Abstract
Revealing the cellular and molecular changes associated with cancer, as they occur in intact living animal models of human neoplastic disease, holds tremendous potential for understanding disease mechanisms and elucidating effective therapies. Since light is transmitted through mammalian tissues, at a low level, optical signatures conferred on tumor cells by expression of reporter genes encoding bioluminescent and fluorescent proteins can be detected externally using sensitive photon detection systems. Expression of reporter genes, such as the bioluminescent enzyme firefly luciferase (Luc) or variants of green fluorescent protein (GFP) in transformed cells, can effectively be used to reveal molecular and cellular features of neoplasia in vivo. Tumor cell growth and regression in response to various therapies have been evaluated non-invasively in living experimental animals using these reporter genes. Detection of Luc-labeled cells in vivo was extremely sensitive with signals over background from as few as 1000 human tumor cells distributed throughout the peritoneal cavity of a mouse with linear relationships between cell number and signal intensity over five logs. GFP offers the strength of high-resolution ex vivo analyses following in vivo localization of the tumor. The dynamic range of Luc detection allows the full disease course to be monitored since disease progression from small numbers of cells to extensive disease can be assessed. As such, therapies that target minimal disease as well as those designed for late stage disease can be readily evaluated in animal models. Real time spatiotemporal analyses of tumor cell growth can reveal the dynamics of neoplastic disease, and facilitate rapid optimization of effective treatment regimens. Thus, these methods improve the predictability of animal models of human disease as study groups can be followed over time, and can accelerate the development of therapeutic strategies.
Keywords: luciferase, photon counting, non-invasive, imaging, in vivo, CCD
Introduction
Development of neoplastic disease is a stepwise process involving loss of genetic regulation, changes in cellular physiology, and failure of effective immune surveillance [1–9]. The progression of neoplastic states to disseminated disease requires interactions between the transformed cells and normal tissues as occurs in invasion, extravasation, migration and neovascularization [10–12]. Although individual steps in these processes may be modeled in correlative cell culture assays, evaluation of the dynamic and interactive processes of the complete disease requires the context of living animals. However, animal models have typically served as “black boxes” where well-defined signals can be applied, but the ultimate evaluation is performed outside of the animal via ex vivo assays. Sophistication of these ex vivo assays has increased dramatically with the development of confocal and two-photon microscopy for the interrogation of thick tissues, reporter proteins with diverse optical signatures, multiparameter flow cytometry and DNA microarrays [13–20].
Tremendous sensitivity can be achieved using amplification methods such as polymerase chain reaction (PCR) to detect tumor cell DNA or mRNA levels, but these assays are time-consuming and severely limited by the size of the sample that can be reasonably evaluated. Thus, the fraction of a given tissue that can be analyzed is limited [21,22]. Studies employing these assays may, therefore, be subject to sampling biases such that results may not represent overall expression levels in the target tissue or organ. In the absence of a signal that can be detected in the intact animal, or even the intact organ, targeting specific tissues for analyses is also difficult, and all tissues cannot be evaluated. Thus, only a small fraction of a few selected tissues is studied when ex vivo assays are employed. To achieve statistical significance with temporal studies, large numbers of animals are used with groups being sacrificed at each time point. Thus, analyses using ex vivo assays are both limited and require large numbers of animals.
The limitations of the ex vivo assays indicate that accessible, versatile and sensitive assays that can rapidly reveal cellular and molecular changes as they occur in intact animal models of human neoplasia are necessary to advance our understanding of disease processes and accelerate the development of effective intervention strategies. In particular, refinement of animal models to include markers for imaging the multiple stages of cancer development in vivo would complement current ex vivo assays and would dramatically accelerate and enrich the analyses of animal models of human neoplastic disease by revealing cellular and molecular changes in vivo and directing the ex vivo assays to critical target tissues.
A variety of imaging strategies designed to reveal the physiologic changes associated with neoplasia and response to therapy have been described. The modalities utilized in these approaches include fluorescence imaging, magnetic resonance imaging (MRI), single photon emission computed tomography (SPECT), and positron emission tomography (PET) [23–34]. Contrast agents that enhance MRI are being developed and molecular reporter genes for MRI have enabled cellular and molecular analyses [35,36]. Similarly, fluorescent dyes that are concentrated at the tumor site or that can be activated by the target cell have been described and utilized to localize tumor cells in animal models [33,37,38]. PET is, by design, a method of revealing metabolic changes and can be used to localize cells and molecules labeled with radioactive tracers. However, many of these imaging strategies are not well-suited for small animals and can be encumbered by long scan times and expensive instrumentation for detection and support. In addition, some of these methods can be difficult to quantify and lack sensitivity.
Sensitive in vitro molecular assays have been developed using light-emitting enzymes, luciferases, as reporters [39–42]. Luciferases comprise a family of photoproteins that can be isolated from a wide variety of species including bacteria and a large number of eukaryotic organisms [43]. Luciferase from the firefly, Photinus pyralis, is the most commonly used photoprotein in molecular biology studies and has been used to evaluate gene expression in transformed cell lines in culture [44,45], and to monitor tumor growth and response of tumors to antineoplastic therapy in animal models of human disease [21,46]. This reporter gene is not only the most widely used member of the photoprotein family of enzymes; it is also the most-studied and has been modified such that it is well suited for studies of neoplastic disease. These modifications include mutations for optimal mammalian codon usage and increased expression due to removal of a peroxisome targeting site [41,42]. These features of the firefly enzyme make it an ideal choice for in vivo monitoring of tumor cell growth. Another luciferase that has been used as a reporter in mammalian cells includes that from the sea pansy (Renilla reniformis). The wavelength of emission from this enzyme (blue, with a peak at 460 nm) and its use of a substrate other than luciferin have led to its use in dual-reporter assays [47,48]. Many luciferases from bioluminescent marine organisms, including that of R. reniformis, use the high-energy compound, coelenterazine, as a substrate. Analyses of these photoproteins as indicators of gene expression have, in the past, required removal of the tissues and assessment of enzymatic activity in cell lysates. This type of assay has many of the same limitations as other ex vivo assays; however, the fact that light can pass through mammalian tissues and the absence of bioluminescence from mammalian cells, suggested that it may be possible to use these reporter genes for the in vivo analysis of neoplasia.
It has previously been demonstrated that internal bioluminescent signals from bacteria and cells of transgenic mice can be externally detected in vivo, even when located at deep tissue sites [49,50]. Using this technology, it has been possible to non-invasively monitor infection, gene expression, and passive transfer of mammalian cells [51]. This approach has recently been applied to the study of tumor progression and response to therapy in living animal models [52,53]. The reporter genes used in these studies included the modified firefly luciferase for eukaryotic gene expression and a bacterial luciferase from the soil bacterium, Photorhabdus luminescens, for prokaryotic expression. Tagging biological processes with reporter genes that are propagated along with the labeled cells permit monitoring cell growth, and transcriptional events without the problem of dilution or loss of signal with cell division. The ability to monitor labeled processes from an external vantage point provides a tremendously powerful tool.
