevents virus replication, while other mutations at S262 and S282 were nonlethal (Fig. 1) (24). We have not yet examined the effect of the above mutations in W233 on the replication of RSV.
Various complementation experiments using recombinant HIV-1 and murine leukemia virus IN demonstrated that IN functions as a multimer (17, 19, 22, 45). A study of other mutations introduced into various domains of recombinant RSV IN suggested that there are notable differences in 3′ OH processing and half-site integration activities between RSV IN mutations (34) and their HIV-1 IN analogs, as shown by other labs (16, 35). In this report, the recombinant RSV P115S IN expressed wt activities prior to heating, while the corresponding mutation in recombinant HIV-1 IN (P109S) (Fig. 1) rendered it inactive (15, 38). The fact that mutations introduced into monomers of recombinant IN, as well as the assembly of active IN, require a dimer structure for half-site or possibly higher-order multimers for full-site integration complicates simple interpretations of introduced mutations. The capabilities of some recombinant retrovirus IN monomers to assemble active dimers in solution may be either nonexistent or inefficient, possibly preventing proper protein-protein interactions. Therefore, even though this present report suggests that a better understanding of full-site integration can be achieved by using recombinant IN, caution must be extended as to how these mutations may be simply affecting protein-protein interactions in solution prior to the pivotal interactions of IN at the LTR att sites.
Acknowledgments
This work was supported by a National Cancer Institute grant (CA16312).
We thank M. Sudduth and V. P. Kurenok for their guidance with the mathematical models and S. Sinha and A. C. Vora for their discussions and reading of the manuscript.
REFERENCES
- 1.Appa, R. S., C.-G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and α helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 276:45848-45855. [DOI] [PubMed] [Google Scholar]
- 2.Asante-Appiah, E., and A. M. Skalka. 1997. A metal-induced conformational change and activation of HIV-1 integrase. J. Biol. Chem. 272:16196-16205. [DOI] [PubMed] [Google Scholar]
- 3.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. J. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Brown, H. E., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the HIV-1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujacz, G. M., J. Jaskolski, A. Alexandratos, A. Woldawer, G. Merkel, R. A. Katz, and A. M. Skalka. 1995. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 253:333-346. [DOI] [PubMed] [Google Scholar]
- 6.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. C.-H., J. Krucinski, L. J. W. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., S. Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intrasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z. G., Y. Yan, S. Munshi, Y. Li, J. Zugay-Murphy, B. Xu, M. Witmer, P. Folock, A. Wolfe, V. Sardana, E. A. Emini, D. Hazuda, and L. C. Kuo. 2000. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-239)-an initial glance of the viral DNA binding platform. J. Mol. Biol. 296:521-533. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, R., and D. P. Grandgenett. 2000. Avian retrovirus DNA internal attachment site requirements for full-site integration in vitro. J. Virol. 74:8292-8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman, J., S. Eaton, G. Merkel, A. M. Skalka, T. Lane. 1997. Characterization of the self association of avian sarcoma virus integrase by analytical ultracentrifugation. J. Biol. Chem. 274:32842-32846. [DOI] [PubMed] [Google Scholar]
- 13.Craigie, R. 2001. Retroviral DNA integration, p. 613-630. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 14.Deprez, E., P. Tauc, H. Leh, J. F. Mouscadet, C. Auclair, M. E. Hawkins, and J. C. Brochon. 2001. DNA binding induces dissociation of the multimeric form of HIV-1 integrase: a time-resolved fluorescence anisotropy study. Proc. Natl. Acad. Sci. USA 98:10090-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drelich, M., M. Haenggi, and J. Mous. 1993. Conserved residues Pro-109 and Asp-116 are required for interaction of the human immunodeficiency virus type 1 integrase protein with its viral DNA substrate. J. Virol. 67:5041-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engleman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-426. [DOI] [PubMed] [Google Scholar]