The use of external detection of an internal bioluminescent signal differs from other optical imaging strategies and offers some distinct advantages. Optical imaging typically involves the use of external light sources to either interrogate the inherent optical properties of tumor tissue, or to assess the concentration of exogenous dyes that accumulate or are activated at tumor sites [33,37,38,54]. In contrast, photons originating from photoproteins in labeled cells can serve as internal biological sources of light that transmit through mammalian tissues to reveal spatial and temporal information in the near absence of background bioluminescence [51]. The weak bioluminescent signal emitted from the labeled cells and transmitted through the animal tissue can be detected and quantified using low light imaging systems such as intensified and ultra-cooled charge-coupled device (CCD) cameras [50]. Bioluminescent reporters may offer greater versatility than fluorescent or other types of markers in mammalian tissues due to the nearly complete absence of spontaneous emission of light from mammalian cells. The use of outside light sources for fluorescence markers can result in tissue autofluorescence, creating background that is greater than the signal. As reporter genes are integrated into the chromosomes of the tumor cells, they are replicated with cell division, which is a distinct advantage over other labeling techniques using dyes that can be diluted out as the cells divide. Published research has demonstrated that real time, non-invasive analyses of pathogenic events, pharmacological monitoring and assessment of promoter activity can be performed in vivo [49–51]. These studies indicate that reporter genes can provide a window through which biological processes can be viewed in living animals, and thus may be useful in illumination of the temporal and spatial distribution of tumor growth and metastasis in vivo.
Detection of bioluminescence from beetle luciferases expressed in mammalian cells either in culture or in vivo requires exogenous delivery of the substrate for the enzymatic reaction. It has been determined that the substrate for the firefly luciferase, luciferin (d-(-)-2-(6′-hydroxy-2′-benzothiazolyl)thiazone-4-carboxylic acid), can be added to cell culture medium such that expression in living cells can be monitored [50]. This contrasts what has been observed for coelenterazine, the substrate used by luciferases from marine eukaryotes (e.g., R. luciferase from R. reniformis), which appears to have significant autoluminescence in the presence of serum proteins (Zhang and Contag, unpublished results). The ability of luciferin to enter into cells is likely due to its small size (280.33 g/mol) and its zwitter ionic nature. Typical studies utilizing luciferase genes as transcriptional reporters require cell lysis with enzymatic activity being analyzed in a luminometer with the attendant loss of temporal information. For detection in living animals, the substrate, luciferin, can be supplied via intraperitoneal injection and luciferase activity can be evaluated at many different tissue sites simultaneously [50], including the central nervous system and fetal tissues in utero (C.H.C., unpublished results). The small size of luciferin also makes it a poor antigen and immune responses to luciferin are unlikely; this will be useful as the models move from xenografts in immunodeficient mice to syngeneic and autologous cell transfers.
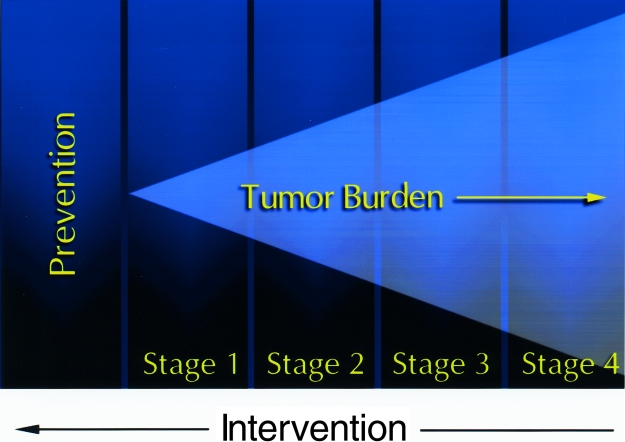
Many of the assays for tumor cell growth and regression in animal models of neoplastic disease rely on changes in the volume of large superficial tumors where changes in three orthogonal diameters can be determined using calipers. These tumor models resemble late stages in human disease, and thus, therapies developed using these models are most applicable to late stage disease. In contrast, therapies that target minimal residual disease either after removal of the tumor, via surgery or therapy, or early in the disease course, cannot readily be developed using these conventional assays. Real time accurate assays that employ reporter genes for rapid detection of minimal disease in animal models will change the paradigm of drug development, and therapies that effectively treat small numbers of transformed cells will be developed (Figure 1).
Figure 1.
New developments in cancer therapy. With the advancement of diagnostics and therapy, there is a need to develop new therapies that target small numbers of tumor cells to prevent both initiation of disease and relapse after treatment. Therefore, development of animal models that represent minimal disease is necessary to evaluate new therapeutic strategies that target these conditions.
Luciferase as a Reporter for Tumor Cell Growth In vivo
The bioluminescent signal from human tumor cells expressing luciferase can be used to follow their growth in mouse xenograft models of human disease. This has been demonstrated using several human tumor lines. For example, growth of a highly metastatic human prostate cancer cell line (PC-3M) [55] has been followed over time in severe combined immunodeficient (SCID) mice (Figure 2). The PC-3M cells were transfected with the SV40-driven modified luciferase gene and stable clonal transfectants were selected by light emission and antibiotic resistance to generate the line designated PC-3M-luc. The luciferase in this construct was optimized for expression in mammalian cells removed [41,42]. Populations of cells arising from single cell clones were generated. Selected clones were used to initiate tumors at subcutaneous sites in the hind flanks of SCID mice, and growth of these cells in vivo was followed over time using an intensified CCD camera to generate whole body images (Figure 2). Light emission was detectable within minutes of injecting the tumor cells, and signals continued to increase logarithmically over a 2-week time period. These data indicate that the bioluminescent light from the transformed cells was detectable externally and that tumor growth can be followed non-invasively. Studies are underway to correlate tumor size with photon counts in this prostate cancer model and to assess the minimal detectable cell number in vivo.
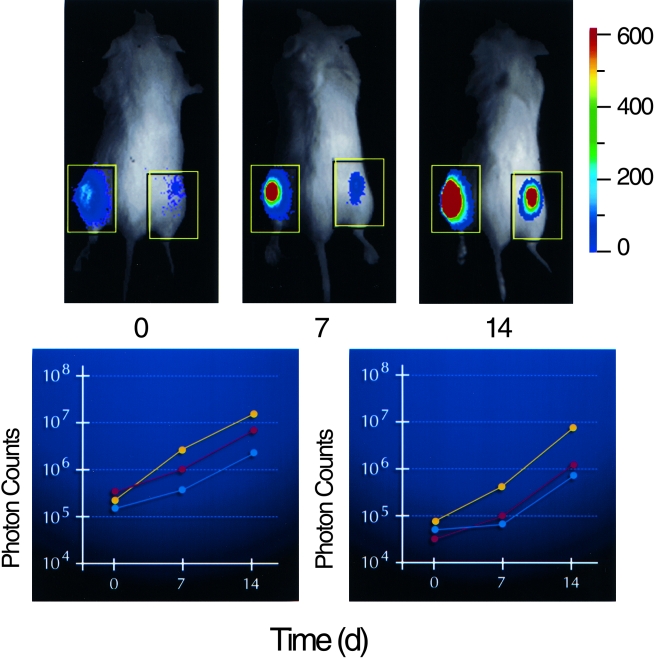
Figure 2.