- 17.Engleman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interactions. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, K., S, C. Butler, and F. D. Bushman. 2001. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 20:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandgenett, D. P., A. C. Vora, and R. Schiff. 1978. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology 89:119-132. [DOI] [PubMed] [Google Scholar]
- 21.Harper, A. L., L. N. Skinner, M. Sudol, and M. Katzman. 2001. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J. Virol. 75:7756-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuer, T., and P. O. Brown. 1998. Photo-crosslinking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37:6667-6678. [DOI] [PubMed] [Google Scholar]
- 23.Hippenmeyer, P. J., and D. P. Grandgenett. 1985. Mutants of the Rous sarcoma virus reverse transcriptase gene are non-defective in early replication events. J. Biol. Chem. 260:8250-8256. [PubMed] [Google Scholar]
- 24.Horton, R., S. R. Mumm, and D. P. Grandgenett. 1991. Phosphorylation of the avian retrovirus integration protein and proteolytic processing at its carboxyl terminus. J. Virol. 65:1141-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katzman, M., and M. Sudol. 1995. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 69:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. Mack, and A. M. Skalka. 1991. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, T. H., T. P. Quinn, D. P. Grandgenett, and M. C. Walsh. 1989. Secondary structural analysis of retrovirus integrase: characterization by circular dichroism and empirical prediction methods. Proteins 5:156-165. [DOI] [PubMed] [Google Scholar]
- 30.Lutzke, R. A., and R. H. Plasterk. 1998. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 72:4841-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCord, M., R. Chiu, A. C. Vora, and D. P. Grandgenett. 1999. Retrovirus DNA termini bound by integrase communicate in trans for full-site integration in vitro. Virology 259:392-401. [DOI] [PubMed] [Google Scholar]
- 32.McCord, M., J. Stahl, T. C. Mueser, C. C. Hyde, A. C. Vora, and D. P. Grandgenett. 1998. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Expr. Purif. 14:167-177. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreau, K., C. Faure, G. Werdier, and C. Ronfort. 2002. Analysis of conserved and non-conserved amino acids critical for ALSV (avian leukemia and sarcoma viruses) integrase functions in vitro. Arch. Virol. 147:1761-1778. [DOI] [PubMed] [Google Scholar]
- 35.Neamati, N., C. Marchand, and Y. Pommier. 2000. HIV-1 integrase inhibitors: present, past, and future. Adv. Pharmacol. 49:147-165. [DOI] [PubMed] [Google Scholar]
- 36.Quinn, T. P., and D. P. Grandgenett. 1988. Genetic evidence that the avian retrovirus DNA endonuclease domain of pol is necessary for viral integration. J. Virol. 62:2307-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinn, T. P., and D. P. Grandgenett. 1989. Avian retrovirus integration protein: structural-functional analysis of viable mutants. Virology 173:478-488. [DOI] [PubMed] [Google Scholar]
- 38.Taddeo, B., F. Carlini, Veranin, P., and A. Engelman. 1996. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J. Virol. 70:8277-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vora, A. C., and D. P. Grandgenett. 1995. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J. Virol. 69:7483-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vora, A., and D. P. Grandgenett. 2001. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J. Virol. 75:3556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vora, A. C., M. McCord, G. Goodarzi, S. J. Stahl, T. C. Mueser, C. C. Hyde, and D. P. Grandgenett. 1997. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J. Biol. Chem. 272:23938-23945. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J.-Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, S. Q., K. Mizuuchi, and R. Craigie. 1998. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Pro. Natl. Acad. Sci. USA 95:10535-10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Z.-N., T. C. Mueser, F. D. Bushman, and C. C. Hyde. 2000. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 296:535-548. [DOI] [PubMed] [Google Scholar]
- 45.Yang, F., and M. J. Roth. 2001. Assembly and catalysis of concerted two-end integration events by Moloney murine leukemia virus integrase. J. Virol. 75:9561-9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, H., G. J. Rainey, S. K. Wong, and J. M. Coffin. 2001. Substrate sequence selection by retroviral integrase. J. Virol. 75:1359-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]