Monitoring tumor growth at subcutaneous sites. PC-3M cells labeled with constitutive expression of a modified luciferase gene (PC-3M-luc) were injected at subcutaneous sites on each hind flank of three animals and growth of the cells was monitored by photon emission over a 14-day time course. A pseudocolor image representing light intensity is superimposed over a grayscale reference image of representative mice from each group of three (upper). Time (day) is indicated below each image. The color bar indicates average signal intensity per pixel represented by a color scheme used in the pseudocolor image. Total signal intensity over the tumor sites (boxes) was determined and plotted with respect to time for each group of three (lower). Inoculum was 1x106 cells in the left flank, n=3 (left); and 1x105 cells in the right flank, n=3 (right).
A human cervical carcinoma cell line (HeLa) has also been used in similar studies, which have indicated that tumor growth and inhibition in response to several different therapeutic agents could be monitored in living mice (Figure 3) [52,53]. Similar to the PC-3M-luc cells, a stable reporter cell line of HeLa cells (HeLa-luc) was generated through selection of an integrated reporter construct consisting of a portion of the viral promoter from SV-40 driving expression of the modified luciferase gene. HeLa-luc cells were introduced into animals via subcutaneous, intraperitoneal and intravenous inoculation, and the kinetics of cellular proliferation of labeled cells in irradiated SCID mice was non-invasively assessed [53]. The signals from luciferase-labeled cells in these 5-minute images were used to rapidly localize tumors in anesthetized animals and to quantify tumor cell proliferation. In this manner, as few as 1000 cells could be quantitatively detected following injection into immunodeficient mice [52,53]. The dynamic range of this assay permitted the study of disease progression from early (non-palpable tumors) to late stages, indicating that this reporter gene system could be used to evaluate therapies directed at various stages in the disease course including minimal disease states.
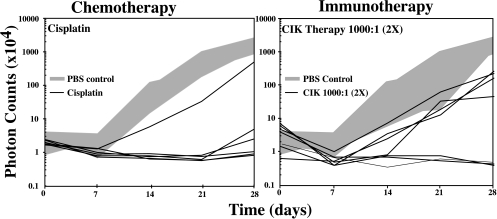
Figure 3.
Effects of chemotherapy and immunotherapy on HeLa-luc cell growth in vivo. HeLa-luc cells were injected into the peritoneal cavity (time 0) at 1x104 cells per animal. Groups of animals were either not treated (n=7), or treated with cis-platinum (n=6) or CIK cells at an effector to target ratio of 1000:1 (n=7). Tumor cell growth, as indicated by transmission of bioluminescent light, was assessed at weekly intervals and plotted for each animal with respect to time. The range of tumor cell growth in the untreated control animals is represented by the gray area.
Detection of Minimal Disease and Evaluation of Therapy in Animal Models
Advancements in imaging that increase the sensitivity of tumor cell detection will have a dramatic effect on the study of minimal disease models where few cells exist early in diagnosis, or remain after therapeutic intervention. Effective intervention strategies that target the small numbers of neoplastic cells that persist after therapy remain major treatment challenges [56,57]. The observation, that while a majority of cancer patients enter a complete remission following chemotherapy or autologous transplantation, a significant proportion go on to relapse, underscores the importance of controlling minimal residual disease (MRD). The limited ability to detect neoplastic cells at these stages and to predict response to therapy in the absence of reliable assays has hampered progress.
Advancements in the treatment of MRD have been limited by a lack of adequate animal models where small numbers of tumor cells can be reliably and quantitatively detected. In some situations, the ability of the tumor cells to grow in SCID mice has been a powerful predictor of the clinical course of the patient [58]. However, detection of neoplastic cells in current animal models of human disease requires a large number of target cells and such models typically use the therapeutic endpoints of gross tumor growth or death of the animal. Molecular techniques, such as DNA amplification using PCR, have been used, but are hampered by sampling limitations, and the need to sacrifice the animal subjects [21,22]. Several other surrogate markers of tumor growth that can be detected in serum and do not require sacrifice of the animal have been used (e.g., human lactate dehydrogenase (LDH) isoenzymes or human nuclear matrix protein 41/7 (NMP 41/7) in sera of SCID mice) [59]. Even though human-derived serologic markers in xenograft models may provide predictable indicators of tumor cell growth, these are indirect assays of tumor cell growth, are only detectable at a week to several weeks after introduction of the tumor cells, and require blood sampling and processing. Thus, further advancements in non-invasive detection and quantification of tumor burden in xenograft models are desirable.
Employing a luciferase-based methodology, conventional chemotherapies and a novel immune cell therapy, utilizing cytokine induced killer (CIK) T cells, have been evaluated in minimal disease xenograft models (Figure 4). The chemotherapeutic agents included cyclophosphamide, 5-fluorouracil, and cis-platinum. These three agents had different effects on the growth of HeLa-luc cells in vivo, with cyclophosphamide being the least effective and cis-platinum the most effective at reducing the signals in tumor-bearing animals [52]. Following treatment responses to therapy could be evaluated in as few as 24 to 72 hours. Long-term evaluation of the animals was also readily accomplished [52].
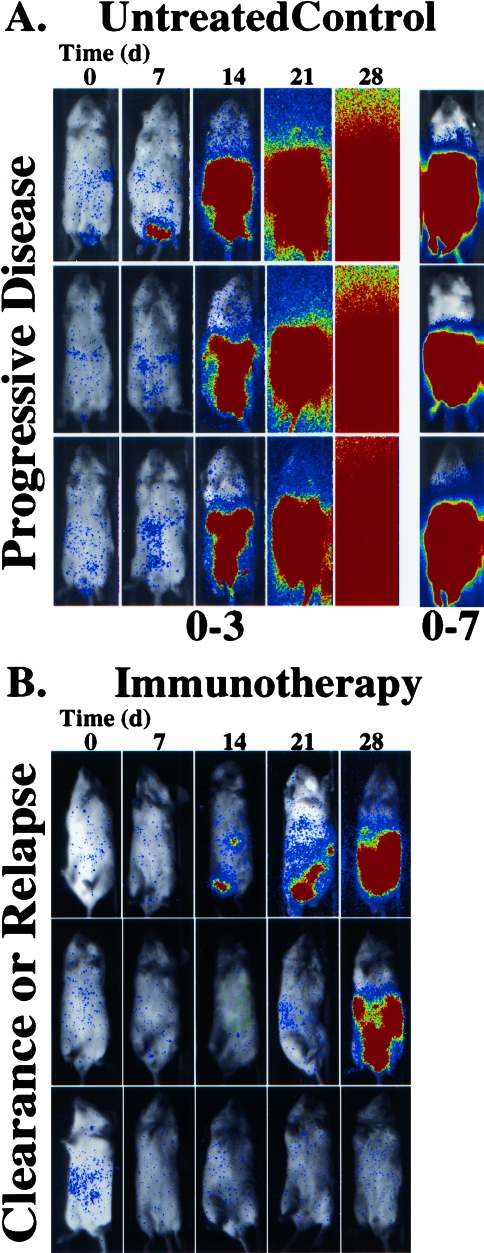
Figure 4.
Effects of immunotherapy on HeLa-luc cell growth in vivo. SCID mice bearing human xenografts, HeLa-luc cells, were either untreated (A) or treated with CIK cells at an effector to target ratio of 1000:1 (B). The growth of the tumor was followed over a 28-day time course. The pseudocolor images represent light intensity collected with a 5-minute integration time, and are all presented at the same display range (0–3 bits) with the exception of the 28-day time point for the untreated controls which is also shown at the 0–7 bit range to reveal spatial information.
The immune cytotherapy arms of this study utilized CIK cells. These cells were generated by culturing peripheral blood mononuclear cells in the presence of IFN-γ, followed 1 day later by IL-2 and an anti-CD3 monoclonal antibody [60–62]. This incubation resulted in the dramatic expansion of effector cells which have cytolytic activity against a broad array of tumor cell targets including multidrug resistant cell lines and autologous fresh tumor isolates [63–65]. The cell type with the most robust anti-tumor activity expresses the surface marker CD3, a general T cell marker, and the natural killer (NK) cell marker, CD56. Initially, an effector to target ratio of 100:1 was used; by 2 weeks, only a modest effect was noted — thus, second and third treatment arms were tested using ratios of 1000:1. In these treatment arms, the signals were reduced to background in many of the animals indicating effective treatment. These studies demonstrated that the non-invasive assays could be used for the rapid development of effective doses and schedules of therapy [52]. The evaluation of novel additional approaches that may enhance the effects of CIK therapy, such as with the use of bi-specific antibodies which crosslink the effector cells with tumor markers, will be accelerated through the use of the luciferase imaging approach.
In these studies, the number of animals required to reach statistically meaningful endpoints was five to seven per treatment group. Differences among the different arms of the study were apparent at 7 days post-treatment with statistically significant results being obtained at 14 days. In conventional tumor models, based upon animal survival, experiments are conducted over several months. Therefore, using this quantitative in vivo assay where a single group of animals can be followed over time and differences between treatment groups can be evident in the first week, a greater number of different treatment strategies can be evaluated.
Luciferase Reporters in Metastatic Models
Detection of metastatic tumor growth in animals has been largely dependent on ex vivo analysis of excised tissues. Histological analyses have been traditionally used to localize and document neoplastic and metastatic growth. These assays, however, suffer from the limitation of sampling selection and extensive labor involvement. GFP expression in tumor cell lines has facilitated directed analyses of tissue samples in these models and represents first steps toward linking the in vivo study of metastasis to tissue culture correlates [66,67]. In these studies, fluorescing metastatic colonies of cells could be detected on murine lung tissue via the GFP tag, and selected tissue samples could then be excised and cultured using a matrix-supported histoculture method to model in vivo tumor colonization. Throughout the studies, GFP expression permitted constant measurement and a means of detection that facilitated manipulation of both the in vivo model and the culture system. This approach demonstrated the strength of using an optical signature to track cells and monitor their growth both in vivo and in culture. Extending this approach to monitoring disease progression in three dimensions in living animal models is the goal of many imaging strategies.
As a step in the direction of real time, three-dimensional tumor cell imaging, luciferase-based methods offer the opportunity of whole body imaging and scanning for tumor growth with fairly short integration times (5 minutes per image). Furthermore, the ability to detect small numbers of luciferase-positive tumor cells enhances the potential to analyze experimental or spontaneous metastatic growth in vivo. Following intravenous injection of HeLa-luc cells, Edinger et al. [53] were able to detect microcolonies at a variety of tissue sites non-invasively. These microcolonies were detectable at 7 days after introduction of the tumor cells and appeared to be located in bone, lung and other tissues. The sensitivity of this assay and the ability to detect microcolonies using a whole body analysis indicated that this approach would be particularly useful in the detection of metastases at sites that are difficult to assay, such as bone or brain. The short time required for the imaging would allow quick screening of large numbers of animals and may permit imaging from multiple angles.
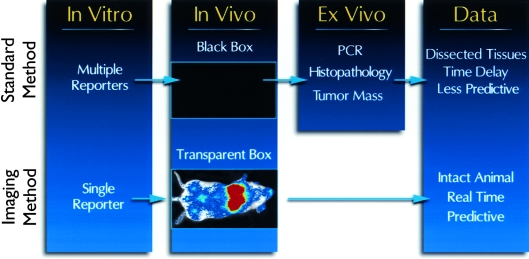
Since reporter gene expression can be monitored in living cells in culture, in animals and in cell lysates derived from tumor tissue, each step in the experimental design can be monitored with a single reporter gene. Examples of this are the studies by Chishima et al. [66] and Edinger et al. [53] and where either GFP or luciferase, respectively, was used both in culture and in animal models. Use of a single reporter gene at multiple steps in an experiment results in a more streamlined approach, and the added advantages of being quantitative and rapid make the use of these methods more efficient and informative than conventional studies. This is especially relevant to the study of metastasis where localizing microcolonies in the whole animal is tantamount to looking for a needle in a haystack. Since the in vivo data can be used to direct ex vivo analyses (i.e., localization of metastatic lesions for subsequent analyses), the labor-intensive assays such as PCR, expression profiling via DNA microarrays and histopathological assays can be performed on targeted tissues as directed by the in vivo analysis. This refinement to the animal models for neoplastic disease will result in more predictive models and improved clinical studies (Figure 5).
Figure 5.
Refinement of animal models using photoprotein reporters. Standard methods of monitoring tumor cell growth utilize multiple reporters and indicators to reveal the effects of experimental therapies on tumor cell growth in correlative cell culture assays. These assays often cannot be applied to in vivo analyses and assessing tumor growth in vivo is limited to measuring tumor size at superficial sites. Thus, ex vivo assays such as PCR, histological examination and weighing tumors have been necessary to assess tumor growth. These ex vivo assays often require sacrifice of the animals at multiple time points. Imaging tumor cell growth in culture and in vivo can be performed using a single reporter gene and signals for this reporter gene can be used to direct the ex vivo assays such that times and tissues can be targeted for analyses. This approach results in animal models and preclinical data that can be more predictive of human disease states.
Green Fluorescent Protein (GFP) as an Indicator of Tumorigenesis
A wide variety of tumor cells have been labeled with GFP including human xenografts and syngeneic transfers. The gene encoding GFP from the jellyfish, Aequeora victoria, has been mutated for optimal mammalian expression and alteration of its excitation and emission wavelengths. A family of genes encoding GFP variants that excite and emit at wavelengths other than the green wildtype excitation and emission of 397 and 509 nm offer versatility for gene expression studies [68–74]. Some of these variants have been used to tag tumor cells and monitor growth and metastasis in animal models [66,67]. The methods used to visualize the fluorescent proteins in living tissues include transillumination of the living animal [75] or intravital microscopy [76,77]. Transillumination of intact living animals produced detectable signals from tumors labeled with GFP, and metastases were visualized in the lung, liver, bones, and brain [75,78,79]. The detection limits were improved by utilizing nude mice or removing fur to reduce the light scatter. Maximum sensitivity was obtained following sacrifice of the animal and removal of the skin and other tissues. Following laparotomy, it was possible to obtain single-cell level resolution [79]. These steps enhanced the ability to detect tumors in mice; however, this approach eliminates temporal analyses and is not easily applied to larger-scale studies.
Orthotopic transplantation of cancer cells that express GFP has been used to visualize metastasis and angiogenesis [66,67,79–81]. Using intravital microscopy, it is also possible to detect single cells labeled with GFP and monitor their migration through blood vessels [77]. While this method allows for temporal analyses, it requires removal of the tissue over the tumor in order to make direct observations of the labeled cells. The excised tissue is replaced with a transparent window to permit repeated analyses. Studies using this technique have revealed migration patterns for immune cells and cancer cells at the tumor site. Studies utilizing intravital microscopy have also revealed expression patterns using the promoter for vascular endothelial growth factor (VEGF) [77].
The utility of fluorescent reporter genes in these types of analyses will be greatly enhanced by the introduction of red fluorescent proteins since the longer wavelength of red light transmits tissues more efficiently than green light. Weissleder et al. [33] have demonstrated the utility of red fluorescence for tumor imaging using an autoquenched red fluorescent dye (Cy5.5) that accumulates at the tumor site and is activated by lysosomal proteases of tumor cells. Quenched fluorochromes were bound to a copolymer consisting of poly-l-lysine and methoxypolyethylene glycol succinate. Following intravenous injection, the conjugates accumulated in solid xenograft tumors presumably after passing through the neovasculature of the tumor. The quenched fluorochrome was released in the tumor by lysosomal proteases in the tumor cells such that the signal was increased 12-fold at the tumor site and submillimetersized tumors were detectable subcutaneously. Fluorescent proteins that excite and emit at longer wavelengths may permit detection of labeled cells several millimeters or centimeters within mammalian tissue. Such reporter proteins have recently been described [16]. However, these orange-red fluorescent proteins have not yet been used in studies of in vivo tumor cell growth.
As transcriptional reporters, GFP and luciferase can be indicative of gene expression when a tumor-specific promoter is linked to the coding sequence, and when constitutively expressed, these reporters can be used to indicate the sites of tumor growth. In vivo assays that utilize reporter genes can also be set up as indicators of other steps in neoplastic disease progression, such as microvessel density (MVD) as an indicator of neovascularization of tumors. Moore et al. [83] have used a 9L cell line constitutively expressing GFP for assessing MVD. In this study using fluorescence microscopy, non-fluorescent regions at the borders of the tumor tissue were interpreted as vascularized regions lacking tumor cells. This was confirmed by utilizing conventional histological methods, anti-CD31 immunohistology, and Hoechst 33258 dye exclusion. MVD allowed quantification of tumor angiogenesis in tissue specimens, thus providing additional information about tumor growth and regression in animal models. Such analysis holds promise in evaluating the efficacy of anti-angiogenic therapies.
Multifunctional Reporters
The various optical reporter genes, including luciferase and GFP, have unique properties that make each useful for particular experimental applications. Luciferase is a real time indicator of transcription; due to its relatively short half-life, it indicates diminution of expression unlike the longer-lived GFP protein. In contrast, GFP can be used for higher resolution imaging in cells. Combining these reporter genes into a single gene could provide additional tools for the analysis of cancer cells in vivo and ex vivo. A recombinant protein consisting of the coding regions for GFP and luciferase would provide the sensitivity of fluorescence and the ease and real-time imaging of bioluminescence. Such a dual-function reporter gene was created and the single encoded protein was shown to be fluorescent and bioluminescent [48]. The GFP portion of the protein allowed for analyses of single living cells expressing the chimeric protein within a population by fluorescence microscopy, and the luciferase activity could be detected from the same living cells.
Luciferases comprise a family of enzymes that use different substrates and biochemical mechanisms to emit light of different wavelengths and that use different biochemical mechanisms [41–43,84–86]. In addition, directed and random mutageneses of firefly genes have resulted in shifts in the wavelength of emission [84,88], offering the possibility of multiparameter in vivo assays. Sets of bioluminescent enzymes with different biochemical requirements and different emission spectra have been used in in vitro and culture-based assays to assess expression of two different genes, usually one as control and the other as the inducible test gene [47,89]. Combining the various fluorescent proteins to the wide range bioluminescent enzymes increases the potential for assays and models to evaluate high throughput multiparameter analyses of oncogenesis.
Summary and Conclusions
Advancing our understanding of neoplastic disease requires the ability to effect biological processes in vivo and evaluate the changes non-invasively at the molecular and cellular levels. A basic premise of biology is that the investigator specifically perturbs a biological process and then studies the effects of this perturbation on the entire system to learn about interactive processes, mechanisms and pathways. A large number of biological events only occur in the context of intact organ systems that cannot be modeled in culture. For this reason, animal models are routinely utilized in biomedical research. However, the ability to interrogate cellular and molecular processes in living animal models has been limited, and thus evaluating the effects of various stimuli on complex biological processes in vivo was not previously possible. In the study of neoplastic disease, many of the molecular and cellular changes that are linked to the initiation and progression of disease only occur in vivo and may be largely undiscovered due to our inability to access these changes in the living animal. With advancements in molecular and cellular imaging, in vivo perturbation and evaluation of steps in interwoven pathways may be possible.
Similarly, improvements and modifications to animal model systems are needed to further understand various mechanisms of immune containment of neoplasia and to develop new therapies to prevent and treat small numbers of neoplastic cells present in minimal disease states. Since as few as 1000 luciferase-labeled tumor cells distributed throughout the peritoneal cavity were reliably detected and quantified in living animals, and the signals were proportional to tumor cell inocula over a broad dynamic range spanning at least five orders of magnitude, the kinetics of tumor growth from early stages of minimal disease to late stage disease could be evaluated over time [52,53]. The ability to monitor kinetics of tumor cell growth and localize micrometastases non-invasively will reveal novel features of these processes as well as elucidate cellular and tissue responses to tumor growth.
The demonstrated ability to non-invasively detect small numbers of tumor cells in living animals will permit the assessment of therapies designed to treat minimal disease states. Minimal residual disease that persists after elimination of the initial tumor mass often results in relapse of disease and remains a major therapeutic challenge [57]. In addition, as neoplastic diagnostic tests improve, therapies that are directed at minimal disease states early in the disease course would need to be developed. The approaches reviewed here can accelerate development of therapeutic strategies by providing a rapid in vivo assay for efficacy that is inexpensive and accessible. In vivo imaging approaches, in general, will improve the predictability of animal models of human neoplastic disease by providing spatiotemporal information, and the ease and versatility of photoprotein imaging can additionally be applied in high throughput animal models for drug discovery. In addition, information about the kinetics of tumor cell growth and effects of endogenous immunological mechanisms can be directly assessed; thus, more information is provided despite the rapid nature of the assay.
In vivo imaging of optical reporter genes is a broadly applicable tool for the study of neoplasia. This approach can provide real time data that indicate the spatial distribution of tumor cells at multiple time points during the disease course. Use of optical reporter genes provides information with relatively short scan times; thus, rapid assessment of a potential intervention in a fully quantitative manner is possible. Since groups of animals can be followed over time, far fewer animal subjects are required to obtain statistically meaningful results. The stress on the animals in these studies is dramatically reduced since whole body imaging replaces the use of death as an endpoint and the models can be evaluated in minimal disease states where the tumor burden is small. Rapid whole body imaging and the use of repeated measurements on groups of animals improve the data set, accelerate the study and reduce stress to the animals; taken together, these serve to generally refine animal studies of neoplastic disease.
This method may also extend to assessing the tissue distribution of host immune cells that recognize and eliminate tumor cells. With these molecular tools for in vivo studies, investigators can begin to analyze the mechanisms of immune surveillance of neoplastic disease in living animal models of human disease. Theoretically, other cell populations or biological functions that can be suitably labeled are also amenable for study in living animal models. Further modifications to the photoprotein reporters used in the imaging strategy may permit the development of multiparameter or dual color assays for monitoring tumor progression and interaction of the host response in the same animal. Studies thus far using optical reporter gene imaging have demonstrated that it is possible to track the fate of tumor cells in vivo.
Advances
Developments in optical reporter gene technologies have been dramatic since the discovery of GFP as an optical tag to indicate promoter activity, intracellular trafficking (by creating fusion proteins with GFP), or cell migration in small nearly transparent animals [90,91]. The lessons learned from the modifications of GFP can be applied to other optical reporter genes such as luciferase. For example, shifting of the emission wavelength of luciferase reporters from primarily blue and green emission to those that are more readily transmitted through tissues will greatly enhance their utility for in vivo analyses [69,70,92]. The barriers to transmission of light through tissues include scattering and absorption. Absorption is largely due to hemoglobin at shorter wavelengths up to 600 nm; above 800 nm, absorption is due to water. Reporter genes that emit within this window would be ideal for in vivo detection.
Advancements in imaging technologies will revolutionize the study, treatment and prevention of neoplastic disease by providing a wealth of critical information that has not been previously available. Continued commitment to imaging research will lead to additional tools to reveal in vivo biological events leading to essentially transparent animal models, and the ability to assess cellular and molecular events non-invasively. Further advancement of non-invasive in vivo methods to study neoplasia will facilitate rapid development of novel therapeutics against new molecular targets, and enable assessment of therapeutic outcomes. Development of multimodality imaging approaches will greatly improve our ability to obtain structural and functional data in animal models, and these advancements will likely have direct bearing on clinical imaging and treatment of human cancers.
Acknowledgements
PC3M cells were kindly provided to Xenogen and Novartis Pharmaceuticals by J.I. Fidler from the M.D. Anderson Medical Center in Houston, TX and the prostate model was evaluated in collaboration with Novartis Pharmaceuticals.
Footnotes
This work was funded, in part, through grants from a Translational Award from The Leukemia Society (609099), the National Cancer Institute (R01 CA80006), and unrestricted gifts from the Mary L. Johnson and Hess Research Funds.
References
- 1.Clemens MJ, Bommer UA. Translational control: the cancer connection. Int J Biochem Cell Biol. 1999;31:1–23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trend Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 3.Devereux TR, Risinger JI, Barrett JC. Mutations and altered expression of the human cancer genes: what they tell us about causes. IARC Sci Publ. 1999:19–42. [PubMed] [Google Scholar]
- 4.Holt SE, Shay JW. Role of telomerase in cellular proliferation and cancer. J Cell Physiol. 1999;180:10–18. doi: 10.1002/(SICI)1097-4652(199907)180:1<10::AID-JCP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson S, Pillus L. Modifying chromatin and concepts of cancer. Curr Opin Gen Dev. 1999;9:175–184. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 6.Markiewicz MA, Gajewski TF. The immune system as anti-tumor sentinel: molecular requirements for an anti-tumor immune response. Crit Rev Oncogen. 1999;10:247–260. [PubMed] [Google Scholar]
- 7.McBurney MW. Gene silencing in the development of cancer. Exp Cell Res. 1999;248:25–29. doi: 10.1006/excr.1999.4454. [DOI] [PubMed] [Google Scholar]
- 8.Kavanaugh DY, Carbone DP. Immunologic dysfunction in cancer. Hem/Oncol Clin N Am. 1996;10:927–951. doi: 10.1016/s0889-8588(05)70376-2. [DOI] [PubMed] [Google Scholar]
- 9.Whiteside TL, Herberman RB. The role of natural killer cells in immune surveillance of cancer. Curr Opin Immunol. 1995;1:704–710. doi: 10.1016/0952-7915(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 10.Christofori G, Hanahan D. Molecular dissection of multistage tumorigenesis in transgenic mice. Sem Cancer Biol. 1994;5:3–12. [PubMed] [Google Scholar]
- 11.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 12.Coussens LM, Hanahan D, Arbeit JM. Genetic predisposition and parameters of malignant progression in K14-HPV16 transgenic mice. Am J Pathol. 1996;149:1899–1917. [PMC free article] [PubMed] [Google Scholar]
- 13.Bigos M, Baumgarth N, Jager GC, Herman OC, Nozaki T, Stovel RT, Parks DR, Herzenberg LA. Nine color eleven parameter immunophenotyping using three laser flow cytometry. Cytometry. 1999;36:36–45. doi: 10.1002/(sici)1097-0320(19990501)36:1<36::aid-cyto5>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 14.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Gen. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 15.Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 16.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from non-bioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 17.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piston DW. Imaging living cells and tissues by two-photon excitation microscopy. Trends Cell Biol. 1999;9:66–69. doi: 10.1016/s0962-8924(98)01432-9. [DOI] [PubMed] [Google Scholar]
- 19.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genomewide analysis of DNA copy-number changes using cDNA microarrays. Nat Gen. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 20.Potter SM. Vital imaging: two photons are betterthan one. Curr Biol. 1996;6:1595–1598. doi: 10.1016/s0960-9822(02)70782-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Hellstrom KE, Chen L. Luciferase activity as a marker of tumor burden and as an indicator of tumor response to antineoplastic therapy in vivo. Clin Exp Metastasis. 1994;12:87–92. doi: 10.1007/BF01753974. [DOI] [PubMed] [Google Scholar]
- 22.Negrin RS, Blume KG. The use of the polymerase chain reaction for the detection of minimal residual malignant disease. Blood. 1991;78:255–258. [PubMed] [Google Scholar]
- 23.Bogdanov A, Jr, Weissleder R. The development of in vivo imaging systems to study gene expression. Trends Biotechnol. 1998;16:5–10. doi: 10.1016/S0167-7799(97)01150-5. [DOI] [PubMed] [Google Scholar]
- 24.Chenevert TL, McKeever PE, Ross BD. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clin Cancer Res. 1997;3:1457–1466. [PubMed] [Google Scholar]
- 25.Ross B, Zhao Y-J, Neal E, Stegman L, Ercolani M, Ben-Yoseph O, Chenevert T. Contributions of cell kill and posttreatment tumor growth rates to the repopulation of intracerebral 9L tumors after chemotherapy: an MRI study. Proc Natl Acad Sci USA. 1998;95:7012–7017. doi: 10.1073/pnas.95.12.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galons J-P, Altbach M, Paine-Murrieta G, Taylor C, Gillies R. Early increases in breast tumor xenograft water mobility in response to paclitaxel therapy detected by non-invasive diffusion magnetic resonance imaging. Neoplasia. 1999;1:113–117. doi: 10.1038/sj.neo.7900009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambhir SS, Barrio JR, Herschman HR, Phelps ME. Imaging gene expression: principles and assays. J Nucl Cardiol. 1999;6:219–233. doi: 10.1016/s1071-3581(99)90083-1. [DOI] [PubMed] [Google Scholar]
- 28.Gambhir SS, Barrio JR, Phelps ME, Iyer M, Namavari M, Satyamurthy N, Wu L, Green LA, Bauer E, MacLaren DC, Nguyen K, Berk AJ, Cherry SR, Herschman HR. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs A, Dubrovin M, Hewett J, Sena-Esteves M, Tan C-W, Slack M, Sadelain M, Breakefield X, Tjuvajev J. Functional coexpression of HSV-1 thymidine kinase and green fluorescent protein: implications for non-invasive imaging of transgene expression. Neoplasia. 1999;1:154–161. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjuvajev JG, Finn R, Watanabe K, Joshi R, Oku T, Kennedy J, Beattie B, Koutcher J, Larson S, Blasberg RG. Non-invasive imaging of herpes virus thymidine kinase gene transfer and expression: a potential method for monitoring clinical gene therapy. Cancer Res. 1996;56:4087–4095. [PubMed] [Google Scholar]
- 31.Tjuvajev JG, Avril N, Oku T, Sasajima T, Miyagawa T, Joshi R, Safer M, Beattie B, DiResta G, Daghighian F, Augensen F, Koutcher J, Zweit J, Humm J, Larson SM, Finn R, Blasberg R. Imaging herpes virus thymidine kinase gene transfer and expression by positron emission tomography. Cancer Res. 1998;58:4333–4341. [PubMed] [Google Scholar]
- 32.Uehara H, Miyagawa T, Tjuvajev J, Joshi R, Beattie B, Oku T, Finn R, Blasberg R. Imaging experimental brain tumors with 1-aminocyclopentane carboxylic acid and alpha-aminoisobutyric acid: comparison to fluorodeoxyglucose and diethylenetriaminepentaacetic acid in morphologically defined tumor regions. J Cereb Blood Flow Metab. 1997;17:1239–1253. doi: 10.1097/00004647-199711000-00013. [DOI] [PubMed] [Google Scholar]
- 33.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 34.Tjuvajev JG, Joshi A, Callegari J, Lindsley L, Joshi R, Balatoni J, Finn R, Larson SM, Sadelain M, Blasberg RG. A general approach to the non-invasive imaging of transgenes using cis-linked herpes simplex virus thymidine kinase. Neoplasia. 1999;1:315–320. doi: 10.1038/sj.neo.7900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harisinghani MG, Saini S, Weissleder R, Hahn PF, Yantiss RK, Tempany C, Wood BJ, Mueller PR. MR lymphangiography using ultrasmall superparamagnetic iron oxide in patients with primary abdominal and pelvic malignancies: radiographic-pathologic correlation. AJR Am J Roentgenol. 1999;172:1347–1351. doi: 10.2214/ajr.172.5.10227514. [DOI] [PubMed] [Google Scholar]
- 36.Josephson L, Tung CH, Moore A, Weissleder R. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconjugate Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds JS, Troy TL, Mayer RH, Thompson AB, Waters DJ, Cornell KK, Snyder PW, Sevick-Muraca EM. Imaging of spontaneous canine mammary tumors using fluorescent contrast agents. Photochem Photobiol. 1999;70:87–94. [PubMed] [Google Scholar]
- 38.Nioka S, Yung Y, Shnall M, Zhao S, Orel S, Xie C, Chance B, Solin L. Optical imaging of breast tumor by means of continuous waves. Adv Exp Med Biol. 1997;411:227–232. doi: 10.1007/978-1-4615-5865-1_27. [DOI] [PubMed] [Google Scholar]
- 39.de Wet JR, Wood KV, Helinski DR, DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wet J, Wood K, DeLuca M, Helinski D, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherf B, Wood K. Firefly luciferase engineered for improved genetic reporting. Promega Notes Mag. 1994;49:14–21. [Google Scholar]
- 42.Wood KV. Marker proteins for gene expression. Curr Opin Biotechnol. 1995;6:50–58. doi: 10.1016/0958-1669(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 43.Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- 44.Willard ST, Faught WJ, Frawley LS. Real-time monitoring of estrogen-regulated gene expression in single, living breast cancer cells: a new paradigm for the study of molecular dynamics. Cancer Res. 1997;57:4447–4450. [PubMed] [Google Scholar]
- 45.Chen H, Biel MA, Borges MW, Thiagalingam A, Nelkin BD, Baylin SB, Ball DW. Tissue-specific expression of human achaete-scute homologue-1 in neuroendocrine tumors: transcriptional regulation by dual inhibitory regions. Cell Growth Differ. 1997;8:677–686. [PubMed] [Google Scholar]
- 46.Takakuwa K, Fujita K, Kikuchi A, Sugaya S, Yahata T, Aida H, Kurabayashi T, Hasegawa I, Tanaka K. Direct intratumoral gene transfer of the herpes simplex virus thymidine kinase gene with DNA-liposome complexes: growth inhibition of tumors and lack of localization in normal tissues. Jpn J Cancer Res. 1997;88:166–175. doi: 10.1111/j.1349-7006.1997.tb00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 48.Day RN, Kawecki M, Berry D. Dual-function reporter protein for analysis of gene expression in living cells. Biotechnology. 1998;25:848–850. doi: 10.2144/98255bt02. [DOI] [PubMed] [Google Scholar]
- 49.Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 50.Contag C, Spilman S, Contag P, Oshiro M, Eames B, Dennery P, Stevenson D, Benaron D. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 51.Contag PR, Olomu IN, Stevenson DK, Contag CH. Bioluminescent indicators in living mammals. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 52.Sweeney T, Mailander V, Tucker A, Olomu AB, Zhang W, Negrin RS, Contag CH. Visualizing tumor cell clearance in living animals. Proc Natl Acad Sci USA. 1999;96:12044–12049. doi: 10.1073/pnas.96.21.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edinger M, Sweeney TJ, Tucker AA, Adesuwa B, Olomu AB, Negrin RS, Contag CH. Non-invasive assessment of tumor cell proliferation in animal models. Neoplasia. 1999;1:303–310. doi: 10.1038/sj.neo.7900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chance B, Luo Q, Nioka S, Alsop DC, Detre JA. Optical investigations of physiology: a study of intrinsic and extrinsic biomedical contrast. Philos Trans R Soc London B Biol Sci. 1997;352:707–716. doi: 10.1098/rstb.1997.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koslowski JM, Fidler IJ, Campbel D, Xu Z, Kaigh ME, Hart IR. Metastatic behavior of human tumor cell lines grown in the nude mouse. Cancer Res. 1984;44:3522–3529. [PubMed] [Google Scholar]
- 56.Cave H, van der Werff ten Bosch J, Suciu S, Guidal C, Waterkeyn C, Often J, Bakkus M, Thielemans K, Grandchamp B, Vilmer E. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer — Childhood Leukemia Cooperative Group. New Engl J Med. 1998;339:591–598. doi: 10.1056/NEJM199808273390904. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch-Ginsberg C. Detection of minimal residual disease: relevance for diagnosis and treatment of human malignancies. Annu Rev Med. 1998;49:111–122. doi: 10.1146/annurev.med.49.1.111. [DOI] [PubMed] [Google Scholar]
- 58.Uckun FM, Sather H, Reaman G, Shuster J, Land V, Trigg M, Gunther R, Chelstrom L, Bleyer A, Gaynon P. Leukemic cell growth in SCID mice as a predictor of relapse in high-risk B-lineage acute lymphoblastic leukemia. Blood. 1995;85:873–878. [PubMed] [Google Scholar]
- 59.Arguello F, Sterry JA, Zhao YZ, Alexander MR, Shoemaker RH, Cohen HJ. Two serologic markers to monitor the engraftment, growth, and treatment response of human leukemias in severe combined immunodeficient mice. Blood. 1996;87:4325–4332. [PubMed] [Google Scholar]
- 60.Schmidt-Wolf IGH, Negrin RS, Kiem HP, Blume KG, Weissman IL. Use of a SCID mouse/human lymphoma model to evaluate cytokine-induced killer cells with potent antitumor cell activity. J Exp Med. 1991;174:139–149. doi: 10.1084/jem.174.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu PH, Negrin RS. A novel population of expanded human CD3+CD56+ cells derived from T cells with potent in vivo antitumor activity in mice with severe combined immunodeficiency. J Immunol. 1994;153:1687–1696. [PubMed] [Google Scholar]
- 62.Ochoa AC, Gromo G, Alter BJ, Sondel PM, Bach FH. Long-term growth of lymphokine-activated killer LAK cells: role of anti-CD3, beta-IL 1, interferon-gamma and -beta. J Immunol. 1987;138:2728–2733. [PubMed] [Google Scholar]
- 63.Schmidt-Wolf IG, Lefterova P, Johnston V, Scheffold C, Csipai M, Mehta BA, Tsuruo T, Huhn D, Negrin RS. Sensitivity of multidrug-resistant tumor cell lines to immunologic effector cells. Cell Immunol. 1996;169:85–90. doi: 10.1006/cimm.1996.0094. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt-Wolf IG, Lefterova P, Mehta BA, Fernandez LP, Huhn D, Blume KG, Weissman IL, Negrin RS. Phenotypic characterization and identification of effector cells involved in tumor cell recognition of cytokine-induced killer cells. Exp Hematom. 1993;21:1673–1679. [PubMed] [Google Scholar]
- 65.Hoyle CH, Bangs CD, Cahng P, Kamel O, Mehta BA, Negrin RS. Expansion of Philadelphia-chromosome-negative CD3+CD56+ cytotoxic cells from CML patients: in vitro and in vivo efficacy in SCID mice. 1998 Submitted. [PubMed] [Google Scholar]
- 66.Chishima T, Yang M, Miyagi Y, Li L, Tan Y, Baranov E, Shimada H, Moossa AR, Penman S, Hoffman RM. Governing step of metastasis visualized in vitro. Proc Natl Acad Sci USA. 1997;94:11573–11576. doi: 10.1073/pnas.94.21.11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chishima T, Miyagi Y, Wang X, Tan Y, Shimada H, Moossa A, Hoffman RM. Visualization of the metastatic process by green fluorescent protein expression. Anticancer Res. 1997;17:2377–2384. [PubMed] [Google Scholar]
- 68.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci USA. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 70.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 72.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 73.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 74.Ito Y, Suzuki M, Husimi Y. A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem Biophys Res Commun. 1999;264:556–560. doi: 10.1006/bbrc.1999.1541. [DOI] [PubMed] [Google Scholar]
- 75.Yang M, Jiang P, Sun F, Hasegawa S, Baranov E, Chishima T, Shimada H, Moossa A, Hoffman R. A fluorescent orthotopic bone metastasis model of human prostate cancer. Cancer Res. 1999;59:781–786. [PubMed] [Google Scholar]
- 76.Naumov G, Wilson S, MacDonald I, Schmidt E, Morris V, Groom A, Hoffman R, Chambers A. Cellular expression of green fluorescent protein, coupled with high-resolution in vivo videomicroscopy, to monitor steps in tumor metastasis. J Cell Sci. 1999;112:1835–1842. doi: 10.1242/jcs.112.12.1835. [DOI] [PubMed] [Google Scholar]
- 77.Fukumura D, Xavier R, Sugiura T, Chen Y, Park EC, Lu N, Selig M, Nielsen G, Taksir T, Jain RK, Seed B. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94:715–725. doi: 10.1016/s0092-8674(00)81731-6. [DOI] [PubMed] [Google Scholar]
- 78.Yang M, Hasegawa S, Jiang P, Wang X, Tan Y, Chishima T, Shimada H, Moossa A, Hoffman R. Widespread skeletal metastatic potential of human lung cancer revealed by green fluorescent protein expression. Cancer Res. 1998;58:4217–4221. [PubMed] [Google Scholar]
- 79.Hoffman R. Orthotopic transplant mouse models with green fluorescent protein-expressing cancer cells to visualize metastasis and angiogenesis. Cancer Met Rev. 1999;17:271–277. doi: 10.1023/a:1006188412324. [DOI] [PubMed] [Google Scholar]
- 80.Hofmann B, Bogdanov A, Jr, Marecos E, Ebert W, Semmler W, Weissleder R. Mechanism of gadophrin-2 accumulation in tumor necrosis. J Magn Reson Imaging. 1999;9:336–341. doi: 10.1002/(sici)1522-2586(199902)9:2<336::aid-jmri28>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 81.Fillmore HL, Shurm J, Furqueron P, Prabhu SS, Gillies GT, Broaddus WC. An in vivo rat model for visualizing glioma tumor cell invasion using stable persistent expression of the green fluorescent protein. Cancer Lett. 1999;141:9–19. doi: 10.1016/s0304-3835(99)00053-1. [DOI] [PubMed] [Google Scholar]
- 82.MacDonald TJ, Tabrizi P, Shimada H, Zlokovic BV, Laug WE. Detection of brain tumor invasion and micrometastasis in vivo by expression of enhanced green fluorescent protein. Neurosurgery. 1998;43:1437–1442. doi: 10.1097/00006123-199812000-00101. [DOI] [PubMed] [Google Scholar]
- 83.Moore A, Marecos E, Simonova M, Weissleder R, Bogdanov A., Jr Novel gliosarcoma cell line expressing green fluorescent protein: a model for quantitative assessment of angiogenesis. Microvase Res. 1998;56:145–153. doi: 10.1006/mvre.1998.2102. [DOI] [PubMed] [Google Scholar]
- 84.Kajiyama N, Nakano E. Isolation and characterization of mutants of firefly luciferase which produce different colors of light. Protein Eng. 1991;4:691–693. doi: 10.1093/protein/4.6.691. [DOI] [PubMed] [Google Scholar]
- 85.Wood KV, Lam YA, Seliger HH, McElroy WD. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- 86.Wood KV, Lam YA, McElroy WD. Introduction to beetle luciferases and their applications. J Biolumin Chemilumin. 1989;4:289–301. doi: 10.1002/bio.1170040141. [DOI] [PubMed] [Google Scholar]
- 87.Wood KV. Luc genes: introduction of colour into bioluminescence assays. J Biolumin Chemilumin. 1990;5:107–114. doi: 10.1002/bio.1170050206. [DOI] [PubMed] [Google Scholar]
- 88.Eames BF, Benaron DA, Stevenson DK, Contag CH. Construction and in vivo testing of a red-emitting firefly luciferase; Proceedings of SPIE Annual Meeting; SPIE. 1999. pp. 36–39. [Google Scholar]
- 89.Stables J, Scott S, Brown S, Roelant C, Burns D, Lee MG, Rees S. Development of a dual glow-signal firefly and Renilla luciferase assay reagent for the analysis of G-protein-coupled receptor signalling. J Recept Signal Transduction Res. 1999;19:395–410. doi: 10.3109/10799899909036660. [DOI] [PubMed] [Google Scholar]
- 90.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 91.Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 92.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]